Background: 3CLpro protease is required for coronaviral polyprotein processing and is only active as a dimer.
Results: MERS-CoV 3CLpro is a weakly associated dimer requiring ligand binding for dimer formation.
Conclusion: Ligand-induced dimerization is a key mechanism for regulating the enzymatic activity of MERS-CoV 3CLpro during polyprotein processing.
Significance: Activation via ligand-induced dimerization may add complexity for the development of MERS-CoV 3CLpro inhibitors as antivirals.
Keywords: analytical ultracentrifugation, enzyme inactivation, enzyme inhibitor, enzyme kinetics, viral protease, X-ray crystallography, β-CoV, MERS-CoV 3CLpro, ligand-induced dimerization, monomer-dimer equilibrium
Abstract
All coronaviruses, including the recently emerged Middle East respiratory syndrome coronavirus (MERS-CoV) from the β-CoV subgroup, require the proteolytic activity of the nsp5 protease (also known as 3C-like protease, 3CLpro) during virus replication, making it a high value target for the development of anti-coronavirus therapeutics. Kinetic studies indicate that in contrast to 3CLpro from other β-CoV 2c members, including HKU4 and HKU5, MERS-CoV 3CLpro is less efficient at processing a peptide substrate due to MERS-CoV 3CLpro being a weakly associated dimer. Conversely, HKU4, HKU5, and SARS-CoV 3CLpro enzymes are tightly associated dimers. Analytical ultracentrifugation studies support that MERS-CoV 3CLpro is a weakly associated dimer (Kd ∼52 μm) with a slow off-rate. Peptidomimetic inhibitors of MERS-CoV 3CLpro were synthesized and utilized in analytical ultracentrifugation experiments and demonstrate that MERS-CoV 3CLpro undergoes significant ligand-induced dimerization. Kinetic studies also revealed that designed reversible inhibitors act as activators at a low compound concentration as a result of induced dimerization. Primary sequence comparisons and x-ray structural analyses of two MERS-CoV 3CLpro and inhibitor complexes, determined to 1.6 Å, reveal remarkable structural similarity of the dimer interface with 3CLpro from HKU4-CoV and HKU5-CoV. Despite this structural similarity, substantial differences in the dimerization ability suggest that long range interactions by the nonconserved amino acids distant from the dimer interface may control MERS-CoV 3CLpro dimerization. Activation of MERS-CoV 3CLpro through ligand-induced dimerization appears to be unique within the genogroup 2c and may potentially increase the complexity in the development of MERS-CoV 3CLpro inhibitors as antiviral agents.
Introduction
Coronaviruses (CoVs)3 are enveloped, positive-strand RNA viruses that infect a variety of vertebrates, including bats, livestock, pets, poultry, and humans (1–3). Although human CoVs cause respiratory illnesses of mild to moderate severity (4–9), two recently emerged CoVs, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have demonstrated their potential to become a serious threat to public health. MERS-CoV emerged late in 2012, and unlike its predecessor SARS-CoV, MERS-CoV continues to exhibit up to a 35% fatality rate (10–12).
Based on the sequence analysis of seven genes of the replicase domain, MERS-CoV has been classified as a β-CoV genogroup 2c member, along with closely related bat coronaviruses HKU5 (Pipistrellus bat) and HKU4 (Tylonycteris bat) (13, 14). Increasing evidence suggests that bats may serve as zoonotic reservoirs for MERS-CoV (15, 16). Evidence presented by recent studies also supports the local zoonotic transmission of MERS-CoV from dromedary camels to humans (17, 18). Alarmingly, human-to-human transmission during close contact, especially in elderly or patients with underlying health conditions, has also been reported for MERS-CoV (19–22). In the wake of the recent upsurge in the laboratory-confirmed cases of MERS-CoV, including two recently identified cases in the United States (23), there is an urgent need to study and characterize the properties of important drug targets of MERS-CoV for the development of effective therapeutics.
Coronaviruses express a >800-kDa replicase polyprotein, which is processed by viral 3CLpro protease (or nsp5) at 11 distinct cleavage sites to yield intermediate and mature nonstructural proteins (nsp) responsible for many aspects of virus replication (3, 24–26). Because of its indispensable role in the virus life cycle, 3CLpro is an important target for therapeutic intervention against coronavirus infections (27–33).
A number of kinetic, biophysical, and x-ray structural studies have demonstrated that SARS-CoV 3CLpro is only active in vitro as a tightly associated dimer with a dimer dissociation constant (Kd) in the low nanomolar range (34–38). The addition or deletion of amino acids, e.g. His6 affinity tags, at either the N or C terminus drastically reduces the enzymatic rate and decreases the ability of SARS-CoV 3CLpro to dimerize (37). Although cellular evidence for the auto-cleavage mechanism (cis versus trans) of 3CLpro is lacking, models for how 3CLpro cleaves itself from the polyprotein to form the mature dimer have been proposed based on in vitro studies using purified 3CLpro (34, 39, 40). A current model posits that two inactive 3CLpro molecules within two separate polyproteins recognize each other and form an immature dimer capable of cleaving the nsp4↓nsp5 and nsp5↓nsp6 sites in trans, followed by formation of an active and mature dimer that can then rapidly process other cleavage sites and multiple polyproteins. It has also been proposed that substrate-induced dimerization regulates the enzymatic activity of SARS-CoV 3CLpro during virus replication; however, no experimental evidence of this has ever been demonstrated in infected cells (40). Although our knowledge of SARS-CoV 3CLpro is extensive, the dimerization properties of 3CLpro from MERS-CoV and other coronaviruses, as well as the factors regulating their enzymatic activity, remain largely unknown.
To understand the properties of MERS-CoV 3CLpro, we conducted a series of kinetic, biophysical and x-ray structural studies. Here, we report a detailed kinetic and biophysical analysis of MERS-CoV 3CLpro activity and dimerization. These kinetic and biophysical studies provide evidence for a weakly associated MERS-CoV 3CLpro dimer. In addition, we utilized our previous knowledge on the design of potent SARS-CoV 3CLpro peptidic inhibitors to design a series of inhibitors of MERS-CoV 3CLpro that exhibit low micromolar potency. We demonstrate that MERS-CoV 3CLpro requires the binding of a ligand for dimer formation, indicating that ligand-induced dimerization is likely a key mechanism in the regulation of MERS-CoV 3CLpro activity during virus infection.
Experimental Procedures
Construct Design and Expression of MERS-CoV 3CLpro
The gene encoding 3CLpro protease of MERS-CoV (amino acid residues 3248–3553 in the replicase polyprotein, GenBankTM accession number AHC74086.1) was codon-optimized for optimal expression in E. coli (BioBasic Inc). The gene was subcloned into pET-11a expression vector with an N-terminal His6 tag followed by the nsp4↓nsp5 auto-cleavage site using the forward primer 5′-ATATACATATGCACCACCACCACCACCACAGCGGTGTTCTGCAGTCTGGTC-3′ and the reverse primer 5′-GACGGATCCTTACTGCATCACAACACCCATGATCTGC-3′. The construct was verified by DNA sequencing at the Purdue University Genomics Core Facility. This construct results in the expression of MERS-CoV 3CLpro without any N- or C-terminal extensions. MERS-CoV 3CLpro was expressed through auto-induction in Escherichia coli BL21-DE3 cells in the presence of 100 μg/ml carbenicillin as described previously (41). Cells were harvested by centrifugation at 5000 × g for 20 min at 4 °C, and the pellets were stored at −80 °C until further use.
MERS-CoV 3CLpro Purification
Frozen pellets from 4 liters of bacterial cell culture were thawed on ice and resuspended in 250 ml of Buffer A (20 mm Tris, pH 7.5, 0.05 mm EDTA, 10% glycerol, and 5 mm β-mercaptoethanol (BME)), containing 500 μg of lysozyme and a small amount of DNase. Cells were then lysed using a single pass through a French press at 1200 p.s.i., and cell debris was removed from the cleared lysate by centrifuging at 29,000 × g for 30 min. Solid ammonium sulfate was added to the cleared lysate to a final concentration of 1 m through gradual mixing on ice.
Hydrophobic Interaction Chromatography
The cleared lysate, mixed with ammonium sulfate, was loaded at a flow rate of 3 ml/min onto a 60-ml phenyl-Sepharose 6 fast-flow high-sub column (Amersham Biosciences) equilibrated with Buffer B (50 mm Tris, pH 7.5, 1 m ammonium sulfate, 0.05 mm EDTA, 10% glycerol, and 5 mm BME). The column was then washed with 5× column volume (300 ml) of Buffer B at a flow rate of 4 ml/min. Protein was eluted using a 5× column volume (300 ml) linear gradient to 100% Buffer A. Fractions (12 ml) were collected, and those containing MERS-CoV 3CLpro, as judged through SDS-PAGE analysis and specific activity measurements, were pooled (120 ml) and exchanged into 2 liters of Buffer A via overnight dialysis in a 10,000 molecular weight cutoff SnakeSkin® dialysis tubing (Thermo Scientific).
DEAE Anion-exchange Chromatography
Dialyzed sample from the previous step was loaded at a flow rate of 3 ml/min onto a 120- ml DEAE anion-exchange column (Amersham Biosciences) equilibrated with Buffer A. The column was then washed with 2× column volume (240 ml) of Buffer A at a flow rate of 4 ml/min. A linear gradient (total volume 480 ml) to 40% Buffer C (50 mm Tris, pH 7.5, 1 m NaCl, 0.05 mm EDTA, 10% glycerol, and 5 mm BME) was used to elute the protein. Fractions (6 ml) were collected, and those containing MERS-CoV 3CLpro were pooled (66 ml) and dialyzed for 4 h in 4 liters of Buffer D (20 mm MES, pH 5.5, 0.05 mm EDTA, 10% glycerol, and 5 mm BME).
Mono S Cation-exchange Chromatography
Following dialysis, the pH of the sample was manually adjusted to 5.5 using 1 m solution of MES, pH 5.5, and any precipitated protein was removed by filtering through a 0.22-μm pore size Millex-GP filter (Millipore). The filtered sample was then loaded at a flow rate of 2 ml/min onto an 8-ml Mono S 10/100 column (Amersham Biosciences) equilibrated in Buffer D. The column was then washed with 5× column volume (40 ml) of Buffer D at a flow rate of 2 ml/min. Protein was eluted using a 25× column volume (200 ml) and a linear gradient to 50% Buffer E (50 mm MES, pH 5.5, 1 m NaCl, 0.05 mm EDTA, 10% glycerol, and 5 mm BME). Fractions (2 ml) were collected, and those containing MERS-CoV 3CLpro were pooled (22 ml) and concentrated to ∼5 mg/ml.
Gel Filtration Chromatography
As the final purification step, the concentrated protein sample was loaded onto the preparation grade Superdex 75 26/60 gel filtration column (Amersham Biosciences) equilibrated with Buffer F (25 mm HEPES, pH 7.5, 10% glycerol, 2.5 mm dithiothreitol (DTT)). Protein was eluted isocratically at a flow rate of 1 ml/min with Buffer F. Fractions (2 ml) containing MERS-CoV 3CLpro were pooled (total volume of 34 ml) and concentrated to ∼5 mg/ml. For final storage of the purified MERS-CoV 3CLpro enzyme, 300-μl protein aliquots were placed into 1-ml screw-cap vials, flash-frozen under liquid nitrogen, and then stored at −80 °C until further use.
Purification of SARS-CoV, HKU4-CoV, and HKU5-CoV 3CLpro
SARS-CoV 3CLpro and HKU5-CoV 3CLpro with authentic N and C termini were expressed and purified as described previously (37, 42). HKU4-CoV 3CLpro was purified utilizing a modified protocol from Ref. 42. Final protein yield was calculated based on the measurement of total activity units (μm product/min), specific activity (units/mg), and milligrams of protein obtained (Bio-Rad protein assay) after each chromatographic step.
Synthesis of Compounds 1–11
The peptidomimetic compounds with Michael acceptor groups (compounds 1-9, Table 3) were synthesized via very similar methods to those published previously (30, 43). Synthesis of noncovalent peptidomimetic compounds 10 and 11 (Table 3) has been described previously (33).
TABLE 3.
Chemical structures and inhibitory activity of compounds 1 to 11 against MERS-CoV 3CLpro
The Michael acceptor group for compound 1 is shaded to highlight this group for all the compounds. The stereochemistry at the benzyl stereocenter of compound 5 is a 1:1 mixture of enantiomers (racemic); therefore, the compound was tested as a mixture of diastereomers.
