Abstract
Flexible filamentous plant viruses are responsible for more than half the viral crop damage in the world, but are also potentially useful for biotechnology. Structural studies began more than 75 years ago but have failed due to the virion’s extreme flexibility. We have used cryo–EM to generate an atomic model for bamboo mosaic virus revealing flexible N– and C–terminal extensions that allow deformation while still maintaining structural integrity.
The flexible filamentous plant viruses1 are single–stranded positive–sense RNA viruses that are widely found, and responsible for more than half the viral crop damage in the world2. Due to their low toxicity they are viewed as potentially useful for biotechnology, such as in vaccines3,4 and biomaterials for drug delivery or imaging5. Filamentous plant viruses are broadly classified into the rigid rod–like, such as tobacco mosaic virus (TMV), and the flexible viruses. While ten rigid filamentous plant viruses have been found in substantial quantities in human stool samples6, the flexible filamentous plant viruses were conspicuously absent, suggesting that they can be metabolized while the rod–like viruses cannot. In addition, the flexible filamentous viruses are potentially valuable for recombinant protein production in plants7. But all of these applications have been hampered by the absence of atomic structures. Published structural studies of these viruses date from 19418, but no atomic model has been possible due to the fact that the viruses cannot be crystallized and have proven to be too flexible for high–resolution x–ray fiber diffraction or electron cryo–microscopy (cryo–EM). TMV, the first virus to be discovered9, is a rigid filamentous plant virus that has been a model system in structural biology and virology. Atomic models of TMV have been produced by both x–ray diffraction10 and cryo–EM11. In contrast to the rigid viruses, the flexible filamentous viruses, including potexviruses such as potato virus X (PVX), could not generate high–quality fiber diffraction patterns. It was suggested, based upon low resolution x–ray fiber diffraction, that all potexviruses may share a common architecture, with slightly less than nine protein subunits per helical turn12. This conclusion was subsequently strengthened using both x–ray diffraction and various forms of EM1,13. A number of low–resolution models of the flexible plant viruses have been generated1,13,14, all implicitly assuming that the virions have a right–handed helical pitch as found in TMV10.
BaMV belongs to the genus Potexvirus, family Alphaflexiviridae. It has a single–stranded RNA genome15 of about 6.4 kb and has a flexible filamentous morphology with a length of 490 nm and diameter of 15 nm16, built mainly from a single protein CP (coat protein). Previous research showed that up to 35 residues of the N–terminus of CP can be deleted with no effect on virus replication and assembly17. Thus, BaMV has been developed as a plant–expression vector for vaccine production by replacing the N–terminal 35 residues with foreign peptides from either foot–and–mouth disease virus (FMDV)4 or infectious bursal disease virus (IBDV)3.
We set out to determine an atomic structure for a flexible filamentous plant virus. We imaged both the wildtype BaMV (wt) and a virion containing a deletion of 35 N–terminal residues of CP, BaMV–Nd35 (Nd35), using cryo–EM with a direct electron detector (Fig. 1). Starting with power spectra from the filaments (Supp. Fig. 1), the symmetry was found by trial–and–error until recognizable secondary structure elements (rod–like features from α–helices) were seen18. We have found no change in the symmetry between the wt and the Nd35, with both having a pitch of ~ 35 Å with ~ 8.8 subunits per turn. Using the crystal structure14 (4DOX.PDB) of a large fragment of the PapMV CP, which has 28% sequence identity with the corresponding region of BaMV, it became obvious that the ~ 35 Å pitch helix must be left–handed (two enantiomorphic reconstructions can be generated that are equally consistent with the images, one right–handed and one left–handed, but the crystal structure could only fit into the left–handed one). The reconstructions of both the wt and Nd35 were improved by using a classification approach to remove the large variability in twist and rise, and a more homogeneous set of ~ 54k segments for the wt and ~ 50k segments for the Nd35 were used for the final reconstructions of each. The absence of any significant structural differences between the two volumes allowed us to use these completely independent reconstructions for an estimate of resolution, which yielded a value of 5.6 Å (Supp. Fig. 2). We then combined the two sets to generate a reconstruction (Fig. 1b) used for modeling. Due to the intrinsic variability of the structure, the resolution of the combined reconstruction was not significantly better than either of the individual reconstructions. A comparison between the surface of the combined reconstruction (Fig. 2a) and the atomic model that we have built, filtered to 5.6 Å (Fig. 2b), shows that our estimate of the resolution is very reasonable.
