We demonstrate a system whereby DNA nanostructure tiles play an active role in their own self-assembly by initiating a binding event that produces a cascading assembly process. We present DNA tiles that have a simple but powerful property: they respond to a binding event at one end of the tile by passing a signal across the tile to activate a binding site at the other end. This action allows sequential, virtually irreversible self-assembly of tiles and enables local communication during the self-assembly process. This localized signal-passing mechanism provides a new element of control for autonomous self-assembly of DNA nanostructures.
A wide variety of nanostructures and devices can self-assemble from DNA[1]. The programmability of DNA as a material extends to both structure and dynamics, where dynamic behavior can be programmed via toehold-mediated isothermal DNA strand displacement[2]. The design of reconfigurable and active nanostructures is an important challenge in DNA nanotechnology[3], but so far there are not many examples that merge structure with dynamics[4].
Self-assembly can be a computational process, as shown by an abstract model called the abstract Tile Assembly Model (aTAM)[5] DNA tiles made from a small double-crossover motif known as DX[6] were the first to be used in the formation of 2D arrays[7] and have been used in algorithmic assembly[8] as modeled by the aTAM. In these previous demonstrations, cooperation between two or more (available) binding sites yields algorithmic self-assembly. Signal-passing in tile self-assembly was proposed as an augmentation to the Tile Assembly Model that brings a new form of communication during assembly and coordination of self-assembly that does not have to depend on passive binding sites cooperativity[9]. Tiles with active binding sites were originally proposed as a method to decrease error rates during self-assembly[10], and active, signal-passing tiles were proposed to coordinate hierarchical recursive assembly[8a]. Other forms of active assembly are being explored in various theoretical models[11], but the mechanism presented here sets up a domino–like cascade[12] that expands upon the hybridization chain reaction[2c] and other triggered self-assembly methods[2e] by producing finite assemblies of specific length, and by incorporating stiff structural motifs into the product assemblies.
We demonstrate the self-assembly of DNA nanostructures with a DNA strand exchange signaling mechanism through a sequence of five DX tiles, four of which contain the full signal-passing machinery; this process is active self-assembly, in contrast to passive self-assembly in un-triggered systems. A binding event occurring at one end of a DX tile releases a previously tethered signal sequence, allowing it to activate a binding site ~18 nm away on the other end of the same DX tile in response. The tethered DNA strand transmits the information locally within the fixed and stiff[13] DX structure, so that each individual assembly grows sequentially by a single tile addition, while the global population remains asynchronous. We have used fluorescent-labeling and non-denaturing PAGE analysis to show that tile assembly occurs sequentially and have verified by atomic force microscopy (AFM) that the expected products are formed.
The signal-passing mechanism is shown in Figure 1. The sequential binding of four tiles to the initiator tile occurs by a toehold-mediated[2a] strand exchange cascade (Figure 1). At the start, there is only one active strand, cBa, (where sequence motifs are listed in 5’ to 3’ order) on Tile A, which we refer to as a “glue strand.” The first binding event is shown in Figure 1b, where Tile A has bound to Tile B via the exposed complementary sequences c and c*, which we refer to as “toeholds” displacing the signal strand. This strand displacement translates the binding event into a signal, carried by the tethered sequence element dBa. The signal is transmitted when the tethered signal strand, dBa in Figure 1b, attaches to the toehold a* on the cover strand, a*B*d*. Here, the cover strand is the strand that functions to protect a portion of the output strand, including its toehold, so that when the cover strand is removed, as in Figure 1c, the output strand, eDb, has been activated. This definition differs slightly from a previous definition of ‘cover strand’[14] The removal of the cover strand translates the signal into a binding site activation. Figure 1c-1f shows the stepwise addition of tiles via this mechanism.
Figure 1.