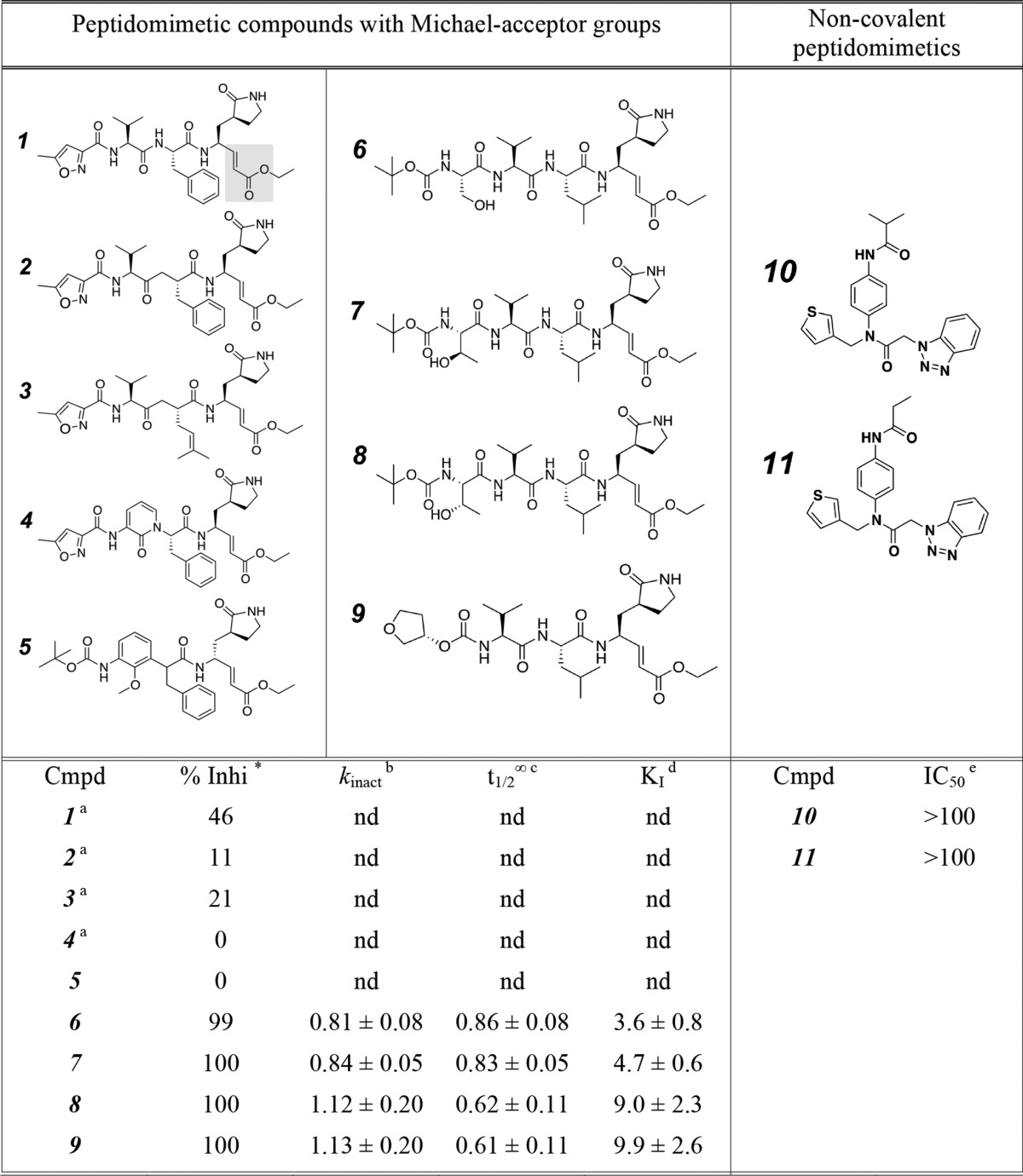
* % inhibition was measured as the % loss in enzymatic activity after 20 min of incubation of 500 nm MERS-CoV 3CLpro with 50 μm of the compound.
a As compounds 1–5 showed <50% inhibition of MERS-CoV 3CLpro, values of kinact, t1/2∞ and KI were not determined (nd) for these compounds.
b kinact is ×10−3 s−1.
c t1/2∞ is × 103 s.
d KI is in μm.
e IC50 values for compounds 10 and 11 were calculated from a dose- response curve determined after 10 min of incubation of 1 μm MERS-CoV 3CLpro with varying concentrations of compounds. IC50 is in μm.
Fluorescence-based Kinetic Assays
The enzymatic activity of 3CLpro was measured using the following custom-synthesized peptide: HilyteFluorTM-488-ESATLQSGLRKAK-(QXLTM-520)-NH2 (AnaSpec, Inc.). The HilyteFluorTM-488 fluorescence group was internally quenched by QXLTM-520 dye. This substrate works as a generic peptide substrate for 3CLpro enzymes and was designed based on the nsp4↓nsp5 cleavage sequence for many coronavirus 3CLpro enzymes. The rate of enzymatic activity was determined at 25 °C by following the increase in fluorescence (λexcitation = 485 nm, λemission = 528 nm, bandwidths = 20 nm) of Hilyte FluorTM-488 upon peptide hydrolysis by the enzyme as a function of time. Assays were conducted in black, half-area, 96-well plates (Corning Glass) in assay buffer (50 mm HEPES, pH 7.5, 0.1 mg/ml BSA, 0.01% Triton X-100, and 2 mm DTT) using a final reaction volume of 100 μl. The resulting florescence was monitored using a BioTek Synergy H1 plate reader. The rate of the reaction in arbitrary fluorescence units/s (AFU/s) was determined by measuring the initial slope of the progress curves, which were then converted to units of micromolars of product produced per min (μm/min) using experimentally determined values of fluorescence “extinction coefficient” as described previously (37). All reactions were carried out in triplicate.
Determination of Enzymatic Efficiency
The apparent enzymatic efficiency for each of the 3CLpro enzymes was determined by measuring the rate of enzymatic activity as a function of varying substrate concentration in 100-μl reactions. Reactions were initiated by the addition of enzyme to the wells of an assay plate containing varying concentrations of substrate. The final substrate concentrations varied over a range from 0 to 2 μm. The final enzyme concentrations for each 3CLpro studied were as follows: MERS-CoV 3CLpro at 1 μm, SARS-CoV 3CLpro at 100 nm, HKU5-CoV 3CLpro at 250 nm, and HKU4-CoV 3CLpro at 200 nm. Because 3CLpro enzymes cannot be saturated with this substrate at a substrate concentration that would still allow accurate fluorescent measurements without the inner filter effect, only the apparent kcat/Km values can be determined from the slope of the line that results from a plot of the enzymatic activity (y axis), normalized for the total enzyme concentration, against the substrate concentration (x axis).
Influence of Dimerization on the Activity of 3CLpro Enzymes
The dependence of the enzymatic activity on the total enzyme concentration was determined using the FRET-based assay described above. The final enzyme concentrations were varied over a concentration range from 2 μm to 100 nm for MERS-CoV 3CLpro, 500 to 10 nm for SARS-CoV 3CLpro, 250 to 0.6 nm for HKU5-CoV 3CLpro, and 200 to 10 nm for HKU4-CoV 3CLpro. Reactions were initiated by the addition of substrate, at a final concentration of 2 μm, to the assay plates containing varying enzyme concentrations in the assay buffer. Initial rates were determined from the initial slopes of the progress curves at each enzyme concentration.
The rates of the 3CLpro-catalyzed reactions measured over a range of enzyme concentrations can be fit to either Equation 1 or 2 to determine the values of the dissociation constant for the monomer-dimer equilibrium as well as the turnover numbers. Nonlinear regression and the program TableCurve 2D version 4.0 were used to fit the data to either Equation 1 or 2 below (44).
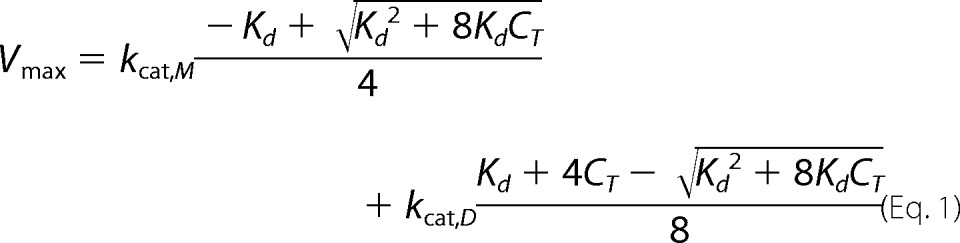 |
In Equation 1, Vmax is the rate of the enzymatic activity calculated at each enzyme concentration (CT); Kd is the monomer-dimer equilibrium dissociation constant, and kcat, M and kcat, D are the turnover numbers for the monomer and the dimer, respectively.
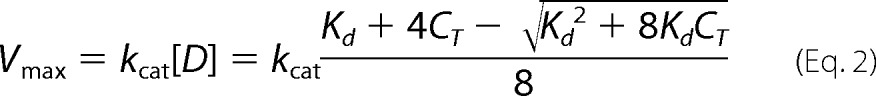 |
In Equation 2, Vmax, CT, and Kd have been described previously, and kcat is the turnover number for the dimer only.
Inhibition Assays
To determine the percent inhibition for compounds 1–9, the total concentration of the substrate was fixed at 1.0 μm, and the enzymes were fixed at 250 nm for SARS-CoV 3CLpro, HKU5-CoV 3CLpro, HKU4-CoV 3CLpro, and at 500 nm for MERS-CoV 3CLpro. DMSO stocks (100×) of the compounds were diluted a hundred-fold to a final concentration of 50 μm in 80 μl of the enzyme solution and incubated for 20 min. After 20 min, the enzymatic activity was measured as initial slope of the progress curve, obtained by initiating the reaction with 20 μl of 5 μm substrate. % inhibition was calculated using Equation 3.
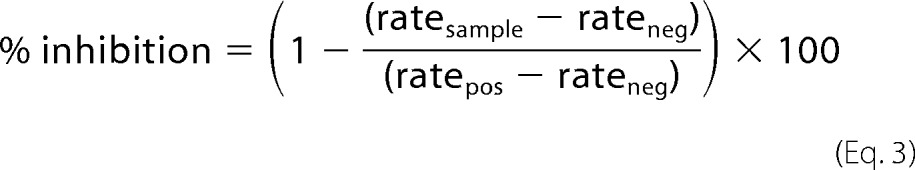 |
In Equation 3, ratesample is the initial slope of the progress curve in AFU/s measured in the presence of the compound; ratepos is the initial slope measured in the absence of any compound, and rateneg is the baseline substrate hydrolysis calculated in the absence of enzyme. All the reactions were carried out in triplicate and contained a final DMSO concentration of 1%. For compounds displaying more than 50% inhibition, a more extensive characterization of the inactivation kinetics was performed through progress curve analysis. To the reaction well, 20 μl of 5 μm substrate was added to a final concentration of 1 μm, and the total inhibitor concentration [I]total was varied from 0 to 50 μm. The reaction was initiated with the addition of 80 μl of MERS-CoV 3CLpro to a final concentration of 500 nm. Fluorescence intensity was then measured over time as AFUt for a period of 70 min. Equation 4 describes the resulting time course of reaction.
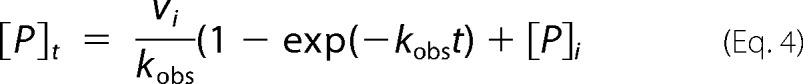 |
In Equation 4, vi is the initial velocity of the reaction; kobs is the observed first-order rate constant for the reaction in the absence and presence of inhibitor; t is the time in minutes; [P]t is the concentration of product produced at time t, and [P]i is the initial product concentration, which is zero. Product concentrations were calculated from the values of AFUt, using the experimentally determined fluorescence extinction coefficient. The resulting values of [P]t were then plotted against time t, and the data were fit to Equation 4 with [P]i = 0 using the nonlinear regression program TableCurve 2D to derive the fitted parameters vi and kobs and their associated errors Δvi and Δkobs.
Values for each kobs were then plotted against [I]total and the data were fit to Equation 5.
 |
In Equation 5, kinact defines the maximum rate of inactivation at infinite inhibitor concentration, and KI defines the concentration of inhibitor that yields a rate of inactivation equal to ½kinact. The half-life of inactivation at infinite inhibitor concentration, which is a measure of inactivation efficiency, is defined as t½∞ = 0.693/kinact.
AUC Analysis
To determine the oligomeric state of MERS-CoV 3CLpro, sedimentation velocity experiments were performed at 20 °C on the Beckman-Coulter XLA ultracentrifuge using varying concentrations of MERS-CoV 3CLpro (4–23 μm) in 25 mm HEPES, pH 7.5, 50 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine at 50,000 rpm. To characterize the effect of the ligand on the monomer-dimer equilibrium of MERS-CoV 3CLpro, sedimentation velocity experiments were conducted on the Beckman-Coulter XLI instrument using different stoichiometric ratios of MERS-CoV 3CLpro with compounds 6 and 10. Samples were prepared by mixing 25 μm MERS-CoV 3CLpro with 25, 50, and 100 μm compound 6 or 10 and incubating the mixture overnight at 4 °C before performing the experiments. Absorbance optics (280 nm) and interference optics were utilized for protein detection. Solvent density, viscosity, and partial specific volumes were calculated using SEDNTERP. SEDPHAT was used to fit the data to the monomer-dimer self-association model to estimate the sedimentation coefficients (s), apparent molecular weights, and Kd and koff values from size distribution analysis. To obtain exact molecular weights, sedimentation equilibrium experiments were performed at concentrations of 3 and 17 μm MERS-CoV 3CLpro. The experiments were done at 20 °C utilizing a two-channel centerpiece and run at multiple speeds (8100, 13,800 and 24,000 rpm) in a AN-60 Ti rotor.
MERS-CoV 3CLpro Activation and Inhibition by a Noncovalent Inhibitor
The rates of the MERS-CoV 3CLpro-catalyzed reactions were determined at final enzyme concentrations of 0.5, 1.0, and 2.0 μm and in the absence and presence of varying concentrations (0.1–60 μm) of compound 10. The substrate concentration was fixed at 2.0 μm. DMSO stocks (100×) of compound 10 were diluted a hundred-fold in 80 μl of enzyme solution and incubated for 10 min. At the same time, a zero-inhibitor control reaction was set up by mixing DMSO to a final concentration of 1% into 80 μl of enzyme solution. After 10 min, the rate of the enzymatic activity was measured as the initial slope of the progress curve, obtained by initiating the reaction with 20 μl of 10 μm substrate. Equation 6 was utilized to calculate the percent activity.