Figure 1.

The BaMV helical capsid structure. (a) Micrographs of the wildtype (left) and Nd35 (right) capsids. The scale bar is 1,000 Å. (b) The 5.6 Å resolution combined reconstruction, showing three full helical turns of the capsid.
Figure 2.
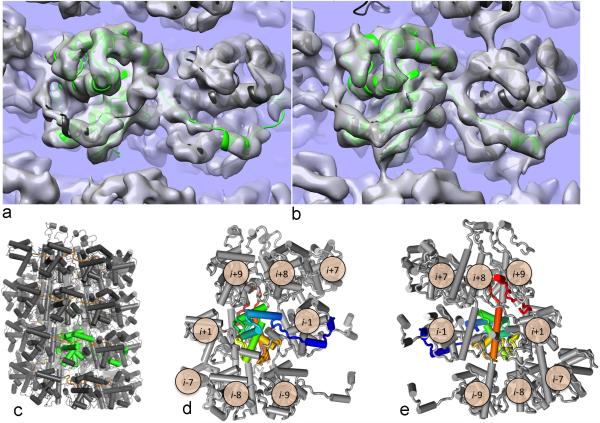
BaMV cryoEM reconstruction allows building and refinement of atomic models. (a) The final refined model of the capsid docked into density shows good agreement with the experimental data (transparent grey surface); one asymmetric unit is highlighted in green. (b) When the model is converted to density and filtered to 5.6 Å resolution (transparent grey surface), it compares extremely well to the actual density map in (a). (c) A cartoon representation of the complete capsid model. One asymmetric unit is indicated in green. (d,e) A cartoon schematic showed the highly interconnected nature of the BaMV capsid, with the asymmetric unit protein colored from blue (N–terminus) to red (C–terminus). Looking from the outside (d), we see the N–terminus wraps around the i–1 subunit, placing a short helix into a hydrophobic groove on the surface of the protein. Looking from the inside (e), we see that the C–terminus makes contact with three subunits from the helical turn above before terminating facing the inside of the capsid where a contact is made between the C–terminus and a subunit from a helical turn below.
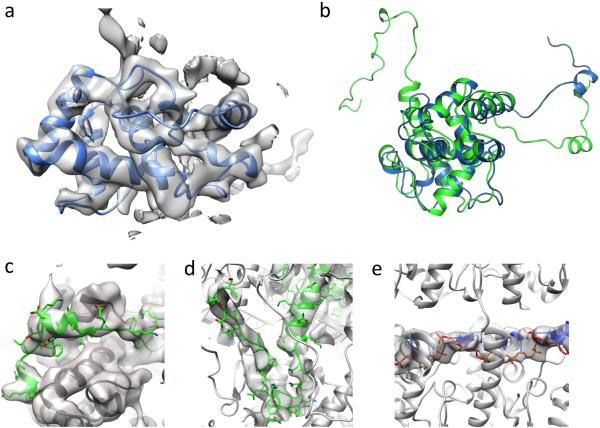
The existence of the crystal structure of the PapMV fragment14 (4DOX.PDB) allowed us to use this as an intial template for building a full atomic model of BaMV (Fig. 2, Supp. Fig. 3). The template was initially docked into density and the core (residues 33–174 in PapMV, corresponding to residues 62–201 in BaMV) showed good agreement to the experimental data, with real–space correlation of 0.61 over these 143 residues comprising the compact core of the crystal structure (Fig. 3a,b). However, this model failed to account for 61 N–terminal BaMV residues (18 of which were present in the template model but were likely stabilized by crystal contacts and poorly fit the experimental density) and 42 C–terminal residues, which were truncated for crystallization14. Continuous density was clearly visible for both the missing N–terminal and C–terminal residues on the outside and the inside of the capsid, respectively.
