Abstract
A genome-wide association study of prostate cancer in Kaiser Permanente health plan members (7,783 cases, 38,595 controls; 80.3% non-Hispanic white, 4.9% African-American, 7.0% East Asian, 7.8% Latino) revealed a new independent risk indel rs4646284 at the previously-identified locus 6q25.3 that replicated in PEGASUS (N=7,539) and MEC (N=4,679) (p=1.0×10−19, OR=1.18). Across the 6q25.3 locus, rs4646284 exhibited the strongest association with expression of SLC22A1 (p=1.3×10−23) and SLC22A3 (p=3.2×10−52). At the known 19q13.33 locus rs2659124 (p=1.3×10−13, OR=1.18) nominally replicated in PEGASUS. A risk score of 105 known risk SNPs was strongly associated with prostate cancer (p<1.0×10−8). Comparing the highest to lowest risk score deciles, the OR was 6.22 for non-Hispanic Whites, 5.82 for Latinos, 3.77 for African-Americans, and 3.38 for East Asians. In non-Hispanic whites, the 105 risk SNPs explained ~7.6% of disease heritability. The entire GWAS array explained ~33.4% of heritability, with a 4.3-fold enrichment within DNaseI hypersensitivity sites (p=0.004).
Keywords: Prostate cancer, genome-wide association study
Introduction
Prostate cancer (PCa) is the second most common cancer diagnosed in men worldwide. PCa family history greatly increases risk; roughly 10–15% of men with PCa have an affected relative (1,2), and familial risk increases roughly two-fold for first-degree male relatives of affected individuals (3). In addition, twin studies indicate PCa is among the most heritable cancers (4–6). It is essential to identify genetic risk factors to fully-characterize disease burden.
Eight years of genome-wide association studies (GWAS) identified at least 105 risk variants for PCa (7–28); most are common with modest effects, and alone explain little heritability. However, an overall genetic risk score combining these variants could be substantially more predictive of disease and explain a reasonable heritability proportion, although there still remain undiscovered loci. How many and where best to search remains unclear.
To further characterize PCa’s genetic basis, we undertook a GWAS using multiple study cohorts within Kaiser Permanente (KP), a fully integrated health care delivery system: the Research Program on Genes, Environment and Health (RPGEH) cohort (dbGaP phs000674.p1), the ProHealth Study, and the California Men’s Health Study (CMHS) (29). This diverse population has not been included in any prior PCa GWAS, allowing for both novel risk variant discovery and assessment of the replication, prediction, and heritability explained by previously-reported risk variants across multiple ethnic populations. We first searched for novel PCa risk SNPs by conducting genome-wide scans using high throughput genotyping arrays optimized for four major race/ethnicity groups—non-Hispanic white, Latino, East Asian, and African-American (30,31)—and meta-analyzing the corresponding results. We then tested the novel findings from this GWAS for independent replication in PEGASUS non-Hispanic whites and African-Americans comprised mostly of the Multiethnic cohort (MEC) (14). Next, we looked at 105 previously-identified risk variants to assess how well they replicate in KP overall and within ethnic subgroups, and their ability to predict PCa. Finally, we evaluated the heritability explained in the largest race/ethnicity group (non-Hispanic whites) by the known SNPs, how much heritability remains unaccounted for, and which genomic regions are most likely to contain additional risk SNPs.
Results
Study population
Table 1 presents descriptive information for KP by PCa status (by study, Table S1), 7,783 men diagnosed with PCa and 38,595 men free from PCa. Approximately 80% were non-Hispanic white, and this ethnic group had a higher case percentage than controls, as did the African-Americans. Most men were over age 60, with an increased percentage of cases diagnosed between ages 60–70. On average, prostate specific antigen (PSA) levels were much higher in cases than controls, as PSA was used for annual PCa screening in KP. Most cases have Gleason scores of six or seven.
Table 1.
Descriptive factors for KP study population used in genome-wide association study of prostate cancer.
| Cases | Controls | |||
|---|---|---|---|---|
|
| ||||
| N | % | N | % | |
|
| ||||
| Total | 7783 | 38595 | ||
| Race/ethnicity: | ||||
| non-Hispanic white | 6406 | 82.3% | 30866 | 80.0% |
| African-American | 601 | 7.7% | 1650 | 4.3% |
| Asian | 288 | 3.7% | 2938 | 7.6% |
| Latino | 488 | 6.3% | 3141 | 8.1% |
|
| ||||
| Age: | ||||
| Age < 50 | 86 | 1.1% | 4619 | 12.0% |
| 50 ≤ Age < 60 | 1224 | 15.7% | 7536 | 19.5% |
| 60 ≤Age < 70 | 3604 | 46.3% | 12240 | 31.7% |
| 70 ≤ Age | 2869 | 36.9% | 14200 | 36.8% |
|
| ||||
| PSA* | ||||
| Mean (SD) | 11.6 | (53.4) | 2.5 | (5.5) |
| Median (MAD) | 6.4 | (3.0) | 1.5 | (1.2) |
| Interquartile Range | 4.8–9.7 | 0.8–2.8 | ||
|
| ||||
| Gleason: | ||||
| ≤5 | 167 | 2.3% | --- | --- |
| 6 | 3067 | 53.8% | --- | --- |
| 7 | 1641 | 33.1% | --- | --- |
| 8 | 292 | 5.8% | --- | --- |
| 9 | 174 | 3.5% | --- | --- |
| 10 | 27 | 0.5% | --- | --- |
PSA is given as the latest measure before diagnosis for cases, and the latest measure available to us from electronic medical records for controls. MAD, median absolute deviation.
GWAS Findings
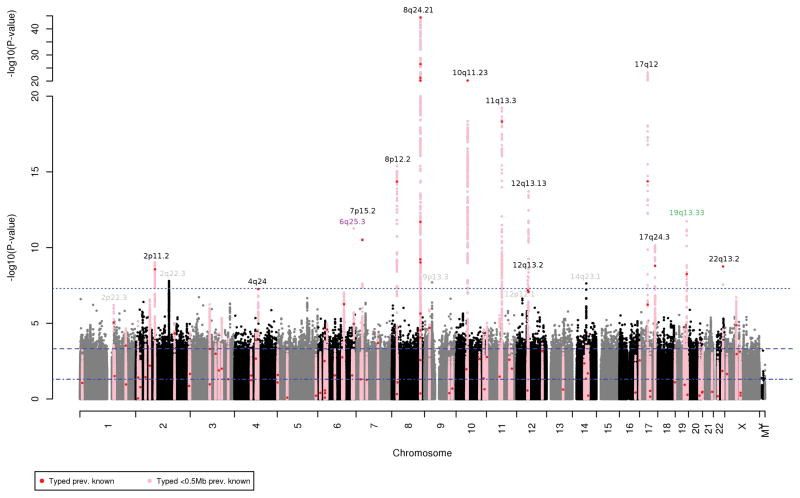
Our GWAS discovery stage detected 16 loci containing Pca-associated variants at genome-wide significance (p<5×10−8) (Figure 1, Table S2). The genomic inflation factor was 1.052, suggesting our findings were not due to systematic bias. Figure S1.1–1.36 shows Manhattan and Q-Q plots for the initial analysis of each race/ethnicity group, in addition to further analyses results conditioning on the genome-wide significant SNPs in KP. Conditional analyses at known loci identified 10 additional independent secondary genome-wide significant variants (Table S2; conditioning round given in first column). Of the 16 original plus 10 conditional genome-wide significant variants, 18 clearly replicated previous GWAS findings (discussed further below). Of the remaining eight variants, four were from our original GWAS: 2q22.3 (rs13016083), 6q25.3 (rs4646284), 14q23.1 (rs34582366), and 19q13.33 (rs2659124); and four were from the conditional analysis: 2p22.3 (rs36004513), 8q24.21 (rs77541621), 9p13.3 (b37 9:33975799), 12p12.1 (b37 12:25430787) (Table S2). While some of these loci contain previously-reported PCa risk SNPs, there was generally limited linkage disequilibrium (LD) between them and the eight significant SNPs that we detected. We assessed whether these eight SNPs were independent novel risk hits by undertaking analyses conditional on known risk SNPs at their corresponding loci and by replication in the PEGASUS and MEC studies.
Figure 1.
Results from a genome-wide association study (GWAS) of prostate cancer in Kaiser Permanente population (8,399 cases and 38,745 controls), highlighting key chromosomal regions. P-values are for variant associations with prostate cancer from trans-ethnic fixed-effects meta-analysis of four race/ethnicity GWAS (non-Hispanic white, Latino, African-American, East Asian), each adjusted for age and ancestry principal components. Horizontal dashed lines indicated genome-wide statistical significance (p<5×10−8), Bonferroni-corrected significance for 105 know prostate cancer risk variants (p<0.05/105), and nominal significance (p<0.05). Red points denote our findings for the 105 known risk SNPs, and pink indicate results for SNPs within a 0.5Mb window around these SNPs. The new 6q25.3 indel rs4646284 is colored magenta, and the 19q13.33 prostate cancer or PSA SNP rs2659124 is noted in green. Those loci containing previously-reported variants that we replicated at genome-wide significance are noted in black text. Loci with variants that were novel in in the KP GWAS but failed to replicate are noted in gray text.