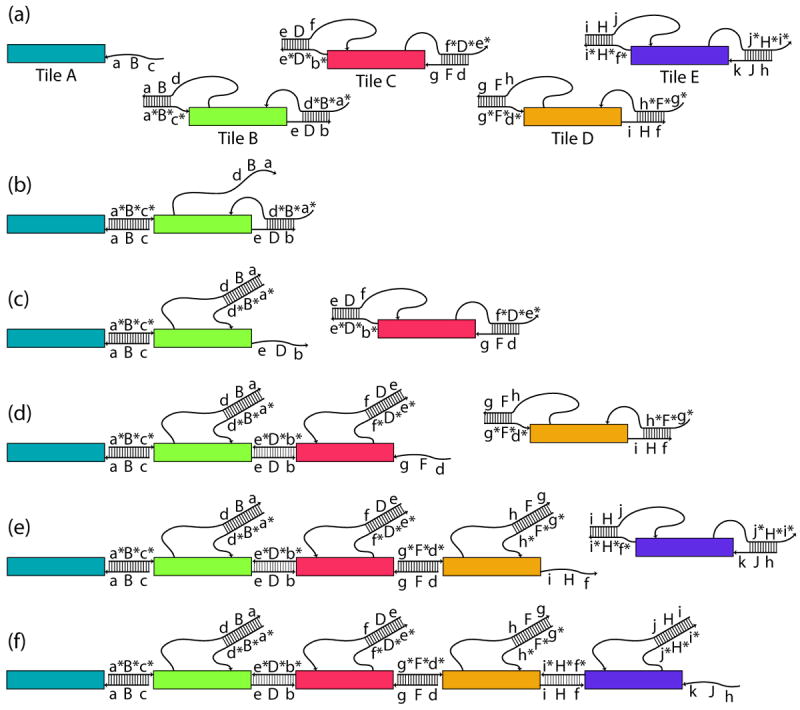
Schematic of DNA strand exchange mechanism for sequential addition of tiles. Rectangles represent the body of the tile, thin lines represent relevant DNA strands in the mechanism with an arrow indicating the 3’ end and a ladder showing base pairs between strands. Sequence segments of 5 nucleotides are labeled with lowercase letters, uppercase denotes 6-base sequences that are a one base augmentation of a 5-base sequence of the same letter, and complementary sequences are starred. a) A schematic representation of the five tiles tested in this paper. Augmented 6-base sequences are such that dB = Db, dF = Df, hF = Hf, and hJ = Hj. b) Binding is initiated at a 5-nucleotide toehold, c*, on the second tile (green). Strand exchange releases the tethered signal strand, allowing it to reach the 5-nucleotide toehold on the cover strand. c) The second strand exchange event removes the cover strand to release the sticky end, activating it. The activated 16-nucleotide glue retains only a 6-nucleotide sequence from the previous glue. On the next tile (two steps away from the initiator) the activated glue sequence will be completely independent of the initial glue sequence. d) Three-tile assembly after the addition of Tile C and the activation of its glue strand. e) Four-tile assembly. f) Schematic of the final assembly of five tiles. Note that in a complete assembly, all bases are paired except for the last output strand of 16 bases.
The tiles are composed of a DX motif consisting of two DNA double helices held together via two crossovers (Figure 2), where one helix, the “active helix”, contains all the signaling machinery and the other helix is passive. The active portions of each tile consist of two sticky ends (input and output), a signal strand and a cover strand (Figure 2). There are 21 nucleotide pairs in the internal loop, and 16-nucleotide sticky ends on the active helix that meet to form a total of 32 nucleotide pairs between crossovers on tiles that are bound together. The passive helix contains 2-nucleotide stabilizing single-stranded overhangs at both ends that do not initiate binding between tiles. To send a signal locally across a tile, the signal sequence remains tethered to the tile via an oligo-dT DNA strand long enough to reach the other end of the ~18 nm-long tile. The oligo-dT DNA was chosen to minimize the chance of interference with the signal strand while remaining easy and inexpensive to synthesize. To decrease potential interference, we replaced the oligo-dT DNA with an alternating dT-PEG6 linker containing the same number of bonds; it did not improve the processing, so we did not pursue this avenue further. Confining the signal strand locally to the tile, where it is expected that in most events, intramolecular interactions will occur far more frequently than intermolecular events, is a design feature that helps avoid unintended inter-tile interactions. The strand displacement mechanism confers on each tile the ability to respond to a binding event at the input end with the activation of a binding site on the other end of the tile by the uncovering of the output strand[14].
Figure 2.