Figure 3.
The refined model reveals atomic details. (a) Model–building was largely guided by a crystal structure of a fragment of the PapMV capsid protein, which showed good agreement with the density data. (b) The final refined model (green) superimposed on the PapMV crystal structure (blue) shows good agreement over the core of the protein, but with large differences in the N–terminus. (c,d,e) Portions of the model not present in the crystal structure that were built into density also show good agreement with the experimental data: the N–terminus (c), C–terminus (d), and RNA (e).
We used Rosetta19 to build the backbone of the missing C–terminus and rebuild the N–terminus (Fig. 3c,d). Due to the length, the relatively low resolution of the local density, and the low apparent secondary structure content of the insertions, building or rebuilding these termini proved especially challenging. Ultimately, we used a novel enumerative backbone sampling protocol, described in the methods section. Sampling these terminal conformations revealed reasonable convergence of the top–scoring models (Supp. Fig. 4). An unbroken tube of density remained unexplained by the models, which seemed likely to correspond to the single stranded RNA. The resolution of the data was unfortunately insufficient to build RNA models de novo with any degree of confidence. Knowing the length of the virus (~ 490 nm), the size of the genome (6.4 kb), and the rise per subunit (4.0 Å), one can estimate ~ 5.2 bases per subunit, which yields 5 as the nearest integer. We docked and refined a 5–nucleotide sequence from rift valley fever virus (4H5O.PDB), as the nucleotide chain in this structure had a very similar radius of curvature to that observed in the density (~23 Å in the crystal structure versus ~30 Å in the density map), and a 5–nucleotide stretch (nucleotides 3–7) showed good agreement when docked into the experimental data (Fig. 3e).
Finally, all–atom refinement of the symmetric full–length model against the experimental density data was carried out in Rosetta. After refinement, the lowest–energy structures were selected and compared. These structures showed relatively tight convergence (Supp. Fig. 4), however, convergence was noticeably worse in the C–terminus, making identification of specific sidechain interactions stabilizing this long loop ambiguous. Regions of the model that used the crystal structure as a starting point were very well converged, and the final model showed only very modest deviation from the initial crystal structure, with a Cα rmsd of 3.1 Å; differences between the two are largely limited to several loops interacting with the single–stranded RNA. Comparison to an independent dataset (Supp. Fig. 2) shows similar agreement to the map used for fitting, indicating the model is not over–refined.
The final structure shows a highly intertwined topology (Fig. 2c), where each subunit makes direct contact with 8 other subunits (Fig. 2d,e). Looking from the outside of the capsid (Fig. 2d), the N–terminus of each subunit wraps around the i–1 subunit, forming a short helix that packs into a hydrophobic cleft on the surface with Phe45 buried in a small pocket on the surface. An N–terminal loop continues wrapping around the structure with Trp41 and Trp68 forming a stacking interaction. The first 38 N–terminal residues are disordered in our model; this is validated by the striking similarity between the wt and Nd35 reconstructions. The C–terminus (Fig. 2e) wraps through the core of the capsid, following a continuous tube of density, and forms contacts with three subunits in the turn above (i+7, i+8, and i+9), before pointing toward the center of the virion where the extreme C–terminal residues contact subunit i–7 in the turn below.
While the resolution of the data does not permit us to draw conclusions on the nature of the protein–RNA interactions, the model suggests that residues Arg99, Lys132, Lys157, Lys202, and Lys213 all potentially make protein–RNA contacts. Among them, Arg99 was found20 to be part of the potential RNA binding motifs in BaMV CP.