The single base pair indel rs4646284 between SLC22A1 and SLC22A2, the site of a previously-reported locus at 6q25.3 was associated with PCa in the KP, PEGASUS and MEC populations (overall meta OR=1.18, p=1.0×10−19; Table 2a). Within KP, the smallest p-value was observed among the non-Hispanic whites (p=6.7×10−11) as expected in light of it being the largest race/ethnic group. We observed nominal significance in African-Americans (p=0.029), and while not significant in Latinos, the estimated magnitude of rs4646284’s effect on PCa was similar to that in non-Hispanic whites and African-Americans; rs4646284 did not exhibit any association among East Asians (Table 2a). This SNP replicated at genome-wide significance in the PEGASUS study of non-Hispanic whites (p=1.4×10−8) and nominally in the MEC study of African-Americans (p=0.0094).
Table 2.
Prostate cancer genome-wide association study results for new risk indel rs4646284 at 6q25.3 and for risk SNP rs2659124 at 19q13.33. These variants were detected in the KP study population and tested for replication in the PEGASUS and MEC cohorts. (a) Single variant and conditional analyses for 6q25.3 indel rs4646284 and the previous prostate cancer risk SNPs rs9364554 and rs651164. (b) Single variant and conditional analyses for 19q13.33 SNP rs2659124 detected here and the previous prostate cancer risk SNP rs2735839. Meta-analysis is given for the fixed effects (FE) and random effects (RE) analysis. Chromosome positions are given in b37. The risk allele frequency (RAF) is the pooled frequency across both cases and controls.
| (a)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No Conditioning | Conditioning on other 6q25.3 SNPs | ||||||||
|
|
|||||||||
| Variant | Risk Allele | Ref. Allele | Group | Risk AF | OR (95% CI) | p | OR (95% CI) | p | r2info |
| Indel: rs4646284 | TG (indel) | T | KP NH white | 0.298 | 1.17 (1.12, 1.23) | 6.7×10−11 | 1.16 (1.10, 1.23) | 3.3×10−7 | 0.91 |
| KP Latino | 0.250 | 1.13 (0.94, 1.36) | 0.20 | 1.06 (0.85, 1.31) | 0.61 | 0.89 | |||
| KP Asian | 0.324 | 1.03 (0.84, 1.27) | 0.76 | 0.91 (0.68, 1.23) | 0.56 | 0.89 | |||
| KP Af-Amer | 0.365 | 1.18 (1.02, 1.37) | 0.029 | 1.21 (1.03, 1.42) | 0.020 | 0.85 | |||
| KP Meta FE | - | 1.17 (1.12, 1.22) | 5.5×10−12 | 1.15 (1.09, 1.21) | 8.5×10−8 | - | |||
| KP Meta RE | - | 1.17 (1.12, 1.22) | 5.5×10−12 | 1.15 (1.07, 1.22) | 7.7×10−5 | - | |||
| MEC | 0.361 | 1.13 (1.03, 1.25) | 0.0094 | 1.13 (1.03, 1.25) | 0.014 | 0.81 | |||
| PEGASUS | 0.278 | 1.27 (1.17, 1.37) | 1.4×10−8 | 1.20 (1.09, 1.32) | 0.00024 | 0.80 | |||
| Overall Meta FE | - | 1.18 (1.14, 1.22) | 1.0×10−19 | 1.16 (1.11, 1.21) | 5.4×10−12 | - | |||
| Overall Meta RE | - | 1.18 (1.13, 1.23) | 9.8×10−17 | 1.16 (1.11, 1.21) | 5.4×10−12 | - | |||
|
| |||||||||
| Previous SNP: rs9364554 | T | C | KP NH white | 0.279 | 1.10 (1.05, 1.15) | 6.6×10−5 | 1.05 (1.00, 1.10) | 0.070 | 1.00 |
| KP Latino | 0.227 | 1.16 (0.97, 1.29) | 0.1 | 1.13 (0.93, 1.36) | 0.21 | 1.00 | |||
| KP Asian | 0.319 | 1.06 (0.87, 1.29) | 0.59 | 1.05 (0.86, 1.28) | 0.63 | 1.00 | |||
| KP Af-Amer | 0.079 | 1.14 (0.89, 1.47) | 0.3 | 1.11 (0.85, 1.43) | 0.45 | 1.00 | |||
| KP Meta FE | - | 1.10 (1.06, 1.15) | 1.1×10−5 | 1.06 (1.01, 1.11) | 0.019 | - | |||
| KP Meta RE | - | 1.10 (1.06, 1.15) | 1.1×10−5 | 1.06 (1.01, 1.11) | 0.019 | - | |||
| MEC | 0.074 | 1.21 (1.02, 1.41) | 0.024 | 1.19 (1.00, 1.40) | 0.041 | 1.00 | |||
| PEGASUS | 0.293 | 1.22 (1.13, 1.31) | 9.3×10−8 | 1.16 (1.07, 1.25) | 0.00022 | 1.00 | |||
| Overall Meta FE | - | 1.14 (1.10, 1.18) | 6.3×10−12 | 1.09 (1.05, 1.13) | 1.9×10−5 | - | |||
| Overall Meta RE | - | 1.15 (1.09, 1.20) | 6.1×10−8 | 1.10 (1.05, 1.15) | 0.00014 | - | |||
|
| |||||||||
| Previous SNP: rs651164 | A | G | KP NH white | 0.308 | 0.94 (0.90, 0.98) | 0.0088 | 1.01 (0.96, 1.06) | 0.79 | 1.00 |
| KP Latino | 0.380 | 0.88 (0.75, 1.04) | 0.12 | 0.91 (0.76, 1.09) | 0.29 | 1.00 | |||
| KP Asian | 0.515 | 0.91 (0.76, 1.10) | 0.35 | 0.85 (0.65, 1.11) | 0.24 | 1.00 | |||
| KP Af-Amer | 0.264 | 1.02 (0.88, 1.18) | 0.82 | 1.08 (0.93, 1.28) | 0.30 | 1.00 | |||
| KP Meta FE | - | 0.94 (0.90, 0.98) | 0.0037 | 1.00 (0.96, 1.05) | 0.96 | - | |||
| KP Meta RE | - | 0.94 (0.90, 0.98) | 0.0037 | 1.00 (0.93, 1.07) | 0.90 | - | |||
| MEC | 0.307 | 0.98 (0.90, 1.08) | 0.73 | 1.03 (0.93, 1.13) | 0.60 | 1.00 | |||
| PEGASUS | 0.306 | 0.91 (0.84, 0.97) | 0.0065 | 0.99 (0.92, 1.07) | 0.82 | 1.00 | |||
| Overall Meta FE | - | 0.94 (0.91, 0.97) | 0.00019 | 1.00 (0.97, 1.04) | 0.89 | - | |||
| Overall Meta RE | - | 0.94 (0.91, 0.97) | 0.00019 | 1.00 (0.97, 1.04) | 0.89 | - | |||
| (b)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No Conditioning | Conditioning on other 19q13.33 SNPs | ||||||||
|
|
|||||||||
| SNP | Risk Allele | Ref. Allele | Study Group | Risk AF | OR (95% CI) | p-value | OR (95% CI) | p-value | r2info |
| Risk SNP: rs2659124 | T | A | KP NH white | 0.854 | 1.22 (1.14, 1.30) | 2.2×10−9 | 1.13 (1.04, 1.23) | 0.0043 | 0.99 |
| KP Latino | 0.809 | 1.31 (1.07, 1.61) | 0.0099 | 1.36 (1.00, 1.86) | 0.053 | 0.99 | |||
| KP Asian | 0.587 | 1.15 (0.95, 1.40) | 0.15 | 1.03 (0.62, 1.71) | 0.91 | 0.98 | |||
| KP Af-Amer | 0.847 | 1.28 (1.05, 1.57) | 0.013 | 1.29 (1.05, 1.59) | 0.014 | 0.95 | |||
| KP Meta FE | - | 1.22 (1.15, 1.29) | 1.9×10−12 | 1.16 (1.07, 1.25) | 9.2×10−5 | - | |||
| KP Meta RE | - | 1.22 (1.15, 1.29) | 1.9×10−12 | 1.16 (1.07, 1.25) | 9.2×10−5 | - | |||
| MEC | 0.839 | 1.06 (0.94, 1.20) | 0.33 | 1.09 (0.97, 1.23) | 0.16 | 0.89 | |||
