Abstract
The molecular underpinnings that drive the heterogeneity of KRAS-mutant lung adenocarcinoma (LUAC) are poorly characterized. We performed an integrative analysis of genomic, transcriptomic and proteomic data from early-stage and chemo-refractory LUAC and identified three robust subsets of KRAS-mutant LUAC dominated, respectively, by co-occurring genetic events in STK11/LKB1 (the KL subgroup), TP53 (KP) and CDKN2A/B inactivation coupled with low expression of the NKX2-1 (TTF1) transcription factor (KC). We further reveal biologically and therapeutically relevant differences between the subgroups. KC tumors frequently exhibited mucinous histology and suppressed mTORC1 signaling. KL tumors had high rates of KEAP1 mutational inactivation and expressed lower levels of immune markers, including PD-L1. KP tumors demonstrated higher levels of somatic mutations, inflammatory markers, immune checkpoint effector molecules and improved relapse-free survival. Differences in drug sensitivity patterns were also observed; notably, KL cells showed increased vulnerability to HSP90-inhibitor therapy. This work provides evidence that co-occurring genomic alterations identify subgroups of KRAS-mutant LUAC with distinct biology and therapeutic vulnerabilities.
Keywords: KRAS, co-mutations, lung adenocarcinoma, STK11, HSP90
Introduction
The identification of subsets of LUAC with oncogenic drivers has transformed the treatment of non-small cell lung cancer (NSCLC), particularly for patients whose tumors harbor activating mutations in EGFR or oncogenic fusions involving the ALK, RET and ROS1 kinases (1, 2). Despite these advances, the goal of developing specific therapeutic strategies for the 25-30% of LUACs that bear activating mutations in KRAS, the most common oncogenic driver in NSCLC, has thus far proven elusive. Both efforts to directly target RAS oncoproteins with small molecules as well as alternative approaches focused on inhibiting RAS post-translational modifications or downstream signaling pathways have been employed with only limited success; recently, however, some promising results have been reported (3-5).
A formidable challenge to the development of effective therapies for KRAS-mutant LUACs is heterogeneity in their biology and therapeutic responsiveness. Unlike NSCLC patients with tumors bearing EGFR activating mutations or ALK fusions, diseases for which targeted agents typically achieve objective responses in 60—80% of cases (1, 2, 6), clinical testing of agents targeting downstream pathways such as MEK and PI3K/AKT in patients with KRAS-mutant tumors has typically yielded response rates of less than 20% (7). This suggests that there is greater molecular diversity in KRAS-mutant tumors compared with those initiated by other known driver events. Preclinical studies provide further support for this notion, and in fact markers designed to predict the varying degrees of KRAS dependence have been developed (8). However, the underlying mechanisms that drive this divergent biological and clinical behavior are not well understood. Previously, we demonstrated that different KRAS amino acid substitutions-particularly common hydrophobic alterations such as G12C and G12V compared with hydrophilic alterations such as G12D- differ in their patterns of downstream signaling and response to targeted agents, suggesting that specific KRAS alleles may account for at least part of this heterogeneity (9). Other reports suggested that specific KRAS codon-12 alleles are not predictive of response to adjuvant therapy (10). Preclinical studies have indicated that Kras-mutant lung tumors bearing Trp53 or Stk11/Lkb1 co-mutations differ in their response to docetaxel with or without selumetinib, suggesting that co-mutations may also impact treatment responsiveness (11).
Here, in order to systematically address the heterogeneity exhibited by KRAS-mutant LUACs, we describe an integrative analysis that incorporates transcriptional, mutational, copy number and proteomic data from cohorts of both chemotherapy-naïve and heavily pre-treated KRAS-mutant tumors and identify three distinct and robust disease subsets. Significantly, we uncover co-occurring genetic events as major determinants of signaling diversification downstream of mutant KRAS and highlight subtype-selective dependencies that can be exploited therapeutically with agents currently undergoing clinical development.
Results
Unsupervised NMF clustering identifies three robust and reproducible subsets of KRAS-mutant LUAC
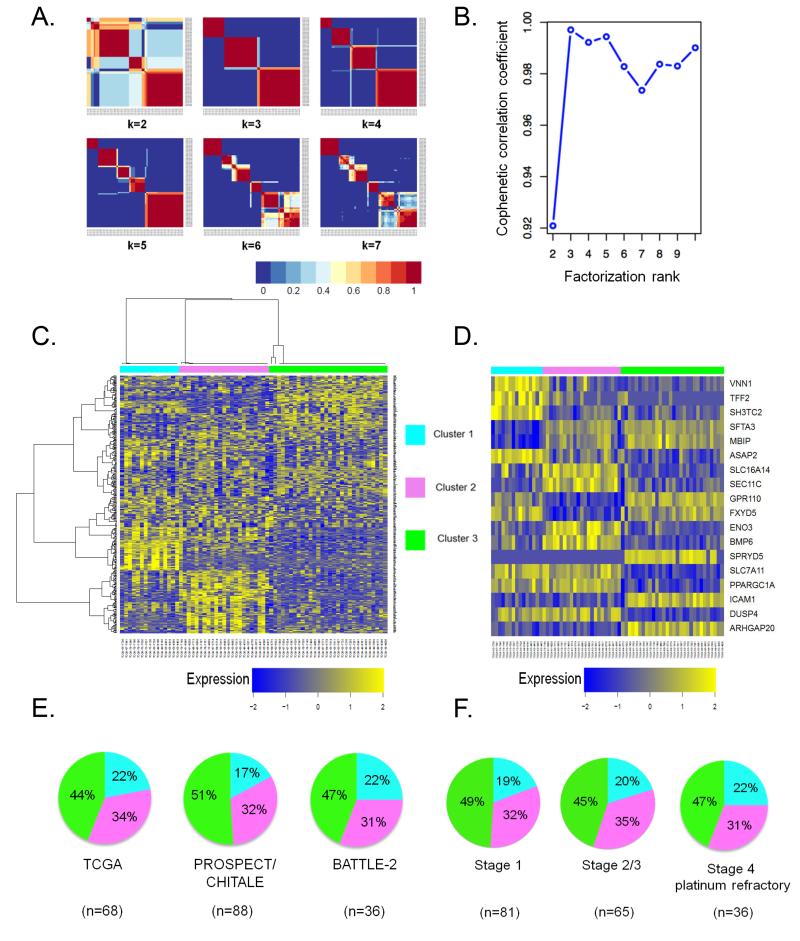
Expression profiling can capture the heterogeneous behavior of complex biological systems and has been successfully applied for the molecular stratification of human tumors. We thus initially interrogated RNA-Seq expression data from a training set of 68 KRAS-mutant LUACs from The Cancer Genome Atlas (12). In order to identify naturally occurring biological patterns, we employed non-negative matrix factorization (NMF) – an unsupervised approach – followed by consensus clustering, an iterative process that assesses the stability of partitioning over a number of clustering runs (13, 14). Application of consensus NMF revealed three to five robust clusters of KRAS-mutant LUAC with cluster stability peaking for k=3, as evidenced by the maximal value of the cophenetic correlation co-efficient, a quantitative measure of consensus matrix dispersion (Figure 1A and 1B). A heatmap depicting expression levels of 384 genes selected for the NMF algorithm is shown in Figure 1C. Potential functional modules within the 384 genes are highlighted in Supplementary Figure S1.
Figure 1. Consensus NMF clustering identifies three robust and reproducible subsets of KRAS-mutant LUAC.
(A) Consensus matrices of 68 KRAS-mutant LUACs from the TCGA dataset, computed for k=2 to k=7.
(B) Cophenetic correlation coefficient plot reveals peak cluster stability for k=3 ranks.
(C) Heatmap depicting mRNA expression levels of 384 genes selected for the NMF algorithm.
(D) Relative expression levels of individual genes that comprise the 18-gene cluster assignment signature in LUACs from the TCGA dataset.
(E) Cluster composition is preserved across distinct datasets of chemotherapy-naïve (PROSPECT, CHITALE) and platinum-refractory (BATTLE-2) KRAS-mutant LUACs.
(F) Subgroup representation is unaffected by increasing disease stage. Stage 4 platinum refractory tumors in this analysis represent the BATTLE-2 clinical cohort.
We next sought to validate our expression-based classification using independent datasets. In order to enable class prediction, we first derived an 18-gene signature consisting of 6 genes whose expression correlated with each subtype in the TCGA cohort using a nearest centroid-based classifier (15) (Figure 1D) (See also Supplementary Methods). For validation, we assembled two clinically distinct collections of KRAS-mutant LUACs with available mRNA expression data: 1) a combined surgical set of 88 chemotherapy–naïve, mostly early-stage or locally advanced tumors consisting of 41 tumors prospectively collected at MD Anderson Cancer Center (PROSPECT dataset) and 47 previously reported LUACs (16) and 2) 36 metastatic, platinum-refractory KRAS-mutant LUACs from the BATTLE-2 clinical trial with tumor re-biopsy performed at trial entry (17). Critically, the three expression clusters retained similar representation in the training and validation datasets (Figure 1E). Cluster composition also remained stable as a factor of increasing disease stage (Figure 1F).
Co-occurring genetic events are major determinants of the biological heterogeneity of KRAS-mutant LUAC
We sought to determine whether alterations in the tumor genome could account for the robustness and reproducibility of cluster assignment. Previously we have reported that individual KRAS codon 12 mutations that result in distinct amino acid substitutions differentially engage downstream effector pathways (9). We compared the distribution of the three most common mutant KRAS alleles (KRASG12C, KRASG12V and KRASG12D and other grouped together) in the expression clusters but found no evidence for enrichment of specific amino acid substitutions (p=0.3, Fisher’s exact test). This suggests that specific KRAS amino acid substitutions are not primary drivers of the molecular diversity of KRAS-mutant LUACs.
Next, we investigated whether somatic mutations or other genomic alterations outside of KRAS itself are dominant determinants of expression cluster membership. In order to formally address this possibility, we identified prevalent somatic mutations [present in ≥ 14.7% (10/68) of evaluated tumors] that were significantly enriched in the three cohorts using a FDR of 0.05 as statistical cutoff. This analysis yielded 11 genes with non-silent somatic mutational events (Supplementary Figure S2).