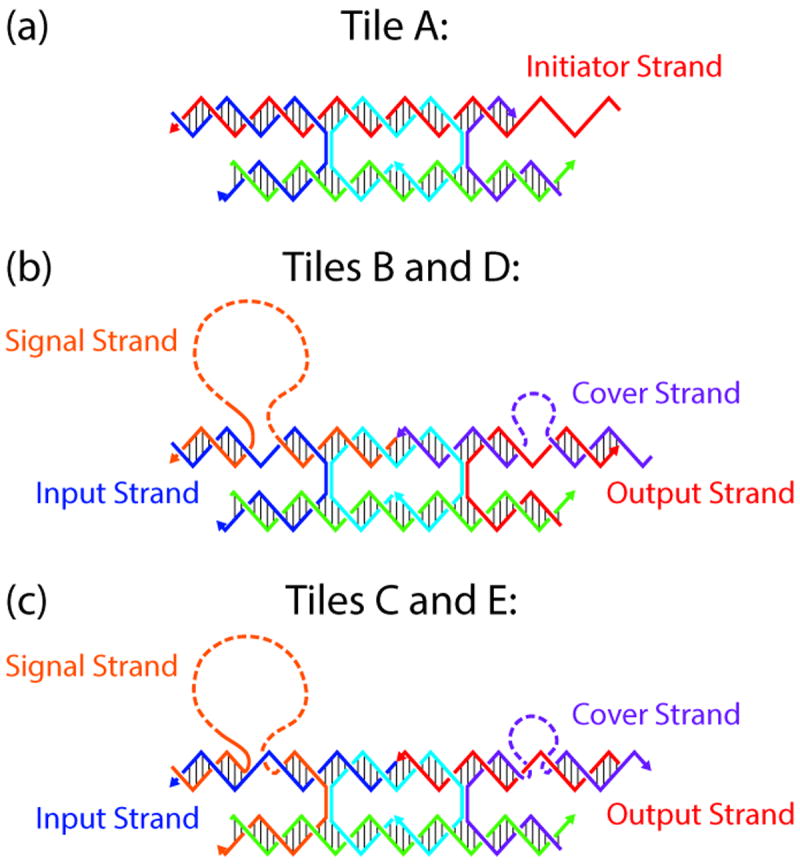
Schematic of the DX signal-passing tile structures. Each tile shares the same core strand sequence (cyan). a) Tile A is the initiator of the assembly signal cascade, while tiles B, C, D, and E each have the complete signaling machinery with a signal strand on the upper right tethered by a poly-T sequence. The upper right of each DX tile has a cover strand (colored purple) that protects the output strand from hybridizing until it has been removed by the signal strand when the signal strand is displaced by a binding event on the input end of the tile. b) Tiles B and D have a 5’ input sticky end and 3’ output sticky end, while these strand polarity orientations are reversed for Tiles C and E in panel c). The lower helix plays a passive role and has 2-nucleotide sticky ends designed to facilitate orientation of the DX tiles, but these interactions are not strong enough to form base pairs at ambient temperatures and concentrations.
The driving force for the cascade of tile additions is a gain of 5 nucleotide pairs per tile. Prior to binding and signal passing, a tile has 22 nucleotide pairs on the glue strands when configured as in Figure 2. On an individual tile, this number would drop to 11 if the signal strand and cover strand were to bind one another in the absence of binding partners for the glue strands, making the pre-activation state energetically more favorable in the absence of other elements. This idea is supported by the fact that each individual tile is produced by annealing in a one-pot reaction with all strands present. Once a tile is bound on both sides and the signal strand is bound to the cover strand, there are no more unpaired nucleotides on that tile aside from the 16-base output strand sequence, the oligo-dT tether and the dinucleotide stabilizers. After signal passing and incorporation into the linear assemly, the total number of nucleotide pairs per tile is 27, if we consider each glue strand to be shared with its neighbour. Thus, there is a net gain of 5 nucleotide pairs of binding free energy with each signaling event. The tether length was chosen to span the length of the tile so the signal strand could reach the cover strand toehold, as well as to maintain a physically unstrained structure after signal-passing. An estimate of tether length is given in the Supplement, section 6.
Non-denaturing PAGE of fluorescently labeled tiles (Figure 3) shows that the reaction proceeds as designed, and can neither progress beyond a missing tile, nor start without the initiator sequence. Figure 3 shows the lengths of the products at which the reaction stops when a tile is missing. We also labeled each tile individually and compared with the initiator to show that the tiles assemble the correct complexes at the correct time (Supp. Figure S4). These experiments show that the tiles add sequentially with an “if A then B” logic. AFM (Figure 4) reveals linear structures of the expected dimensions for this 5-tile linear complex. The expected 16-base duplexes between signal and cover strands that should be tethered to the 5-tile complex are difficult to resolve in the AFM image. The images cannot reveal which end contains the initiator tile, but they show that the assemblies are the correct length.