| PEGASUS | 0.868 | 1.16 (1.05, 1.28) | 0.0027 | 1.15 (1.01, 1.31) | 0.031 | 1.00 | |||
| Overall Meta FE | - | 1.18 (1.13, 1.24) | 1.3×10−13 | 1.14 (1.08, 1.21) | 4.3×10−6 | - | |||
| Overall Meta RE | - | 1.18 (1.12, 1.24) | 3.0×10−10 | 1.14 (1.08, 1.21) | 4.3×10−6 | - | |||
|
| |||||||||
| Previous SNP: rs2735839 | T | C | KP NH white | 0.150 | 1.20 (1.13, 1.28) | 1.6×10−8 | 1.11 (1.02, 1.23) | 0.016 | 1.00 |
| KP Latino | 0.214 | 1.21 (1.00, 1.47) | 0.052 | 0.97 (0.72, 1.30) | 0.83 | 1.00 | |||
| KP Asian | 0.419 | 1.15 (0.95, 1.40) | 0.15 | 1.13 (0.68, 1.88) | 0.63 | 1.00 | |||
| KP Af-Amer | 0.302 | 1.02 (0.88, 1.18) | 0.70 | 0.98 (0.84, 1.14) | 0.75 | 1.00 | |||
| KP Meta FE | - | 1.17 (1.11, 1.24) | 5.7×10−9 | 1.07 (1.00, 1.15) | 0.056 | - | |||
| KP Meta RE | - | 1.16 (1.07, 1.25) | 0.00012 | 1.07 (1.00, 1.15) | 0.056 | - | |||
| MEC | 0.311 | 0.93 (0.85, 1.02) | 0.11 | 0.91 (0.83, 1.00) | 0.058 | 1.00 | |||
| PEGASUS | 0.140 | 0.90 (0.82. 0.99) | 0.038 | 0.99 (0.88, 1.13) | 0.92 | 0.99 | |||
| Overall Meta FE | - | 0.88 (0.85, 0.92) | 2.0×10−10 | 1.01 (0.96, 1.06) | 0.76 | - | |||
| Overall Meta RE | - | 0.89 (0.84, 0.93) | 3.5×10−6 | 1.00 (0.92, 1.09) | 0.99 | - | |||
Pairwise correlations of these three SNPs in European, Latino, East Asian and African-Americans, respectively (KP data):
| r2rs4646284,rs9364554 | = 0.15, 0.090, 0.00028, 0.017; |
| r2rs4646284,rs651164 | = 0.19, 0.19, 0.53, 0.10; |
| r2rs4646284,rs9364559 | = 0.00047, 0.0088, 0.0018, 0.0010. |
Pairwise correlations of these two SNPs in European, Latino, East Asian and African-Americans, respectively (KP data): r2rs2659124, rs2735839 = 0.50, 0.58, 0.85, 0.055.
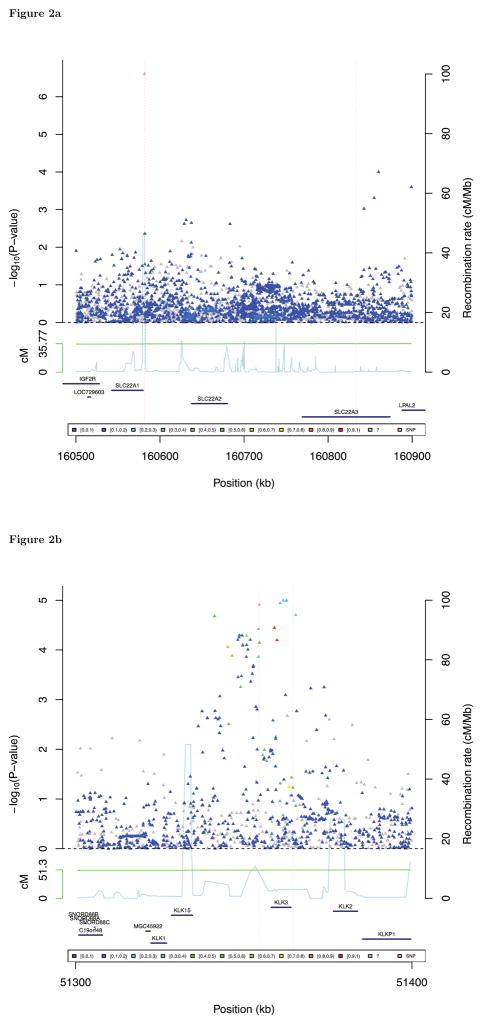
The indel rs4646284 is in weak LD with two previously-reported PCa risk SNPs at 6q25.3: rs9364554 and rs651164 (pairwise r2 of these three SNPs was less than 0.20 in all race/ethnicity groups, except r2rs4646284,rs651164=0.55 in East Asians, see Table 2a). To verify that rs4646284 is associated with PCa independently of these two SNPs, we fit joint (conditional) models containing all three variants. Here, rs4646284 remained strongly associated with PCa overall (OR=1.16, p=5.4×10−12; Table 2a). The conditional analysis slightly weakened the association for rs9364554 (overall p-value went from 6.3×10−12 to 1.9×10−5 after conditioning), and completely attenuated the result for rs651165 (overall p-value decreased from 1.9×10−4 to 0.89 with conditioning). A 6q25.3 regional plot of association p-values conditioning on rs9364554 and rs651165 for KP shows that rs4646284 is a single hit, with very limited surrounding LD and no other strongly associated risk variants nearby (Figure 2a, each race/ethnicity group Figure S2.1–2.24).
Figure 2.
Regional fixed-effects meta-analysis plots from GWAS in Kaiser Permanente population of the two risk variants that replicated: (a) 6q25.3 (rs4646284), (b) 19q13.33 (rs2659124). The color code for the points represents the r2 of each SNP with the risk variant (ranges defined in the legend). The dotted vertical lines pass through the risk variants and the other significant SNPs in the region, on which analyses were conditioned.
Our cis-eQTL analysis detected an association between rs4646284 and decreased expression of SLC22A1, SLC22A2, and SLC22A3 in prostate tissue (Table S3). In the Mayo Clinic data we observed extremely significant associations between the rs4646284 insertion and lower expression of SLC22A1 (effect size=−0.42, p=1.3×10−23) and SLC22A3 (coefficient=−0.68, p=3.2×10−52). Regional eQTL analysis of all variants within 1.1Mb of these two genes—including previously-reported PCa risk SNPs—indicated that the rs4646284 indel was clearly the strongest predictor of expression at SLC22A1 and SLC22A3 in the Mayo Clinic samples, with eQTL p-values for surrounding SNPs orders of magnitude larger (Figure S3.1–3.2). Our replication analyses in the PHS+HPFS normal/tumor and TCGA tumor tissues also showed reduced expression in SLC22A1 (overall coefficient=−0.07, one-sided p=0.046) and even more so in SLC22A3 (overall coefficient=−0.32, one-sided p=0.0012). The PHS+HPFS normal tissue drove the limited replication for SLC22A1 expression (Table S3); in that study the expression array did not perform very well for SLC22A1 (90% expression mark=3.4, where >5 is desirable). This allele may also be associated with lower expression of lipoprotein(a)-like 2 (LPAL2), a pseudogene structurally similar to the gene that encodes lipoprotein(a) (LPA), but produces mRNA with a premature stop codon (32). Taken together, these results indicate rs4646284 is an independent risk indel for PCa that improves upon the previously-reported findings for the 6q25.3 locus.
Another genome-wide significant SNP in KP, rs2659124 at 19q13.33, replicated in the PEGASUS study. This SNP is near the 5′ UTR terminus of KLK3 (kallikrein-related peptidase 3), which encodes PSA. The KP meta-analysis yielded p=1.9×10−12, with similar ORs observed in the non-Hispanic whites, African-Americans, and Latinos (Table 2b). rs2659124 replicated in PEGASUS with p=0.0027 (overall pooled p=1.3×10−13, OR=1.18). The rs2659124 association was slightly attenuated but remained suggestive (pooled p=4.3×10−6) after adjusting for the previously-known risk SNP at 19q12.33 (rs2735839). In contrast, the conditional analysis completely attenuated the rs2735839 association with PCa (Table 2b). Figure 2b gives a regional plot for this locus, showing that rs2735839 is the strongest risk SNP.
