Background: Bacterial ADP-ribosylating toxin C3 has long been used to study the diverse regulatory functions of Rho GTPases.
Results: The complex structures of C3-RhoA(GTP) and C3-RhoA(GDP) were revealed.
Conclusion: C3 recognizes RhoA via the switch I, switch II, and interswitch regions.
Significance: The structures presented explain RhoA recognition by C3 including the ARTT loop and provide insight into the ART reaction.
Keywords: ADP-ribosylation, bacterial toxin, CDC42, protein structure, protein-protein interaction, Rho (Rho GTPase), signal transduction, RalA, RhoA, C3 exoenzyme
Abstract
C3 exoenzyme is a mono-ADP-ribosyltransferase (ART) that catalyzes transfer of an ADP-ribose moiety from NAD+ to Rho GTPases. C3 has long been used to study the diverse regulatory functions of Rho GTPases. How C3 recognizes its substrate and how ADP-ribosylation proceeds are still poorly understood. Crystal structures of C3-RhoA complex reveal that C3 recognizes RhoA via the switch I, switch II, and interswitch regions. In C3-RhoA(GTP) and C3-RhoA(GDP), switch I and II adopt the GDP and GTP conformations, respectively, which explains why C3 can ADP-ribosylate both nucleotide forms. Based on structural information, we successfully changed Cdc42 to an active substrate with combined mutations in the C3-Rho GTPase interface. Moreover, the structure reflects the close relationship among Gln-183 in the QXE motif (C3), a modified Asn-41 residue (RhoA) and NC1 of NAD(H), which suggests that C3 is the prototype ART. These structures show directly for the first time that the ARTT loop is the key to target protein recognition, and they also serve to bridge the gaps among independent studies of Rho GTPases and C3.
Introduction
Rho GTPases (∼20 kDa) are key regulators of cytoskeletal dynamics, affecting such processes as morphogenesis, cell migration, neuronal development, and cell division and adhesion (1, 2) and functioning as molecular switches that control signal transduction pathways from plasma membrane receptors to the cytoskeleton. The switching mechanism is thought to be mediated by conformational changes in two switch regions (I and II). Switching between an inactive GDP-bound conformation and an active GTP-bound one (*Rho(GDP) or Rho(GTP) indicates the Rho-bound nucleotide) (3–5) is tightly controlled by three regulatory proteins: (i) the guanine nucleotide exchange factor (GEF)2 activates Rho GTPase by exchanging GDP for GTP; (ii) the GTPase-activating protein (GAP) inactivates Rho GTPase by increasing its intrinsic rate of GTP-hydrolysis; and (iii) the guanine nucleotide dissociation inhibitor (GDI) sequesters isoprenylated Rho GTPase in the cytosol away from the membrane.
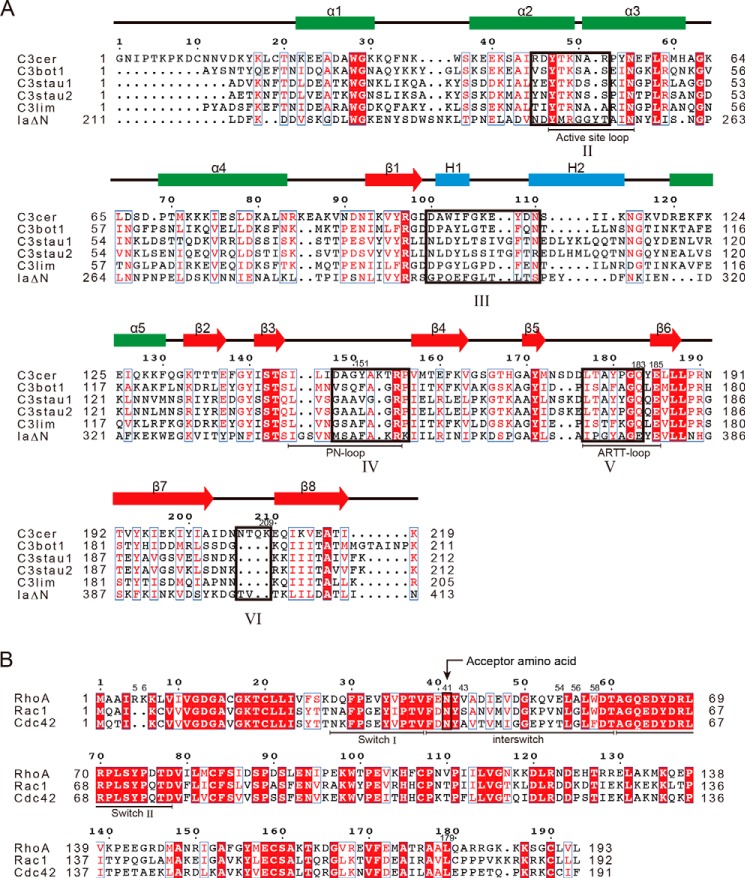
ADP-ribosylation is an important post-translational protein modification known to be catalyzed by bacterial toxins as well as eukaryotic endogenous ADP-ribosyltransferases (ARTs). Based on their target specificity, bacterial ARTs are traditionally classified into several types. Interestingly, however, there is a prominent structural similarity between the Rho GTPase-specific C3 exoenzyme and the binary toxin enzymatic units Ia (6), C2I (7), VIP2 (8), and CDTa (9), even though they have different substrate specificities (10); C3-like ARTs (25 kDa) catalyze the ADP-ribosylation of Rho GTPases on Asn-41 (11) and Ia-like ARTs modify actin on Arg-177 (6). C3 exoenyzme has been used extensively to examine the function of Rho GTPases (12–16). C3-like ARTs are expressed by various Gram-positive bacteria and have been characterized on the basis of their substrate specificity. So far, seven C3-like isoforms have been described: two isoforms of C3 from Clostridium botulinum (C3bot1 and C3bot2) (17–21), C3lim from Clostridium limosum (22), C3 from Bacillus cereus (C3cer) (23), and three isoforms of C3 from Staphylococcus aureus (C3stau1, C3stau2, and C3stau3) (24–27). C3bot, C3lim, and C3cer preferentially ADP-ribosylate RhoA, RhoB, and RhoC but not Rac1 or Cdc42 (28). ADP-ribosylation of Rho GTPases by C3-like ARTs prevents GEF binding, thereby inhibiting subsequent binding of Rho GTPases to effector proteins (29). Moreover, ADP-ribosylation increases the ability of RhoA to complex with GDI (30), so that ADP-ribosylated RhoA is found exclusively in the cytosolic fraction of C3-treated cells. This C3-induced sequestration most likely blocks Rho-dependent signaling (31).