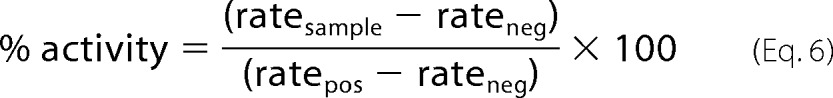 |
The ratesample, ratepos, and rateneg are as described above for Equation 3.
MERS-CoV 3CLpro Crystallization, X-ray Data Collection, and Structure Determination
Purified MERS-CoV 3CLpro was concentrated to 1.6 mg/ml in 25 mm HEPES, pH 7.5, and 2.5 mm DTT. Inhibitor complexes of MERS-CoV 3CLpro with compounds 6 and 11 were formed by incubating MERS-CoV 3CLpro with the compounds in a 1:3 stoichiometric ratio at 4 °C overnight. After iterative rounds of optimization of the crystallization conditions based on the initial hits obtained from high throughput screening of Qiagen Nextel Screens, crystals of MERS-CoV 3CLpro inhibitor complexes suitable for x-ray diffraction were grown by the hanging-drop, vapor diffusion method at 20 °C in 0.2 m sodium acetate, 0.1 m BisTris, pH 7.0, and 20% PEG-3350 for the MERS-CoV 3CLpro and 6 complex, and 0.2 m ammonium acetate, 0.1 m BisTris, pH 5.5, 12% PEG-3350 for the MERS-CoV 3CLpro and 11 complex. For x-ray data collection, crystals were flash-cooled in liquid nitrogen after dragging the crystals through a cryo-solution that contained the crystallization solution supplemented with 15% 2-methyl-2,4-pentanediol.
X-ray diffraction data were collected for MERS-CoV 3CLpro and 6 and MERS-CoV 3CLpro and 11 complexes at the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) Sector 31 and the Life Sciences Collaborative Access Team (LS-CAT) Sector 21 at the Advanced Photon Source, Argonne National Laboratory, respectively. Data were processed and scaled using Mosflm version 7.0.5 (45) and HKL2000 version 706 (46). The method of molecular replacement was used to obtain initial phases using the program PHASER-MR in Phenix suite version 1.8.4 (47). For MERS-CoV 3CLpro and 6 complex, the x-ray structure of SARS-CoV 3CLpro (PDB code 3V3M) was used as a phasing model (32). The final MERS-CoV 3CLpro and 6 complex structure was then used to calculate the initial phases for the MERS-CoV 3CLpro and 11 complex model. Automated model building using Autobuild in Phenix was initially used to build a preliminary model of the MERS-CoV 3CLpro and 6 inhibitor complex. Each structure was then refined using iterative cycles of refinement using Phenix Refine coupled to manual model building using COOT (48) based on Fo − Fc and 2Fo − Fc maps. Coordinates and molecular library files for inhibitor molecules were built using the program eLBOW in the Phenix suite. Water molecules were added to peaks in residual (Fo − Fc) density maps that were greater than 3σ using the “Find Water” function in COOT. MolProbity was used to assess structural quality of the final model (49). The measured structure factor amplitudes and the atomic coordinates for the final structures were deposited in the Protein Data Bank with accession codes 4RSP (MERS-CoV 3CLpro and 6 complex) and 4YLU (MERS-CoV 3CLpro and 11 complex), respectively. Structural superposition was performed using the method of least squares fitting of C-α atoms in COOT. PyMOL was used to generate figures of all the structures (50).
Results
Production of MERS-CoV 3CLpro with Authentic N and C Termini
Insertion of the nsp4↓nsp5 cleavage site between the N-terminal His6 tag and the coding region for MERS-CoV 3CLpro results in autoprocessing of the His tag and overexpression of MERS-CoV 3CLpro without any N-terminal extension in E. coli BL21-DE3 cells. MERS-CoV 3CLpro was purified to high purity and an overall yield of 10% using four sequential chromatographic steps. A summary of the percent enzyme yield, total activity units, and the fold-purification after each chromatographic step is summarized in Table 1. Approximately 12 mg of highly pure MERS-CoV 3CLpro can be obtained per liter of bacterial cell culture.
TABLE 1.
Purification summary of MERS-CoV 3CLpro per liter of E.coli BL21-DE3 cells
| Sample | Protein | Total activity units | Specific activity | Fold purification | % yield |
|---|---|---|---|---|---|
| mg | units/mg | ||||
| Lysate | 1102 | 1168 | 1 | 1 | 100 |
| Phenyl-Sepharose | 219 | 185 | 1 | 1 | 16 |
| DEAE | 22 | 189 | 8 | 8 | 16 |
| Mono S | 15 | 142 | 9 | 9 | 12 |
| Superdex 75 | 12 | 114 | 10 | 10 | 10 |
To verify the production of the enzyme with correct N and C termini, the molecular mass of purified MERS-CoV 3CLpro was determined by MALDI to be 33.4 kDa, which is close to the theoretical molecular mass of 33.3 kDa for the authentic/mature MERS-CoV 3CLpro monomer. Western blot analysis of purified MERS-CoV 3CLpro using an anti-His6 antibody also confirmed the absence of the N terminus His6 tag associated with the expression plasmid (data not shown). These results demonstrate that the N-terminal His6 tag is auto-catalytically removed by MERS-CoV 3CLpro during its expression in E. coli, indicating MERS-CoV 3CLpro is enzymatically active when expressed in E. coli.
MERS-CoV 3CLpro Hydrolyzes a Fluorescent Peptide Substrate with Lower Efficiency than Other 3CLpro Enzymes
A FRET-based peptide substrate was used to measure the enzymatic activity of MERS-CoV 3CLpro as a function of substrate concentration over a substrate concentration range from 0 to 2.0 μm (Fig. 1A). We observed that MERS-CoV 3CLpro cannot be saturated by the substrate over this concentration range, which is typical for other coronavirus 3CLpro enzymes because the Km values for peptide substrates approach 1 mm (51–54). Therefore, the slope of the kinetic response of MERS-CoV 3CLpro to increasing substrate concentration was determined to derive an apparent (kcat/Km) value, which is a measure of enzymatic efficiency. We also determined and compared the apparent (kcat/Km) values for 3CLpro enzymes from SARS-CoV, HKU5-CoV, and HKU4-CoV under similar experimental conditions (Fig. 1B). MERS-CoV 3CLpro is able to hydrolyze the peptide substrate; however, the enzymatic efficiency of MERS-CoV 3CLpro (kcat/Km = 3.1 ± 0.03 × 10−2 μm−1 min−1) is noticeably lower than other 3CLpro enzymes tested. Specifically, MERS-CoV 3CLpro was 5-fold less efficient at processing the peptide substrate when compared with SARS-CoV 3CLpro. Even among the β-CoVs from the same 2c genogroup (MERS, HKU5, and HKU4), MERS-CoV 3CLpro was the least efficient enzyme.
FIGURE 1.
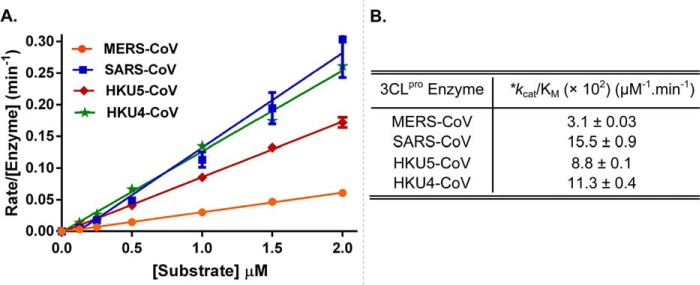
Comparison of enzymatic efficiencies (kcat/Km) of 3CLpro enzymes from different CoVs. A, rates for the enzymatic activity, normalized to the total enzyme concentration, are plotted as a function of varying substrate concentrations. Total concentration of each enzyme in the final reaction is as follows: MERS-CoV 3CLpro at 1 μm; SARS-CoV 3CLpro at 100 nm; HKU5-CoV 3CLpro at 250 nm; and HKU4-CoV 3CLpro at 200 nm. Slope of the line represents the apparent value of kcat/Km. Error bars represent the standard deviation for triplicate data. B, *, apparent value of kcat/Km for the nonsaturable substrate, calculated as the slope of the linear plot from panel A.
MERS-CoV 3CLpro Is a Weakly Associated Dimer
Because a dimer has consistently been shown to be the catalytically active form of all 3CLpro enzymes studied to date, we tested the hypothesis that the lower enzymatic efficiency of MERS-CoV 3CLpro is a result of the reduction in its ability to dimerize. Therefore, we determined the dependence of the enzymatic activity of MERS-CoV 3CLpro on the total enzyme concentration and compared it with other 3CLpro enzymes from HKU4, HKU5, and SARS coronaviruses (Fig. 2).
FIGURE 2.
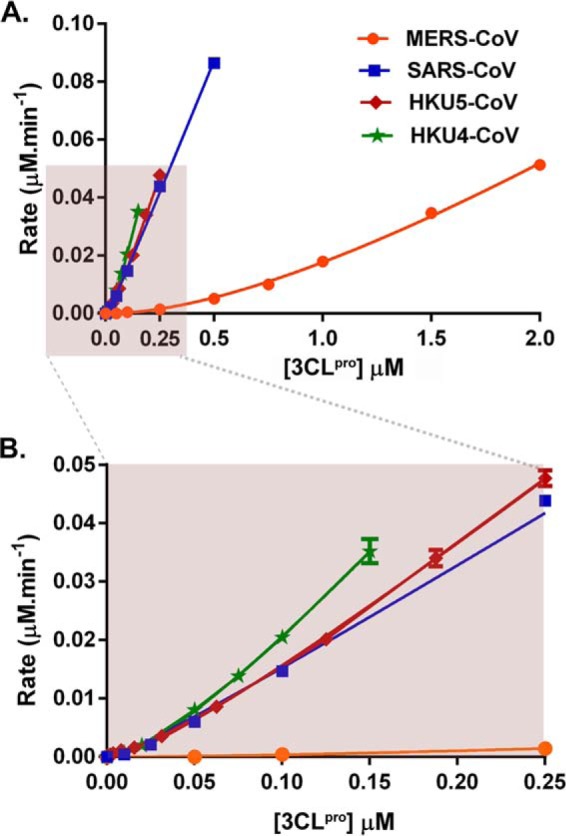
Dependence of the enzymatic activity of MERS-CoV, HKU4-CoV, HKU5-CoV, and SARS-CoV 3CLpro on the total enzyme concentration. A, kinetic response of each CoV 3CLpro to increasing enzyme concentration is plotted along with the resulting fit of the data to Equation 2. Resulting values for the apparent turnover number, kcat, and the monomer-dimer equilibrium constant, Kd, are shown in Table 2. Final enzyme concentrations varied over the concentration ranges of 2 μm to 100 nm for MERS-CoV 3CLpro, 500 to 10 nm for SARS-CoV 3CLpro, 250 to 0.6 nm for HKU5-CoV 3CLpro, and 200 to 10 nm for HKU4-CoV 3CLpro. Final substrate concentration was fixed at 2 μm. Experiments were done in triplicate. Error bars represent the standard deviation for triplicate data. Shaded box represents the data that are plotted in B. B, enlarged view of the fitted data at low total enzyme concentrations, marked in shaded box in A, illustrating the nonlinear dependence of enzymatic activity on the total concentrations of 3CLpro from SARS-CoV, HKU5-CoV, and HKU4-CoV.
It is immediately apparent from the data plotted in Fig. 2 that the response of MERS-CoV 3CLpro enzymatic activity to an increasing enzyme concentration is nonlinear. The strong curvature suggests that a dimer is either the most active form or the only active form of MERS-CoV 3CLpro. To determine the mechanism of dimerization, the data in Fig. 2 were first fit to Equation 1 (see “Experimental Procedures”), which describes a model where both the monomer and the dimer are active. A fit of the data to Equation 1 yielded a negative turnover value for the monomer (kcat, M), suggesting the monomer is inactive and that the dimer is the only active form of the enzyme. Therefore, the data were fit to Equation 2 (see “Experimental Procedures”), which considers only the dimer as the active form of the enzyme. The kinetic data for all four 3CLpro enzymes, MERS-CoV, HKU4-CoV, HKU5-CoV, and SARS-CoV, fit well to this model, and the resulting values for the monomer-dimer equilibrium dissociation constant, Kd, and apparent turnover number, kcat, for each enzyme are provided in Table 2.
TABLE 2.
Comparison of the apparent turnover number, kcat, and the monomer-dimer dissociation constant, Kd, for 3CLpro from different CoVs
| 3CLpro | Nonlinear fitting
of kinetic dataa |
|
|---|---|---|
| kcatb | Kd | |
| min−1 | μm | |
| MERS-CoV | 0.2 ± 0.02 | 7.8 ± 1.3 |
| SARS-CoV | 0.47 ± 0.03 | 0.06 ± 0.01 |
| HKU5-CoV | 0.53 ± 0.02 | 0.06 ± 0.01 |
| HKU4-CoV | 0.84 ± 0.07 | 0.1 ± 0.03 |
a Values were determined through nonlinear fitting of the kinetic data to Equation 2.
b kcat represents the apparent turnover number.
The lower kcat value for MERS-CoV 3CLpro, when compared with other coronavirus 3CLpro enzymes, indicates a moderate reduction (2–4-fold) in its ability to turn over the substrate, which is consistent with the observed lower apparent (kcat/Km) value. In contrast, there is a substantial reduction in the ability of MERS-CoV 3CLpro to dimerize compared with the other 3CLpro enzymes. Based on the Kd values, the capacity of MERS-CoV 3CLpro to dimerize is ∼78–130-fold weaker than the other enzymes (Table 2). These results indicate that the MERS-CoV 3CLpro dimer is much more weakly associated than the other coronavirus 3CLpro enzymes studied, and these results raise questions as to the structural and mechanistic differences among the 3CLpro enzymes that ultimately regulate protease activity during coronavirus replication.