The remaining six genome-wide significant SNPs in our cohort did not clearly replicate in PEGASUS or MEC (Table S2). With regard to their PCa effects, rs13016083 in 2q22.3 had KP meta-analysis OR=1.13, but PEGASUS OR=1.03 and MEC OR=0.99. For the SNP 9:33975799, the KP OR=0.86 but PEGASUS OR=1.05 and MEC OR=0.94. rs34582366 had KP OR=1.32 but PEGASUS and MEC had ORs in the opposite direction (0.90 and 0.96, respectively). The SNP 12:25430787 had KP OR=0.74 but the effect was in the opposite direction in PEGASUS, OR=1.10; this SNP had too low an allele frequency among African-Americans to be tested in MEC. rs360054513 had KP OR=0.58 but MEC OR=0.98, and was too rare in non-Hispanic whites to test in PEGASUS. Finally, rs77541621 remained significant after conditioning on all 12 8q24 loci in KP non-Hispanic whites (OR=0.61) (it was very rare in African-Americans and Asians). In the PEGASUS cohort, it was genome-wide significant before conditioning (OR=0.51, p=8.6×10−10), and was nominally significant after adjusting for the 12 8q24 loci (OR=0.68, p=0.02). However, since we conditioned on 12 SNPs here, some which were imputed, we suspect this result may reflect incomplete tagging of existing 8q24 loci.
Replication of Known GWAS Results
The remaining 18 of 26 genome-wide significant associations were clear replications of previously-reported findings at 2p11.2, 3p11.2, 4q24, 6q22.1, 7p15.2, 8p21.2, 8q24.21, 10q11.23, 11q13.3, 12q13.12, 12q13.13, 17q12, 17q24.3, 19q13.33, and 22q13.2 (Figure 1) (7–9,9–16,24–28). Our 18 lead risk SNPs did not appear to improve upon or exhibit independence from those previously-reported. Table S2 gives the correlation between these SNPs in non-Hispanic whites.
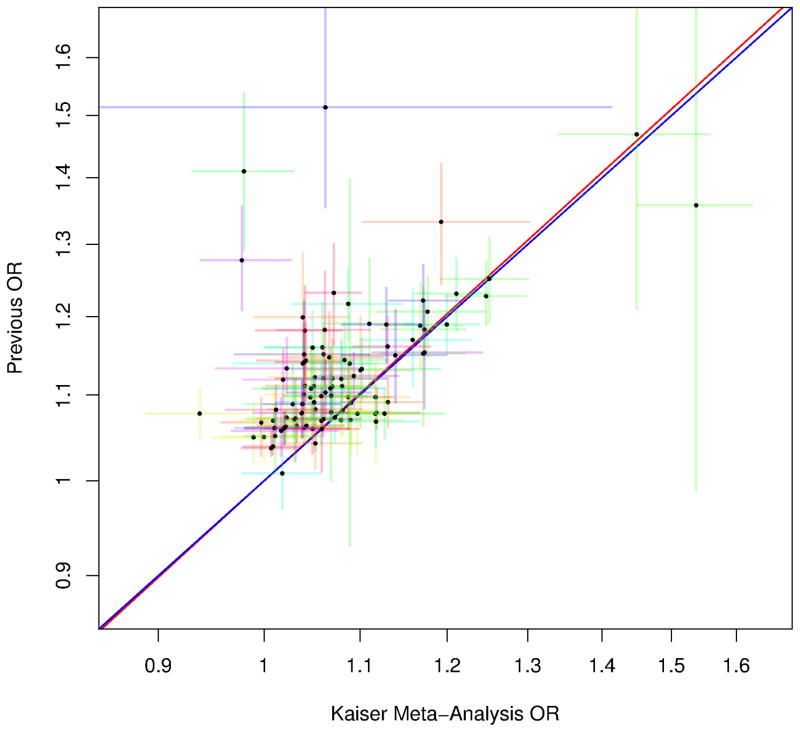
Table S4 presents ORs and p-values for the associations between PCa and these 18 SNPs plus the other previously-reported variants (105 SNPs total) from the previous reports (16) and from KP. These 105 variants exhibited high replication based on the magnitude and direction of their associations with PCa. In particular, we observed excellent agreement between the ORs for variant associations from previous reports and those from the KP GWAS (Meta-analysis in Figure 3; each race/ethnicity group in Figure S4). A large majority of the ORs were larger in previous studies than observed in ours; nevertheless, we saw extremely high agreement: the slope of a line fit to the ORs was almost identical to 1 (Figure 3). Moreover, within ethnic groups, 99/103 of the non-Hispanic white, 67/99 of East Asian, 79/102 of Latino, and 78/105 of African-American ORs were in the same direction as previously found (i.e., ORs>1.0). Only four of the variants with ORs<1.0 were statistically significant: three for Asians; and one for African-Americans.
Figure 3.
Comparison of variant associations from previous reports and results from the KP GWAS meta-analysis for 105 known prostate cancer risk SNPs. Plotted values are the log odds ratios from the previous and current studies. Error lines denote the 95% CIs for the respective studies, colored by chromosome. Blue line is the diagonal and red is regression fit through the points, showing extremely high correlation between the previous and new odds ratios. The SNP rs116041037 appears to be an African-American specific SNP and is an outlier so we left it off of the plot to more easily view the other points; the SNP had a previous OR=2.45 (95% CI=1.65–3.62), and KP OR=2.67 (1.96, 3.64).
The meta-analysis p-values were less than 0.05 for 66 of the 105 SNPs (62.9%) and less than the Bonferroni-corrected alpha-level of 0.00048=0.05/105 for 33 (31.4%). When stratified by ethnic groups, most SNPs that had p-values less than these alpha levels did so in non-Hispanic whites, as expected since this population has been most commonly used for discovery efforts and is the largest ancestry group in the KP study. In particular, the known risk variants had p<0.05 for 62 SNPs (59.0%) in non-Hispanic whites, 11 (10.5%) in Latinos, 13 (12.4%) in East Asians, and 13 (12.3%) in African-Americans. The known risk variants had p<0.00048 for 30 SNPs in non-Hispanic whites, zero in Latinos, one (1.0%) in East Asians, and two (1.9%) in African-Americans.
Prediction with GWAS SNPs
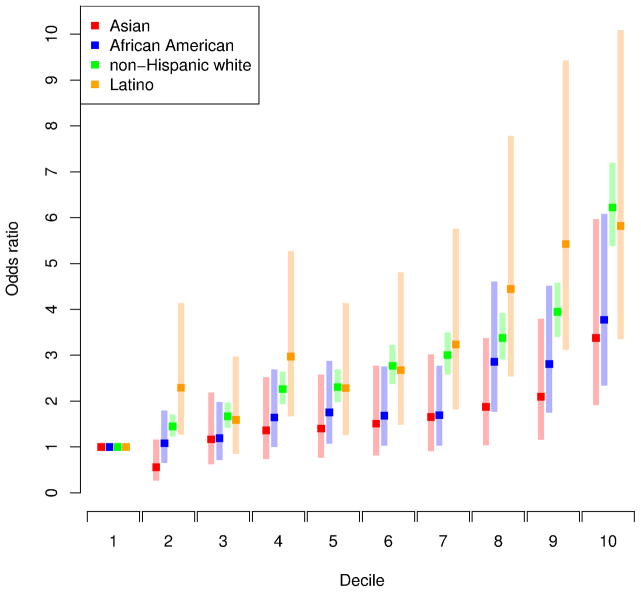
We used risk profile scoring to assess the predictive value of the 105 known GWAS risk SNPs for PCa in KP (which is independent of the populations in which these risk SNPs were discovered). We combined the 105 SNPs into a single score by applying the previously estimated ORs to our genotype data (details in Methods). The risk score was highly statistically significant for all four major ethnic groups: non-Hispanic white p=1.0×10−211, Latino p=3.5×10−16, East Asian p=1.0×10−8, and African-American p=1.1×10−15. To see how the magnitude of the associations with risk scores varied across race/ethnic groups, we calculated PCa ORs for corresponding to increasing deciles of risk scores, using the lowest decile as the referent (Figure 4). All four race/ethnic populations exhibited clear trends of increasing risk score ORs across increasing deciles, but the non-Hispanic white and Latino groups had substantially higher ORs than the African-Americans, and the East Asians always had the lowest ORs (Figure 4). Comparing the highest to lowest risk score deciles, the association with PCa for non-Hispanic whites OR=6.22 (95% CI=5.38–7.19), for Latinos OR=5.82 (95% CI=3.36–10.1), for African-Americans OR=3.77 (95% CI=2.34–6.08), and for East Asians OR=3.38 (95% CI=1.91–5.97). While the risk score was highly predictive across groups, it was much less predictive for the African-Americans and East Asians. This may in part reflect lower LD in these ethnic groups.
Figure 4.
Impact of increasing deciles of risk profile scores based on 105 known risk SNPs across different race/ethnicity groups. Risk scores generated by combining the 105 SNPs into a single score, applying the logs odds ratio estimated by previous studies to our KP study population genotype data. Odds ratios for effect of risk profile scores on prostate cancers calculated within each decile of the scores, using the lowest decile of risk profile scores as the referent category. The non-Hispanic white and Latino groups had substantially higher ORs than the African-Americans, and the East Asians always had the lowest ORs.