The first structural details of a C3-like ART were obtained from C3bot1 in a NAD+-free state using x-ray crystallography (10). Han and Tainer (32) proposed that the bipartite ADP-ribosylating toxin turn-turn (ARTT) loop, which consists of turns 1 and 2, is responsible for substrate recognition and is thus crucial for the ARTase activity of C3-like exoenzymes and binary toxins. This finding has prompted the examination of the conformation and significance of the ARTT loop within the NAD+-bound and -free structures of C3 (33, 34). However, recognition of RhoA by the ARTT loop has never been directly verified, because the structure of a C3-like ART in complex with a protein substrate has never been determined. On the other hand, several RhoA amino acid residues involved in recognition by C3bot have been identified through mutational analysis (35).
The complex structure C3-RhoA is a long sought after piece of this C3 field study. How C3-like ART recognizes Rho GTPase and how the ADP-ribosylation of asparagine occurs are major issues to be addressed. Here we have reported the crystal structures of apo(NAD+-free)-C3-RhoA(GTP), NADH-C3-RhoA(GTP), and NADH-C3-RhoA(GDP). These structures provide the first direct evidence that the ARTT loop is essential for target protein recognition and not only explain the recognition between C3 and RhoA but also provide insight into the asparagine ART reaction.
Experimental Procedures
Preparation of C3cer
His-tagged C3cer (UniProt ID: Q8KNY0) was designed and subcloned into the pRham vector, after which the plasmid was transformed into DH10B cells using electroporation. Cells containing pRham-C3cer plasmid were selected on LB agar plates containing kanamycin. A single colony was inoculated into 1.5 liters of LB medium (containing 25 μg/ml kanamycin, 0.2% rhamnose, and 0.05% glucose) and cultured overnight at 37 °C with vigorous shaking. The cells were then harvested by centrifugation and stored at −80 °C. The frozen cells were later thawed, resuspended with 50 ml of buffer CA (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 5 mm imidazole), disrupted using a French press, and centrifuged at 180,000 × g for 40 min. The resultant supernatant was loaded onto a Ni-NTA agarose column. After washing the column with buffer CA to remove the unbound residues, the His-tagged sample was eluted using buffer CB (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 300 mm imidazole). The His tag was then cleaved using His-tagged TEV (tobacco etch virus) protease overnight at 10 °C. The resultant mixture was concentrated, and the buffer in the mixture was exchanged for buffer CA without imidazole using an Amicon Ultra filter (Millipore). To remove the His tag and His-tagged TEV protease, the sample was run on a nickel-nitrilotriacetic acid-agarose column, after which the concentrated sample was loaded onto a Superdex 75 size-exclusion column (GE Healthcare) and eluted with buffer CC (10 mm Tris-HCl (pH 7.4) and 100 mm NaCl). The C3cer fractions were collected and concentrated to ∼20 mg/ml and stored at −80 °C.
Preparation of RhoA
The C-terminal 14 residues of RhoA (UniProt ID: P61586) were truncated, and Phe25 was substituted with Asn as described elsewhere (36). Truncated and mutated RhoA was subcloned into the pGEX 4T-1 vector, after which the plasmid was transformed into BL21(DE3) cells using heat shock. Cells containing pGEX 4T-1-RhoA plasmid were selected on LB agar plates containing ampicillin, and a single colony was inoculated into a small volume of LB medium and precultured overnight at 37 °C. The preculture was transferred into 1.5 liters of LB medium containing 100 μg/ml ampicillin, and the cells were cultured for 4 h at 37 °C with vigorous shaking. After inducing expression with isopropyl-1-thio-β-d-galactopyranoside at a final concentration of 0.5 mm, the culture was incubated for an additional 22 h at 20 °C. The cells were then harvested by centrifugation and stored at −80 °C. The frozen cells were later thawed, resuspended with 50 ml of buffer RA (50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, and 5 mm DTT) containing complete EDTA-free (Roche Applied Science), passed through a French press, and centrifuged at 17,000 × g for 50 min at 4 °C. The supernatant was loaded onto a glutathione-Sepharose 4B column (GE Healthcare), which was then washed with buffer RA to remove the unbound residues. The GST tag was cleaved in buffer RA containing 2.5 mm CaCl2 and 350 units of thrombin for 18 h at room temperature on the Sepharose 4B column. The sample and the thrombin mixture were then eluted and concentrated using an Amicon Ultra filter, after which the concentrated mixture was loaded onto a Superdex 75 size-exclusion column and eluted with buffer RC (10 mm Tris-HCl (pH 8.0), 1 mm MgCl2, and 1 mm DTT). RhoA fractions were collected, concentrated to ∼20 mg/ml, and stored at −80 °C.
Preparation of Cdc42
Cdc42 (UniProt ID: P60953-1) in which the C-terminal 14 residues were truncated was subcloned into pGEX 4T-1 vector, after which the plasmid was transformed into BL21(DE3) cells using heat shock. Protein expression was the same as described in RhoA.
Preparation of RhoA and Cdc42 Mutants
RhoA and Cdc42 mutants were constructed by site-directed mutagenesis using a PrimeSTAR mutagenesis basal kit (TaKaRa Bio Inc.) according to the manufacturer's instructions. Protein expression was the same as described for RhoA.
Crystallization and NADH Soaking
To obtain apo-C3cer-RhoA crystals, C3cer and RhoA samples were mixed at a 1:1 molar ratio before the addition of GTPγS or GDP to a final concentration of 1 mm. Crystallization was carried out using the hanging drop vapor diffusion method against a reservoir solution containing 100 mm MES (pH 6.4) and 20% PEG 1500 at 4 °C. The drop was composed of equal volumes of the protein and reservoir solutions. In 4 to 6 weeks, rod-like crystals appeared in a cluster for both C3cer-RhoA(GTP) and C3cer-RhoA(GDP). NADH cannot be an ADP-ribosylation substrate of C3. To obtain crystals of the NADH-bound C3cer-RhoA(GTP or GDP) complex, a small portion of the clustered crystals was soaked in buffer containing 10 mm NADH with 30% ethylene glycol as a cryoprotectant for 1 h at 4 °C.
Data Collection
The crystals were plunged into a stream of nitrogen gas at 100 K. Data collection for the apo- and NADH-soaked C3cer-RhoA(GDP) was performed at 100 K using an x-ray wavelength of 1.0 Å with an ADSC Quantum 210r detector system on beamlines BL-5A and AR-NW12A, respectively, at the KEK Photon Factory. The data for NADH-soaked C3cer-RhoA(GTP) were collected in-house using an x-ray wavelength of 1.54 Å with a MicroMax-007 HF generator and RAXISVII (Rigaku). A total of 360 frames of each of the data sets were collected for each crystal with 0.5° oscillations. The best data set was collected at 1.8 Å for apo-C3cer-RhoA(GTP). The datasets of NADH-C3cer-RhoA(GTP) and NADH-C3cer-RhoA(GDP) were collected at 2.5 Å, respectively. The diffraction images for all crystals were indexed, integrated, and scaled using the programs DENZO and SCALEPACK from the HKL2000 suite (37). The space group for all crystals was P32, and the asymmetric unit contained one C3cer and one RhoA(GTP or GDP). Data collection statistics and cell constants are summarized in Table 1.
TABLE 1.