Figure 3.
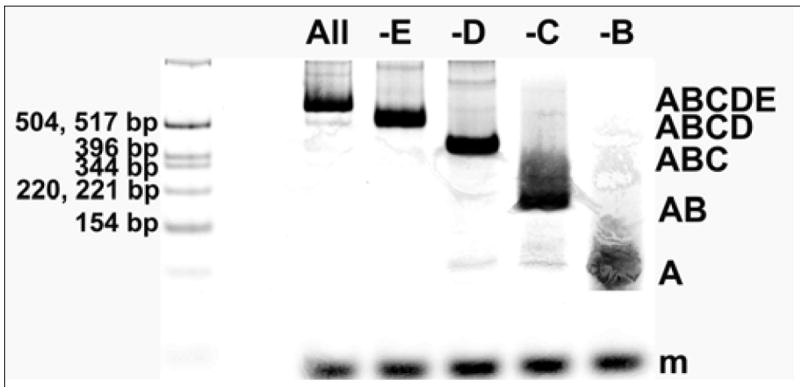
Native PAGE on tile reaction mixtures where Tile A is fluorescently labeled with ATTO633. The leftmost lane is a fluorescein-labeled molecular weight marker and each lane contains a 70 base-pair scaling marker labeled with fluorescein. Tiles were mixed in TAE/10.5 mM Mg2+ buffer and incubated 16 hours at 16° C. Major product complexes of expected size formed in each case when a tile was omitted or with the full tile set. Complexes formed are indicated at the right side of the image. Lane contents are as follows, where an underscore marks the addition of buffer in place of a missing tile, and the percentage of total lane contents according to gel analysis using ImageQuant.net software: (1) molecular weight marker fluorescein labeled (2) ABCDE (3) ABCD_ (4) ABC_E (5) AB_DE (6) A_CDE. This gel is highly enhanced to show minor products. The fluorescence intensity of the product band in each lane is adjusted by the intensity of the scaling marker in the same lane and the percentages of products formed (from an average of 2 reactions) are the following: Lane 2, ABCDE: 70.5%, Lane 3, ABCD: 80.5%, Lane 4, ABC: 87.5%, Lane 5, AB: 90.5%, Lane 6, A: 100%. Average step-yield, 91.6%.
Figure 4.
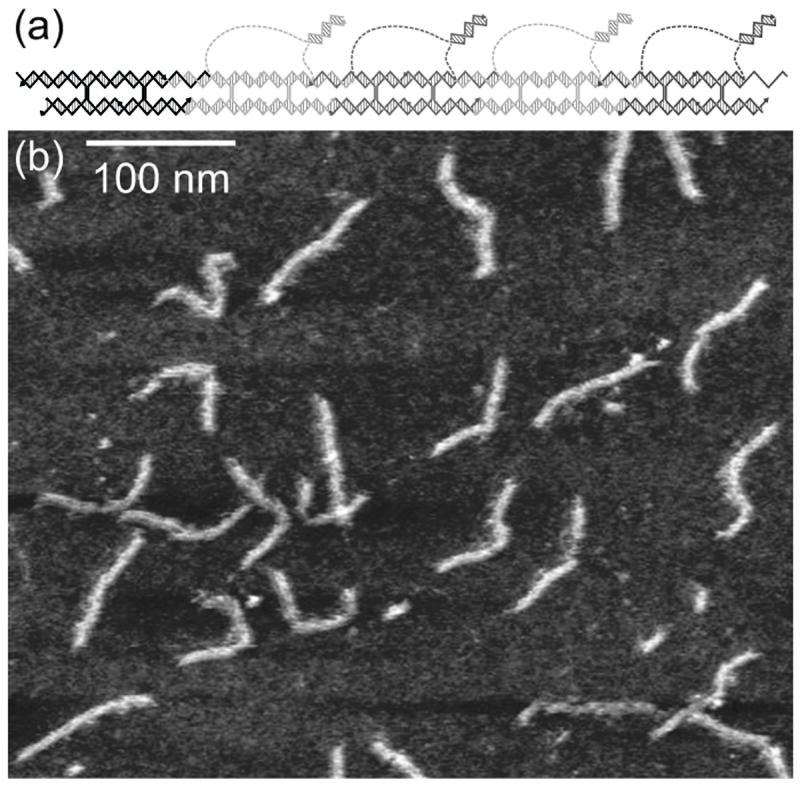
Atomic force microscopy of the 5-tile assembly. a) Schematic of the expected 5-tile assembly. b) Atomic force micrograph of the 5-tile assembly product, purified by native PAGE. Linear assemblies of expected length near 93 nm are seen
Material that does not eventually become incorporated into the 5-tile complex may consist of tiles that have participated in unintended side reactions. In the literature on nucleic acid strand displacement systems, there are instances of so-called “leakage”[2c, 2e], where a small portion of strands participate in strand displacement in the absence of triggering. In some cases the intended reaction simply proceeds without the trigger, and in some cases complexes of unintended size are observed. In this last case, the leakage reactions produce unintended aggregates. Due to the necessity of repeating certain 5 to 6-nucleotide segments in the glue strand, signal strand, and cover strand sequences, there are matching sequences between strands that are not intended to bind one another in the designed mechanism (Figure 2), but which may do so via sequence domains that are shorter than the intended binding regions. These may be the source of the unintended aggregates that migrate more slowly than the 5-tile product seen on the fluorescently labeled native gels (Figure 3). While we appear to lose some material to uncharacterized aggregation as the reaction proceeds, a 24-hour incubation of tile mixtures lacking the initiator at 16° C no visible leakage products of the first kind in an ethidium bromide stained native gel (see Supp. Figure S5) That is to say, the linear polymerization of the tiles appears not to proceed in the absence of initiator. In this regard, this system is robust to false triggering.