We also wanted to see how well we could predict PCa using the genetic risk score versus other covariates (i.e., body mass index (BMI), age, ancestry principal components (PCs)). For each race/ethnicity group, we split the cohort in half for training/testing to estimate the non-genetic covariate coefficients. Results for different combinations of genetic risk score and covariates are shown in Figure S5. Relative to the other covariates alone, we generally observed an increase in AUC of approximately 5% with the genetic risk score. Table S5 presents the variance in PCa explained by BMI, age, and ancestry covariates, compared to that explained by also including the risk score. For non-Hispanic whites, including the risk score increased the variance explained from 0.077 to 0.127. The increase was similar in the other groups, though the overall variance explained was lowest in African-Americans. Ignoring the covariates and restricting the risk score to only include those SNPs with nominal (p<0.05), replication-wide (p<0.00048), and genome-wide (p<5×10−8) significance level in the non-Hispanic whites gave variance explained of 0.122, 0.122, and 0.114 respectively.
Heritability: GWAS Array and Functional Categories
We calculated the narrow sense heritability (i.e., the additive genetic component of the phenotypic variance) explained by variants typed and tagged by the GWAS array in the non-Hispanic whites. The estimated heritability for the genotyped SNPs was h2=0.201 (standard error, SE=0.041). When including the imputed SNPs this substantially increased to h2=0.334 (SE=0.060). When we partitioned the data into the 105 previously-known genotyped SNPs versus the rest of the genome, the heritability estimates were 0.076 (SE=0.012) and 0.215 (SE=0.058), respectively. These approximately sum to the entire study’s heritability, and are close to the increase in variability explained.
We then calculated the narrow sense heritability explained by variants within functional regions of the genome, and estimated whether certain regions explained a disproportionate amount of this heritability in comparison with their size (33). We found that the DNaseI hypersensitivity sites (DHSs) exhibited 4.3-fold increased enrichment (p=0.0039) and the intergenic regions had a 0.2-fold decreased enrichment (p=0.0058) (Table 3). We also found non-significant enrichment in coding (4.6-fold), UTR (3.7-fold), and intronic (0.4-fold) regions (Table 3). These results suggest that the SNPs underlying PCa are more likely located in the coding, DHS, and UTR regions, less likely located in the promoter regions, and least likely located in the intronic and intergenic regions (Table 3). Looking at the previously-reported risk SNPs (102 autosomal) we found a similar but slightly weaker pattern of enrichment: coding, 2.4-fold; UTR, 0.7-fold, DHS, 2.5-fold; intron, 0.7-fold; and intergenic, 0.7-fold. For the two key variants reported here, rs4646284 is in a DHS and rs2659124 is intergenic. Note that the smaller percentage of the genome in the promoter category observed here, in contrast with (33) reflects our use of a more accurate promoter definition from the Eukaryotic Promoter Database new v003 (34).
Table 3.
Array heritability enrichment of the coding, UTR, promoter, DHS, intron, and intergenic partitions for non-Hispanic whites using the imputed data. Results are from fitting each partition separately (univariate), and then subsequent results are from the joint multivariate fit, Kaiser Permanente Study Population.
| Univariate | Multivariate fit | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Region | hg2 (SE) | hg2 (SE) | % hg2 | # SNPs | % Genome | Enrichment (95% CI) | Enrichment P |
| Coding | 0.029 (0.014) | 0.013 (0.014) | 3.9% | 18,608 | 0.85% | 4.6 (0.0, 13.5) | 0.45 |
| UTR | 0.048 (0.019) | 0.016 (0.020) | 4.7% | 27,778 | 1.27% | 3.7 (0.0, 12.2) | 0.56 |
| Promoter | 0.001 (0.003) | 0.001 (0.003) | 0.2% | 3.347 | 0.15% | 1.3 (0.0, 10.8) | 0.94 |
| DHS | 0.299 (0.049) | 0.243 (0.065) | 70.9% | 361,426 | 16.56% | 4.3 (2.0, 5.6) | 0.0039 |
| Intron | 0.193 (0.045) | 0.039 (0.055) | 11.5% | 699,518 | 32.04% | 0.4 (0.0, 1.2) | 0.14 |
| Intergenic | 0.199 (0.051) | 0.030 (0.060) | 8.7% | 1,072,335 | 49.12% | 0.2 (0.0, 0.8) | 0.0058 |
| Total | 0.334 (0.060) | 0.342 (0.060) | 100.0% | 2,183,012 | 100.00% | 1.0 | - |
Discussion
In this large, ethnically diverse, and previously unstudied cohort, we detected two risk variants for PCa: an indel (rs4646284 at 6q25.3) and a SNP that may be involved with PSA (rs2659124 at 19q13.33). We also replicated a large majority of the known risk SNPs, which taken together as a risk score in a polygenic model were very strongly associated with PCa, albeit with substantial variation across ethnic groups. In addition, we estimated that approximately 65% of the heritability assayed by the GWAS array remains unexplained by the 105 known risk SNPs, and that heritability is overall enriched in the DHS regions. This indicates that substantial genetic variation in PCa remains to be uncovered.
The novel intergenic indel we identified (rs4646284) is in a recombination hotspot, and is positioned roughly 1.8kb downstream of the 3′ UTR terminus of SLC22A1 and 56kb upstream from SLC22A2. Previous work identified other PCa susceptibility variants at 6q25.3, including rs9364554 within intron 5 of SLC22A3 (9) and, 250kb upstream, rs651164 outside the 3′UTR of SLC22A1 (9); these SNPs appear to reflect independent susceptibility alleles (23). The rs651164 risk SNP is only 170 base pairs from the novel indel (rs4646284). In our study, joint models including all three variants at 6q25 indicated that rs4646284 is more explanatory than the other two risk SNPs (rs651164 and rs9364554). Furthermore, our cis-eQTL analysis found that the rs4646284 indel was strongly associated with decreased expression of SLC22A1 and SLC22A3, substantially more than any other variant at the 6q25 locus, including the two previous GWAS-identified PCa risk SNPs. This rs4646284 indel is within binding site signals from ENCODE ChIP-seq data for several transcription factors, and appears closer than rs651164 to a local binding site peak for c-Myc across several cell lines and to peaks for CTCF in both androgen treated and androgen untreated prostate adenocarcinoma (LNCaP) cells (35, 36). While rs4646284 is a common (insertion frequency ~30% in non-Hispanic whites) and highly statistically significant risk variant, previous work may not have detected this indel because it is not in HapMap and is in low LD with neighboring SNPs, making it difficult to cover with early-generation genotyping arrays. Our finding highlights the importance of studying indels in GWAS and that these can be imputed with high confidence with appropriate reference panels.
The suggestive SNP rs2659124 is 3.3kb from the 5′ UTR terminus of KLK3, which encodes PSA and is one of 15 kallikrein genes located sequentially on 19q13.33-41. rs2659124 is 10kb away from rs2735839, a previously-reported risk SNP for both PCa (9) and PSA levels (35,36). rs2659124 and rs2735839 are on the opposite sides of KLK3, and the latter SNP is <1kb from the 3′ UTR terminus (r2=0.50 between these SNPs in KP non-Hispanic whites). There is evidence that the association between rs2735839 and PCa risk may depend on PSA levels (37) or disease aggressiveness (35,38). rs2659124 was previously-reported as part of a fine mapping study of the KLK3 locus conducted among men from the Cancer Genetic Markers of Susceptibility Project (CGEMS)—which is included in the PEGASUS sample—with nominal statistical significance (p=0.02) (35).
We replicated a large proportion of the 105 known risk SNPs, especially when considering the ancestry group in which the SNPs were discovered. The “winner’s curse,” that effect sizes are often larger in the populations in which they are discovered, may be one reason why some SNPs failed to replicate, and why ORs were generally smaller in our cohort than previously found (39). It is also possible that control misclassification yielded more conservative estimates at these and all SNPs. Nevertheless, the aggregate risk scores combining information across all 105 known SNPs were highly significant in all ethnic groups, although the magnitude of this association varied substantially by ethnicity. The variance in PCa risk explained by SNPs in KP increased as we relaxed the SNP inclusion criteria and incorporated larger numbers of variants: r2=0.112 for the risk SNPs exhibiting genome-wide significance; r2=0.122 for those replicating; and r2=0.127 for all 105 previously-reported risk SNPs. We also showed an increase of roughly 5% in the AUC when adding a genetic risk score to BMI, age, and PCs of ancestry. Finally, we showed that these aggregate risk scores had large ORs when comparing the upper to lower deciles. In these results, East Asians and African-Americans had much lower ORs for the highest decile than the non-Hispanic whites and Latinos. This may be due to different allele frequencies, LD patterns, or causal risk alleles in these populations, since the previous discovery cohorts were largely of European ancestry (Table S4). For example, African-Americans and East Asians have higher average absolute risk allele frequency differences from non-Hispanic whites (0.16 and 0.15, respectively) than Latinos (0.05). As more information becomes available for ethnic-specific PCa risk SNPs, ethnic-specific risk scores should improve prediction.