MERS-CoV 3CLpro Inhibition by Designed Peptidomimetic Compounds
In an effort to develop potent inhibitors of MERS-CoV 3CLpro, we designed and synthesized nine peptidomimetic compounds containing a Michael acceptor group, i.e. an α,β-unsaturated carbonyl, capable of irreversibly reacting with the active site cysteine of MERS-CoV 3CLpro (Table 3). These compounds were designed and synthesized based on our understanding and knowledge of the interactions of similar inhibitor molecules with SARS-CoV 3CLpro (30, 31). At a concentration of 50 μm, compounds 6–9 displayed more than 50% inhibition of MERS-CoV 3CLpro and were further evaluated for their ability to inactivate the enzyme in a time- and concentration-dependent manner (Fig. 3). Data from the kinetic progress curve for compound 6 (Fig. 3), as well as for compounds 7-9 (data not shown), were fit to the appropriate equations (see under “Experimental Procedures”) to obtain the kinetic parameters, kinact, t½∞, and KI, and the resulting values are provided in Table 3.
FIGURE 3.
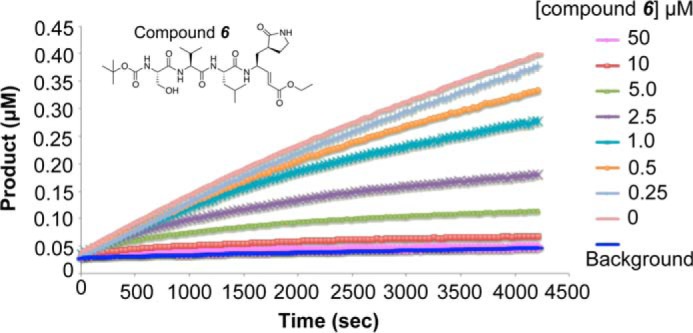
Progress curves for the MERS-CoV 3CLpro-catalyzed reaction in the presence of compound 6. Time-dependent hydrolysis of 1 μm substrate catalyzed by 500 nm MERS-CoV 3CLpro was measured over a time period of 70 min and at fixed variable concentrations of compound 6 ranging from 0 to 50 μm. Values for the inactivation kinetic parameters kinact, t½∞, and KI were calculated by fitting the progress curve data to Equations 4 and 5. Chemical structure of compound 6 is shown in the inset.
We identified four compounds, 6–9, as micromolar inhibitors of MERS-CoV 3CLpro with KI values less than 10 μm (Table 3). Analysis of structure-activity relationships of these compounds suggests that the S2 subsite pocket of MERS-CoV 3CLpro is small and can only accommodate a smaller P2-isobutyl substituent (compounds 6-9) but not bigger substituents such as P2-benzyl or P2-isobutylenyl (compounds 1-5). It was also observed that replacing the P4-ethoxy (compound 6) with P4-isopropoxy (compounds 7 and 8) had no effect on the inhibitory activity of the compounds. Finally, these compounds provide an excellent chemical scaffold to study the molecular details of interactions of substrate-like compounds with the enzyme and to develop more potent inhibitors of MERS-CoV 3CLpro for therapeutic intervention.
To evaluate broad spectrum specificity of these compounds, we also calculated % inhibition of SARS-CoV 3CLpro, HKU5-CoV 3CLpro, and HKU4-CoV 3CLpro after 20 min of incubation in the presence of 50 μm compounds 6–9. Except for compound 9, which inhibited SARS-CoV 3CLpro by 76%, we observed 100% inhibition of all other enzymes in the presence of compounds 6-9. Furthermore, we performed progress curve analysis of HKU5-CoV 3CLpro and HKU4-CoV 3CLpro in the presence of varying concentrations of compounds 6–9. The KI values of compounds 6–9 for HKU5-CoV 3CLpro are 0.49 ± 0.16, 0.60 ± 0.21, 1.30 ± 0.53, and 0.47 ± 0.06 μm, respectively. The KI values of compounds 6–9 for HKU4-CoV 3CLpro are 0.39 ± 0.14, 0.50 ± 0.17, 0.85 ± 0.33, and 0.64 ± 0.25 μm, respectively. These data suggest that peptidomimetic compounds 6–9 have the potential to be developed as coronavirus 3CLpro inhibitors with broad spectrum specificity.
Weak Association of the MERS-CoV 3CLpro Dimer Is Supported by AUC Studies
To further explore the mechanism of MERS-CoV 3CLpro dimerization, we performed analytical ultracentrifugation sedimentation velocity (AUC-SV) studies at varying concentrations of MERS-CoV 3CLpro (Fig. 4A). Unlike enzyme kinetics, AUC allows determination of the monomer-dimer equilibrium constant (Kd) in the absence of substrate. MERS-CoV 3CLpro displayed a continuous size distribution at different protein concentrations. Two distinct peaks corresponding to monomer (2.9 S) and dimer (3.9 S) species are observed, with the dimer peak becoming more pronounced at higher enzyme concentrations (Fig. 4A). We fit the AUC data to a monomer-dimer equilibrium model to determine the values for Kd and koff, where Kd is the equilibrium dissociation constant for a monomer from the dimer, and koff is the rate constant for dissociation of the monomer from the dimer. The resulting best fit value for Kd is 52 ± 5 μm and that for koff is 10−4 s−1. The Kd value of 52 μm for MERS 3CLpro is dramatically different from SARS-CoV 3CLpro, which has reported Kd values ranging from low nanomolar up to 10 μm depending on the enzyme construct used and the experimental conditions and methods utilized to determine the dissociation constant (37). The dimer affinity of MERS-CoV 3CLpro is substantially weaker than that for SARS-CoV 3CLpro, when comparing the same enzyme construct, i.e. the enzyme without any N- or C-terminal modifications. The AUC-SV calculated Kd value for MERS-CoV 3CLpro is ∼150,000 times higher than the value of 0.35 nm determined for SARS-CoV 3CLpro (34).
FIGURE 4.
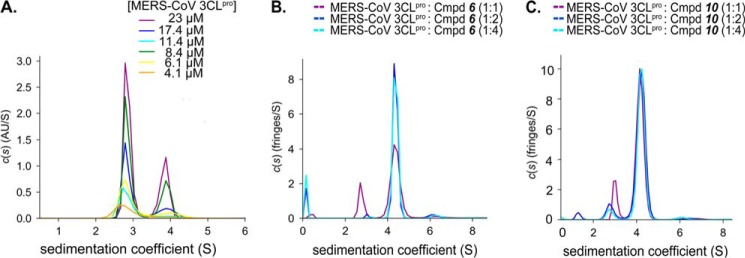
AUC-SV analyses of ligand-induced dimerization of MERS-CoV 3CLpro. A, sedimentation coefficient distribution for varying concentrations of MERS-CoV 3CLpro (4.1 to 23 μm) with sedimentation coefficient values of 2.9S and 3.9S for the monomer and the dimer, respectively. The best fit value for AUC-SV-calculated Kd is 52 ± 5 μm. B, sedimentation coefficient distribution of MERS-CoV 3CLpro (25 μm) in the presence of different stoichiometric ratios of compound 6 (25, 50, and 100 μm). C, sedimentation coefficient distribution of MERS-CoV 3CLpro (25 μm) in the presence of different stoichiometric ratios of compound 10 (25, 50, and 100 μm). A significant shift in the 2.9S peak (monomer) to a 4.1S peak (dimer) is detected upon addition of increasing concentrations of compounds 6 and 10.
The AUC results (Fig. 4A) show that the monomer peak at ∼2.9S does not gradually shift peak position toward the dimer peak at ∼3.9S with increasing concentrations of MERS-CoV 3CLpro; rather, the two peaks change in area, which is indicative of very slow monomer-dimer exchange rate (koff ∼10−4 s−1) and the formation of hydrodynamically stable monomer and dimer species (55). This koff value is 1000 times slower than the koff value (10−1 s−1) reported for SARS-CoV 3CLpro indicating that the SARS-CoV enzyme has a significantly more rapid monomer-dimer exchange rate (56). These observations support a model whereby the MERS-CoV 3CLpro dimer is weakly associated, suggesting the enzyme exists mainly as a monomer in solution.
MERS-CoV 3CLpro Undergoes Extensive Ligand-induced Dimerization
The weak association of MERS-CoV 3CLpro monomers engenders the following questions. “Are higher levels of expression of 3CLpro in MERS-CoV-infected cells necessary to allow formation of active dimer?” “Are other mechanisms such as substrate- or ligand-induced dimerizations involved in activating 3CLpro?” To explore the latter question of ligand-induced dimerization of MERS-CoV 3CLpro, we performed AUC experiments in the presence of compound 6, which acts as a substrate mimetic and mechanism-based inhibitor, also known as a suicide substrate. Peptidomimetic compounds such as compound 6, which contains a Michael acceptor group, interact and react with the active site cysteine of cysteine proteases to covalently modify them. We utilized compound 6 to form a covalent MERS-CoV 3CLpro and inhibitor 6 complex that is stable over long periods of time, making it amenable to analysis by AUC-SV experiments. In contrast, incubation of a normal peptide substrate with the enzyme would lead to immediate hydrolysis of the substrate and dissociation of the products from the enzyme, confounding AUC experiments and subsequent data analysis.
MERS-CoV 3CLpro was incubated with varying concentrations of compound 6 in stoichiometric ratios of 1:1, 1:2, and 1:4. The modified enzyme was then subjected to AUC studies to determine the influence of compound 6 on the monomer-dimer equilibrium (Fig. 4B). A significant shift in the area under 2.9S peak (monomer) to 4.1S peak (dimer) is detected upon addition of increasing concentrations of compound 6. We obtained similar results when AUC studies were performed utilizing a complex of MERS-CoV 3CLpro with a noncovalent peptidomimetic inhibitor (compound 10, Figs. 4C). The transition of MERS-CoV 3CLpro from monomer to dimer in the presence of compounds 6 and 10 suggests that the enzyme undergoes extensive dimerization upon substrate binding.
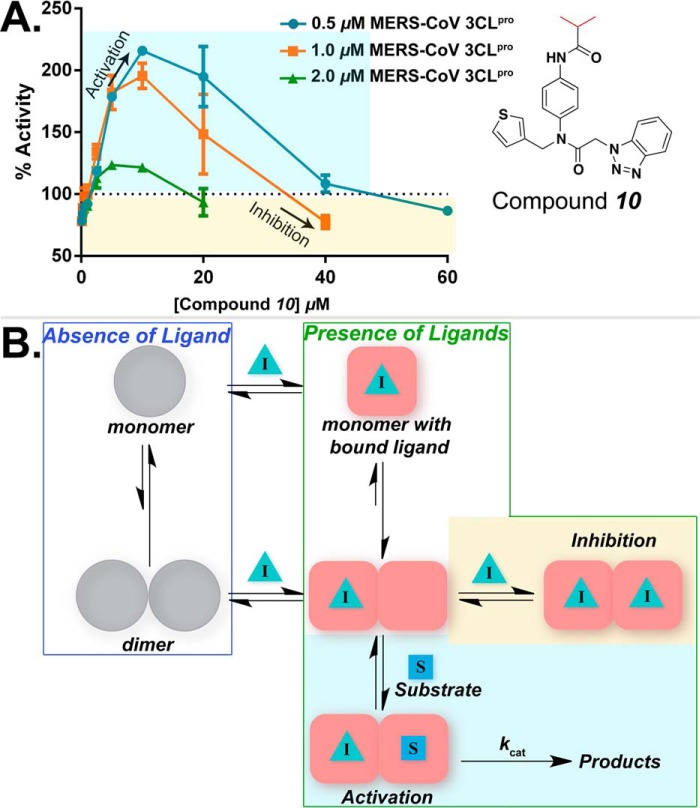
MERS-CoV 3CLpro Is Activated by Ligand-induced Dimerization
The observed ligand-induced dimerization of MERS-CoV 3CLpro, as demonstrated through AUC studies, prompted us to investigate whether or not the enzymatic activity of MERS-CoV 3CLpro could be increased at low concentrations of a compound via ligand-induced dimerization. To do so, we chose to use a noncovalent peptidomimetic compound (compound 10, Fig. 5A) that we previously identified as an inhibitor of SARS-CoV 3CLpro. Because of the time-dependent, irreversible nature of the reaction between compound 6 and MERS-CoV 3CLpro, use of compound 6 was not ideal for these kinetic studies as it would further complicate kinetic data analysis.
FIGURE 5.
Activation of MERS-CoV 3CLpro via ligand-induced dimerization. A, enzymatic activity of 0.5, 1.0, and 2.0 μm MERS-CoV 3CLpro was measured in the absence and presence of varying concentrations of compound 10. Substrate concentration was fixed at 2.0 μm. % activity, normalized to zero inhibitor enzymatic activity, was plotted as a function of increasing inhibitor concentrations. Error bars represent the standard deviation for triplicate data. Increase in enzymatic activity (highlighted in cyan-shaded box) is observed in the presence of low concentrations of compound 10. Inhibition of enzymatic activity is observed at higher inhibitor concentrations (highlighted in yellow-shaded box). B, kinetic model describing the equilibrium between different species of MERS-CoV 3CLpro that are formed in the absence (blue box) and presence (green box) of a ligand is shown. Based on the AUC-calculated Kd value of ∼ 52 μm, MERS-CoV 3CLpro primarily exists as a monomer in solution in the absence of a ligand. Upon ligand binding (inhibitor I in our case) to the monomer, the monomer-dimer equilibrium shifts toward dimer formation. Next, under lower inhibitor concentrations (cyan-shaded box), substrate (S) binds in the second active site and catalysis takes place. However, under higher inhibitor concentrations (yellow-shaded box), inhibitor directly competes with the substrate for the second active site, and inhibition of the enzymatic activity is observed.