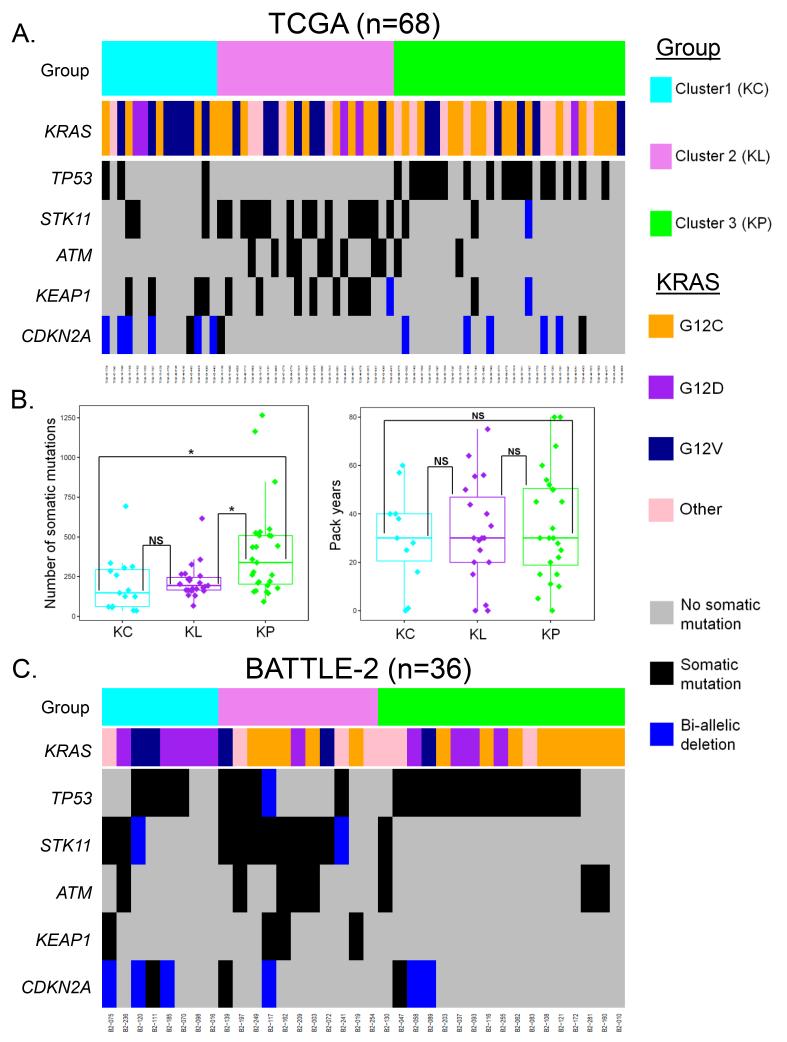
Co-mutations in TP53 (P=3.8e−06) and STK11/LKB1 (P=1.03 e−05) were the most significantly enriched genetic events in cluster 3 and cluster 2, (henceforth referred to as the KP and KL clusters), respectively (Figure 2A) and were largely non-overlapping in the context of chemotherapy-naïve disease. We confirmed this in a second cohort of KRAS-mutant tumors from TCGA as well as in a larger merged dataset of 176 previously untreated (mostly early stage) KRAS-mutant LUACs with available somatic mutation data, where only 4% of KRAS-mutant tumors harbored co-mutations in both TP53 and STK11 (Supplementary Figure S2) (16, 18-20). The occurrence of triple-mutant tumors (KRAS;STK11;TP53, referred to as KPLm in order to distinguish from the expression-based clusters) was significantly less frequent than expected by chance in both the original TCGA set (n=68) (P=0.0018, permutation test based on 10000 permutations) as well as in the second set of 77 KRAS-mutant TCGA LUACs analyzed since (P=0.01692, permutation test based on 10000 permutations), and of borderline significance in the merged validation cohort (n=176) (P=0.0693, permutation test based on 10000 permutations). Other prominent genes with non-random mutation patterns across the three subsets included two that were enriched in the KL subgroup: ATM (P=0.002), a gene encoding an apical kinase in the DNA damage response and KEAP1 (P=0.006), a gene encoding a ubiquitin ligase that functions as a negative regulator of NRF2 (also known as NFE2L2)-mediated transcription (Figure 2A). For a list of individual mutations in KRAS, STK11/LKB1, TP53, ATM and KEAP1 see Supplementary Table S1.
Figure 2. Co-occurring genetic events in pivotal tumor suppressor genes are differentially represented in the three KRAS-mutant LUAC subgroups.
(A) Co-mutation plot for 68, predominantly early-stage, KRAS-mutant LUACs from the TCGA dataset.
(B) Comparison of overall non-synonymous somatic mutation rate (left panel) and cumulative exposure to smoking (expressed in pack years) (right panel) in the KC, KL and KP subgroups. ANOVA was used for the three group comparison, and Tukey’s post-test was applied to all pair-wise comparisons. Asterisks denote statistical significance at P≤0.05.
(C) Co-mutation plot for 36 metastatic, platinum-refractory, KRAS-mutant LUACs with available somatic mutation data from the BATTLE-2 trial.
Interestingly, the mean overall number of non-silent somatic mutations per tumor also differed between the three subgroups, with KP tumors harboring a significantly higher overall mutational load (Figure 2B, left panel) despite comparable exposure to smoking (measured in pack years) (Figure 2B, right panel), suggesting increased mutation tolerance or genomic instability in this subset.
Marked genomic copy number aberrations can alter gene function and are considered functionally relevant genetic events. We thus computed the distribution of focal bi-allelic deletions and high copy number gains across the genome in the three clusters using GISTIC2.0-derived deletion and amplification peaks generated from Affymetrix SNP 6.0 array hybridization data (21). This analysis revealed bi-allelic deletions of CDKN2A (encoding for the p16 tumor suppressor) and CDKN2B (encoding for the p15 tumor suppressor), both located at 9p21.3, as significantly enriched events in the first cluster (henceforth referred to as KC) (P=0.004 and P=0.002 respectively, Fisher’s exact test; Figure 2A and Supplementary Figure S3). It is worth noting, however, that many tumors in the KC cluster did not have observable copy numbers alterations in these genes. When combined with somatic mutations, alterations in CDKN2A were present in 7/15 tumors in the KC cluster compared to 1/23 and 5/30 tumors in the KL and KP clusters, respectively. Among other components of the G1/S transition machinery, somatic mutations in RB1 were more frequent in the KL cluster (P=0.044), although this was based on a small number of events (n=3) (Supplementary Figure S4). Inclusion of both somatic mutations and bi-allelic deletions in STK11, TP53, KEAP1, ATM and CDKN2A in the analysis of co-occurring genetic events did not alter the statistical conclusions (STK11: P=0.000049, TP53: P=0.0000038, KEAP1: P=0.01258, ATM: P=0.0015, CDKN2A: P=0.00648, Fisher’s exact test).
A direct corollary of the conjecture that key co-mutations dictate signaling diversification downstream of mutant KRAS is the prediction that these genetic events are likely to be clonal in nature and occur early during the process of branched tumor evolution. Indeed, clonality analysis utilizing an algorithm that incorporates variant allele counts with estimates of sample purity and tumor ploidy concluded that the overwhelming majority of reported somatic mutation events in KRAS (98.53%), STK11/LKB1 (100%), TP53 (95.24%) and KEAP1 (100%) in the TCGA dataset were likely to be clonal (Supplementary Figure S5) (22). In contrast, ATM mutations were clonal in only 81.82% of mutant LUACs. Overall, 67.64% of all identified somatic mutations in this analysis were reported to be clonal, in agreement with recent work from our group and others utilizing deep multi-region sequencing of localized, chemotherapy-naïve LUACs (23, 24).
We then tested these findings using other datasets and confirmed that the expression cluster enrichment in STK11/LKB1 and TP53 co-mutations was recapitulated in the two independent validation cohorts. Among 66 KRAS-mutant LUACs with available somatic mutation data from the combined Chitale and PROSPECT dataset, the distribution of KRAS;LKB1, KRAS;TP53 and KRAS;LKB1;TP53 genotypes differed significantly between the three expression clusters (P=0.047, Fisher’s exact test), with LKB1 somatic mutations accumulating, as predicted, in the KL cluster (P=0.036, Fisher’s exact test) (Supplementary Figure S6). Lack of copy number data precluded evaluation of CDKN2A/CDKN2B inactivation in this cohort. Consistent with TCGA results, the distribution of the KRASG12D, KRASG12V and KRASG12C alleles was not different across the three subgroups in this independent set of LUACs (P=0.338, Fisher’s exact test).
We next examined whether similar patterns were seen among tumors from platinum-refractory, metastatic LUAC, assessed as part of the BATTLE-2 trial (Figure 2C). Consistent with the analysis from early stage, treatment-naïve tumors, highly significant association between co-occurring genetic events in STK11/LKB1 and TP53 (incorporating both somatic mutations and bi-allelic deletions) and subtype allocation was also evident among 36 metastatic, platinum-refractory KRAS-mutant LUACs from the BATTLE-2 trial (P<0.001, Fisher’s exact test), with striking enrichment for LKB1-inactivating events in the KL cluster (P<0.001, Fisher’s exact test). Genetic abrogation (somatic mutation or bi-allelic loss) of CDKN2A was more frequent in the KC cluster (50% versus 18.2% versus 17.6%), mirroring the relative frequencies observed in the TCGA dataset, but this result was not statistically significant likely due to the small number of events observed (P=0.278, Fisher’s exact test). Interestingly, there was a trend for enrichment of somatic mutations and/or bi-allelic deletions involving KEAP1 among tumors in the KL cluster, in agreement with data from TCGA (P= 0.056, Fisher’s exact test).
Despite their overall concordance, some notable differences could also be appreciated between the early stage, treatment-naïve cohorts and the BATTLE-2 cohort. The prevalence of mutations/bi-allelic deletions in TP53 was higher overall in the refractory BATTLE-2 cohort, and there was increased frequency of concurrent mutations/bi-allelic deletions in both TP53 and LKB1, which clustered predominantly with the KL subgroup (Figure 2C and Supplementary Figure S2). The distribution of ATM mutations was not different among the three subgroups in the heavily pre-treated BATTLE-2 patient population (P=0.449, Fisher’s exact test). Finally, in this cohort, the distribution of mutant KRAS alleles appeared non-random with tumors harboring the KRASG12D allele occurring more frequently in the KC cluster (P=0.047, Fisher’s exact test). This suggests that patterns of co-occurring genomic events may evolve as tumors metastasize and/or develop platinum resistance.