This highly intertwined structure of a flexible filamentous virus is in sharp contrast to the highly compact architecture of TMV. The asymmetric unit of TMV is similar in size to that of BaMV: 158 residues in TMV versus 204 residues in BaMV (ignoring the disordered 38 N–terminal residues), with an accessible surface area of 8,787 Å2 per subunit for TMV versus 12,922 Å2 for BaMV (8,075 Å2 not including the extended termini). TMV however, forms much more extensive contacts with neighboring subunits, with 4,499 Å2 (or about 51%) of surface forming the interface in the assembled capsid. On the other hand, BaMV’s compact core is only making modest contacts between subunits, with 1,806 Å2 (22%) of surface contacting neighboring subunits; it is only when including the N– and C–terminal extensions, connected to the core of the subunit by very flexible linkers, that the contacting surface increases to 5,752 Å2 (45%), in line with TMV. It is this architecture that allows extensive non–covalent interactions with many surrounding subunits to be maintained in the flexible filamentous viruses as the structures deform due to mechanical forces.
Online Methods
BaMV preparation
The plasmid pCB is a full–length cDNA infectious clone of BaMV–S (GenBank accession number AF018156) in a pCass2 vector as described previously21. The pCB–Nd35 was derived from pCB by deletion of the N–terminal 35 amino acid sequence of CP. The plasmids (1–2 μg) were used to inoculate Chenopodium quinoa. The inoculated leaves with local lesions were collected at 7–10 days post inoculation. BaMV were extracted and purified as described previously22.
Cryo–EM and image processing
The sample (3 μL, 1–2μg/μl) was applied to lacey carbon grids that were plasma cleaned (Gatan Solarus) and vitrified in a Vitrobot Mark IV (FEI, Inc.). Grids were imaged in a Titan Krios at 300 keV, and recorded with a Falcon II direct electron detector at 1.05 Å per pixel, with seven “chunks” per image. Each chunk, containing multiple frames, represented a dose of ~ 20 electrons per Å2. A total of 914 images (each 4k x 4k) from the wt and 560 images from the Nd35 sample were selected that were free from drift or astigmatism, and had a defocus less than 3.0 μm. The program CTFFIND323 was used for determining the Contrast Transfer Function (CTF) and the range used was from 0.6 to 3.0 μm. The SPIDER software package24 was used for most subsequent steps. The CTF was corrected by multiplying each image by the theoretical CTF, both reversing phases where they need to be reversed and improving the Signal–to–Noise ratio. The program e2helixboxer within EMAN225 was used for boxing long filaments from the micrographs, and 5,099 and 3,400 such boxes of varying length were generated from the wt and Nd35 samples, respectively. Overlapping boxes, 384 px long with an 8 px shift between adjacent boxes (98% overlap) were extracted from these long filaments, yielding 236,726 segments from the wt and 217,773 segments from Nd35. The CTF determination and particle picking came from the integrated images (all seven chunks), while the segments used for the initial alignments and reconstruction came from the first two chunks.
The determination of the helical symmetry was by trial and error, searching for a symmetry which yielded recognizable secondary structure 18. The IHRSR algorithm26 was used for the helical reconstructions, starting from a solid cylinder as an initial model. Once the correct symmetry was determined (an axial rise of ~ 4.0 Å and a rotation of ~ −40.9° per subunit) a preliminary reconstruction was used for both eliminating segments with large out–of–plane tilt (greater than 9°) and as a basis for sorting by axial rise and twist. The sorting was done by generating reconstructions with variations in both twist and rise, and these were used as multiple references in a classification. The final data sets contained 54,547 segments for the wt and 49,886 segments for the Nd35. After determining that there were no significant differences between the two reconstructions, the sets were combined into a total set containing 104,433 segments. The final reconstruction was generated by imposing the helical parameters found for each segment using the first two chunks on segments containing only the first chunk (~ 20 electrons per Å2) and using these for the back–projection in SPIDER. The Fourier Shell Correlation (FSC) was generated by comparing two completely independent reconstructions: the wt and the Nd35 (Supp. Fig. 2), and the FSC=0.143 criterion27 was used.
Building and refining atomic models into density
Model building began by docking a crystallized fragment from PapMV (PDB id: 4DOX) into the experimental density data using Chimera’s “dock into density” tool. The fragment was truncated to the core residues (residues 33–174) that showed good agreement to the density data. However, this model failed to account for 58 N–terminal residues and 42 C–terminal residues. Continuous density was clearly visible for both the missing N–terminal and C–terminal residues at the outside and the inside of the capsid, respectively.