The kinetic response of MERS-CoV 3CLpro to increasing concentrations of compound 10 was first measured at a single enzyme concentration of 1.0 μm (Fig. 5A). Interestingly, an increase in the activity of MERS-CoV 3CLpro, as high as 195%, was observed in the presence of low inhibitor concentrations (0.1 to 20 μm). Inhibition of enzymatic activity was observed only at higher inhibitor concentrations (40 μm or greater). These results suggest that at low concentrations, compound 10 binds to a monomer and induces the formation of a dimer. The resulting dimer then has one free active site that is capable of processing the substrate. At higher concentrations of inhibitor, the substrate and inhibitor directly compete for the free active site.
The model of activation and inhibition suggested by the data at 1 μm enzyme would predict that at higher enzyme concentrations less activation by a compound would be observed at lower inhibitor concentrations, and the inhibition of activity would be detected at lower inhibitor concentrations because the equilibrium would be pushed toward dimer formation. In contrast, lower enzyme concentrations would result in higher activation by compounds, and inhibition by the compound would occur at significantly higher compound concentrations. Therefore, we further measured the activity of MERS-CoV 3CLpro at two additional enzyme concentrations (0.5 and 2.0 μm) in the presence of varying concentrations of compound 10. Remarkably, we observed that the activation effect was most pronounced at the lowest MERS-CoV 3CLpro concentration tested (0.5 μm), and the effect decreased as the enzyme concentration was increased (1.0 and 2.0 μm) (Fig. 5A). Moreover, inhibition by compound 10 occurred at lower compound concentrations when higher concentrations of enzyme were used. These observations further support a model whereby enzyme activation can occur through ligand-induced dimerization.
The activation and inhibition of MERS-CoV 3CLpro by compound 10 can be explained by a simple kinetic model depicted in Fig. 5B. The MERS-CoV 3CLpro monomer exists in equilibrium with the dimer, and their relative concentrations depend on the total enzyme concentration. In the absence of substrate or compound, the Kd value is 52 μm, and the equilibrium is represented by the gray spheres (blue box) in Fig. 5B. The monomer is unable to hydrolyze the substrate and is therefore inactive. Binding of inhibitor (Fig. 5B, green triangle) to the monomer results in monomer to dimer switch leading to the formation of a dimer that contains inhibitor bound in one of the active sites. Once the dimer is formed, the substrate binds in the second active site and catalysis takes place. Under high inhibitor concentrations, however, the inhibitor molecule directly competes with substrate for the free dimer active site, and inhibition of the enzymatic activity is observed as a result.
We would also expect to observe induced dimerization and activation in the presence of the substrate. Indeed, the monomer-dimer kinetic studies performed in Fig. 2 were performed at a fixed concentration of substrate at 2 μm. In this experiment, the Kd value for the MERS-CoV 3CLpro dimer was determined to be 7.8 μm, which is lower than the Kd value determined in the absence of substrate using AUC, thereby supporting substrate-induced dimerization. Given the high Km value of 3CLpro for the peptide substrate (51–54), even higher substrate concentrations would be required to observe substrate activation in a plot of catalytic activity versus substrate concentration. However, we are limited to use our FRET-based substrate only at low concentrations due to a significant inner filter effect at higher concentrations of substrate. Therefore, a compound that both mimics substrate and has higher binding affinity can act as a useful surrogate for the substrate, allowing the observation of ligand-induced dimerization and activation even at low substrate concentrations.
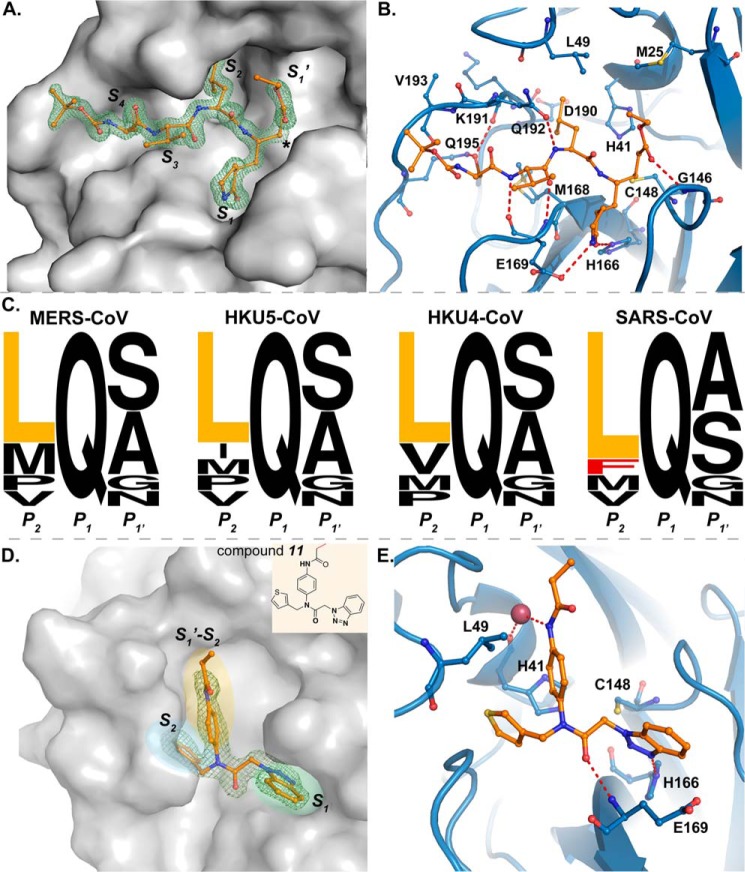
X-ray Structure of MERS-CoV 3CLpro in Complex with Compound 6
To gain atomic level detail and molecular insight into the mechanism for substrate-induced dimerization of MERS-CoV 3CLpro, we attempted to crystallize and determine the x-ray structures of the unliganded MERS-CoV 3CLpro monomer and the MERS-CoV 3CLpro covalently modified with compound 6. Unfortunately, we were unable to crystallize the unliganded MERS-CoV 3CLpro monomer after multiple attempts, but we were able to crystallize and determine the x-ray structure of MERS-CoV 3CLpro in complex with compound 6 to a resolution of 1.6 Å. The statistics for x-ray data collection, processing, and refinement are summarized in Table 4. The MERS-CoV 3CLpro and 6 complex crystallized as a biologically relevant, symmetrical dimer in space group C2 with one monomer in the asymmetric unit. Electron density for the entire protein was clearly visible and strong electron density (Fo − Fc >4σ) was present for compound 6 within the active site (Fig. 6A).
TABLE 4.
X-ray data collection and refinement statistics
| MERS-CoV 3CLpro·6 | MERS-CoV 3CLpro·11 | |
|---|---|---|
| Beamline | LRL-CAT sector 31 ID-D | LS-CAT sector 21 ID-G |
| Data collection | ||
| Wavelength (Å) | 0.9793 | 0.9786 |
| Resolution range (Å) | 19.35–1.62 (1.68–1.62)a | 50.00–2.10 (2.14–2.10)a |
| Protein monomers in asymmetric unit | 1 | 4 |
| Space group | C2 | P21 |
| Unit cell dimensions | ||
| a, b, c (Å) | 106.49, 57.31, 48.88 | 63.44, 114.93, 92.34 |
| α, β, γ (°) | 90, 112.78, 90 | 90, 90.89, 90 |
| Total no. of reflections | 63,855 | 816,216 |
| No. of unique reflections | 32,851 | 76,865 |
| Multiplicity | 1.9 (1.9)a | 2.2 (2.2)a |
| Completeness (%) | 95.0 (93.8)a | 96.8 (93.8)a |
| Mean I/σI | 5.2 (1.3)a | 11.17 (1.83)a |
| Rmerge (%))b | 8.3 (67.2)a | 8.8 (58.6)a |
| Refinement | ||
| Resolution range (Å) | 19.35–1.62 | 42.59–2.10 |
| No. of reflections in working set | 30824 | 76623 |
| No. of reflections in test set | 2026 | 2019 |
| Rwork (%)c | 17.8 | 15.91 |
| Rfree (%)c | 21.7 | 21.51 |
| No. of non-hydrogen atoms | ||
| Protein/water | 2380/208 | 9383/995 |
| r.m.s.d.,d bond lengths (Å) | 0.007 | 0.013 |
| r.m.s.d., bond angles (°) | 1.09 | 1.35 |
| Ramachandran favored (%) | 99 | 98 |
| Ramachandran outliers (%) | 0 | 0 |
| Molprobity clash score | 3.3 | 1.94 |
| Average B-factor (Å2) | 20.4 | 33.1 |
| Protein | 19.8 | 32.5 |
| Ligands | 16.6 | 41.1 |
| Solvent | 27.7 | 37.9 |
a Values in parentheses are for highest resolution shell.
b Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
c Rwork and Rfree = h(|F(h)o| − | F(h)c|)/h | F(h)o| for reflections in the working and test sets, respectively.
d r.m.s.d. is root mean square deviation.
FIGURE 6.
X-ray crystal structure of MERS-CoV 3CLpro in complex with inhibitors. A, solvent-accessible surface (gray-shaded surface) of MERS-CoV 3CLpro and compound 6 complex. Compound 6 is displayed in ball and stick model with atoms colored as follows: carbons (orange), nitrogens (blue), and oxygens (red). Electron density associated with compound 6 is shown as an Fo − Fc electron density difference map contoured to 3σ (green mesh). Substrate binding pockets S4-S′1 are labeled, where asterisk indicates the electrophilic carbon of compound 6 that forms a C–S covalent bond with the active site cysteine Cys-148. B, MERS-CoV 3CLpro and compound 6 complex with the MERS-CoV 3CLpro backbone represented as a ribbon model and relevant amino acids that interact with compound 6 represented as ball and sticks. MERS-CoV 3CLpro carbon atoms are colored blue, and compound 6 carbon atoms are colored orange. Nitrogen atoms are colored blue, and oxygen atoms are colored red. Catalytic residues Cys-148 and His-41 are also shown. Hydrogen bonds are depicted as red dashed lines. C, sequence logos showing amino acid conservation for the 11 polyprotein cleavage sites of different 3CLpro enzymes (MERS-CoV, HKU5-CoV, HKU4-CoV, and SARS-CoV), generated using the WebLogo server (63). Residues P2-P′1 are shown. Height of each letter corresponds to the amino acid conservation at that position. D, solvent-accessible surface (gray-shaded surface) of MERS-CoV 3CLpro and compound 11 complex. Compound 11 is displayed in ball and stick model. Electron density associated with compound 11 is shown as a 2Fo − Fc electron density difference map contoured to 1.5σ (green mesh). Functional groups of compound 11 with their corresponding binding pockets are highlighted in yellow, green, and blue ellipses. Chemical structure of compound 11 is shown in the inset. E, interactions between MERS-CoV 3CLpro and compound 11 are illustrated. Catalytic residues Cys-148 and His-41 are also shown. Hydrogen bonds are depicted as red dashed lines.
MERS-CoV 3CLpro Has a Smaller S2 Pocket than SARS-CoV 3CLpro
The active site of MERS-CoV 3CLpro bound with compound 6 is shown in Fig. 6, A and B. Compound 6 is covalently bound to the active site cysteine (Cys-148) via a 1.8 Å bond between the γ-sulfur and the electrophilic β-carbon of the Michael acceptor. The P′1-ethyl ester carbonyl, which mimics the carbonyl of the scissile bond in a substrate, forms a hydrogen bond with the backbone NH of Gly-146 that forms part of the oxyanion hole (Fig. 6B). Within the S1 subsite, the P1-lactam carbonyl, which is a surrogate for the amide of P1-glutamine of substrates, participates in a hydrogen bonding interaction with the imidazole ring of His-166, and the P1-lactam NH forms a hydrogen bond with the carboxylate oxygen of Glu-169. The P2-backbone amide NH forms a hydrogen bond with the side chain carbonyl of Gln-192 (Fig. 6B). The P2-leucine side chain atoms of the inhibitor make hydrophobic contacts with the side chains of Met-168 and Leu-49 that line the S2 subsite pocket. Moreover, compared with the equivalent residue Thr-25 in SARS-CoV 3CLpro, Met-25 in the S2 pocket of MERS-CoV 3CLpro is expected to reduce the size of the hydrophobic pocket, which is supported by our observed SAR described above.
The smaller size of the S2 pocket in MERS-CoV 3CLpro is also consistent with the preference for a smaller leucine residue at the P2 position of cleavage sites instead of a bulkier phenylalanine or methionine residue. Indeed, analysis of the preference for leucine or phenylalanine at the P2 position for the 11 3CLpro cleavage sites within the polyprotein of MERS-CoV shows that none of the 11 cleavage sites contain a phenylalanine residue at this position (Fig. 6C). Leucine is the predominantly favored residue at this position followed by methionine. Analysis of the cleavage sites from SARS-CoV, HKU4-CoV, and HKU5-CoV shows that none of the 11 cleavage sites from group 2c members (MERS-CoV, HKU4-CoV, and HKU5-CoV) contain a phenylalanine residue at the P2 position; however, the SARS-CoV nsp5↓nsp6 cleavage site contains a phenylalanine residue at this position.