Data collection and structure refinement statistics of C3cer-RhoA complexes
| apo-C3cer-RhoA(GTP) | NADH-C3cer-RhoA(GTP) | NADH-C3cer-RhoA(GDP) | |
|---|---|---|---|
| Data collection | |||
| group | P32 | P32 | P32 |
| Cell dimensions | |||
| a, b, c (Å) | 50.75, 50.75, 135.87 | 50.42, 50.42, 136.67 | 50.48, 50.48, 136.67 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| X-ray source | PF BL-5A | Rigaku MicroMax-007 HF | PF-AR NW-12A |
| Wavelength (Å) | 1.00 | 1.54 | 1.00 |
| Resolution range (Å) | 50.00–1.80 (1.86–1.80) | 50.00–2.50 (2.59–2.50) | 50.00–2.50 (2.59–2.50) |
| Observed reflections | 205,280 | 73,141 | 63,158 |
| Rmeasa | 0.074 (0.859) | 0.090 (0.912) | 0.127 (1.336) |
| Rpimb | 0.031 (0.367) | 0.038 (0.428) | 0.059 (0.628) |
| CC½c | 0.998 (0.823) | 0.998 (0.703) | 0.994 (0.449) |
| I/σI (%) | 21.4 (2.2) | 21.0 (2.6) | 17.4 (2.0) |
| Completeness (%) | 100.0 (100.0) | 99.9 (99.9) | 99.6 (100.0) |
| Redundancy | 5.7 (5.4) | 5.5 (4.4) | 4.7 (4.4) |
| Refinement | |||
| Resolution (Å) | 45.29-1.80 | 45.56-2.50 | 45.56-2.50 |
| Rwork/Rfree (%)d | 17.7 / 22.3 | 19.6 / 25.4 | 21.1 / 24.6 |
| Overall B-factors (Å2)e | 32.8 | 49.7 | 45.9 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.008 | 0.010 | 0.020 |
| Bond angles (°) | 1.34 | 1.25 | 1.74 |
| Ramachandran plot | |||
| Favored regions (%) | 91.2 | 89.4 | 93.5 |
| Allowed regions (%) | 8.8 | 10.6 | 6.5 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| PDB ID | 4XSG | 4XSH | 5BWM |
a Rmeas = Σhkl{N(hkl)/[N(hkl) − 1]}1/2Σi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) are the observed intensities.
b Rpim = Σhkl{1/[N(hkl) − 1]}1/2Σi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) are the observed intensities, 〈I(hkl)〉 is the average intensity, and N(hkl) is the multiplicity of reflection hkl.
c CC½ = percentage of correlation between intensities from random half-data sets. CC½ is calculated according to the formula found in Ref. 54.
d Rwork = Σhkl‖Fobs | − |Fcalc‖/Σhkl|Fobs|. Rfree is the cross-validation R-factor for the test set (5%) of reflections omitted from model refinement.
e Obtained by TLS refinement in Refmac5 of CCP4i.
Model Building and Refinement
The structure of apo-C3cer-RhoA(GTP) was determined using the molecular replacement method with the Phaser-MR program in the Phenix suite (38). The structures of C3lim (PDB code: 3BW8 (39)) and RhoA (PDB code: 1A2B (3)) served as templates. The structure was refined iteratively using Phenix.refine in the Phenix suite (40), REFMAC5 in the CCP4i suite (41), and Coot (42). A comparison of the structures and the preparation of the figures were done using PyMOL (43). Finally, the structure was refined and validated using the PDB_REDO Web server (44). For both NADH-bound C3cer-RhoA(GTP) and NADH-bound C3cer-RhoA(GDP), using the resolved apostructure served as the template, and the structures were determined and improved using the same methods used for the apostructure. The final model statistics for all structures are summarized in Table 1.
ADP-ribosylation
The crystal assay was conducted as follows. Crystals of the C3-RhoA complex were washed with mother liquor twice. Biotin-NAD+ (50 μm) was then added to the mother liquor containing the C3-RhoA complex, and the mixture was kept at room temperature for 4 h. These samples were then subjected to SDS-PAGE. The gel was washed twice with PBS, stained with streptavidin-FITC (250 nm) overnight, washed twice with PBS again, and scanned using a Typhoon FLA9000 laser scanner (General Electric). The solution assay was conducted as follows. In the experiment shown in Fig. 2, C3cer (0.3 μm) and GST-RhoA (3 μm) or C3cer (0.3 μm) and non-tagged RhoA (3 μm) were mixed together in buffer RC. In the mutational experiment (Fig. 5), C3cer (0.03 μm) and GST-RhoA (3 μm) or C3cer (0.3 μm) and GST-Cdc42 (mutants) (3 μm) were mixed together in buffer RC. Biotin-NAD+ (3.5 μm) was then added to the mixture, which was kept at 37 °C for 10 min. Thereafter, the same protocol used for the crystal assay was applied.
FIGURE 2.
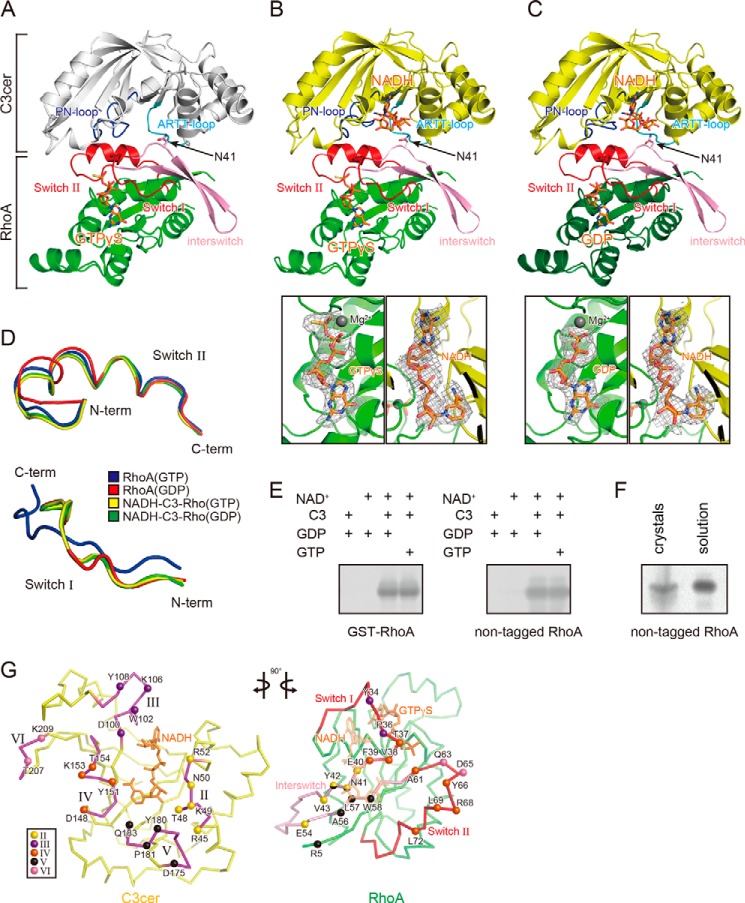
Crystal structures of the C3cer-RhoA complex. A, overall structure of the apo-C3cer-RhoA. B, overall structure of the NADH-bound C3cer-RhoA(GTP) (upper panel) and electron density maps for GTPγS and NADH (lower panels). C, overall structure of the NADH-bound C3cer-RhoA(GDP) (upper panel) and electron density maps for GDP and NADH (lower two panels). The apo-form of C3cer is shown in white; the other forms are in yellow. In A–C, all RhoA are in green, and their switch regions and interswitch regions are in red and pink, respectively. Nucleotides (NADH, GTPγS, and GDP) are shown in stick form and colored orange. The ARTT loops and PN loops are in cyan and blue, respectively. The Fo − Fc simulated annealing omit maps are drawn at 1.0 σ, and the clipping distances are 30 Å. D, conformations of the main chains in the switch I (residues 28–38 (left panel)) and switch II (residues 61–78 (right panel)) regions of Rho: the GTP form (PDB code: 1A2B), GDP form (PDB code: 1FTN), NADH-bound C3cer-RhoA(GTP), and NADH-bound C3cer-RhoA(GDP) are colored blue, red, yellow, and green, respectively. E, ADP-ribosylation of GST-tagged and non-tagged of RhoA(GDP) and RhoA(GTP) by C3cer. F, ADP-ribosylation of crystal and solution of C3-RhoA(GTP). G, butterfly representation of the recognition residues in C3cer and RhoA. Roman numerals (II–VI) show the five binding regions in C3cer. In the left panel, the RhoA recognition residues in the five regions are shown as spheres. In the right panel, the C3cer recognition residues of RhoA are shown as spheres colored as in the left panel. The switch I and II regions are shown in red. In both panels NADH is shown in stick form and colored orange. The definitions of each region are provided in Fig. 1.