As expected, our estimated heritability among non-Hispanic whites (0.334, SE=0.060) was lower than that from twin studies (0.58) (5), but higher than previous array heritability estimates (0.204, SE=0.056) that used a slightly smaller PCa lifetime risk of 0.09 (40). The heritability estimate for the 105 known risk SNPs was 0.077 (SE=0.058); there is still an estimated heritability of 0.215 (SE=0.058) that remains unexplained. Our heritability estimate is unbiased from winner’s curse since the known risk SNPs were previously discovered. This heritability appears enriched within the DHS regions, and possibly in the coding, UTR, and promoter regions. These enrichment result pattern is roughly similar to those found for the non-autoimmune phenotypes in (33). Further refining these regions may be a promising area of future research.
In summary, we were able to detect new risk variants for PCa, confirm many previously-reported associations at various levels across ethnic groups, and provide independent evidence that additional risk SNPs are still to be found. Since a large amount of narrow-sense genetic array heritability remains to be explained, larger analyses or meta-analyses may uncover further genetic variants associated with disease. Additional advances may be possible by applying to existing data novel analytic approaches such as Bayesian models that incorporate local heritability estimates or prior biological knowledge, or by undertaking scans for pleiotropic effects that leverage data across multiple cancers. Additionally, we showed that the existing GWAS results are robust and predictive of PCa risk, which may have important implications for using risk SNPs to guide individualized screening for this common but complex disease.
Materials and Methods
Participants, Phenotype, and Genotyping
Our primary analyses used cases and controls from three KP studies: RPGEH, ProHealth, and CMHS. RPGEH participants included men in the RPGEH Genetic Epidemiology Research on Aging (GERA) cohort (dbGaP phs000674.v1.p1), as well as PCa cases with a DNA sample in the RPGEH biorepository but who were not part of GERA. These studies have been previously described (29–31,41) (dbGaP phs000674.v1.p1). Briefly, the ProHealth study focused on ascertaining KP Northern California African-American PCa cases. The CMHS is a prospective cohort study of KP California men. PCa cases in these cohorts were identified from the KP Northern California Cancer Registry (KPNCCR), the KP Southern California Cancer Registry (KPSCCR) or through review of clinical electronic health records through the end of 2012. The Cancer Registries capture data on all PCa cases newly diagnosed or treated at KP facilities. The Cancer Registries conform to standards of the North American Association of Central Cancer Registries and the National Cancer Institute’s Surveillance, Epidemiology and End Results (SEER) program. Controls were all men in GERA without PCa diagnosis; who could have had other cancers. Our analyses included 7,783 cases and 38,595 controls after exclusions described here. These men were genotyped at over 650,000 SNPs on four race/ethnicity-specific Affymetrix Axiom arrays optimized for individuals of European (EUR), African-American (AFR), East Asian (EAS), and Latino (LAT) race/ethnicity. Specific details of array designs including estimated genome-wide coverage have been previously published (30,31). Briefly, the proportion of common (MAF>0.05) 1000 Genomes SNPs (Interim Phase I release, 1,092 subjects) covered by the genome-wide array with r2>0.8 equals 0.93, 0.89, 0.93, and 0.95 for non-Hispanic whites, African-Americans, Asians, and Hispanics, respectively; the proportion of less common (0.01<MAF≤0.05) variants covered is 0.73, 0.65, 0.80, and 0.80, respectively. The arrays were designed using human genome b36, but the probesets were remapped to b37, used for all SNP locations reported here. For the EUR and EAS arrays, we used dbSNP v130, and for the LAT and AFR arrays we used dbSNP v132. All of the EAS and most of the EUR arrays (more details below) were processed using the Axiom v1 reagent kit, while the other arrays were processed on the Axiom v2 reagent kit. We note that a small number of participants in CMHS from Southern California overlap with MEC because these studies had overlapping geographic areas of recruitment (14,42). We identified and removed 107 subjects who overlapped between these studies using a set of 1,000 random non-AT/GC SNPs with MAF>5%. We also excluded any first-degree relatives based on the same analysis. The Kaiser Permanente and University of California Institutional Review Boards approved this project. Informed Consent was obtained, and the studies were conducted in accordance with the Declaration of Helsinki.
GWAS Pre-imputation Quality Control
Genotype quality control (QC) procedures for the original GERA cohort assays were performed on an array-wise basis, as described previously (dbGap phs000674.v1.p1). This process was repeated including ProHealth, CMHS, and additional genotyped individuals not in the originally genotyped GERA cohort. Because we genotyped an additional set of individuals who were much more enriched for cases than the original GERA cohort, we additionally employed the following filters to control for batch effects: we removed SNPs with minor allele frequency (MAF)<0.01, call rate <95%, or Hardy-Weinberg equilibrium (HWE) p-value among homogeneous control groups <1×10−5. This completed QC for EAS and LAT, resulting in 653,943 and 678,790 SNPs, respectively. EAS individuals were genotyped using the same reagent kit, as were LAT individuals. In addition, the cases and controls were randomly distributed among the genotype packages to control for any potential batch effects.
On the EUR array, 3,843 men were run on a different reagent kit (Axiom v2 versus v1). To adjust for any potential kit effects, we undertook GWAS of the association between each SNP and reagent kit separately among cases and controls, adjusting for PCs. We removed kit-associated SNPs (p<1×10−4). We also genotyped each of the new PCa sample plates (those not genotyped with the original set of GERA individuals) with 12 other plates from the originally genotyped GERA cohort, and removed SNPs with >13/1,268 (1%) mismatches. This resulted in 604,255 SNPs.
Similarly, we addressed potential plate batch issues for 1,308 men genotyped with the AFR array through a GWAS of the association between SNPs and whether a subject was typed in the originally genotyped GERA cohort versus later in additional PCa batches separately among cases and controls, adjusting for PCs. Here, we removed batch-associated SNPs at p<0.05. We used a stronger batch filter threshold on the AFR array than on the EUR array because fewer individuals were analyzed on the AFR array, resulting in lower power to detect batch effects. As before, we also genotyped the individuals in packages with just the new plates, re-genotyped them with some of the previous plates, and removed SNPs with >2/78 (2.6%) mismatches. This resulted in 568,496 SNPs.
We were able to accurately impute (see below) many SNPs removed by the QC steps. Specifically, of those genotyped SNPs that failed QC (MAF>0.01), we imputed with accuracy r2≥0.3 (and r2≥0.8) a total of 58,333/63,863 or 91.3% (82.3%), 44,390/51,180 or 87.5% (77.6%), 30,273/35,765 or 84.6% (58.2%), and 305,303/312,202 or 97.8% (91.5%) of the SNPs among non-Hispanic whites, Latinos, East Asians, and African-Americans, respectively. The larger decrease in coverage with higher r2 values for East Asians may reflect having designed the array with only a greedy SNP selection as opposed to the hybrid greedy/imputation-based approach of the AFR and LAT arrays. Although the EUR array was also designed with exclusively greedy SNP selection, this population may have higher r2 than EAS because of a larger sample size (giving more accurate phasing) and stronger LD.
GWAS Genomic Imputation
Imputation was also performed on an array-wise basis. First genotypes were pre-phased with Shape-it v2.5 (43), including cryptic relatives to improve phasing. Variants were then imputed from the 1000 Genomes Project October 2014 release with 2,504 samples with singletons removed (which impute terribly) and as a cosmopolitan reference panel with Impute2 v2.3.1 (44–46). The estimated QC rinfo2 metric given here is the info metric from Impute2, which estimates the correlation between the true and imputed genotype (47). Our GWAS analysis used 10,109,774; 9,283,528; 10,776,138; and 17,141,436 SNPs with r2≥0.3 and MAF≥0.01 for non-Hispanic white, East Asian, Latino, and African-American men, respectively (19,977,088 unique SNPs).
GWAS Analysis and Covariate Adjustment
We first analyzed each of the four race/ethnicity groups (non-Hispanic White, Latino, East Asian, and African-American) separately. Within these groups, each SNP was modeled using additive dosages to account for imputation uncertainty, which works well in practice (48). Each SNP association with Pca was tested via a logistic regression model adjusting for age and ancestry (described below). Age is given at diagnosis for cases, and at last PSA measured for controls. For computational efficiency, we initially regressed the phenotype on all covariates excluding the SNP. We then computed the sum of the estimated beta coefficients times the original covariates to create a single covariate, and tested each SNP in a logistic regression model with this single covariate.