Other interactions are also observed to play a significant role in stabilizing the MERS-CoV 3CLpro-compound 6 complex. The P3-carbonyl and P3-NH participate in hydrogen bonding interactions with the backbone NH and carbonyl of Glu-169. The P4-serine side chain is within hydrogen bonding distance of the side chain carboxamide of Gln-195 and the backbone carbonyl of Lys-191.
X-ray Structure of MERS-CoV 3CLpro in Complex with a Noncovalent Inhibitor
We were also able to obtain diffraction quality crystals of MERS-CoV 3CLpro in complex with compound 11, which has an almost identical chemical structure as that of compound 10 (Fig. 6D). We previously showed that compounds similar to 10 and 11 act as potent noncovalent inhibitors of 3CLpro from SARS-CoV (33). The x-ray structure of compound 11 bound to MERS-CoV 3CLpro was determined to a resolution of 2.1 Å and the x-ray data collection, processing, and refinement statistics are summarized in Table 4. The MERS-CoV 3CLpro and 11 complex crystallized in space group P21 with two biologically relevant dimers in the asymmetric unit. The overall root mean square deviation between the C-α atoms of the four chains was less than 1 Å, with the highest C-α root mean square deviation of 0.719 Å between chains C and D. Strong electron density (Fo − Fc >4σ) was present for compound 11 within all the four active sites of the two dimers (Fig. 6D).
The binding orientation for compound 11 in the active site of MERS-CoV 3CLpro is similar to the binding orientation of related compounds in the active site of SARS-CoV 3CLpro (PDB code 4MDS). The benzotriazole group binds in the S1 subsite; phenyl propionamidyl occupies the S′1-S2 subsite, and the thiophene group binds in the S2 subsite. Compound 11 also forms two direct and one water-mediated hydrogen bond interactions with amino acids in the MERS-CoV 3CLpro active site (Fig. 6E). The N3 of the benzotriazole ring forms a hydrogen bond with the side chain ϵ-nitrogen of conserved His-166, and the central acetamide oxygen forms a hydrogen bond with the backbone NH of conserved Glu-169. The NH of the phenyl propionamidyl group interacts with backbone carbonyl oxygen of the catalytic His-41 residue through a water-mediated hydrogen bond, and the imidazole ring of His-41 engages with the phenyl ring of phenyl propionamidyl group through T-shaped π stacking. The phenyl ring also form hydrophobic contacts with Leu-49.
Interactions at the 3CLpro Dimer Interface
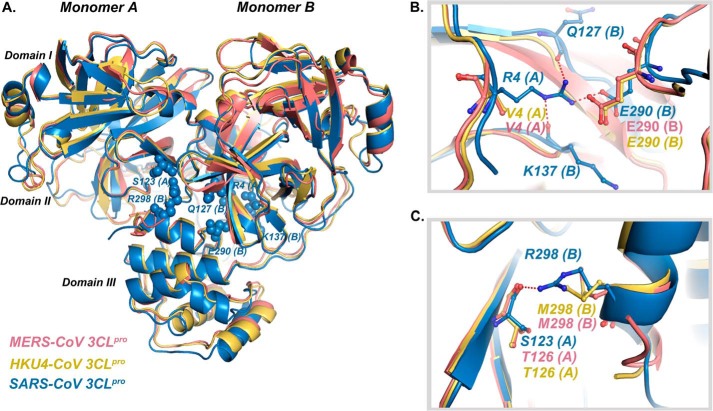
Analysis of the MERS-CoV 3CLpro and 6 and MERS-CoV 3CLpro and 11 crystal structures reveals key differences between the dimer interface of MERS-CoV and SARS-CoV 3CLpro (PDB code 2ALV) (Fig. 7) (30). Two arginine residues, Arg-4 and Arg-298 (Fig. 7, A–C), form some of the key interactions at the dimer interface of SARS-CoV 3CLpro, and mutation of either of these amino acids results in a drastic loss of dimerization in SARS-CoV 3CLpro (36, 38). Interestingly, these two arginine residues (Arg-4 and Arg-298) are substituted in MERS-CoV 3CLpro by two hydrophobic residues (Val-4 and Met-298) that are unable to participate in the formation of hydrogen bonds or salt bridges. Therefore, we initially thought that the loss of these key interactions might simply explain the >100,000-fold weaker dimerization observed for MERS-CoV 3CLpro compared with SARS-CoV 3CLpro. Surprisingly, however, structural analysis of the dimer interface from the available x-ray structure of HKU4-CoV 3CLpro (PDB code 2YNB; Fig. 7, B and C), and primary sequence alignment of 3CLpro from MERS-CoV, HKU5-CoV, HKU4-CoV and SARS-CoV (Fig. 8) revealed that Val-4 and Met-298 are conserved between all the β-CoV 2c members studied here. Substantial differences between the ability of MERS-CoV 3CLpro and HKU4/HKU5-CoV 3CLpro to dimerize, despite their high sequence identity, led us to the hypothesis that nonconserved residues between MERS-CoV and other β-CoV 2c members that are remote from the dimer interface may play a significant role in dimer formation.
FIGURE 7.
Comparison of x-ray crystal structures of 3CLpro dimers from MERS-CoV, HKU4-CoV, and SARS-CoV. A, superposition of dimers of MERS-CoV 3CLpro (pink color), HKU4-CoV 3CLpro (yellow color, PDB code 2YNB), and SARS-CoV 3CLpro (blue color, PDB code 2ALV). For SARS-CoV 3CLpro, residues Arg-4 and Ser-123 from monomer A, and residues Gln-127, Lys-137, Glu-290, and Met-298 from monomer B are represented as spheres. B, for SARS-CoV 3CLpro, interactions between the side chain of Arg-4 from monomer A and Gln-127, Glu-290, and Lys-137 residues from monomer B are shown. The corresponding residues in MERS-CoV 3CLpro and HKU4-CoV 3CLpro are Val-4 in monomer A and Glu-290 in monomer B, which do not interact at the dimer interface. C, for SARS-CoV 3CLpro, Ser-123 from monomer A engages in hydrogen bonding with Arg-298 from monomer B across the dimer interface. The corresponding residue in monomer B of MERS-CoV 3CLpro and HKU4-CoV 3CLpro is Met-298, which does not participate in any interaction with Thr-126 from monomer A across the dimer interface.
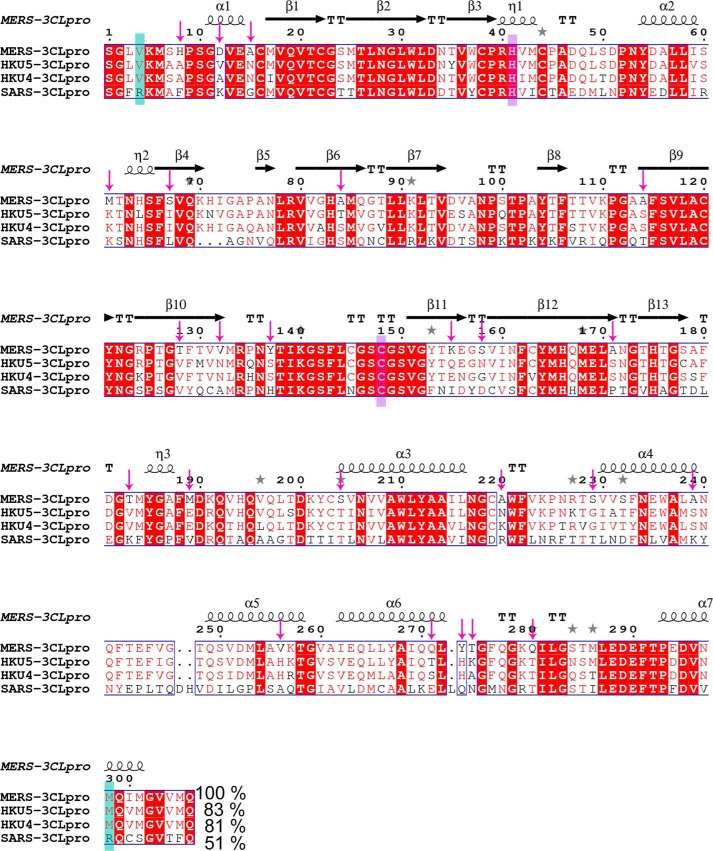
FIGURE 8.
Sequence alignment of 3CLpro enzymes from MERS-CoV, HKU5-CoV, HKU4-CoV, and SARS-CoV. Programs MultAlin (64) and ESPript (65) were used for the sequence alignment and visualization. Secondary structural elements of MERS-CoV 3CLpro are represented as spirals for α-helix, arrows for β-strands, η for 310 helix, and T for β-turns. Residues Val-4 and Met-298 in MERS-CoV, HKU5-CoV, HKU4-CoV 3CLpro, and Arg-4 and Arg-298 in SARS-CoV are shown in a green box; catalytic residues His-41 and Cys-148 are highlighted in a purple box. The nonconserved residues of MERS-CoV 3CLpro are marked with pink arrows. % identity with MERS-CoV 3CLpro is shown.
Analysis of Nonconserved Residues of MERS-CoV 3CLpro
Analysis of our current crystal structures does not reveal a clear mechanism for the monomer to dimer switch of MERS-CoV 3CLpro upon ligand binding. Therefore, we attempted to identify the nonconserved residues in MERS-CoV 3CLpro that might affect enzymatic activity due to their proximity to key residues involved in substrate binding and/or dimer formation.
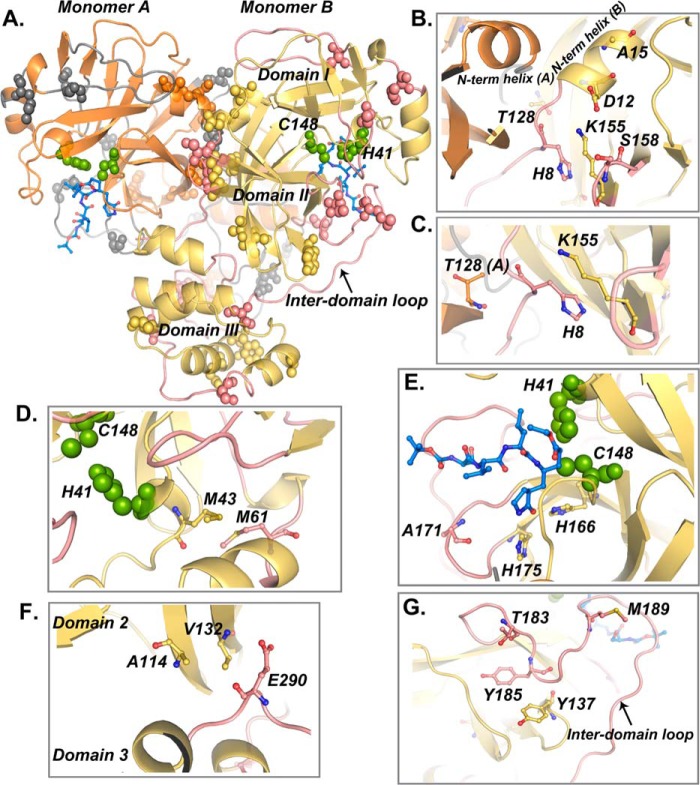
Based on a sequence alignment, MERS-CoV 3CLpro contains ∼24 nonconserved amino acids (pink arrows in Fig. 8). Upon analyzing the position of these amino acids in the crystal structure, we observed that a remarkable number of these amino acids are present in the loop regions. Fig. 9A illustrates the nonconserved residues present in the loop regions as gray (monomer A) and pink (monomer B) spheres. Interestingly, we also observed that there are hot spots in the protein structure where most of these amino acids are clustered. These hot spots include the N-terminal region, the active site region, the inter-domain loop (loop between the catalytic fold and domain III), and the domain III. In MERS-CoV 3CLpro, nonconserved amino acid His-8, which forms van der Waals contacts with Lys-155 of the same monomer and Thr-128 of the other monomer, is present at the end of the N-terminal finger (Fig. 9, B and C), whereas amino acids Asp-12 and Ala-15 are part of the N-terminal helix (Fig. 9B). Additionally, amino acids Thr-128, Lys-155, and Ser-158 are present within 6 Å of the N-terminal region (Fig. 9B). Substitution to these amino acids in MERS-CoV 3CLpro might have changed the protein dynamics in a way that only ligand binding populates the monomer conformation, which is more amenable to dimer formation.
FIGURE 9.
Analysis of the nonconserved amino acids of MERS-CoV 3CLpro. A, representation of MERS-CoV 3CLpro dimer with monomers A and B colored in orange and yellow, respectively. Nonconserved residues that are present in the loop regions are shown as spheres in gray and pink for monomers A and B, respectively. Other nonconserved residues are represented as spheres with the corresponding chain color. Domains I–III and the inter-domain loop are labeled. Catalytic residues His-41 and Cys-148 are shown as green spheres. Inhibitor molecule is shown in both active sites in blue sticks. B–G, residues of monomer B are shown (yellow and pink), unless otherwise labeled. B, clustering of some of the nonconserved amino acids, His-8, Asp-12, Ala-15, Thr-128, Lys-155, and Ser-158, near the N-terminal region is shown. N-terminal helices for both monomers are labeled. C, His-8 from the N-terminal region forms van der Waals contacts with Lys-155 of the same monomer and Thr-128 of the other monomer in the dimer. D, nonconserved residue Met-61 forms hydrophobic contacts with the Met-43 residue, which is in close proximity to the catalytic residue His-41. E, loop containing the nonconserved residue Ala-171 forms the S1 pocket along with residues His-166 and His-175. F, Val-132 forms hydrophobic contacts with a residue within the same domain (Ala-114), as well as Glu-290 from domain III. G, nonconserved residue Tyr-137 makes hydrophobic contacts with Tyr-185; Tyr-185 along with two other nonconserved residues Thr-183 and Met-189 are present on the inter-domain loop.