FIGURE 5.
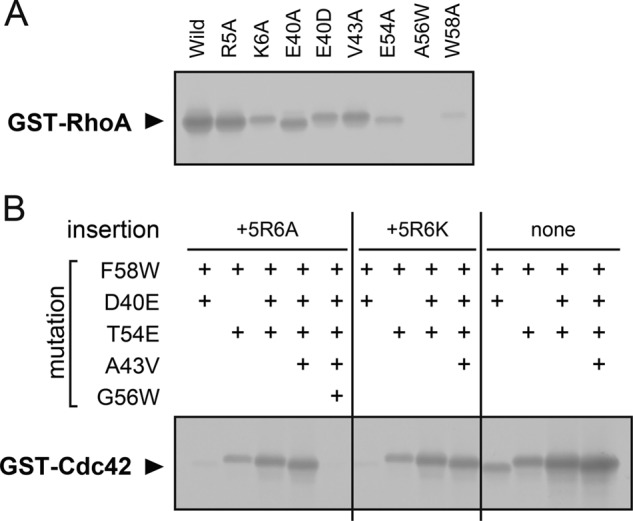
ADP-ribosylation of RhoA and Cdc42 mutants by C3cer A, ADP-ribosylation of RhoA single point mutations. B, ADP-ribosylation of Cdc42 combined mutations. Wild type and mutants of RhoA and Cdc42 were purified as GST-tagged protein.
Results
Crystal Structures of C3cer-RhoA Complex
we used C3cer for crystallization of the complexes. The structure of C3cer is unknown, but its sequence shows identity with C3bot1 (38%), C3bot2 (36%), C3lim (36%), C3stau1 (29%), and C3stau2 (29%) (Fig. 1A). To investigate how C3cer recognizes RhoA, we determined the crystal structure of apo(NAD+-free)-C3cer complexed with human RhoA(GTP) at 1.8 Å resolution (Fig. 2A and Table 1). Then, by soaking the apo-C3-RhoA(GTP) crystal with NADH, we obtained the structure of NADH-bound C3-RhoA(GTP) at 2.5 Å resolution (Fig. 2B). Using the same approach with C3-RhoA(GDP), we obtained the structure of NADH-C3-RhoA(GDP) at 2.5 Å resolution (Fig. 2C). The overall structure of C3cer is similar to those of other C3 exoenzymes (i.e. a mixed α/β-fold with a β-sandwich core). Structural alignment of C3cer with C3bot1 shows that the main differences are restricted to two 310-helices in C3cer (Fig. 1A). There was no large conformational change between apo-C3-RhoA(GTP) and NADH-C3-RhoA(GTP), except in the phosphate nicotinamide (PN) loop, which is one of the RhoA binding regions. Only in the PN loop, which includes Tyr-151, does a large conformational change from the apo to the NADH structure occur. On the other hand, a comparison between NADH-C3-RhoA(GTP) and NADH-C3-RhoA(GDP) showed that there was no large conformational change except for GTP/GDP binding in RhoA. It is important to note that the density of both GTP and GDP is very clear in both forms (Fig. 2, B and C). Interestingly, within the structures of the NADH-C3-RhoA(GTP) and NADH-C3-RhoA(GDP) complexes, the switch I and II regions adopted the GDP and GTP conformations, respectively (Fig. 2D). This corresponds to the fact that C3-like ARTs ADP-ribosylate both the GTP- and GDP-bound forms of RhoA (Fig. 2E). However, the nucleotide-dependent time course of the ADP-ribosylation also revealed that the GDP form is the optimal substrate and that the binding of GTP analogs reduces the reaction rate by a factor of 5 but does not change the total amount of ADP-ribosylation (35). The structures of the complexes of C3-RhoA explain why RhoA(GDP) is a better substrate. Although the C3 binding to RhoA(GTP) or RhoA(GDP) induces conformational change in the switch I and II regions, the conformational change in the switch I region (r.m.s.d. 2.46 Å) is much larger than that in the switch II region (r.m.s.d. 1.08 Å). Within the NADH-C3-RhoA ternary complex, RhoA(GTP) needs to change the switch I region, whereas RhoA(GDP) needs to change the switch II region. Consequently, it is easier for RhoA(GDP) to form a ternary complex than for RhoA(GTP). This suggests that the conformational change in RhoA is the rate-limiting step during ADP-ribosylation by C3-like ARTs. Judged from the fact of ADP-ribosylation occurring in the C3-RhoA(GTP) crystal, it is important to note that the structure presented here is the active complex (Fig. 2F).
FIGURE 1.
Sequence alignments of C3 toxins and Rho GTPases. A, comparison of C3 toxins and IaΔN-(211–413). B, comparison of Rho GTPases. Secondary structures are shown over the alignments. H, 310-helix. The five interaction regions (II–VI) are outlined in black boxes. The structural features (loops and switches) are shown below the alignments. Alignments were carried out using T-Coffee.