To adjust for genetic ancestry, we performed PC analysis using Eigenstrat v4.2 (49) on each of the four race/ethnicity subgroups. We used a subset of 28,174 SNPs with CR>99% common to all arrays (dbGaP phs000674.p1). For the largest race/ethnicity group (non-Hispanic whites), we performed the PC analysis on 20,000 random individuals, projecting the remaining individuals into the same space, as has been shown to work very well in practice (dbGaP phs000674.v1.p1). The PC analysis are nearly identical to that previously been shown for GERA (dbGaP phs000674.p1). The top 10 eigenvectors were included in each logistic regression model. The genomic inflation factor (50) was very modest for all GWAS analyses (all ≤1.065; exact values given for each analysis in Figure S1.1–1.36).
We undertook both random effects (RE) and fixed effects (FE) meta-analysis to combine the results of the four race/ethnicity groups using Metasoft (51). Then we assessed whether conditioning on the observed genome-wide significant results highlighted additional significant findings. Here, we used results from the meta-analysis to group clumps of genome-wide significant SNPs (p<5×10−8) and within 1Mb of another GWAS significant SNP. We chose the most significant SNP in each clump, and completely reran the full genome-wide analysis, adjusting for these SNPs. We iterated these conditional analyses until no additional genome-wide significant SNPs were found. We also looked for additional independent SNPs at loci that previously-known to be associated with PCa. We searched in a 1Mb window around known SNPs at 87 loci, constituting 2.9% of the genome, and so adjusted for multiple comparisons by 5.8×10−8×2.9%=2.1×10−6, but no additional loci were found.
Replication of SNPs in PEGASUS and MEC Cohorts
To determine if any of the eight new genome-wide significant PCa associations from KP replicated, we evaluated them using independent data from PEGASUS and what we refer to as and consists mostly of MEC plus other African-American studies (14)up (dbGaP phs000306.v3.p1). PEGASUS included 4,599 PCa cases and 2,940 controls of non-Hispanic white race/ethnicity, genotyped using Illumina HumanOmni2.5 and imputed using 1000 Genomes Project Phase I data (1,092 individuals). The PEGASUS replication analyses adjusted for statistically significant ancestry PCs. MEC included 2,265 cases and 2,414 controls of African-American race/ethnicity, genotyped using the Illumina Infinium 1MDuo (dbGap phs000306.v3.p1) and imputed using the same 1000 Genomes reference panel as by KP. The MEC replication analyses adjusted for the first 10 ancestry PCs. All individuals from these replication cohorts were independent of KP.
Confirmation of Imputed rs4646284 Indel
We used two approaches to validate that we correctly imputed the rs4646284 variant. First, we Taqman genotyped (Life Technologies) 352 individuals who were also genotyped on our EUR array and imputed with our EUR individuals. We computed the correlation with the imputed genotype, and a bias-corrected and accelerated bootstrap CI (99,999 iterations). The genotyped indel showed high agreement with the imputed indel (r2=0.81, 95% CI=0.75–0.86). Second, we subsetted the 1000 Genomes Project data to the SNPs on the EUR array, and imputed the rs4646284 indel in a leave-one-out manner as described in (31), using the 1000 Genomes Project data as a reference. We then computed the correlation between what was genotyped in 1000 Genomes and this imputed value. This also exhibited high agreement between the actual and imputed indel genotypes (r2=0.84).
eQTL Analysis of Indel and 6q25 Locus
We evaluated the potential effect of the novel risk indel rs4646284 on expression of neighboring genes and pseudogenes (IGF2R, LOC729603, SLC22A1, SLC22A2, SLC22A3, LPAL2, and LPA) in normal and cancerous prostate tissue. This eQTL analysis was undertaken in three studies. First, the Mayo Clinic included normal prostate tissue from 471 men with Gleason ≤7 disease undergoing radical prostatectomy or cystoprostatectomy (52). Surgical hematoxylin and eosin (H&E) sections from fresh frozen materials were reviewed to identify normal (non-cancerous) tissue samples and RNA-seq data was obtained with an Illumina HiSeq 2000. The second eQTL analysis included prostate tumor tissue samples from 99 men and normal prostate tissue from 56 men with incident PCa who participated in the Physicians’ Health Study (PHS) and Health Professionals Follow-up Study (HPFS) (NCBI GEO GSE62872) (53) using genotype data from the Breast and Prostate Cancer Cohort Consortium (BPC3) aggressive PCa GWAS (23). Prostate tissue was collected from archival trans-urethral resection or prostatectomy specimens (formalin-fixed paraffin-embedded). mRNA expression was assayed using the Affymetrix GeneChip Human Gene 1.0 ST microarray. The third study included prostate tumor tissue from 128 participants in The Cancer Genome Atlas (TCGA) (NCBI GEO GSE21032) (54) using Illumina HiSeq 2000 mRNA expression data and genotypes from matched normal using the Affymetrix SNP 6.0 array (55). All three studies successfully imputed the rs4646284 indel for the eQTL analyses (Mayo rallelic2=0.71, PHS rinfo2=0.87, HPFS rinfo2=0.89, TCGA rinfo2=0.84), and used linear regression on the dosage.
Replication Analysis of Previously Detected SNPs
To determine whether the 105 previously-reported variants associated with PCa replicated in our cohort, we used a nominal significance level (0.05) and a Bonferroni-corrected alpha level of 0.05/105=0.00048. We computed the retrospective power with the R package GeneticsDesign (56), using the previously-reported ORs, numbers of cases and controls in KP, alpha level of 0.05/105=0.00048, the r2 estimate from imputation, and a PCa prevalence=0.12, the KP GERA cohort prevalence.
Risk Scores
We constructed a risk score for each man by summing up the additive coding of each SNP previously associated with PCa weighted by the previously-reported log(OR) from (16). In the non-Hispanic white, Latino, and African-American race/ethnicity groups, no two SNPs had r2>0.3, so all were used. In the East Asian race/ethnicity group, rs116041037 and rs7210100 had r2=0.87, and rs1016343 and rs6983562 has r2=0.55; the latter SNP was removed in both circumstances when computing the risk score. To estimate the variance explained by these risk scores, we report Nagelgerke’s pseudo-r2 estimate (57).
GWAS Array Heritability
We estimated the additive array heritability using GCTA v1.24 (58). Array heritability estimates can be more sensitive to artifacts than GWAS results (58). Thus, to limit any potential batch effects, we limited this analysis to the homogeneous group of 30,598 non-Hispanic white men (3,605 cases and 26,993 controls) genotyped with the EUR array with Axiom v1 reagent kit. We also employed a stronger set of filters than used in the GWAS (58). Specifically, in addition to the filters noted above, we excluded SNPs with: HWE p<0.05 (in controls); significant differences in case-control missingness (p<0.05); and absolute MAF differences >0.15 compared to 1000 Genomes Project European ancestry individuals. We also removed 22 outlier individuals who were outside of five standard deviations of the first two PCs, and eliminated individuals such that there were no pairwise relationships with estimated kinship >0.05. We used only the autosomes, as is commonly done for estimating heritability. Finally, we LD-filtered the SNPs such that no two SNPs had a pairwise r2>0.8. This resulted in 26,226 individuals (3,143 cases and 23,083 controls), 402,748 genotyped SNPs, and 2,184,083 imputed SNPs with rinfo2≥0.3 and MAF≥0.01 used in the analysis. We assumed a population prevalence of PCa=0.12 in the liability threshold model.
We further partitioned the genome into several sets using a joint variance components fit in GCTA (59). We first tested the previously-known hits vs. the rest of the genome, and then partitioned into the functional categories, prioritized similarly to (33): coding, UTR, promoter, DHS, intron, and intergenic. SNPs that happened to fall in overlapping regions were assigned to the highest priority category. The coding, UTR, and intron regions were determined from the UCSC Genome Browser known gene database (60). Unlike (33), who defined the promoters as +/−2Kb of a transcription start site, we defined the promoters from the Eukaryotic Promoter Database new v003 (34). The DHSs were determined from (33). For this partitioning, we defined enrichment for each category as the percentage of heritability explained divided by the percentage of genome. CIs and p-values were determined by 108 bootstrap iterations.
Supplementary Material
Significance.
Taken together, our findings of independent risk variants, ethnic variation in existing SNP replication, and remaining unexplained heritability have important implications for further clarifying the genetic risk of prostate cancer. It also suggests that there may be much promise in evaluating understudied variation such as indels and ethnically diverse populations.