We also observe that some of the nonconserved residues in MERS-CoV 3CLpro are located in proximity to the substrate-binding site and might contribute toward ligand-induced dynamic changes favorable for dimer formation. For example, nonconserved amino acid Met-61 forms hydrophobic interactions with Met-43, which in turn is in close proximity to the catalytic residue His-41 (Fig. 9D). Residue Ala-171 is present on a loop, and this loop, along with conserved residues His-166 and His-175, forms the S1 subsite for binding the P1 amino acid of the substrate (Fig. 9E). In addition to its influence on substrate binding, Ala-171 may also contribute toward dimer formation upon substrate binding due to its close proximity with Glu-169. This glutamate residue in SARS-CoV 3CLpro (Glu-166) has been established as a key residue linking the substrate-binding site to the dimer interface (56). Val-132 forms hydrophobic interaction with other nonconserved residue Ala-114 within domain II (Fig. 9F). Additionally, Val-132 is present within van der Waals contact distance of Glul-290 from extra-helical domain III (Fig. 9F). It is noteworthy that Glu-290 forms a salt bridge with Arg-4 across the dimer interface in SARS-CoV 3CLpro. However, this interaction is not formed in MERS-CoV 3CLpro due to the substitution of Arg-4 with Val-4. Tyr-137 forms hydrophobic contacts with the conserved residue Tyr-185 (Fig. 9G).
Besides amino acid Val-132 that connects domains II and III, residue Tyr-185, along with two other nonconserved residues, Thr-183 and Met-189, is present on the inter-domain loop that connects the catalytic fold (domains I and II) with the extra-helical domain III (Fig. 9G). Flexibility within these residues might affect the orientation of domain III required for dimer formation.
Discussion
Model for Regulation of the Enzymatic Activity of MERS-CoV 3CLpro during Polyprotein Processing
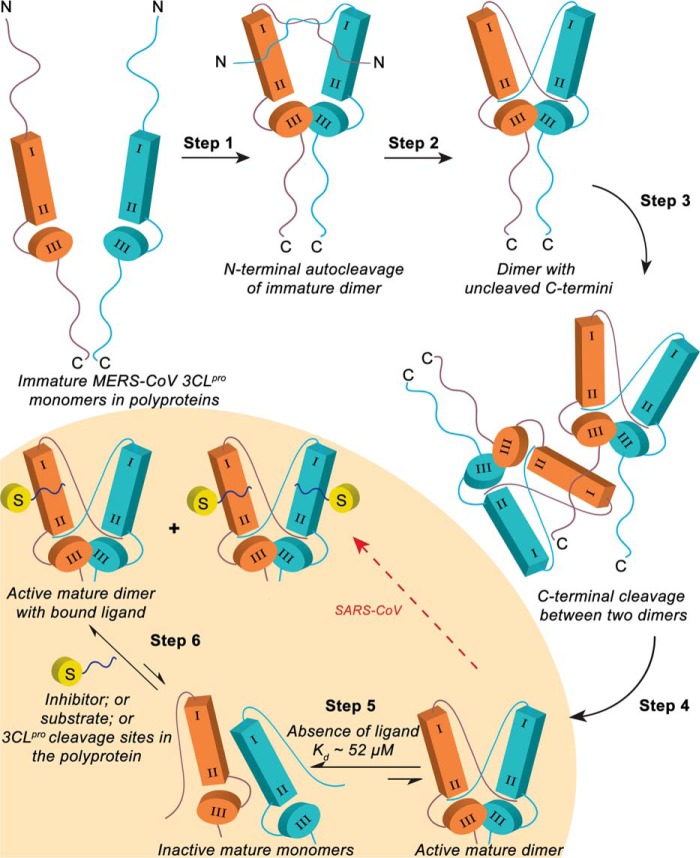
Enzymatic activity of coronavirus 3CLpro is required for the processing of viral polyproteins at 11 distinct cleavage sites, allowing the release of nonstructural proteins that subsequently form a replication complex for virus genome replication. Because of its indispensable role in the virus life cycle, regulation of the enzymatic activity of 3CLpro is instrumental for efficient replication of coronaviruses. Based on our experimental results, we propose a model to explain the mechanism for regulating the enzymatic activity of MERS-CoV 3CLpro in the context of polyprotein processing during virus infection (Fig. 10).
FIGURE 10.
Proposed model for polyprotein processing in MERS-CoV regulated by ligand-induced dimerization of MERS-CoV 3CLpro. MERS-CoV 3CLpro domains I and II are together represented as the rectangular box, and domain III is represented as a cylinder. The N and C termini are labeled, and the yellow cylinder labeled S represents a ligand that can be a peptide inhibitor, peptide substrate, or 3CLpro cleavage sites in the polyprotein. Various steps required for the auto-release of 3CLpro from the polyprotein and subsequent processing of the polyprotein cleavage sites are described in the text. Suggested by our AUC and kinetic studies, the shaded region (steps 5 and 6) highlights the additional steps MERS-CoV 3CLpro would undertake during polyprotein processing and have been described in the kinetic model depicted in Fig. 5B.
A number of in vitro studies performed on SARS-CoV 3CLpro have established the mechanism for 3CLpro auto-release from the polyprotein (34, 39, 40). Based upon these studies and our data on MERS-CoV 3CLpro, we propose the polyprotein processing model in Fig. 10. The steps proposed for auto-release of MERS-CoV 3CLpro from the polyprotein (steps 1–4, Fig. 10) have been adapted from Chen et al. (39), where it is suggested that the N-terminal auto-processing does not require the formation of a mature 3CLpro dimer for SARS-CoV. Based on the differences between the properties of SARS-CoV 3CLpro and MERS-CoV 3CLpro, as highlighted in our studies, we added two additional steps (steps 5 and 6, Fig. 10) that MERS-CoV 3CLpro may need to utilize for efficient polyprotein processing. In Fig. 10, step 1, two immature MERS-CoV 3CLpro monomers in the polyprotein approach each other and form an immature dimer via interactions between domain III, which allows each of the monomers to insert their N termini into the active site of the other monomer. In step 2, the N termini are cleaved, and the dimer with uncleaved C termini adopts a conformation similar to the mature dimer. Our observation of auto-cleavage of the N-terminal His6 tag from MERS-CoV 3CLpro during expression in bacterial cells supports steps 1 and 2, where formation of an immature dimer capable of auto-processing the N terminus occurs. In step 3, two dimers with uncleaved C termini approach each other, followed by insertion of the C terminus from one dimer into one of the active sites of the other dimer. In step 4, the C termini are cleaved and mature dimer is released from the polyprotein.
For SARS-CoV, the 3CLpro dimer formed in step 4 continues to process cleavage sites in the polyprotein, effectively skipping steps 5 and 6 (red arrow in Fig. 10) because the dimer is tightly associated. However, the high Kd value of MERS-CoV 3CLpro dimer suggests that the active and mature dimer may dissociate into inactive, mature monomers in the absence of any ligand (step 5). In order for polyprotein processing to proceed, another step (step 6) must occur. In step 6, a substrate S, e.g. one of the 11 polyprotein cleavage sites, would induce dimer formation and hence activate catalysis and cleavage at the substrate recognition sites. Our AUC results and the kinetic activation studies performed in the absence and presence of inhibitors support steps 5 and 6 where the inactive but mature monomers require binding of a ligand to undergo ligand-induced dimerization and formation of an active, mature dimer that can then process the polyprotein cleavage sites.
Nonconserved Amino Acids of MERS-CoV 3CLpro May Regulate the Dimer Formation
Long range interactions have been reported to modulate dimerization and activity of 3CLpro enzymes. Barrila et al. (57) demonstrated that mutation of a conserved amino acid Ser-147, which is distant from the dimer interface, results in a total loss of dimerization and enzymatic activity of SARS-CoV 3CLpro. Although Ser-147 does not form direct interactions at the dimer interface, disruption of the dimer upon mutation stems from the fact that Ser-147 makes several interactions with other residues involved in forming a hydrogen bonding network within SARS-CoV 3CLpro. Site-directed mutagenesis studies on domain III of SARS-CoV 3CLpro, where N214A and S284A/T285A/I286A mutants were characterized, revealed that despite being present on an entirely different domain, these residues affect catalysis through a network of residues undergoing correlated motions across the entire protease (58, 59). Utilizing 3CLpro temperature-sensitive mutants of MHV, Stobart et al. (60) have also demonstrated that second-site mutation physically distant from the temperature-sensitive mutation suppresses the temperature-sensitive phenotype through long range interactions, thereby regulating 3CLpro enzymatic activity during polyprotein processing and virus replication.
Our studies also suggest that long range interactions among the nonconserved residues can significantly alter the properties of MERS-CoV 3CLpro. A detailed analysis of nonconserved residues of MERS-CoV 3CLpro among β-CoV 2c members identified hot spots, including the N-terminal finger and helix, the active site region, the inter-domain loop, and the domain III, where these residues are clustered. Several studies done on SARS-CoV 3CLpro have demonstrated that amino acids from the N-terminal finger, the N-terminal helix, and domain III significantly contribute toward dimer formation.
In addition to the direct interactions at the dimer interface, correct orientation between the catalytic fold and domain III is also crucial for dimer formation. Wu et al. (61) showed that the most dramatic difference between the crystal structures of monomer and the ligand-bound dimer of the R298A mutant of SARS-CoV 3CLpro was a 33° rotation of domain III (38). This rotation results in a steric clash between domain III from two monomers and would essentially block dimer formation. However, upon addition of a ligand, domain III of the R298A mutant adopts the correct orientation and results in the formation of a dimer structure. Similar to the SARS-CoV 3CLpro R298A mutant, ligand binding into the active site of the MERS-CoV 3CLpro monomer possibly stabilizes the inter-domain loop conformation that maintains domain III in the correct orientation for dimer formation. Most of the nonconserved residues within domain III are present on the surface and also are distant from the dimer interface. These residues may be involved in providing the flexibility required for conformational changes during the monomer to dimer switch.
We have identified several amino acids in MERS-CoV 3CLpro that may contribute to the dimer formation upon ligand binding. However, single amino acid mutagenesis alone is unlikely to reveal significant differences in the dimerization properties. As demonstrated by Myers et al. (62) for ornithine decarboxylase, the response of single amino acid to ligand binding may be limited to only local conformational changes and may not have significant contribution toward dimer stability. However, local conformational changes in a network of residues may propagate larger effects that stabilize dimer formation upon ligand binding. Analysis of the nonconserved residues of MERS-CoV 3CLpro discussed here sets forth a framework to perform systematic single or multiple mutagenesis studies to gain insights into the mechanism for ligand-induced dimerization of the enzyme.
Development of 3CLpro Inhibitors with Broad Spectrum Specificity
Insights into the mechanistic and structural similarities as well as differences between 3CLpro enzymes from different coronavirus subgroups are instrumental for the development of 3CLpro inhibitors with broad spectrum specificity. To evaluate the broad spectrum specificity of our peptidomimetic compounds, we determined their inhibitory activity against 3CLpro from MERS-CoV, SARS-CoV, HKU5-CoV, and HKU4-CoV. Our inhibitory data and KI values clearly show that compounds 6–9 inhibit all the 3CLpro enzymes tested here. The x-ray structure of MERS-CoV 3CLpro in complex with compound 6 revealed that out of eight direct hydrogen bonds formed between compound 6 and MERS-CoV 3CLpro, four of these hydrogen bonds involve interactions with conserved structural elements of the peptide backbone of the enzyme. Furthermore, the amino acids that form hydrogen bonds with compound 6 through side chain interactions are conserved in all the coronavirus 3CLpro enzymes evaluated here, as well as 3CLpro enzymes from other β-coronaviruses like MHV, OC43, and HKU1. These results suggest that canonical structural features exist among the 3CLpro enzymes that can be exploited for structure-based design of broad spectrum inhibitors.
For the noncovalent inhibitor compound 11, the x-ray structure reveals two direct hydrogen bonding interactions between the compound and MERS-CoV 3CLpro. One of the hydrogen bonds forms with the side chain ϵ-nitrogen of conserved His-166, and the second involves the backbone NH of conserved Glu-169. We speculate these interactions remain conserved in other 3CLpro enzymes as well, because His-166 and Glu-169 amino acids are conserved in all 3CLpro enzymes. In fact, the crystal structure of SARS-CoV 3CLpro in complex with an inhibitor similar to compound 11 (PDB code 4MDS) reveals that the interactions of the inhibitor with the amino acids His-166 and Glu-169 are conserved.
The identification of 3CLpro-inhibitor interactions utilizing conserved elements of the protein structure, including the peptide backbone and conserved side chains of active site residues, suggests that the development of broad-spectrum inhibitors of coronavirus 3CLpro is feasible.
Our studies here demonstrate the unique properties of MERS-CoV 3CLpro among β-CoV 2c members, evident from the requirement for a ligand to induce dimerization. Although the peptidomimetic compounds containing a Michael acceptor group (for example, compounds 6–9) induce dimer formation of MERS-CoV 3CLpro, the irreversible nature of their reaction with the active site cysteine ensures complete inhibition of the enzyme at stoichiometric ratios in a time-dependent manner. On the contrary, noncovalent peptidomimetic compounds (for example, compounds 10 and 11) inhibit the enzymatic activity of MERS-CoV 3CLpro only at high compound concentrations. Based on these observations, compounds that irreversibly modify the 3CLpro active site may serve as better candidates for the development of inhibitors for MERS-CoV 3CLpro.