Interaction between C3 and RhoA
The surface area involved in the interface of the C3cer-RhoA heterodimer was extensive (1119 Å2). The structural components of C3cer that mediated RhoA binding consisted of four loops around NAD+ and one additional loop: loop II, composed of residues 45–52 (the active site loop) (the loop numbers are derived from the Ia-actin complex structure (45)); loop III, composed of residues 100–110 (adenine loop); loop IV, composed of residues 148–156 (PN loop); loop V, composed of residues 175–183 (ARTT loop); and loop VI, composed of residues 206–209 (an additional loop in C3cer) (Fig. 2G). In the case of the actin-specific binary toxin Ia, similar loops (II–V) in the C-terminal enzymatic domain as well as an extra loop I in the N-terminal domain, necessary for binding with Ib, are used for actin binding. However, C3cer lacks loop I; instead loop VI assists the binding of C3cer to RhoA. The structure of the complex showed that, with the exception of Arg-5, all RhoA residues involved in recognizing C3cer are within the switch I region (residues 28–38), the switch II region (residues 61–78), or the region linking switch I and II regions (residues 39–60) (interswitch). On the C-terminal side of switch I and the N-terminal side of the interswitch region, Tyr-34, Pro-36, Thr-37, Val-38, Phe-39, Glu-40, Asn-41, Tyr-42, and Val-43 contact C3cer (Figs. 2G and 3A). On the C-terminal side of the interswitch region and the N-terminal side of switch II, Glu-54, Ala-56, Leu-57, Trp-58, Ala-61, Gln-63, Asp-65, Tyr-66, Arg-68, Leu-69, and Leu-72 contact C3cer (Figs. 2G and 3B).
FIGURE 3.
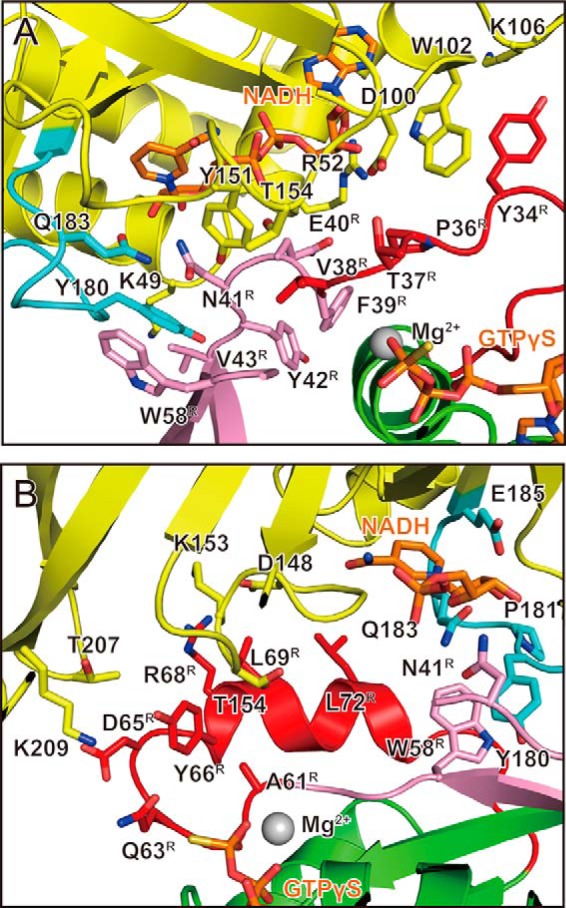
Interactions around switch I and switch II in C3cer-RhoA(GTP) complex. This is a detailed view around switch I (A) and switch II (B). The residues involved in the interaction are labeled. C3cer, RhoA, the switch regions, and the interswitch region are colored yellow, green, red, and pink, respectively. The ARTT loops are shown in cyan (as in Fig. 2B).
RhoA Recognition by ARTT Loop
Within the ARTT loop of C3-like ARTs, a conserved aromatic residue (C3bot1: Phe-169/C3cer: Tyr-180) in turn 1 has been thought to recognize substrate RhoA via a hydrophobic patch around the acceptor amino acid residue in RhoA (Asn-41), and Gln (C3bot1: Gln-172/C3cer: Gln-183) in turn 2 has been thought to be essential for interaction with Asn-41 (Fig. 4A) (10). Actually, within the structure of the complex, Tyr-180 of C3cer interacts with a hydrophobic patch on RhoA composed of Val-43, Ala-56, and Trp-58 (Fig. 4B), and the hydroxyl group of Tyr-180 forms a hydrogen bond with the main-chain carbonyl group of Leu-57. Importantly, Asn-41 forms a hydrogen bond with Gln-183 in the QXE motif within the ARTT loop of C3cer (Fig. 4B). This interaction was conserved in all forms: apo-C3-RhoA, NADH-C3-RhoA(GTP), and NADH-C3-RhoA (GDP).
FIGURE 4.
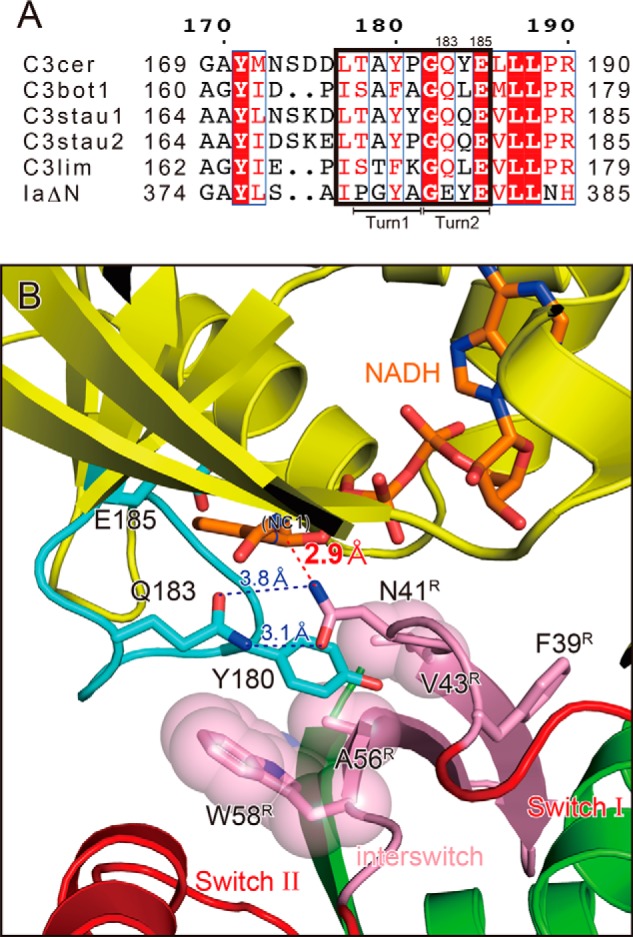
RhoA recognition by the ARTT loop of C3 exoenzyme. A, sequence alignment of the ARTT loop (outlined in black box) in C3 exoenzymes and IaΔN-(211–413). The turn 1 and turn 2 regions are indicated below the alignment. The sequence alignments of all amino acids in C3 exoenzymes and Rho GTPases are provided in Fig. 1. B, detailed view of the active site within C3cer-RhoA(GTP) with NADH. Colors are the same as in Fig. 3. The distances between Asn-41 of RhoA and Gln-183 of C3cer and between Asn-41 of RhoA and the electrophile (NC1 of NAD(H)) are shown as dotted lines.