The signal initiated self-assembly mechanism demonstrated here introduces a new means for an active self-assembly of DNA nanostructures. We have enabled communication and control in the self-assembly process. Two important features of this triggered assembly mechanism are that (1) in principle it can be nucleated from anything to which a 16-nucleotide initiator strand can be attached, such as a gold nanoparticle or a DNA origami structure and (2) it is a controlled cascading assembly process with an “if A then B” action, like a row of dominoes. Control of sequential assembly occurs isothermally, and does not require a carefully chosen annealing temperature or the thermal programming that is often required for achieving cooperation in passive DNA tile assembly. This is a rudimentary mechanism that could be used for signal-passing tile assembly[9a, 9b, 9d], as described by the Signal-passing Tile Assembly Model, (STAM)[9b,9d], a theoretical model of tile self-assembly augmented with a basic communication mechanism. Future directions for this work that would fully implement the STAM theoretical model include glue deactivation and fanout (where one input signal would produce two or more output signals). Signal transmission demonstrated here across four steps is robust, showing a good average step-yield of 91.6%. Our experiments (not included here) show that the system can be extended up to seven tiles with similar yield. For very long signal transmission pathways, we suggest that redundant pathways could be used to ensure that at least one signal reaches the other side of a larger DNA nanostructure. Redundant pathways could utilize orthogonal sequence designs to minimize interference. Given its inherent robustness and potential for redundancy, the mechanism demonstrated here for self-assembly via signal transmission may prove useful in coordinating autonomous self-assembly of DNA nanostructures across a range of size scales. In addition to its capability of facilitating the reliability of algorithmic assembly, the approach described here can be used to initiate structure-forming pathways from a variety of sites at a variety of times, and from a variety of starting materials, ranging from origami to nanoparticles.
Supplementary Material
Footnotes
This research has been supported by grant CCF-1117210 from the National Science Foundation to NCS, NJ, and JEP, NIH grant R01GM-109459-01 to NJ; and the following grants to NCS: GM-29554 from NIGMS, grants CMMI-1120890, and EFRI-1332411 from the NSF, MURI W911NF-11-1-0024 from ARO, grants N000141110729 and N000140911118 from ONR, grant 3849 from the Gordon and Betty Moore Foundation. We would like to thank Yoel Ohayon for technical advice.
Contributor Information
Jennifer E. Padilla, Department of Chemistry, New York University, New York, NY 10003, USA
Ruojie Sha, Email: ruojie.sha@nyu.edu, Department of Chemistry, New York University, New York, NY 10003, USA.
Martin Kristiansen, Email: mk3474@nyu.edu, Department of Chemistry, New York University, New York, NY 10003, USA.