Low expression of TTF1 (NKX2-1) is a defining feature of the KC cluster and drives a distinct, GI-like differentiation program
Given that characteristic co-occurring genomic events could not be identified in many of the tumors in the KC cluster, and recent preclinical data suggesting CDKN2A/B loss may impact the differentiation of LUACs (25), we further interrogated patterns of mRNA, micro-RNA and (phospho)-protein expression in the three KRAS-driven LUAC subsets and probed for potentially relevant expression modules using Gene Set Enrichment Analysis (GSEA) (26).
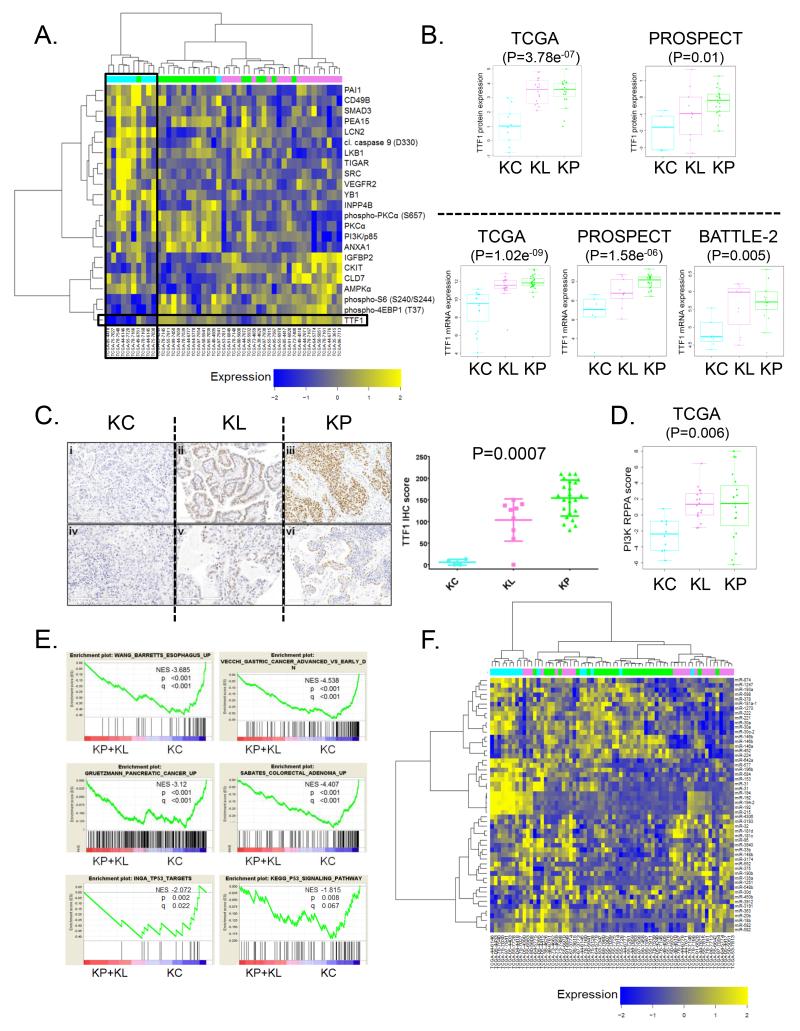
Supervised analysis of reverse phase protein array (RPPA)–derived proteomic (total and phospho-proteins) data revealed marked differences between the three subgroups (Figure 3A). Notably, expression of TTF1 (NKX2-1), a lineage-specific, homeobox-containing transcription factor routinely assessed in diagnostic histopathology and detected in 80-85% of LUACs, was almost uniformly suppressed among tumors in the KC cluster (P=3.78e−07, ANOVA) (Figure 3B). This was also observed at the level of mRNA expression (P=1.02e−09, ANOVA) and was further validated in the PROSPECT (P=1.58e−06, ANOVA) and BATTLE-2 (P=0.005, ANOVA) cohorts (Figure 3B). Importantly, lower TTF1 expression could be demonstrated by immunohistochemical (IHC) analysis of a tissue microarray that included a subset of surgically resected tumors from PROSPECT (P=0.0007, Kruskal-Wallis test) (Figure 3C). Thus, low TTF1 expression resulting in little to no IHC staining is a convergent feature of KRAS-mutant LUACs in the KC cluster and may represent a clinical biomarker facilitating their identification.
Figure 3. Multi-platform profiling identifies low expression of TTF-1 (NKX2-1) as a defining feature of KRAS-mutant LUACs in the KC cluster.
(A) Supervised hierarchical clustering of reverse-phase protein array (RPPA) expression data demonstrates consistently suppressed levels of TTF1 protein in the KC cluster.
(B) Quantitative analysis of TTF1 protein (top panels) and TTF1 mRNA (bottom panels) expression in the TCGA, PROSPECT and BATTLE-2 cohorts. Statistical comparison between the three groups is based on ANOVA.
(C) Immunohistochemical analysis of TTF1 expression in KRAS-mutant LUACs from PROSPECT. Representative images from tumors in the three clusters are shown in the left panel. Scale bar = 200μm. The Kruskal-Wallis test was used for statistical comparison. Error bars represent standard deviation of the mean.
(D) Dot-plot representation of PI3K proteomic (RPPA) score in the three KRAS-mutant LUAC subsets.
(E) GSEA shows enrichment of gene expression signatures reflecting both upper and lower GI neoplastic processes as well as wild-type p53 transcriptional activity in the KC cluster.
(F) Supervised hierarchical clustering reveals distinct patterns of miRNA expression in the three KRAS-mutant LUAC subsets.
In addition to low TTF1 expression, we detected significantly suppressed levels of phospho-S6 (Ser240/Ser244) and phospho-4EBP1 (Thr37) in KC LUACs compared with KL and KP tumors, indicating reduced mTORC1 signaling output (Figure 3A). This was further confirmed using a proteomic (RPPA) “PI3K score” (Figure 3D). It is notable that in this analysis KP LUACs exhibited comparable levels of PI3K-AKT-MTOR pathway activation to KL tumors. Genomic alterations in pathway genes other than STK11/LKB1 may account for this observation (Supplementary Figure S7).
In agreement with a recently reported role for TTF1/NKX2-1 in restraining a latent gastric differentiation program in genetically engineered murine models (GEMMs) of hemi- or homozygous Nkx2-1 deletion in the context of mutant KrasG12D, 6/15 KC LUACs were classified as invasive mucinous carcinomas. This histological subtype was not detected in the KL and KP subgroups (with the exception of a single colloid KL tumor, P=0.000125, Fisher’s exact test). Furthermore, expression of HNF4A (P=3.17e−05, unpaired t-test) and PDX1 (P=6.85e−05, unpaired t-test), two master regulators of GI cell fate, was significantly elevated in tumors from the KC cluster (27, 28) (data not shown). As expected, markers of mucinous differentiation, including CK20, MUC5B and AGR2 were also up-regulated (data not shown) and several gene signatures reflective of both upper and lower GI neoplastic processes were among the most highly enriched in this cluster (Figure 3E). Interestingly, higher average expression of the normally embryonically-restricted chromatin regulator HMGA2 (P=0.001, unpaired t-test) was observed across tumors in this subset consistent with data from the GEMMs (data not shown). Together, these preclinical studies and our data support a role for both CDKN2A/B and TTF1/NKX2-1 in the differentiation of KRAS-mutant LUAC.
Finally, GSEA highlighted several gene signatures associated with elevated wild-type p53 transcriptional output in the KC cluster compared to the KP and KL clusters (Figure 3E). This finding is consistent with wild-type TP53 status in the majority of tumors in this cluster and is further supported by low rates of TP53 copy number loss and quantitative analysis of proteomic markers that represent established p53 targets (for example TIGAR and PAI-1, Figure 3A).
Tumors in the KC subgroup further exhibited a distinct and highly concordant pattern of micro-RNA expression (Figure 3F), characterized by up-regulation of the miRNA-192, miRNA-194 and miRNA-215 cluster, among others, as well as elevated miRNA-31, a validated lung cancer onco-miR (29). Notably, the miRNA-192/194/215 cluster represents an established p53 transcriptional target (30).