Substrate Specificity of C3-like ARTs
C3-like ARTs modify RhoA but not Cdc42 or Rac1, despite their high amino acid identities (Fig. 1B). What accounts for this selectivity? As described above, the interaction between the modified Asn-41 of RhoA and Gln-183 of C3cer is crucial for binding, but Asn-41 does not explain the specificity of the C3-RhoA interaction because RhoA, Rac1, and Cdc42 all contain that asparagine. Among the residues of RhoA engaged in the binding to C3cer, seven residues in the switch II region are strictly conserved among RhoA, Rac1, and Cdc42, which means that other regions (Arg-5 and Glu-40–Trp-58) must provide substrate specificity to C3-like ARTs (Fig. 1B). It is largely the side chains that interact with the toxin, although Phe-39, Tyr-42, and Leu-57 interact with C3cer via their main chains (Fig. 3). The side chains of Phe-39, Tyr-42, and Leu-57 are oriented in the opposite direction from the C3cer interface and seem important to the stabilization of the switch I and II loops. Consequently, based on the complex structure, the substrate specificity of C3-like ARTs were suggested to be provided by Arg-5, Glu-40, Val43, Glu54, and Trp58 whose side chains interact with C3cer and these residues differ among RhoA, Rac1 and Cdc42.
Structure-based Mutational Analysis
Based on the complex structures, we assessed whether one point mutation within C3-RhoA interface would impair the activity of C3 with RhoA (Fig. 5A). E40A, E40D, and V43A lost activity slightly compared with wild type RhoA. On the other hand, E54A and W58A lost considerable activity. In the Ala-56 position, it seems that only a small hydrophobic side chain could fit in that space of the complex. As we expected, RhoA(A56W) lost activity totally. In Arg-5 and Lys-6, the results obtained are the opposite of what we expected. R5K did not change the ADP-ribosylation, but K6A decreased the ADP-ribosylation. Next we assessed whether the combined mutations within the interface would gain the activity of C3 with Cdc42, which is a poor substrate of C3cer (28) (Fig. 5B). (The residue numbers of Cdc42 are based on the numbers of RhoA in Fig. 1B.) A double mutant (D40E/F58W) showed a slight increase in activity compared with the wild type Cdc42. The other double mutant (T54E/F58W) showed more of an increase in activity. Furthermore, triple (D40E/T54E/F58W) and quadruple (D40E/A43V/T54E/F58W) mutants showed increasingly strong activity. To determine the effect of 5RK6, we also assessed two-residue insertions, Cdc42(5RA6) and Cdc42(5RK6). However, all non-insertion mutants (double, triple, and quadruple) showed more activity compared with insertion mutants (Cdc42(5RA6) and Cdc42(5RK6)) (Fig. 5B). Finally, we confirmed that G56W disturbed the activity of the active mutant as shown in Cdc42(5RA6/D40E/A43V/T54E/G56W/F58W).
Discussion
C3-RhoA complex structures imply information not only about binding but also about ADP-ribosylation reaction. The observed structure of the ARTT loop implies that Tyr-180 and Gln-183 of C3cer function as anchors to dock with RhoA. However, the function of Tyr-180 and Gln-183 is not limited to binding RhoA; the two residues are also reportedly very important for the ART reaction. The Y180T and Q183E mutants show normal NADase activity but diminished ARTase activity (28). An earlier structural analysis of RhoA(GTP) reveals that the distance between the indole ring in the side chain of Trp-58 and the carbonyl oxygen of Asn-41 is 3.3–3.5 Å (3), which suggests that the Trp-58 indole ring acts as a strong electron donor, enhancing the nucleophilic properties of the side-chain nitrogen of Asn-41. However, the present structure shows that the Tyr-180 side chain prevents interaction between the indole ring of Trp-58 and the side chain of Asn-41. Alternatively, the flat phenol ring of Tyr-180 interacts in parallel with the flat carbamoyl group of the Asn-41 side chain, suggesting that the phenol ring of Tyr-180 enhances the nucleophilic properties of the side-chain nitrogen of Asn-41 (Fig. 4B).
By mutational analyses of RhoA and Rac1, Wilde et al. (35) report that seven residues are important for the substrate specificity of C3-like ARTs (Arg-5, Lys-6, Glu-40, Val-43, Glu-47, Glu-54, and Trp-58). In our present mutational and structural analysis, we confirmed that four residues particularly contribute to the activity (Trp-58 > Glu-54 > Glu-40 > Val-43), and these four residue combined mutants of Cdc42 could be a good substrate of C3. Furthermore, we also confirmed that a small hydrophobic side chain (Ala in RhoA or Gly in Cdc42) is necessary at residue 56. Within the complex structure, Arg-5 interacts with Asp-175 of C3cer with a distance of 2.7 Å, and Lys-6 does not interact with C3cer. Nevertheless, RhoA(K6A) lost activity. Although we do not understand the exact reason for this, Arg-6 may stabilize the β1 strand of RhoA. The side chain of Glu-40 was invisible within the complex structure due to its flexibility, but actually it contributed to the activity. We initially expected this glutamate to play a role similar to that of Asp-179 in actin, which fixes the N-ribose position in the second oxocarbenium cation intermediate, but this is unlikely based on the structure, and its function remains unknown. Glu-54 binds to Lys-49 of C3cer. Although the residue at position 54 differs among RhoA, Rac1, and Cdc42, the counter-residue, Lys-49, is conserved in all C3-like ARTs (arginine in C3lim), which suggests that Glu-54 is important for substrate specificity. In the complex structure, Glu-47 obviously does not interact with C3cer, although it was suggested to be an important residue for activity in a previous study (35). The residues Val-43 and Trp-58 form a hydrophobic patch for anchoring Tyr-180 of C3cer (Fig. 4B). Based on the structural analysis, we succeeded in altering non-active Cdc42 to an active mutant, Cdc42 (D40E/A43V/T54E/F58W). The contribution of a single mutation was not great, but these combinations affected the activity significantly.