Junghuei Chen, Email: junghuei@udel.edu, Department of Chemistry and Biochemistry, University of Delaware, Newark, DE 19716, USA.
Natasha Jonoska, Email: jonoska@usf.edu, Department of Mathematics, University of South Florida, Tampa, FL, USA.
Nadrian C. Seeman, Email: ned.seeman@nyu.edu, Department of Chemistry, New York University, New York, NY 10003, USA.
References
- 1.a) Edwards A, Yan H. In: Nucleic Acid Nanotechnology. Kjems J, Ferapontova E, Gothelf KV, editors. Vol. 29. Springer Berlin Heidelberg; 2014. pp. 93–133. [Google Scholar]; b) Linko V, Dietz H. Curr Opin Biotechnol. 2013;24:555–561. doi: 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]; c) Krishnan Y, Simmel FC. Angew Chem. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]; d) Seeman NC. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]; b) Yan H, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]; c) Dirks RM, Pierce NA. Proc Natl Acad Sci USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]; e) Yin P, Choi HM, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]; f) Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]; g) Wang F, Lu CH, Willner I. Chem Rev. 2014;114:2881–2941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]; h) Zhang DY, Hariadi RF, Choi HM, Winfree E. Nat commun. 2013;4 doi: 10.1038/ncomms2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinheiro AV, Han D, Shih WM, Yan H. Nat Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Nie Z, Wang P, Tian C, Mao C. Angew Chem. 2014;53:8402–8405. doi: 10.1002/anie.201404307. [DOI] [PubMed] [Google Scholar]; b) Sadowski JP, Calvert CR, Zhang DY, Pierce NA, Yin P. ACS Nano. 2014 doi: 10.1021/nn4038223. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Teichmann M, Kopperger E, Simmel FC. ACS Nano. 2014;8:8487–8496. doi: 10.1021/nn503073p. [DOI] [PubMed] [Google Scholar]; d) Ding B, Seeman NC. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winfree E. PhD Thesis. California Institute of Technology; 1998. [Google Scholar]
- 6.Fu TJ, Seeman NC. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 7.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 8.a) Rothemund PW, Papadakis N, Winfree E. PLoS Biol. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fujibayashi K, Hariadi R, Park SH, Winfree E, Murata S. Nano Lett. 2007;8:1791–1797. doi: 10.1021/nl0722830. [DOI] [PubMed] [Google Scholar]
- 9.a) Padilla JE, Liu W, Seeman NC. Nat Comput. 2012;11:323–338. doi: 10.1007/s11047-011-9268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Padilla JE, Patitz MJ, Schweller RT, Seeman NC, Summers SM, Zhong X. Int J Found Comput S. 2014;25:459–488. [Google Scholar]; c) Hendricks J, Padilla JE, Patitz MJ, Rogers TA. In: DNA Computing and Molecular Programming. Soloveichik D, Yurke B, editors. Vol. 8141. Springer; 2013. pp. 90–104. LNCS. [Google Scholar]; d) Jonoska N, Karpenko D. Int J Found Comput S. 2014;02:141–163. [Google Scholar]
- 10.Majumder U, LaBean TH, Reif JH. In: DNA Computing. Garzon MH, Yan H, editors. Vol. 4848. Springer; 2008. pp. 15–25. LNCS. [Google Scholar]
- 11.a) Chen M, Xin D, Woods D. In: DNA Computing and Molecular Programming. Soloveichik D, Yurke B, editors. Vol. 8141. Springer; 2013. pp. 16–30. LNCS. [Google Scholar]; b) Woods D, Chen H-L, Goodfriend S, Dabby N, Winfree E, Yin P. Proceedings of the 4th Conference on Innovations in Theoretical Computer Science; ACM; 2013. pp. 353–354. [Google Scholar]; c) Dabby N, Chen H-L. Proceedings of the Twenty-Fourth Annual ACM-SIAM Symposium on Discrete Algorithms; SIAM; 2013. pp. 1526–1536. [Google Scholar]
- 12.Omabegho T, Sha R, Seeman NC. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Yang X, Wenzler LA, Qi J, Li X, Seeman NC. J Am Chem Soc. 1998;120:9779–9786. [Google Scholar]; b) Sa-Ardyen P, Vologodskii AV, Seeman NC. Biophys J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong H, Seeman NC. Nano Lett. 2006;6:2899–2903. doi: 10.1021/nl062183e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