Functional inactivation of the LKB1-AMPK axis is common in STK11/LKB1 mutation-negative KL tumors
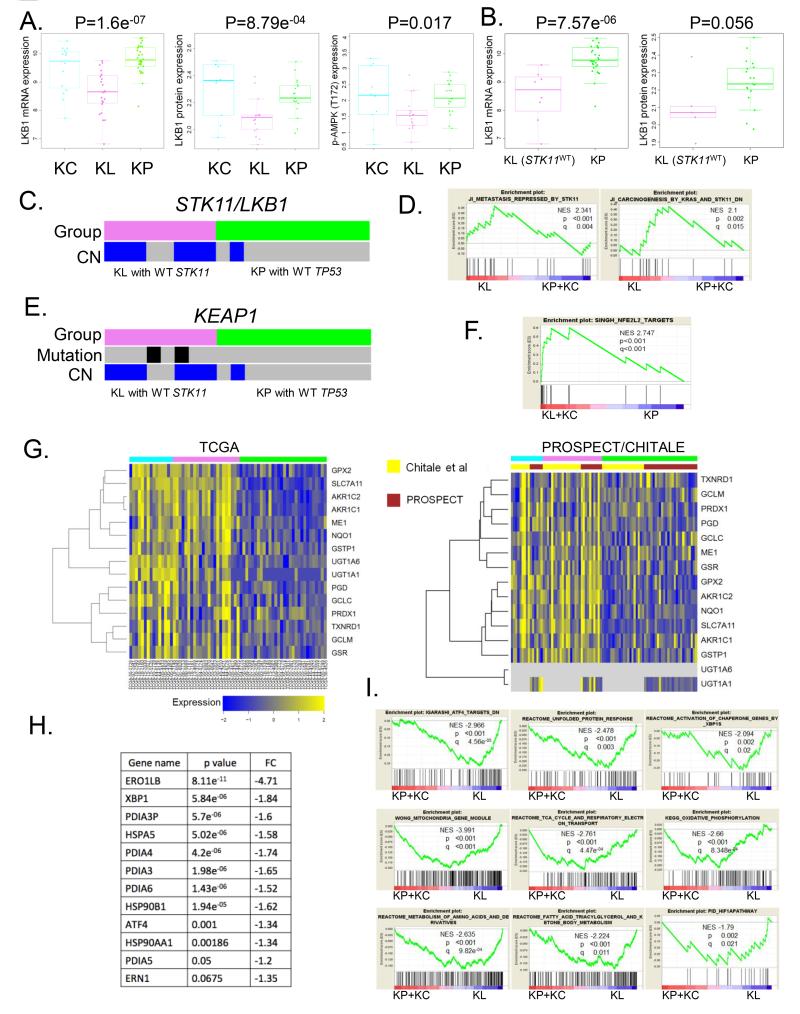
Several lines of evidence point towards functional inactivation of the LKB1-AMPK axis as a hallmark of tumors in the KL cluster, even among cases that lack somatic mutations in STK11/LKB1. First, in keeping with the observed LKB1 mutation spectrum that is dominated by nonsense and frameshift mutations, expression levels of both LKB1 (P=8.79e−04, ANOVA) protein and its direct target, phospho-AMPK Thr172 (P=0.017, ANOVA), were significantly lower in the KL cluster compared with the KP and KC clusters (Figure 4A). Second, LKB1 mRNA expression was significantly suppressed among LKB1-mutation negative cases in the KL cluster compared to KP tumors (P=7.57e−06, unpaired t-test), and this was associated with a trend towards lower LKB1 protein expression by RPPA (P=0.056, unpaired t-test) (Figure 4B). Third, copy-number losses at the LKB1 locus were more prevalent among LKB1-mutation negative KL tumor compared to TP53-mutation negative KP tumors (P=0.006, Fisher’s exact test) (Figure 4C). Finally, GSEA identified “Carcinogenesis by KRAS and STK11” and “Metastasis repressed by STK11”, two gene signatures derived from tumors arising in a genetically engineered murine model of NSCLC following concomitant inactivation of Stk11/Lkb1 and expression of endogenous oncogenic KRASG12D, as significantly enriched in KL LUACs compared to KP and KC combined (Figure 4D).
Figure 4. KL tumors with functional inactivation of the LKB1-AMPK pathway display evidence of adaptation to oxidative, proteotoxic and energetic stress.
(A) Functional inactivation of the LKB1-AMPK axis in KL tumors. Box plot representation of (from left to right) LKB1 mRNA/protein and phospho-AMPK (Thr172) protein expression. Comparison is based on ANOVA.
(B) Suppressed levels of LKB1 mRNA and protein are noted even among STK11/LKB1-wild type tumors in the KL subgroup. The un-paired t-test is used for comparison with KP LUACs.
(C) Frequent single copy number loss at the STK11/LKB1 locus among LKB1 somatic mutation-negative KL tumors.
(D) GSEA reveals enrichment of two STK11/LKB1 related signatures in the KL subgroup.
(E) Frequent genetic abrogation of the KEAP1 locus in LKB1 wild-type KL LUACs.
(F) Significant enrichment of a NRF2 (NFE2L2) expression signature in KL and KC tumors.
(G) Heatmap display of relative mRNA expression levels of several prototypical NRF2 target genes in the TCGA and combined PROSPECT/CHITALE datasets.
(H) Higher expression of several cytoplasmic and ER chaperone proteins and core unfolded protein response components among KL LUACs. P values are based on an unpaired t-test.
(I) GSEA identifies altered cellular bio-energetics, activation of the unfolded protein response and HIF-1α pathway up-regulation as prominent modules among LUACs in the KL cluster.
Adaptation to oxidative and endoplasmic reticulum stress is a hallmark of KRAS-mutant LUACs with functional inactivation of the LKB1-AMPK pathway
We observed significant accumulation of genetic events involving KEAP1 among both LKB1-mutation positive and negative LUACs in the KL cluster (Figure 2A, 2C and Figure 4E). In contrast, KEAP1 mutations were infrequently observed among KP tumors in either the TCGA or BATTLE-2 cohorts, as well as in a separate collection of 49 surgically resected KRAS-mutant LUACs reported previously (18), unless the tumor also harbored a STK11/LKB1 mutation (ie triple-mutant KPLm tumors). KEAP1 mutations and/or somatic copy number losses (mono- or bi-allelic), were also present in the majority of STK11/LKB1-wild type KL tumors (Figure 4E). The significant co-occurrence of KEAP1 and STK11/LKB1 copy number losses is not surprising, as the corresponding genomic loci reside on the short arm of chromosome 19 (at 19p13.2 for KEAP1 and 19p13.3 for STK11).
Notably, 4/15 tumors in the KC subgroup also harbored mutations in KEAP1 (Figure 2A). In two cases, these co-occurred with mutations in STK11/LKB1 whereas one additional tumor with KEAP1 mutation exhibited suppressed levels of phospho-AMPK Thr172.
In keeping with the function of KEAP1 as a regulator of the proteasomal degradation of NRF2, a transcription factor with a pivotal role in the cellular defense against oxidative stress and xenobiotics, a NRF2 gene expression signature was significantly enriched in the KL and KC clusters compared to KP (Figure 4F). Indeed, several prototypical NRF2 target genes were among the most differentially expressed transcripts in both the TCGA and PROSPECT datasets (Figure 4G). Thus, activation of a NRF2-driven anti-oxidant and cyto-protective transcriptional program is a common feature and may be selected for in the evolution of tumors with functional inactivation of the LKB1-AMPK axis. Additional mechanisms of NRF2 upregulation and/or stabilization may account for the prominent upregulation of NRF2-target genes in KC LUACs given the relatively low frequency of KEAP1 somatic mutations in this subgroup. For example, expression levels of miR-200a, a microRNA that has been previously reported to suppress KEAP1 (31), differed significantly between the three clusters (P=0.0065, ANOVA), with higher levels detected in KC LUACs.
In addition to antioxidant genes, several molecular chaperones and components of the cellular proteostasis network were also expressed at higher levels in the KL cluster (Figure 4H). These involved cytoplasmic and ER chaperones, several members of the protein disulfide isomerase family as well as XBP1 and ATF4. LKB1 loss - dependent up-regulation of nodal components of the unfolded protein response (UPR) was further demonstrated in pairs of isogenic LKB1-deficient/proficient NSCLC cell lines (Supplementary Figure S8). Accordingly, XBP1 and ATF4 expression signatures were significantly enriched in the KL cluster (Figure 4I), which also exhibited evidence of altered bio-energetics, in agreement with previous reports (32).
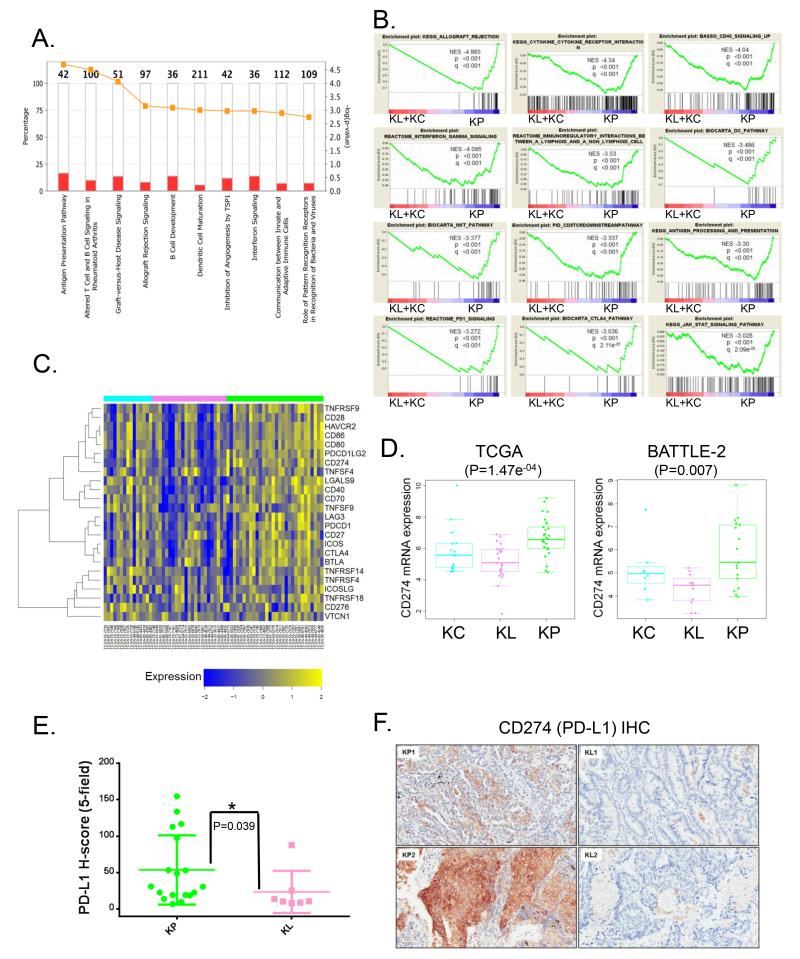
Inflammation and active immuno-editing in KP LUACs suggest potential avenues for therapeutic intervention
GSEA and Ingenuity Pathway Analysis (IPA) highlighted inflammation and several immune-related pathways as prominent modules in the KP cluster. Both gene sets associated with activation of anti-tumor immunity and immune tolerance/escape were enriched in this subset of KRAS-driven LUACs (Figure 5A and 5B). In contrast, KL tumors demonstrated comparative lack of immune system engagement, while KC tumors demonstrated a mixed picture. Notably, several targetable mediators of cell intrinsic co-inhibitory signals were expressed at higher levels in the KP cluster including PD-L1(CD274)(P=0.000147, unpaired t-test), PD-1(CD279)(P=0.01278, unpaired t-test) and CTLA-4 (CD152) (P=0.0269, unpaired t-test) (Figure 5C and 5D) and there was a trend towards more dense infiltration with CD3+, CD8+ and CD45RO+ populations of lymphocytes (Supplementary Figure S9). PD-L1 mRNA expression was also higher among KP LUACs in the BATTLE-2 cohort (P=0.007, un-paired t-test) (Figure 5D). Furthermore, we confirmed the association of KP with elevated PD-L1 expression in the PROSPECT dataset, using an immunohistochemical (Histo)-score (Figure 5E and 5F). In support of these findings, Koyama et al. demonstrated reduced number of tumor-infiltrating lymphocytes and reduced expression of PD-L1 in a genetically engineered murine model (GEMM) of Kras-driven, Lkb1-deficient NSCLC (Kras;Lkb1) compared to Kras-mutant tumors with intact Lkb1 status (Kras) (submitted manuscript). Importantly, Kras;Lkb1 tumors were resistant to therapy with an anti-PD-1 antibody in this GEMM.