We resolved the structure of NADH-C3-RhoA in the pre-ADP-ribosylation state. Previously, we determined the structures of the Ia-actin complex in the pre- and post-ADP-ribosylation states (45, 46) and proposed a strain alleviation model for the ADP-ribosylation of actin caused by Ia (46, 47). Upon scission of the nicotinamide via an Sn1 reaction, the oxocarbenium cation deforms to an alleviation conformation. This gives the electrophile (NC1 of N-ribose) of the oxocarbenium cation access to the nucleophile (Arg-177, the acceptor amino acid of actin). Here, we suggest that the ADP-ribosylation of RhoA proceeds via the same strain alleviation model on the basis of the following similarities. (i) The structure of Ia (C-terminal domain) is similar to C3cer with an r.m.s.d. of 2.2 Å. (ii) The conformations of NADH complexed with Ia and C3cer are similar. In all ARTs, NAD+ exhibits a highly folded structure (strained conformation) (48). (iii) Both Ia and C3cer recognize their substrate via four common loops, and similar substrate recognition mechanisms involving the ARTT loop have been proposed (10). It is noteworthy, however, that there are the following important differences between C3-RhoA and Ia-actin complexes (Fig. 6). (i) Surprisingly, the structural relationship between NC1 of NAD(H) and the target residues (Arg-177 in actin and Asn-41 in RhoA) is totally different. The structure of NADH-C3-RhoA reflects the close relationship among Gln-183 in the QXE motif (C3cer), the modified Asn-41 residue (RhoA), and NC1 of NADH. The distance between NC1 of the N-ribose and ND2 of Asn-41 is only 2.9 Å. Within the NAD+-Ia-actin complex, the distance between the nucleophile (Arg-177) and the electrophile (NC1 of N-ribose) is 8.4 Å (46). This difference between C3-RhoA and Ia-actin reflects the difference in the flexibility of the region in which the target residue is located. Within RhoA, Asn-41 is located at the edge of the flexible switch I region, whereas Arg-177 within actin is located in a β-strand. Although the switch I region adopts a conformation that brings Asn-41 into close proximity to NC1 of NADH within the complex with C3cer, the β-strand containing Arg-177 does not. To reduce the distance between the nucleophile and the electrophile within the Ia-actin structure, we propose a model of strain alleviation via two oxocarbenium cation intermediates (45, 46). Within the C3-RhoA structure, by contrast, the distance is not a problem. As soon as NAD+ binds, ADP-ribosylation can occur without a large conformational change in C3cer, RhoA, or NAD+. In that context, C3cer (C3-like ARTs) appears to be the prototype ART. (ii) In Ia and C3-like ARTs, a common mechanism has been proposed by which the target protein is recognized by the ARTT loop: a turn 2 residue (Glu-378 in the Ia EXE motif and Gln-183 in the C3cer QXE motif) interacts with the target residue (Arg-177 in actin and Asn-41 in RhoA), and an aromatic residue (Tyr-375 in Ia and Tyr-180 in C3cer) in turn 1 interacts with the hydrophobic region of actin or RhoA (10). But recognition by the ARTT loop has not been observed within the Ia-actin complex (45, 46). For Ia, the ARTT loop recognition may occur transiently during ADP-ribosylation and the transition from the pre- to post-ADP-ribosylation state. It is likely that these similarities and differences between C3cer and Ia are also observed in other Rho- and actin-specific ARTs.
FIGURE 6.
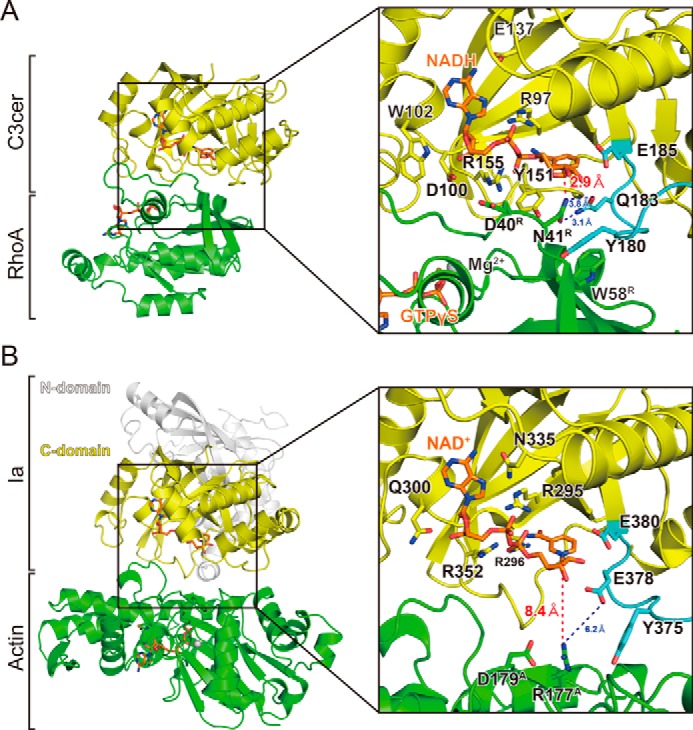
Comparison of RhoA-specific and actin-specific ADP-ribosylating toxins. A, detailed view around the active site in C3cer. Components are colored as described in the legend for Fig. 2B. The distances between Asn-41 of RhoA and Gln-183 of C3cer and between Asn-41 of RhoA and the electrophile (NC1 of NAD(H)) are shown as dotted lines. B, detailed view around the active site in Ia (PDB code: 4H03). The Ia N terminus, Ia C terminus, actin, and NAD+ are shown in white, yellow, green, and orange, respectively. The residues needed for NAD(H) binding are also labeled. The distances between Arg-177 of actin and the electrophile (NC1 of NAD+) are shown as dotted lines.
Rho and related small GTPases are the most common targets for bacterial toxins and are of major importance for the entry of bacteria into mammalian host cells (49). It is well known that various bacterial toxins modify small GTPases through glucosylation and deamidation in addition to ADP-ribosylation. For example, the bacterial toxin Fic (filamentation induced by cyclic AMP) catalyzes the addition of AMP to Rho-GTPases (adenylylation), thereby preventing their interaction with cellular effectors. In complex with the Fic domain of IbpA from Histophilus somni (IbpAFic2) (50), Cdc42 mimics the GDI-bound state of Rho GTPase. Within that structure, most of the interface is within the switch I and II regions, whereas in the structure of C3cer complexed with RhoA, most of the interface is within the switch I and II and interswitch regions. This means that both IbpA and C3cer interact with the same side of their target GTPases, despite their totally different structures and functions, suggesting that interaction with target GTPases via the switch I and II regions is a common bacterial strategy for disrupting GTPase function.
RalA, a Ras-related GTPase, reportedly inhibits C3bot1, C3lim, and C3cer but not C3stau2 (51). Holbourn et al. (52) and Pautsch et al. (53) have reported different structures for the C3-RalA complex; however, the former α4-ARTT model is likely to be a crystallographic artifact, based on crystallographic and mutational results (53). The model proposed by Pautsch et al. (53) is that the loop between helices α3 and α4 (C3bot93–C3bot101) inserts into the groove of the switch I and II regions of RalA. When we compare the model of Pautsch et al. with our C3-RhoA structure, the binding sites of C3 with RalA and RhoA do not overlap each other. It suggests that RalA would not sterically interfere with RhoA. How does RalA binding inhibit ADP-ribosylation of RhoA? One possibility is that a structural change occurs in C3 upon RalA binding. RalA increases the NAD+ glycohydrolase activity of C3bot by about 5-fold (51). This means that a structural change in C3 must occur upon RalA binding, but that concept remains to be addressed in the future.