Initially, these termini were rebuilt using RosettaCM, which combines Monte Carlo sampling of backbone fragments with Cartesian space minimization28. However, due to the length, the relatively low resolution of the local density, and the low apparent secondary structure content of the insertions, backbone conformational sampling in RosettaCM was poorly converged, and the best–scoring models still poorly fit the density, with large segments outside of density and significant amounts of unexplained density remaining. Instead, we used a novel enumerative rebuilding strategy in Rosetta to overcome this sampling issue. Rather then sample the entire backbone segment simultaneously, we iteratively sampled short, three–residue segments of backbone. By only considering three residue segments, we may completely explore the space of backbone conformations given each three amino acid segment. Each iteration, we stored a “beam” containing up to the 50 best solutions; the subsequent iteration attempts to extend each of these solutions, and stores up to the best 50 after the next extension. The density data are used to filter obviously wrong solutions (by throwing out solutions with density agreement significantly worse than the best seen over the same stretch of backbone), and additional filters ensure that models stored each iteration are sufficiently different from one another. Sampling these terminal conformations revealed good convergence of the top–scoring models (Supp. Fig. 4a), when we looked at the final “beam.”
After rebuilding the N– and C– termini, an unbroken tube of density remained unexplained by the models, which presumably corresponded to the single stranded RNA. The resolution of the data was unfortunately insufficient to build RNA models de novo with any degree of confidence. Based on the length of the capsid and genome, we assumed that there were 5 nucleotides in each asymmetric unit of the capsid. We identified a set of structures that had RNA with a similar radius of curvature (PDB ids: 1C9S, 1RMV, 3PDM, 4BKK, and 4H5O), and considered docking and refining every 5–residue segment into the density map. Refinement of the RNA was carried out using the symmetry of the capsid29, with constraints used to ensure that bond geometry was maintained between adjacent asymmetric units. This refinement (25 different RNA stretches) showed that the best agreement to density was observed for residues 3–7 of 4H5O, a crystal structure of rift valley fever virus. The RNA conformation clashed with residues 85–96 using the docked crystal structure, so these were rebuilt with RosettaCM, with the RNA model present.
Finally, all–atom refinement of the symmetric full–length model against the experimental density data was carried out in Rosetta, using a previously described protocol30. A total of 600 refined models were generated. After refinement, the lowest–energy 10 structures were selected and compared. These structures showed relatively tight convergence over most of the structure, with most deviation in the C–terminus; this is unsurprising as this is the region with worst local resolution, possibly due to conformational heterogeneity in this region. All coordinate and B–factor refinement was carried out against the wildtype map reconstruction. The independent Nd35 reconstruction was then used to evaluate overfitting of models to density.
An FSC curve comparing the final model to the wt map (Supp. Fig. 2) shows good agreement between model and map, with FSC=0.5 at a resolution of about 5 Å, in line with the resolution of the data. The agreement of the model with the Nd35 reconstruction shows a similarly good fit, suggesting the refined model is not overfit to the density data.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health GM035269 (to E.H.E), NSC98–2321–B–005–005–MY3 (to Y.H.H), NSC99–2628–B–001–012–MY3 and Academia Sinica (Taipei, Taiwan) (to N.S.L), and Scholarships for Excellent Students to Study Abroad from National Chung Hsing University (Taichung, Taiwan) (to C.C.C). We thank K. Dryden for assistance with the cryo–EM. C.C.C. thanks R. H. Cheng, L. Xing, R. Diaz, Z. H. Zhou and G.G. Liou for their assistance.
Footnotes
Accession Codes
The map and the model have been deposited at the EMDB and the PDB, respectively, with accession codes EMD–3020 and 5A2T.PDB.