Figure 5. KRAS-mutant LUAC subsets exhibit distinct patterns of immune system engagement.
(A) IPA identifies several immune-related modules among the top ten up-regulated pathways in the KP cluster.
(B) GSEA reveals prominent enrichment of signatures relating to inflammation, anti-tumor immunity and immune tolerance/escape in the KP subgroup.
(C) Heatmap representation of relative mRNA expression levels of selected targetable immune checkpoint mediator/effector molecules.
(D) Expression of CD274 (PD-L1) mRNA in KRAS-mutant LUAC subsets. ANOVA is used for statistical comparison between the three groups.
(E) Comparison of PD-L1 H-score between KL and KP tumors in a tissue microarray from PROSPECT. A single confirmed triple mutant tumor (KRAS;TP53;LKB1) in this cohort is excluded from the analysis. Wilcoxon rank-sum test is used for statistical comparison. Error bars represent standard deviation of the mean.
(F) Representative images of CD274 immuno-staining in LUACs from the KL (KL1/KL2) and KP (KP1/KP2) clusters. Scale bar = 200μm.
Prognostic relevance of KRAS-mutant LUAC subgroups
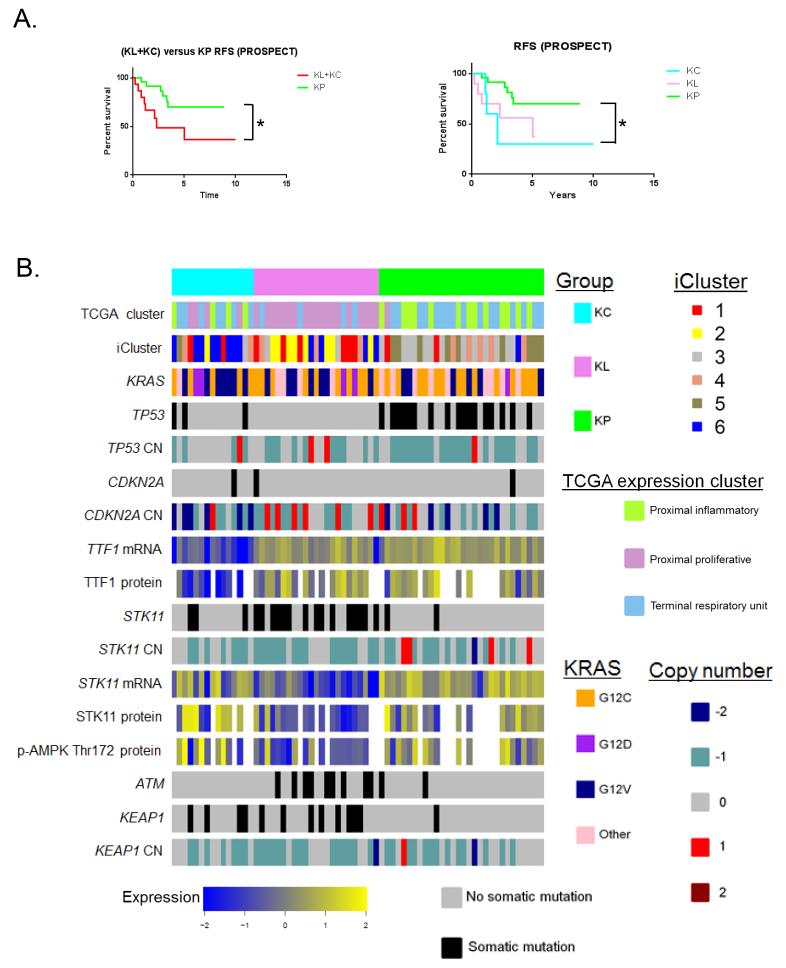
Increased lymphocytic infiltration has been associated with improved clinical outcome after surgical resection in colorectal cancer, that also harbors frequent mutations in KRAS (33). We therefore explored the prognostic utility of the KRAS-mutant LUAC subgroups using relapse-free survival data (RFS) from PROSPECT. This analysis revealed that KP tumors had significantly longer RFS compared to KL and KC LUACs combined (P=0.029, log-rank test) (Figure 6A, left panel) and compared to the KC subgroup alone (P=0.0269, log-rank test) (Figure 6A, right panel). Cluster membership remained a significant independent predictor of RFS on multivariate analysis (incorporating adjuvant therapy and nodal status) [P=0.03 for the KP versus (KL+KC) comparison and P=0.03 for the KP versus KC comparison]. In contrast, overall survival did not differ between the subgroups in a larger merged dataset of KRAS-mutant tumors with available overall survival (OS) data (P=0.3) (Supplementary Figure S10). Due to the limited numbers of tumors with available molecular and clinical data reported here these results require future validation in larger, prospectively collected datasets.
Figure 6. Prognostic utility of KRAS-mutant LUAC subgroups and integrated view of their key genetic, transcriptional and proteomic features.
(A) Kaplan-Meier estimates of relapse-free survival following surgical resection of 40 KRAS-mutant LUACs from PROSPECT (one stage 4 tumor was excluded from the analysis). Comparison of RFS was based on the log-rank test. Asterisk denotes statistical significance at the P≤0.05 level.
(B) Integrated view of key genetic, transcriptional and proteomic features of the three KRAS-mutant LUAC subgroups. The relationship with the TCGA expression (proximal proliferative/proximal inflammatory/terminal respiratory unit) and integrated clusters (iCluster) is also indicated.
An integrated heatmap encompassing the key genomic, transcriptional and proteomic features of the three KRAS-mutant LUAC subgroups, as well as their relationship to the previously defined TCGA and i-Clusters (12) is presented in Figure 6B. Corresponding heatmaps for a higher number of KRAS-mutant LUAC subgroups (k=4 and k=5) based on the NMF algorithm are included in Supplementary Figure S11.
KRAS mutant cell lines with LKB1 loss show enhanced sensitivity to HSP90 inhibition
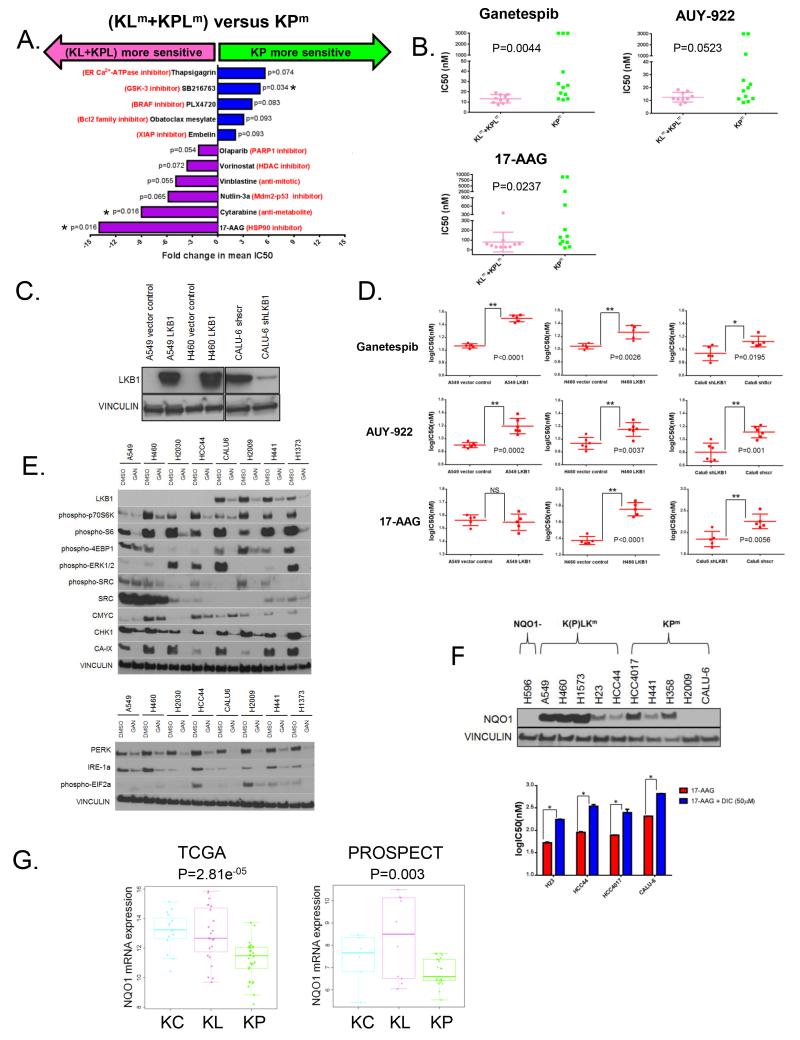
We posited that differences in the underlying biology of KRAS-driven LUACs with co-mutations in STK11/LKB1 or TP53 would result in distinct therapeutic vulnerabilities. In support of this hypothesis, re-analysis of publicly available large-scale cell line drug sensitivity data (Genomics of Drug Sensitivity in Cancer) (34) for KRAS-mutant NSCLC cell lines on the basis of co-mutations in STK11/LKB1 (KLm/KPLm) and TP53 (KPm) yielded divergent patterns of drug sensitivity (Figure 7A).
Figure 7. KRAS co-mutations are associated with distinct therapeutic vulnerabilities.
(A) Re-analysis of drug sensitivity data (34) for 19 KRAS-mutant NSCLC cell lines based on the presence of co-mutations in STK11/LKB1 and TP53.KRAS;LKB1 (KLm) and KRAS;TP53;LKB1 (KPLm) triple mutant lines were grouped together (n=9) for this analysis and compared to KRAS;TP53 (KPm) lines with wild-type STK11/LKB1 status (n=10). Drugs with Wilcoxon rank-sum test-derived P values ≤ 0.1 for the (KLm+KPLm) versus KPm comparison are displayed and fold change in mean IC50 values is plotted on the x axis. Asterisk denotes statistical significance at the P≤0.05 level.