Potential Complexity in the Development of MERS-CoV 3CLpro Inhibitors as Antiviral Agents
Induced dimerization of MERS-CoV 3CLpro, as seen in the presence of peptidomimetic inhibitors, has significant implications in the development of antiviral agents targeting MERS-CoV 3CLpro. As a consequence of enzyme activation, the development of an effective antiviral agent may necessitate the development of a compound that can inhibit the MERS-CoV 3CLpro monomer and stabilize it without inducing dimerization and/or inhibit the active sites of the dimer at low doses, ensuring inactivation of both the active sites within the dimer. On the contrary, it is also possible that the presence of an inhibitor could enhance the activity of MERS-CoV 3CLpro to an extent that results in a complete loss of the temporal and spatial regulation of the enzymatic activity, thereby disrupting viral genome replication. Ramifications of ligand-induced dimerization and activation of MERS-CoV 3CLpro, as seen in the presence of lower concentrations of inhibitor, will need to be further explored in virus-infected cells.
Author Contributions
S. T. and A. D. M. conceived and coordinated the study and wrote the paper. S. T., M. L. J. and S. E. S. J. designed, performed, and analyzed the experiments shown in Figs. 1 and 2. S. T. and L. N. P. designed, performed, and analyzed the experiments shown in Fig. 4. S. T. and M. L. J. designed, performed, and analyzed the experiments shown in Fig. 5. S. T. and A. D. M. determined the crystal structures. H. L. O., P. R. N., and A. K. G. synthesized compounds 1–9. S. E. S. J., A. K. G., and M. R. D. provided substantial contributions to analysis and interpretation of data.
Acknowledgments
Crystallization and DNA sequencing were partially supported by the Purdue Center for Cancer Research Macromolecular Crystallography and DNA Sequencing Shared Resources, which are partially supported by National Institutes of Health Grant P30 CA023168. We acknowledge the LS-CAT and LRL-CAT beam line staff for their help in acquiring x-ray data. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Argonne National Laboratory, was supported by the United States Department of Energy under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and the Michigan Technology Tri-Corridor Grant 085P1000817. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Co., which operates the facility. We thank Yahira Baez-Santos, Dia Beachboard and Sergey Savinov for their helpful suggestions during manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants AI08508 (to A. D. M.) and AI026603 (to A. D. M. and M. R. D.). This work was also supported by the Walther Cancer Foundation (to A. D. M.). All authors reviewed the results and approved the final version of the manuscript. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4RSP and 4YLU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CoV
- coronavirus
- MERS
- Middle East respiratory syndrome
- SARS
- severe acute respiratory syndrome
- nsp
- nonstructural protein
- 3CLpro
- 3-chymotrypsin-like protease
- AUC
- analytical ultracentrifugation
- SV
- sedimentation velocity
- BME
- β-mercaptoethanol
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank.
REFERENCES
- 1. Alenius S., Niskanen R., Juntti N., Larsson B. (1991) Bovine coronavirus as the causative agent of winter dysentery: serological evidence. Acta Vet. Scand. 32, 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barber D. M., Nettleton P. F., Herring J. A. (1985) Disease in a dairy herd associated with the introduction and spread of bovine virus diarrhoea virus. Vet. Rec. 117, 459–464 [DOI] [PubMed] [Google Scholar]
- 3. Perlman S., Netland J. (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McIntosh K., Becker W. B., Chanock R. M. (1967) Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U.S.A. 58, 2268–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cavallaro J. J., Monto A. S. (1970) Community-wide outbreak of infection with a 229E-like coronavirus in Tecumseh, Michigan. J. Infect. Dis. 122, 272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamre D., Procknow J. J. (1966) A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121, 190–193 [DOI] [PubMed] [Google Scholar]
- 7. Woo P. C., Lau S. K., Chu C. M., Chan K. H., Tsoi H. W., Huang Y., Wong B. H., Poon R. W., Cai J. J., Luk W. K., Poon L. L., Wong S. S., Guan Y., Peiris J. S., Yuen K. Y. (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Hoek L., Pyrc K., Jebbink M. F., Vermeulen-Oost W., Berkhout R. J., Wolthers K. C., Wertheim-van Dillen P. M., Kaandorp J., Spaargaren J., Berkhout B. (2004) Identification of a new human coronavirus. Nat. Med. 10, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui L. J., Zhang C., Zhang T., Lu R. J., Xie Z. D., Zhang L. L., Liu C. Y., Zhou W. M., Ruan L., Ma X. J., Tan W. J. (2011) Human coronaviruses HCoV-NL63 and HCoV-HKU1 in hospitalized children with acute respiratory infections in Beijing, China. Adv. Virol. 2011, 129134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 [DOI] [PubMed] [Google Scholar]
- 11. Butler D. (2012) Clusters of coronavirus cases put scientists on alert. Nature 492, 166–167 [DOI] [PubMed] [Google Scholar]
- 12. WHO-Global Alert and Response (2014) Middle East respiratory syndrome coronavirus (MERS-CoV)–Saudi Arabia. World Health Organization, Geneva, Switzerland [Google Scholar]
- 13. van Boheemen S., de Graaf M., Lauber C., Bestebroer T. M., Raj V. S., Zaki A. M., Osterhaus A. D., Haagmans B. L., Gorbalenya A. E., Snijder E. J., Fouchier R. A. (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3, e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan J. F., Li K. S., To K. K., Cheng V. C., Chen H., Yuen K. Y. (2012) Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 65, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Annan A., Baldwin H. J., Corman V. M., Klose S. M., Owusu M., Nkrumah E. E., Badu E. K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y. A., Kalko E. K., Lina P. H., Godlevska E. V., et al. (2013) Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19, 456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Memish Z. A., Mishra N., Olival K. J., Fagbo S. F., Kapoor V., Epstein J. H., Alhakeem R., Durosinloun A., Al Asmari M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A. A., Lipkin W. I. (2013) Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 19, 1819–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reusken C. B., Haagmans B. L., Müller M. A., Gutierrez C., Godeke G. J., Meyer B., Muth D., Raj V. S., Smits-De Vries L., Corman V. M., Drexler J. F., Smits S. L., El Tahir Y. E., De Sousa R., van Beek J., et al. (2013) Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 13, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perera R. A., Wang P., Gomaa M. R., El-Shesheny R., Kandeil A., Bagato O., Siu L. Y., Shehata M. M., Kayed A. S., Moatasim Y., Li M., Poon L. L., Guan Y., Webby R. J., Ali M. A., Peiris J. S., Kayali G. (2013) Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 18, pii=20574 [DOI] [PubMed] [Google Scholar]
- 19. Cauchemez S., Van Kerkhove M. D., Riley S., Donnelly C. A., Fraser C., Ferguson N. M. (2013) Transmission scenarios for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and how to tell them apart. Euro Surveill. 18, 20503. [PMC free article] [PubMed] [Google Scholar]
- 20. Assiri A., McGeer A., Perl T. M., Price C. S., Al Rabeeah A. A., Cummings D. A., Alabdullatif Z. N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J. A., Cotten M., Watson S. J., et al. (2013) Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 369, 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breban R., Riou J., Fontanet A. (2013) Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 382, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cotten M., Watson S. J., Kellam P., Al-Rabeeah A. A., Makhdoom H. Q., Assiri A., Al-Tawfiq J. A., Alhakeem R. F., Madani H., AlRabiah F. A., Al Hajjar S., Al-nassir W. N., Albarrak A., Flemban H., Balkhy H. H., et al. (2013) Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382, 1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Centers for Disease Control (2014) MERS in the U.S. CDC, Atlanta, GA [Google Scholar]
- 24. Lu Y., Lu X., Denison M. R. (1995) Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69, 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai M. M., Cavanagh D. (1997) The molecular biology of coronaviruses. Adv. Virus Res. 48, 1–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ziebuhr J., Snijder E. J., Gorbalenya A. E. (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81, 853–879 [DOI] [PubMed] [Google Scholar]
- 27. Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., Hilgenfeld R. (2003) Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300, 1763–1767 [DOI] [PubMed] [Google Scholar]
- 28. Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G. F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. (2003) The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 100, 13190–13195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang H., Bartlam M., Rao Z. (2006) Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr. Pharm. Des. 12, 4573–4590 [DOI] [PubMed] [Google Scholar]
- 30. Ghosh A. K., Xi K., Ratia K., Santarsiero B. D., Fu W., Harcourt B. H., Rota P. A., Baker S. C., Johnson M. E., Mesecar A. D. (2005) Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 48, 6767–6771 [DOI] [PubMed] [Google Scholar]
- 31. Ghosh A. K., Gong G., Grum-Tokars V., Mulhearn D. C., Baker S. C., Coughlin M., Prabhakar B. S., Sleeman K., Johnson M. E., Mesecar A. D. (2008) Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 18, 5684–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S. A., Chase P., Eggler A., Dawson E. S., Baez-Santos Y. M., Tomar S., Mielech A. M., Baker S. C., Lindsley C. W., Hodder P., Mesecar A., Stauffer S. R. (2013) Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 56, 534–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J. S., Dawson E., Saldanha A., Chase P., Baez-Santos Y. M., Lindsley C. W., Hodder P., Mesecar A. D., Stauffer S. R. (2013) Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 23, 6172–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsu M. F., Kuo C. J., Chang K. T., Chang H. C., Chou C. C., Ko T. P., Shr H. L., Chang G. G., Wang A. H., Liang P. H. (2005) Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 280, 31257–31266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lai L., Han X., Chen H., Wei P., Huang C., Liu S., Fan K., Zhou L., Liu Z., Pei J., Liu Y. (2006) Quaternary structure, substrate selectivity and inhibitor design for SARS 3C-like proteinase. Curr. Pharm. Des. 12, 4555–4564 [DOI] [PubMed] [Google Scholar]
- 36. Chen S., Zhang J., Hu T., Chen K., Jiang H., Shen X. (2008) Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 143, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grum-Tokars V., Ratia K., Begaye A., Baker S. C., Mesecar A. D. (2008) Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 133, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi J., Sivaraman J., Song J. (2008) Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 82, 4620–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen S., Jonas F., Shen C., Hilgenfeld R., Higenfeld R. (2010) Liberation of SARS-CoV main protease from the viral polyprotein: N-terminal autocleavage does not depend on the mature dimerization mode. Protein Cell 1, 59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li C., Qi Y., Teng X., Yang Z., Wei P., Zhang C., Tan L., Zhou L., Liu Y., Lai L. (2010) Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J. Biol. Chem. 285, 28134–28140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Báez-Santos Y. M., Barraza S. J., Wilson M. W., Agius M. P., Mielech A. M., Davis N. M., Baker S. C., Larsen S. D., Mesecar A. D. (2014) X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 57, 2393–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Agnihothram S., Yount B. L., Jr., Donaldson E. F., Huynh J., Menachery V. D., Gralinski L. E., Graham R. L., Becker M. M., Tomar S., Scobey T. D., Osswald H. L., Whitmore A., Gopal R., Ghosh A. K., Mesecar A., et al. (2014) A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. MBio 5, e00047–00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghosh A. K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W., Houser K. V., Baker S. C., Johnson M. E., Mesecar A. D. (2007) Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 17, 5876–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen Y., Chou C. Y., Chang G. G., Tong L. (2006) Is dimerization required for the catalytic activity of bacterial biotin carboxylase? Mol. Cell 22, 807–818 [DOI] [PubMed] [Google Scholar]
- 45. Leslie A. G., Powell H. R. (2007) Processing diffraction data with MOSFLM. NATO Sci. Ser. ii Math. 245, 41–51 [Google Scholar]
- 46. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 47. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLano W. L. (2014) The PyMOL Molecular Graphics System, Version 1.7.2.1, Schrödinger, LLC, New York [Google Scholar]
- 51. Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. (2004) Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 279, 1637–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi J., Wei Z., Song J. (2004) Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 279, 24765–24773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blanchard J. E., Elowe N. H., Huitema C., Fortin P. D., Cechetto J. D., Eltis L. D., Brown E. D. (2004) High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 11, 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., Rupprecht D., Schmuck C., Baumann K., Ziebuhr J., Schirmeister T. (2005) A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 48, 6832–6842 [DOI] [PubMed] [Google Scholar]
- 55. Brown P. H., Balbo A., Schuck P. (2008) Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. Chapter 18, Unit 18.15 [DOI] [PubMed] [Google Scholar]
- 56. Cheng S. C., Chang G. G., Chou C. Y. (2010) Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 98, 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barrila J., Bacha U., Freire E. (2006) Long-range cooperative interactions modulate dimerization in SARS 3CLpro. Biochemistry 45, 14908–14916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi J., Han N., Lim L., Lua S., Sivaraman J., Wang L., Mu Y., Song J. (2011) Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain. PLoS Comput. Biol. 7, e1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lim L., Shi J., Mu Y., Song J. (2014) Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS One 9, e101941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stobart C. C., Lee A. S., Lu X., Denison M. R. (2012) Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. J. Virol. 86, 4801–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu C. G., Cheng S. C., Chen S. C., Li J. Y., Fang Y. H., Chen Y. H., Chou C. Y. (2013) Mechanism for controlling the monomer-dimer conversion of SARS coronavirus main protease. Acta Crystallogr. D Biol. Crystallogr. 69, 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Myers D. P., Jackson L. K., Ipe V. G., Murphy G. E., Phillips M. A. (2001) Long-range interactions in the dimer interface of ornithine decarboxylase are important for enzyme function. Biochemistry 40, 13230–13236 [DOI] [PubMed] [Google Scholar]
- 63. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]