References
- 1.Kendall A, et al. Structure of flexible filamentous plant viruses. Journal of Virology. 2008;82:9546–9554. doi: 10.1128/JVI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López–Moya J, García J. Potyviruses (Potyviridae) Encyclopedia of virology. 1999;3:1369–1375. [Google Scholar]
- 3.Chen TH, et al. Induction of protective immunity in chickens immunized with plant–made chimeric Bamboo mosaic virus particles expressing very virulent Infectious bursal disease virus antigen. Virus research. 2012;166:109–115. doi: 10.1016/j.virusres.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Yang CD, et al. Induction of protective immunity in swine by recombinant bamboo mosaic virus expressing foot–and–mouth disease virus epitopes. BMC biotechnology. 2007;7:62. doi: 10.1186/1472-6750-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shukla S, et al. Biodistribution and clearance of a filamentous plant virus in healthy and tumor–bearing mice. Nanomedicine (London, England) 2014;9:221–235. doi: 10.2217/nnm.13.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS biology. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lico C, Chen Q, Santi L. Viral vectors for production of recombinant proteins in plants. Journal of cellular physiology. 2008;216:366–377. doi: 10.1002/jcp.21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernal JD, Fankuchen I. X–Ray and Crystallographic Studies of Plant Virus Preparations. Iii. The Journal of general physiology. 1941;25:147–165. doi: 10.1085/jgp.25.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beijerinck MW. Over een contagium vivum fluidum als oorzaak van de vlekziekte der tabaksbladen. Versl.Gew.Verg.Wis en Natuurk.Afd. 1898;7:229–235. [Google Scholar]
- 10.Namba K, Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986;231:1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- 11.Ge P, Zhou ZH. Hydrogen–bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc.Natl.Acad.Sci.U.S.A. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson JF, Tollin P, Bancroft JB. The architecture of the potexviruses. Virology. 1981;112:34–39. doi: 10.1016/0042-6822(81)90609-7. [DOI] [PubMed] [Google Scholar]
- 13.Kendall A, et al. A common structure for the potexviruses. Virology. 2013;436:173–178. doi: 10.1016/j.virol.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, et al. Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus. J Mol Biol. 2012;422:263–273. doi: 10.1016/j.jmb.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin NS, et al. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. The Journal of general virology. 1994;75:2513–2518. doi: 10.1099/0022-1317-75-9-2513. Pt 9. [DOI] [PubMed] [Google Scholar]
- 16.Lin M, Kitajima E, Cupertino F, Costa C. Partial purification and some properties of bamboo mosaic virus. Phytopathology. 1977;67:1439–1443. [Google Scholar]
- 17.Lan P, Yeh WB, Tsai CW, Lin NS. A unique glycine–rich motif at the N–terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Molecular plant–microbe interactions : MPMI. 2010;23:903–914. doi: 10.1094/MPMI-23-7-0903. [DOI] [PubMed] [Google Scholar]
- 18.Egelman EH. Helical Ambiguities. eLife. 2014;3:e04969. doi: 10.7554/eLife.04969. doi:10.7554/eLife.04969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leaver–Fay A, et al. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung CJ, et al. Two key arginine residues in the coat protein of Bamboo mosaic virus differentially affect the accumulation of viral genomic and subgenomic RNAs. Molecular plant pathology. 2014;15:196–210. doi: 10.1111/mpp.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen HC, et al. The conserved 5' apical hairpin stem loops of bamboo mosaic virus and its satellite RNA contribute to replication competence. Nucleic acids research. 2012;40:4641–4652. doi: 10.1093/nar/gks030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin N-S, Chen C-C. Association of bamboo mosaic virus(BaMV) and BaMV–specific electron–dense crystalline bodies with chloroplasts. Phytopathology. 1991;81:1551–1555. [Google Scholar]
- 23.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 25.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single–particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal PB, Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single–particle Electron Cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Song Y, et al. High–resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiMaio F, Leaver–Fay A, Bradley P, Baker D, Andre I. Modeling symmetric macromolecular structures in Rosetta3. PloS one. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMaio F, et al. Atomic–accuracy models from 4.5–A cryo–electron microscopy data with density–guided iterative local refinement. Nature methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.