(B) KRAS-mutant NSCLC cell lines with LKB1 inactivation show increased sensitivity to HSP90 inhibition. Scatter plots of median IC50 values (nM) (from three to five independent experiments) of 10 KLm/KPLm and 12 KPm cell lines for three chemically distinct HSP90 inhibitors. The Wilcoxon rank sum test is used for statistical comparison.
(C) Western blot analysis of LKB1 expression in three isogenic pairs of LKB1 deficient/proficient KRAS-mutant NSCLC cell lines.
(D) LKB1-status dependent sensitization of KRAS-mutant NSCLC cell lines to HSP90 inhibitors. Log10IC50 (nM) values from five to six independent experiments for each isogenic pair were compared using the un-paired t-test.* denotes significance at the P≤0.05 and ** at the P≤0.01 level. Error bars represent SD of the mean.
(E) Ganetespib simultaneously destabilizes multiple proteins that support the fitness of KRAS-mutant NSCLC cell lines with LKB1 inactivation.
(F) NQO1-mediated bio-activation contributes to the sensitivity of NSCLC cell lines to 17-AAG. Top panel: Robust expression of NQO1 among KRAS;(TP53);LKB1 lines, which frequently harbor co-occurring mutations in KEAP1[K(P)LKm]. KP lines display variable NQO1 expression (CALU-6 expresses NQO1 after prolonged exposure). The *2NQO1- polymorphic H596 cell line is used as negative control. Bottom panel: Co-treatment with dicumarol, a NQO1-inhibitor, renders KRAS-mutant NSCLC partially resistant to 17-AAG. Un-paired t-test is used for all statistical comparisons. Error bars represent SD of the mean from two independent experiments.
(G) Differential expression of NQO1 mRNA in the three KRAS-mutant LUAC subgroups in the TCGA (left panel) and PROSPECT (right panel) cohorts. ANOVA is used for statistical comparison.
17-AAG, an ansamycin benzoquinone ATP-competitive HSP90 inhibitor emerged as the top hit in this analysis with enhanced activity observed in STK11/LKB1 co-mutant (KLm and KPLm) lines. We therefore further assessed whether HSP90 inhibition represented a novel therapeutic vulnerability for cells bearing KRAS mutation and LKB1 loss.
We first validated and extended these findings by assessing the sensitivity of an extended panel of 22 KRAS-mutant NSCLC cell lines with different LKB1 status (10 KLm/KPLm and 12 KPm) to three distinct HSP90 inhibitors (17-AAG, Ganetespib, AUY922) using a 72-hour luminescent cell viability assay (Cell-Titer Glo®, Promega). Significantly, KLm/KPLm cell lines were more sensitive to Ganetespib and 17-AAG, and showed a trend towards increased sensitivity to AUY922 compared to KPm lines (17-AAG: P=0.0237, Ganetespib: P=0.0044, AUY922: P=0.0523, Mann-Whitney test for all comparisons) (Figure 7B).
LKB1-status dependent sensitization to HSP90 inhibition was further assessed in pairs of isogenic cell lines (Figure 7C). Expression of wild type LKB1 in A549 and H460 (two naturally LKB1-deficient cell lines) and knockdown of LKB1 in Calu-6 (a KPm line that expresses wild-type LKB1 protein) resulted in statistically significant increases in IC50 values to ganetespib, AUY-922 and 17-AAG (with the single exception of 17-AAG in the A549 isogenic pair) (Figure 7D). Thus, sensitivity to HSP90 inhibition in these lines is at least partially mediated by LKB1 inactivation.
Hyper-sensitivity to HSP90 inhibitors is usually attributed to destabilization of oncogenic kinases or other HSP90 client proteins that are required for the survival or proliferation of malignant cells or, alternatively, may result from the induction of irresolvable levels of proteotoxic stress. In order to explain the heightened responsiveness of KRAS;LKB1 co-mutant lines to HSP90 inhibition, we focused our attention on nodal components of signaling networks that are known to regulate cellular fitness in the context of LKB1 inactivation. Following treatment of a panel of 8 KRAS-mutant cell lines with 150nM of ganetespib for 24 hours, we observed reduction in levels of c-MYC as well as carbonic anhydrase 9, a prototypical HIF-1α target gene (Figure 7E, top panel). Both c-MYC and HIF-1 have been previously reported to underpin the metabolic adaptation of LKB1-deficient cells (32, 35). In addition, treatment with ganetespib simultaneously suppressed mTORC1 signaling output (evidenced by low levels of phospho-S6, phospho-p70S6K and phospho-4EBP1 protein), MAPK pathway activation (measured by levels of phospho-ERK1/2) and phosphorylated SRC kinase levels (Figure 7E, top panel), thus emulating the therapeutic combination that has previously been demonstrated to induce regression of Kras;Lkb1 co-mutant tumors in a GEMM model of NSCLC (36). Furthermore, CHK1, a checkpoint protein that recently emerged in a functional genetic screen as synthetic lethal in the context of LKB1 inactivation (37) was markedly depleted in response to HSP90 inhibition (Figure 7E, top panel). Finally, levels of IRE1A and PERK, two nodal components of the proteostasis network, were potently suppressed following HSP90 inhibition in all tested cell lines, in agreement with previous reports (Figure 7E, bottom panel) (38).
For 17-AAG and other chemically related benzoquinone ansamycin HSP90 inhibitors that are subject to NQO1-mediated bio-activation, we further reasoned that robust NRF2-driven up-regulation of NQO1 in LKB1-mutant lines – which frequently, like primary tumors, harbor mutations in KEAP1- would represent an additional mechanism accounting for their hyper-sensitivity to this chemical class of inhibitors (39). As predicted, LKB1-mutant cell lines – and most notably those with concurrent mutations in KEAP1- expressed higher levels of NQO1 protein (Figure 7F). Furthermore, concurrent treatment with dicumarol (50μM), a known inhibitor of NQO1 enzymatic activity, conferred resistance to 17-AAG, supporting a role for NQO1-mediated bio-activation in these cells. Finally, KL tumors expressed higher levels of NQO1 transcripts than KP tumors in two independent datasets (Figure 7G), in-keeping with widespread inactivation of KEAP1 and elevated basal NRF2 transcriptional activity.
Discussion
The development of more effective treatment strategies for LUAC bearing activating mutations in KRAS is hampered by the biological and phenotypic heterogeneity of KRAS-mutant tumors. Here we implement an integrated approach to the discovery of biologically distinct subgroups of KRAS-mutant LUAC. Employing NMF, an algorithm based on “decomposition by parts”, we identify three major expression-based subgroups that are highly reproducible across diverse clinical cohorts of both early-stage, chemotherapy-naïve and metastatic, platinum-refractory tumors. We subsequently explore the molecular underpinnings and potential therapeutic vulnerabilities of these subgroups.
Previously, we and others have demonstrated variable effector pathway engagement downstream of different common mutant KRAS alleles (KRASG12C, KRASG12V, KRASG12D) (9). Based on these observations we initially explored the possibility that specific alleles may be associated with the three subgroups but found generally no consistent association between KRAS alleles and the three expression clusters. We then considered other molecular features and discovered that co-occurring genetic events (non-silent mutations and/or bi-allelic deletions) in distinct tumor suppressor genes were significantly enriched in the three subgroups. Specifically, the second and third expression clusters were dominated, respectively, by genomic alterations in STK11/LKB1 and TP53. Somatic events involving these genes were clonal and largely non-overlapping among chemotherapy-naïve LUACs initiated by oncogenic KRAS, indicating that they likely represent gatekeepers to divergent pathways of tumor evolution. This finding is consistent with prior work in genetically engineered murine models of LUAC initiated by somatic activation of an endogenous KrasG12D allele, whereby concomitant genetic inactivation of either Stk11/Lkb1 or Trp53 sufficed to promote fully invasive and widely metastatic LUAC with shortened latency and complete penetrance, but variable phenotypic characteristics (40). In contrast, in the platinum-refractory BATTLE-2 cohort, increased frequency of triple mutant tumors (KRAS;STK11;TP53 - KPLm) was observed. It is unclear whether this represents expansion of a pre-existing subclone or accumulation of de novo mutations arising during therapy. We are currently investigating this issue using multi-region sequencing as we have recently described (24). It is notable, however, that all KRAS;STK11;TP53 triple-mutant tumors in BATTLE-2 still clustered together in the KL expression subgroup, suggesting that transcriptional fingerprints of LKB1 inactivation are retained in the context of co-occurring mutations in TP53.
Two additional frequently co-mutated genes, ATM and KEAP1, showed predilecton for the KL cluster. Interestingly, somatic mutations in ATM and KEAP1 were also largely non-overlapping in the TCGA, Imielinski and BATTLE-2 tumor collections. The frequent co-occurrence of KEAP1 mutations in KL tumors infers the application of positive selection pressure for up-regulation of a NRF2-mediated antioxidant, cyto-protective and anti-inflammatory transcriptional program. Increased generation of reactive oxygen species from cells undergoing energetic stress, coupled with defective H2O2 detoxification due to impaired NADPH maintenance may provide the impetus for this adaptive response (41, 42). However, it should be noted that a NRF2 expression signature was also enriched in KC LUACs, despite lower rates of KEAP1 somatic mutation, indicating potential alternative mechanisms for NRF2 upregulation and/or stabilization in this subgroup.
ATM mutations were most frequently encountered among KL LUACs that displayed evidence of functional LKB1-AMPK pathway inactivation but retained wild-type STK11/LKB1 genomic sequence. Activation of LKB1 through ATM-dependent phosphorylation at Thr366 in response to genotoxic or oxidative stress has been recently reported and may account for this predilection (43). Therefore, inactivating mutations in ATM, in addition to their effects on the DNA damage signaling cascade, may also promote a functional LKB1-deficient state.