Acknowledgments
Grant support:
This work was supported by National Institutes of Health grants: CA127298, CA088164 and CA112355 (JSW); AG046616 and DC013300 (TJH); R01 GM107427 (MLF); CA136578, CA141298, and CA131945 (KLP, LAM); CA151254 and CA157881 (SNT). This work was also supported by the UCSF Goldberg-Benioff Program in Cancer Translational Biology (JSW), Prostate Cancer Foundation Young Investigator Awards (KLP, LAM), Department Of Defense grant W81XWH-11-1-0261 (SNT), the MEC and the genotyping in AAPC were supported by NIH grants CA63464, CA54281, CA1326792, CA148085 and HG004726, funds from the California Cancer Research Program, grant number 99-86883, and from The Community Benefit Program, Kaiser Permanente Northern California. Support for participant enrollment, survey completion, and biospecimen collection for the RPGEH was provided by the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, the Ellison Medical Foundation, and Kaiser Permanente national and regional community benefit programs. Genotyping of the GERA cohort was funded by a grant from the National Institute on Aging, National Institute of Mental Health, and the National Institute of Health Common Fund (RC2 AG036607 to CAS and NJR). PEGASUS was supported by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to the Kaiser Permanente Northern California members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment, and Health, the ProHealth Study and the California Men’s Health Study. We would like to thank Peter Kraft, David Hunter, Sara Lindstrom, and Constance Chen for the contribution of PHS and HPFS genetic data through the BPC3.
Abbreviations list
- PCa
prostate cancer
- KP
Kaiser Permanente
- RPGEH
Research Program on Genes, Environment, and Health
- MEC
Multiethnic Cohort
- PSA
prostate specific antigen
- LD
linkage disequilibrium
- BMI
body mass index
- DHSs
DNaseI hypersensitivity sites
- CGEMs
Cancer Genemics Markers of Susceptibility Project
- CMHS
California Men’s Health Study
- GERA
Genetic Epidemiology Research on Aging
- KPNCCR
Kaiser Permanente Northern California Cancer Registry
- KPSCCR
Kaiser Permanente Southern California Cancer Registry
- SEER
Surveillance, Epidemiology, and End Results
- EUR
European-optimized array
- EAS
East Asian-optimized array
- LAT
Latino-optimized array
- AFR
African-American-optimized array
- QC
quality control
- MAF
minor allele frequency
- HWE
Hardy-Weinberg Equilibrium
- PCs
Principal components
- H&E
hemetoxylin and eosin
- PHS
Physician’s Health Study
- HPFS
Healthy Professionals Follow-up study
- BPC3
Breast and Prostate Cancer Cohort Consortium
- TCGA
The Cancer Genome Atlas
- FE
fixed effects
- RE
random effects
- CI
confidence interval
Footnotes
Conflict of Interest: No potential conflicts of interest were disclosed.
References
- 1.Hayes RB, Liff JM, Pottern LM, Greenberg RS, Schoenberg JB, Schwartz AG, et al. Prostate cancer risk in U.S. blacks and whites with a family history of cancer. Int J Cancer J Int Cancer. 1995;60:361–4. doi: 10.1002/ijc.2910600315. [DOI] [PubMed] [Google Scholar]
- 2.Whittemore AS, Wu AH, Kolonel LN, John EM, Gallagher RP, Howe GR, et al. Family history and prostate cancer risk in black, white, and Asian men in the United States and Canada. Am J Epidemiol. 1995;141:732–40. doi: 10.1093/oxfordjournals.aje.a117495. [DOI] [PubMed] [Google Scholar]
- 3.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13(Spec No 1):R103–21. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 4.Baker SG, Lichtenstein P, Kaprio J, Holm N. Genetic susceptibility to prostate, breast, and colorectal cancer among Nordic twins. Biometrics. 2005;61:55–63. doi: 10.1111/j.0006-341X.2005.030924.x. [DOI] [PubMed] [Google Scholar]
- 5.Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, Graff RE, et al. The heritability of prostate cancer in the nordic twin study of cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored. Am Soc Prev Oncol. 2014;23:2303–10. doi: 10.1158/1055-9965.EPI-13-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and Heritable Factors in the Causation of Cancer — Analyses of Cohorts of Twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 7.Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, Kubo M, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–9. S1. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 8.Cheng I, Chen GK, Nakagawa H, He J, Wan P, Laurie CC, et al. Evaluating Genetic Risk for Prostate Cancer among Japanese and Latinos. Cancer Epidemiol Biomarkers Prev. 2012;21:2048–58. doi: 10.1158/1055-9965.EPI-12-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eeles RA, Kote-Jarai Z, Olama AAA, Giles GG, Guy M, Severi G, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eeles RA, Olama AAA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–91. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y, Signorello LB, Strom SS, Kittles RA, Rybicki BA, Stanford JL, et al. Generalizability of established prostate cancer risk variants in men of African ancestry. Int J Cancer. 2015;136:1210–7. doi: 10.1002/ijc.29066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, et al. Functional Enhancers at the Gene-Poor 8q24 Cancer-Linked Locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kote-Jarai Z, Olama AAA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven novel prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindstrom S, Schumacher F, Siddiq A, Travis RC, Campa D, Berndt SI, et al. Characterizing Associations and SNP-Environment Interactions for GWAS-Identified Prostate Cancer Risk Markers—Results from BPC3. PLoS ONE. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindström S, Schumacher FR, Campa D, Albanes D, Andriole G, Berndt SI, et al. Replication of Five Prostate Cancer Loci Identified in an Asian Population—Results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2012;21:212–6. doi: 10.1158/1055-9965.EPI-11-0870-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olama AAA, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22:408–15. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, et al. Sequence Variants at 22q13 Are Associated with Prostate Cancer Risk. Cancer Res. 2009;69:10–5. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 26.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–5. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 29.Enger SM, Van Den Eeden SK, Sternfeld B, Loo RK, Quesenberry CP, Rowell S, et al. California Men’s Health Study (CMHS): a multiethnic cohort in a managed care setting. BMC Public Health. 2006;6:172. doi: 10.1186/1471-2458-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–30. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrne CD, Schwartz K, Lawn RM. Loss of a splice donor site at a “skipped exon” in a gene homologous to apolipoprotein(a) leads to an mRNA encoding a protein consisting of a single kringle domain. Arterioscler Thromb Vasc Biol. 1995;15:65–70. doi: 10.1161/01.atv.15.1.65. [DOI] [PubMed] [Google Scholar]
- 33.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreos R, Ambrosini G, Périer RC, Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–6. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikh H, Wang Z, Pettigrew KA, Jia J, Daugherty S, Yeager M, et al. Fine mapping the KLK3 locus on chromosome 19q13.33 associated with prostate cancer susceptibility and PSA levels. Hum Genet. 2011;129:675–85. doi: 10.1007/s00439-011-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J, Tao S, Gao Y, Peng T, Tan A, Zhang H, et al. Genome-wide association study identified novel genetic variant on SLC45A3 gene associated with serum levels prostate-specific antigen (PSA) in a Chinese population. Hum Genet. 2013;132:423–9. doi: 10.1007/s00439-012-1254-3. [DOI] [PubMed] [Google Scholar]
- 37.Ahn J, Berndt SI, Wacholder S, Kraft P, Kibel AS, Yeager M, et al. Variation in KLK Genes, Prostate Specific Antigen, and Risk of Prostate Cancer. Nat Genet. 2008;40:1032–4. doi: 10.1038/ng0908-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Gu J, Strom S, Logothetis CJ, Kim J, Wu X. The prostate cancer susceptibility variant rs2735839 near KLK3 gene is associated with aggressive prostate cancer and can stratify gleason score 7 patients. Clin Cancer Res Off J Am Assoc Cancer Res. 2014;20:5133–9. doi: 10.1158/1078-0432.CCR-14-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraft P. Curses—Winner’s and Otherwise—in Genetic Epidemiology. Epidemiology. 2008;19:649–51. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 40.Zaitlen N, Kraft P, Patterson N, Pasaniuc B, Bhatia G, Pollack S, et al. Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann TJ, Sakoda LC, Shen L, Jorgenson E, Habel LA, Liu J, et al. Imputation of the Rare HOXB13 G84E Mutation and Cancer Risk in a Large Population-Based Cohort. PLoS Genet. 2015;11:e1004930. doi: 10.1371/journal.pgen.1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–57. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 44.Howie B, Marchini J, Stephens M. Genotype Imputation with Thousands of Genomes. G3 Genes Genomes Genet. 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, Li Y, Abecasis GR, Scheet P. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet Epidemiol. 2011;35:102–10. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 2004;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 51.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am J Hum Genet. 2011;88:586–98. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larson NB, McDonnel S, French AJ, Fogarty Z, Cheville J, Middha S, et al. Am J Hum Genet. 2015 doi: 10.1016/j.ajhg.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penney KL, Sinnott JA, Tyekucheva S, Gerke T, Shui IM, Kraft P, et al. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored. Am Soc Prev Oncol. 2015;24:255–60. doi: 10.1158/1055-9965.EPI-14-0694-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q, Stram A, Chen C, Kar S, Gayther S, Pharoah P, et al. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum Mol Genet. 2014;23:5294–302. doi: 10.1093/hmg/ddu228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu W, Lazarus R. GeneticsDesign: Functions for designing genetic studies. R Package version 1.28.0. 2010 [Google Scholar]
- 57.Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–2. [Google Scholar]
- 58.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–70. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.