The up-regulation of several ER chaperone proteins including GRP78 (Glucose-regulated protein – otherwise known as BiP) in KL LUACs indicates that adaptation to endoplasmic reticulum stress may constitute an additional homeostatic mechanism that contributes to cellular fitness in the context of LKB1 deficiency. Elevated rates of protein synthesis driven by constitutively active mTORC1 and high levels of intracellular ROS, coupled with hypoxia and energy/nutrient deprivation that are characteristic of in vivo tumor growth may, under conditions of impaired LKB1 function, promote increased dependency on ER and cytoplasmic chaperone proteins in order to mitigate the consequences of a high proteotoxic load. Activation of the UPR downstream of active mTORC1 has been previously demonstrated in genetically engineered murine models of constitutive TSC1/TSC2 depletion and contributes to several phenotypic characteristics of the tuberous sclerosis clinical syndrome (43). In addition, several nodal components of the UPR, including GRP78 and IRE1a, have been reported to exert context-dependent tumor promoting functions in various experimental models of cancer, and ATF4 can promote cyto-protective autophagy independently of AMPK and mTORC1(44); this latter function may be particularly relevant in the context of KRAS;LKB1 co-mutant cancer cells, which rely on efficient lysosomal degradation of macromolecules for supply of mitochondrial TCA-cycle substrates (45).
Unexpectedly, KP and KL tumors displayed distinct patterns of immune system engagement. KP tumors were characterized by an inflammatory response and their expression profiles showed enrichment for signatures of JAK/STAT pathway activation and interferon signaling. There was further evidence of active immunoediting, as indicated by robust expression of several co-stimulatory (e.g. CD28) and co-inhibitory molecules, including PD-L1, which was further validated immunohistochemically in the PROSPECT dataset. Thus, intrinsic regulation of T-cell function may be crucial for the establishment of functional tolerance towards KP LUACs. Accordingly, these tumors may be particularly amenable to therapeutic strategies that incorporate immune checkpoint blockade with inhibitors of PD-L1, PD-1, CTLA-4 or other checkpoint mediators (46). Recent preclinical studies from our group and others are consistent with this possibility (47). In keeping with the premise that a high somatic mutational load fosters tumor immunogenicity by generating a rich repertoire of neo-antigens, KP LUACs displayed higher global mutation rates than KL tumors in the TCGA cohort, despite similar cumulative exposure to smoking.
Contrary to tumors with somatic TP53 mutations, KL LUACs appeared largely “immune-inert”. The mechanism underlying this phenotype is not clear although a lower rate of somatic mutations may partly account for this relative lack of immune system engagement. It is also notable that secreted lactic acid, a by-product of HIF-1α–mediated metabolic reprogramming of LKB1-deficient cells towards glycolysis (32) is a potent lymphotoxin and may thus actively promote establishment of loco-regional immune privilege. Additionally, anti-inflammatory signals emanating from NRF2 may further oppose and prevent mounting of a significant inflammatory response to developing tumors with pertubations in the LKB1/AMPK pathway. Detailed characterization of the immune contexture of developing and established KL and KP tumors using genetically engineered murine models and large collections of human specimens will likely yield further insights into onco-genotype-specific immunoediting mechanisms.
Integrative analysis identified low expression of the NKX2-1 (TTF1) lineage-specific homeodomain transcription factor as a defining feature among tumors in the KC cluster. In striking resemblance to the lung cancer phenotype of GEMMs expressing oncogenic KRASG12D in the context of hemi- or homozygous Nkx2-1 inactivation, both trans-differentiation towards a GI-like phenotype – mediated by expression of the prototypical gastrointestinal transcription factors HNF4A and PDX1- and de-differentiation towards a more primitive state through derepression of the normally embryonically restricted chromatin regulator HMGA2 were evident among tumors in this group (27, 28). Notably, 6/15 tumors in this subgroup were classified histologically as invasive mucinous carcinomas, a phenotype that was not observed in the KL and KP subgroups. These results are concordant with a separate study that reported a strong association between negative TTF1 immunohistochemical staining and mucinous histology in KRAS-mutant LUACs (48).
The KC subgroup was significantly enriched in tumors bearing bi-allelic loss of CDKN2A and/or CDKN2B. Consistent with this finding, concomitant genetic ablation of Cdkn2a and Cdkn2b in a murine model of Kras-mutant lung cancer resulted in a high percentage of TTF1-negative tumors that frequently overexpressed HMGA2, suggesting that loss of this locus may directly impact the differentiation state of at least some LUACs in the KC subgroup (25). It is also noteworthy- and potentially clinically relevant- that KC tumors displayed evidence of reduced mTORC1 signaling output and expressed high basal levels of wild-type p53 regulated transcripts.
It should be noted that the KRAS -focused analysis presented here affords additional and complementary biological insights to the three expression and six integrated clusters proposed by TCGA based on an un-selected pool of LUACs. For example, the KC subgroup integrates tumors from all three TCGA expression clusters and includes most iCluster6 LUACs, an integrative cluster for which no distinctive features had hitherto been identified. In addition, iCluster2 incorporates most KL LUACs with ATM co-mutations and the majority of CIMP-H (CpG-island methylator phenotype-high) (iCluster3 and iCluster4) KRAS-mutant tumors are classified as KP in our analysis. In contrast, lower levels of p16 (CDKN2A) promoter methylation in KL LUACs are consistent with an intact CDKN2A/CDKN2B genomic locus in the majority of tumors in this subgroup.
Comparison of RFS in the three KRAS-mutant LUAC subgroups in PROSPECT demonstrated improved RFS in KP compared to either KC alone or non-KP (KL and KC) tumors. This result is intriguing in view of the immune phenotype of KP LUACs and previous reports linking lymphocytic infiltration in surgically resected colorectal tumors with reduced likelihood of relapse (33). However, due to the limited number of patients reported here, definitive assessment of the prognostic utility of the proposed classification will require additional, adequately powered, prospective studies.
Finally, we provide evidence that the proposed sub-classification of KRAS-mutant LUACs is therapeutically relevant. By coupling re-analysis of publicly available large scale drug sensitivity data with targeted screening of an extensive panel of comprehensively annotated KRAS;LKB1 (KLm/KPLm) and KRAS;TP53 (KPm) NSCLC cell lines, we uncover differential sensitivity to a number of drugs including enhanced sensitivity of LKB1-mutant (KLm or KPLm) cells to several chemically distinct inhibitors of the HSP90 molecular chaperone. We further demonstrate that in response to ganetespib, an HSP90 inhibitor that is currently undergoing phase 3 clinical evaluation in combination with docetaxel in metastatic LUAC patients following failure of first line therapy, several signaling pathways and adaptive biological responses with an established role in maintenance of cellular fitness in the setting of LKB1 inactivation are concomitantly destabilized. This enhanced sensitivity is therefore likely to be multifactorial and may be at least partly attributed to: 1) Degradation of HIF-1α and MYC, which underpin metabolic adaptation to LKB1 loss by promoting a shift towards glycolysis; 2) Depletion of CHK1, which may be necessary for processing of stalled replication forks in the setting of depleted nucleoside pools; and 3) Inactivation of HSP90-dependent cyto-protective branches of the UPR that facilitate tolerance of a high proteotoxic load. Regardless of the mechanism, however, we find that HSP90 sensitivity can directly be impacted by modulating LKB1 status in vitro, thus supporting a direct role for LKB1. Furthermore, this analysis identifies NRF2-mediated anti-oxidant responses and UPR pathways as potential therapeutic targets for LKB1-deficient KRAS-mutant LUACs.
Taken together, our findings have several important implications for the molecular stratification and precise therapeutic targeting of LUACs initiated by oncogenic KRAS. First, STK11/LKB1 and TP53 appear to control distinct tumorigenic pathways downstream of mutant KRAS. Therefore, clinical trials enrolling patients with KRAS-mutant LUACs should take into account the co-mutation status of individual tumors. Second, it is important to recognize that patterns of co-occurring or mutually exclusive genetic events may evolve under the selective pressures imposed by cancer therapy – as is the case for somatic mutations in STK11/LKB1 and TP53 that rarely co-occur in KRAS-mutant chemo-naïve tumors but frequently do so following development of secondary resistance to cytotoxic agents. Thus, tumors initiated by the same apical oncogenic event may warrant different therapeutic approaches depending on their treatment history. Third, it is evident that even in the absence of identifiable genomic aberrations, a substantial number of LUACs exhibit functional abrogation of the LKB1-AMPK pathway; development of validated assays for the identification of these cases based on immunohistochemical detection of LKB1 or phospho-AMPK Thr172 protein expression (49), serum biomarkers, mRNA-based expression signatures (50) or even “surrogate” somatic mutations in KEAP1 or ATM is therefore a critical step towards appropriate patient stratification. Finally, our analysis highlights dependence on the molecular chaperone machinery as a therapeutic vulnerability in cancer cells with co-mutations in KRAS;LKB1 that can be tackled with HSP90 inhibitors; on the other hand, tolerance of a higher mutation load in tumors with concurrent mutations in TP53 and establishment of a prominent inflammatory response may render this subset particularly susceptible to immune checkpoint blockade and other novel immunotherapy approaches. Thus, our work advances current understanding of KRAS-driven lung carcinogenesis, suggests a framework for the molecular classification of KRAS-mutant LUACs and facilitates the implementation of personalized therapy.
Materials and methods
Clinical cohorts
The TCGA, PROSPECT, Imielinski et al., Chitale et al., Ding et al. and JBR.10 clinical cohorts of un-treated, mostly early-stage LUACs have been previously reported (12, 16, 18-20, 51). BATTLE-2 enrolled patients with advanced NSCLC, following failure of at least one front-line chemotherapy regimen, with fresh tumor biopsy mandated prior to adaptive randomization. 41 KRAS-mutant LUACs from stage 1 of BATTLE-2 were included in the current study, with mRNA expression data available for 36 tumors. In all cases, bio-specimens were obtained following patient informed consent, under protocols approved by Institutional Review Boards at all participating institutions. All human studies were conducted in accordance with the Declaration of Helsinki.
Molecular profiling
Level 3 somatic mutation, copy number (GISTIC 2.0), RNA-Seq, miRNA-seq and RPPA data for TCGA LUACs were accessed through the TCGA portal. Expression profiling of PROSPECT and BATTLE-2 tumors was performed using the Illumina Human WG-6 v3 BeadChip and Affymetrix GeneChip®Human Gene 1.0 ST Array, respectively. Massively parallel sequencing exome capture of 19 KRAS-mutant LUACs from PROSPECT was performed using the NimbleGen Sequence Capture 2.1M human exome array and whole exome sequencing was undertaken on the Illumina HiSeq2000 platform as previously described (52). Sample preparation, basic alignment and downstream analyses were performed according to previously described methods (53-55). Detailed methodology can be found in Supplementary Methods. Targeted sequencing of coding exons and selected introns of cancer-related genes from 41 KRAS-mutant BATTLE-2 tumors and copy number analysis based on Affymetrix SNP 6.0 array profiling was performed by Foundation Medicine, as previously described (56). RPPA analysis was performed as previously reported (12, 57). The ABSOLUTE algorithm was employed for clonality analyses (22). Unsupervised NMF consensus clustering was performed using NMF Version 0.5.06 (R package) (14). Development of the cluster assignment signature was based on the ClaNC algorithm (15). Further details can be found in Supplementary Methods.
mRNA expression profiling
For LUACs included in the TCGA cohort, experimental procedures regarding RNA extraction from tumors, mRNA library preparation, sequencing (on the Illumina HiSeq platform), quality control and subsequent data processing for quantification of gene expression have been previously reported (12). Details regarding sample collection, storage, selection and RNA extraction for PROSPECT tumors have also been previously published (20). Array-based expression profiling of PROSPECT tumors was performed using the Illumina Human WG-6 v3 BeadChip, according to the manufacturer’s protocol. BATTLE-2 LUACs were profiled using the GeneChip®Human Gene 1.0 ST Array from Affymetrix. The tumors reported by Chitale et al were profiled using the HG-U133A and HG-U133A 2.0 arrays from Affymetrix, as previously reported (16). Gene expression data for the PROSPECT cohort have been previously deposited in the GEO repository (GSE42127). Gene expression data for the BATTLE-2 tumors have been deposited in the GEO repository (GSE61913). Tumors from the JBR.10 trial were profiled using the U133A oligonucleotide microarray from Affymetrix as previously described (51) and were accessed through the GEO repository (GSE14814). For cross-platform integration of mRNA expression data, expression levels for each gene were derived by averaging corresponding probesets and standardized within each dataset. Missing values were replaced with 0.
Immunohistochemistry
4μm- thick tissue sections were stained with the indicated antibodies (see Supplementary Table 2) using an automated staining system (Leica Bond Max, Leica Microsystems, Vista, CA, USA), according to standard protocols. The Aperio Image Analysis Toolbox (Aperio, Leica Microsystems) was used for digital analysis of images obtained from scanned slides, as described in Supplementary Methods. The H-score for TTF1 was determined manually, jointly by two thoracic pathologists, as previously described (58).
NSCLC cell line propagation and authentication
NSCLC cell lines were established by Dr J.D.Minna and Dr A. Gazdar at the National Cancer Institute and the University of Texas Southwestern Medical Center (Dallas, TX) or were obtained from ATCC. They were maintained in RPMI-1640 (R8758, Sigma Life Science), supplemented with glutamine, 10% heat-inactivated fetal bovine serum (Gibco) and 1% Penicillin/Streptomycin (Sigma Life Science) in a humidified chamber at 5% CO2. All cell lines were authenticated between 2009 and 2011 using STR profiling (PowerPlex 1.2, Promega, Madison, WI) for at least eight different loci and results were compared to reference STR profiles available through ATCC or provided by Dr Minna at UTSW. Following authentication, cell line stocks were frozen and maintained in liquid nitrogen until they were used in the reported experiments. All cell lines were mycoplasma- tested prior to experiments.
Western blotting and qRT-PCR
qRT-PCR and Western analysis were performed following standard protocols, as detailed under Supplementary Methods. 35μg of total protein lysate was transferred to PVDF membranes and blotted with the indicated antibodies prior to development using picoLUCENT™ PLUS-HRP (G-Bioscience) or ECL (Amersham) detection reagents.
Cell viability assay
Cell viability was determined following 72- hour exposure of NSCLC cells (in triplicate wells) to seven serial fold dilutions of the indicated drugs using the Cell-Titer Glo® Luminescent assay (Promega) according to the manufacturer’s protocol, with minor modifications. For IC50 determination, multiple models were fitted and the best one selected based on residual standard error. Details of the procedures can be found in Supplementary Methods.
Statistical analyses
Statistical analyses were conducted using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, California, USA) or the R system for statistical computing. All reported P values are two-tailed and for all analyses, P≤0.05 is considered statistically significant, unless otherwise specified. Box and whiskers plots indicate median and interquartile range. Kaplan-Meier RFS and OS curves were compared using the log rank test. Multivariate analysis was based on the Cox proportional hazards model and included adjuvant therapy and nodal status as additional co-variates. Further details can be found in Supplementary Methods.
Supplementary Material
Significance.
Co-occurring genetic alterations in STK11/LKB1, TP53 and CDKN2A/B – the latter coupled with low TTF1 expression - define three major subgroups of KRAS-mutant LUAC with distinct biology, patterns of immune system engagement and therapeutic vulnerabilities.
Acknowledgements
We would like to thank Emily Roarty, Ph.D. for critical review of the manuscript and editorial assistance and Drs Ming-Sound Tsao, M.D., Frances A. Shepherd, M.D. and Chang-Qi Zhu, M.D.,Ph.D. for providing information on the KRAS mutation status of LUACs from the JBR.10 clinical trial.
Financial support: This work was supported by The University of Texas Southwestern Medical Center and The University of Texas MD Anderson Cancer Center Lung SPORE grant 5 P50 CA070907; DoD PROSPECT grant W81XWH-07-1-0306; 1 R01 CA155196-01A1 to V.P. and R.S.H.; Lung Cancer Moon Shot Program; M.D. Anderson Immunotherapy Platform; National Institute of Health Cancer Center Support Grant (CA016672); 1R01 CA168484-01 to J.V.H.; V Foundation Grant to J.V.H.; David Bruton, Jr. Endowed Chair to J.V.H.; Ford Petrin Donation; F.S was supported by the Jeanine T. Rainbolt Advanced Scholar Endowment, the Eric and Pat Bodin Lung Cancer Research Fellowship and a Uniting Against Lung Cancer Legacy Program for Advances in Lung Cancer Research grant.
Footnotes
Potential conflicts of interest: V.M. and G.M.F. are employees of and shareholders in Foundation Medicine. P.S. and J.P.A. serve as founders and consultants for Jounce Therapeutics; P.S. is a consultant for Bristol-Myers Squibb and GlaxoSmithKline. G.B.M. is a consultant for or member of the Scientific Advisory Board of AstraZeneca, Blend, Critical Outcome Technologies, HanAl Bio Korea, Illumina, Nuevolution, Pfizer, Provista Diagnostics, Roche, Signalchem Lifesciences, Symphogen, Tau Therapeutics; holds stock/options in Catena Pharmaceuticals, PTV Ventures, Spindle Top Ventures; has received research grant support from Adelson Medical Research Foundation, AstraZeneca, Critical Outcomes Technology, GSK. All other authors declare no conflicts of interest.
References
- 1.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31(8):1105–11. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 6.Berge EM, Doebele RC. Targeted therapies in non-small cell lung cancer: emerging oncogene targets following the success of epidermal growth factor receptor. Semin Oncol. 2014;41(1):110–25. doi: 10.1053/j.seminoncol.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stinchcombe TE, Johnson GL. MEK inhibition in non-small cell lung cancer. Lung Cancer. 2014 doi: 10.1016/j.lungcan.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104(3):228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd FA, Domerg C, Hainaut P, Janne PA, Pignon JP, Graziano S, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31(17):2173–81. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483(7391):613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101(12):4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabney AR. ClaNC: point-and-click software for classifying microarrays to nearest centroids. Bioinformatics. 2006;22(1):122–3. doi: 10.1093/bioinformatics/bti756. [DOI] [PubMed] [Google Scholar]
- 16.Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28(31):2773–83. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadimitrakopoulou V, Wistuba II, Lee JJ, Tsao AS, Kalhor N, Fossella FV, et al. BATTLE-2 program: A biomarker-integrated targeted therapy study in previously treated patients with advanced non-small cell lung cancer (NSCLC) ASCO Meeting Abstracts. 2013;31(15_suppl):TPS8118. doi: 10.1200/JCO.2015.66.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19(6):1577–86. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster K, Venkateswaran N, Rabellino A, Girard L, Pena-Llopis S, Scaglioni PP. Nullifying the CDKN2AB locus promotes mutant K-ras lung tumorigenesis. Mol Cancer Res. 2014;12(6):912–23. doi: 10.1158/1541-7786.MCR-13-0620-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50(2):185–99. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda Y, Tsuchiya T, Hao H, Tompkins DH, Xu Y, Mucenski ML, et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest. 2012;122(12):4388–400. doi: 10.1172/JCI64048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68(24):10105–12. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 31.Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem. 2011;286(47):40725–33. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A. 2014;111(7):2554–9. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. The Journal of pathology. 2010;222(4):350–66. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai LH, Wu JY, Cheng YW, Chen CY, Sheu GT, Wu TC, et al. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene. 2014 doi: 10.1038/onc.2014.118. [DOI] [PubMed] [Google Scholar]
- 36.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17(6):547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3(8):870–9. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1alpha. Mol Cell Biol. 2002;22(24):8506–13. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel D, Yan C, Ross D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem Pharmacol. 2012;83(8):1033–40. doi: 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 41.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–58. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–5. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107(9):4153–8. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122(12):4621–34. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–66. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quezada SA, Peggs KS, Simpson TR, Allison JP. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol Rev. 2011;241(1):104–18. doi: 10.1111/j.1600-065X.2011.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nature communications. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol. 2013;26(10):1307–19. doi: 10.1038/modpathol.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakada Y, Stewart TG, Pena CG, Zhang S, Zhao N, Bardeesy N, et al. The LKB1 tumor suppressor as a biomarker in mouse and human tissues. PLoS One. 2013;8(9):e73449. doi: 10.1371/journal.pone.0073449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, et al. LKB1 Loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J Thorac Oncol. 2014;9(6):794–804. doi: 10.1097/JTO.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28(29):4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao S, Choi M, Overton JD, Bellone S, Roque DM, Cocco E, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci U S A. 2013;110(8):2916–21. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi M, Scholl UI, Yue P, Bjorklund P, Zhao B, Nelson-Williams C, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331(6018):768–72. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–90. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res. 2013;19(23):6556–65. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.