We have shown the structural basis for the binding of a bacterial C3 exotoxin to substrate RhoA. The structures of the complexes directly demonstrate that the ARTT loop recognizes RhoA. The interaction of residues of Rho GTPase with C3cer was confirmed by mutagenesis. Furthermore, we successfully changed Cdc42 to an active substrate by introducing four combined mutations (D40E/A43V/T54E/F58W). In summary, these findings are also useful for understanding the interaction of other ARTs with their substrates. We believe that these findings may pave the way for the identification of new inhibitors and may also contribute to the development of novel ARTs specific for other Rho GTPases (Rac1 and Cdc42), which could serve as novel tools for biochemistry.
Author Contributions
A. T. and H.T. designed the research; A. T., T. T., and Y. T. performed the research; A. T., T. T., and H. T. analyzed the data; and A. T., T. T., T. Y., and H. T. wrote the article.
Acknowledgments
This paper is dedicated to Prof. Alan Hall. We thank the staff at the KEK Photon Factory for data collection. We also thank Masataka Oda (Niigata University) and Masahiro Nagahama (Tokushima Bunri University) for discussion.
This work supported in part by a Strategic Research Foundation Grant-aided project for private universities from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and by Grants-in-aid for Scientific Research 15K08289 and 25121733 from MEXT of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4XSG, 4XSH, and 5BWM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GEF
- guanine nucleotide exchange factor
- ART
- ADP-ribosyltransferase
- GDI
- guanine nucleotide dissociation inhibitor
- ARTT
- ADP-ribosylating toxin turn-turn
- PN
- phosphate nicotinamide
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Heasman S. J., Ridley A. J. (2008) Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 2. Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 3. Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273, 9656–9666 [DOI] [PubMed] [Google Scholar]
- 4. Wei Y., Zhang Y., Derewenda U., Liu X., Minor W., Nakamoto R. K., Somlyo A. V., Somlyo A. P., Derewenda Z. S. (1997) Crystal structure of RhoA-GDP and its functional implications. Nat. Struct. Biol. 4, 699–703 [DOI] [PubMed] [Google Scholar]
- 5. Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J. (1997) Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature 389, 758–762 [DOI] [PubMed] [Google Scholar]
- 6. Tsuge H., Nagahama M., Nishimura H., Hisatsune J., Sakaguchi Y., Itogawa Y., Katunuma N., Sakurai J. (2003) Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J. Mol. Biol. 325, 471–483 [DOI] [PubMed] [Google Scholar]
- 7. Schleberger C., Hochmann H., Barth H., Aktories K., Schulz G. E. (2006) Structure and action of the binary C2 toxin from Clostridium botulinum. J. Mol. Biol. 364, 705–715 [DOI] [PubMed] [Google Scholar]
- 8. Han S., Craig J. A., Putnam C. D., Carozzi N. B., Tainer J. A. (1999) Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 6, 932–936 [DOI] [PubMed] [Google Scholar]
- 9. Sundriyal A., Roberts A. K., Shone C. C., Acharya K. R. (2009) Structural basis for substrate recognition in the enzymatic component of ADP-ribosyltransferase toxin CDTa from Clostridium difficile. J. Biol. Chem. 284, 28713–28719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han S., Arvai A. S., Clancy S. B., Tainer J. A. (2001) Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J. Mol. Biol. 305, 95–107 [DOI] [PubMed] [Google Scholar]
- 11. Sekine A., Fujiwara M., Narumiya S. (1989) Asparagine residue in the Rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 264, 8602–8605 [PubMed] [Google Scholar]
- 12. Aktories K., Hall A. (1989) Botulinum ADP-ribosyltransferase C3: a new tool to study low molecular weight GTP-binding proteins. Trends Pharmacol. Sci. 10, 415–418 [DOI] [PubMed] [Google Scholar]
- 13. Ridley A. J., Hall A. (2004) Snails, Swiss, and serum: the solution for Rac 'n' Rho. Cell 116, Suppl. 2, S23–S25 [DOI] [PubMed] [Google Scholar]
- 14. Ridley A. J., Hall A. (1992) The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 15. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 16. Nobes C. D., Hall A. (1995) Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 17. Aktories K., Weller U., Chhatwal G. S. (1987) Clostridium botulinum type C produces a novel ADP-ribosyltransferase distinct from botulinum C2 toxin. FEBS Lett. 212, 109–113 [DOI] [PubMed] [Google Scholar]
- 18. Ohashi Y., Narumiya S. (1987) ADP-ribosylation of a Mr 21,000 membrane protein by type D botulinum toxin. J. Biol. Chem. 262, 1430–1433 [PubMed] [Google Scholar]
- 19. Rubin E. J., Gill D. M., Boquet P., Popoff M. R. (1988) Functional modification of a 21-kilodalton G protein when ADP-ribosylated by exoenzyme C3 of Clostridium botulinum. Mol. Cell. Biol. 8, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popoff M., Boquet P., Gill D. M., Eklund M. W. (1990) DNA sequence of exoenzyme C3, an ADP-ribosyltransferase encoded by Clostridium botulinum C and D phages. Nucleic Acids Res. 18, 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nemoto Y., Namba T., Kozaki S., Narumiya S. (1991) Clostridium botulinum C3 ADP-ribosyltransferase gene: cloning, sequencing, and expression of a functional protein in Escherichia coli. J. Biol. Chem. 266, 19312–19319 [PubMed] [Google Scholar]
- 22. Just I., Mohr C., Schallehn G., Menard L., Didsbury J. R., Vandekerckhove J., van Damme J., Aktories K. (1992) Purification and characterization of an ADP-ribosyltransferase produced by Clostridium limosum. J. Biol. Chem. 267, 10274–10280 [PubMed] [Google Scholar]
- 23. Just I., Selzer J., Jung M., van Damme J., Vandekerckhove J., Aktories K. (1995) Rho-ADP-ribosylating exoenzyme from Bacillus cereus: purification, characterization, and identification of the NAD-binding site. Biochemistry 34, 334–340 [DOI] [PubMed] [Google Scholar]
- 24. Inoue S., Sugai M., Murooka Y., Paik S. Y., Hong Y. M., Ohgai H., Suginaka H. (1991) Molecular cloning and sequencing of the epidermal cell differentiation inhibitor gene from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 174, 459–464 [DOI] [PubMed] [Google Scholar]
- 25. Wilde C., Just I., Aktories K. (2002) Structure-function analysis of the Rho-ADP-ribosylating exoenzyme C3stau2 from Staphylococcus aureus. Biochemistry 41, 1539–1544 [DOI] [PubMed] [Google Scholar]
- 26. Yamaguchi T., Hayashi T., Takami H., Ohnishi M., Murata T., Nakayama K., Asakawa K., Ohara M., Komatsuzawa H., Sugai M. (2001) Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 69, 7760–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilde C., Chhatwal G. S., Aktories K. (2002) C3stau, a new member of the family of C3-like ADP-ribosyltransferases. Trends Microbiol 10, 5–7 [DOI] [PubMed] [Google Scholar]
- 28. Wilde C., Vogelsgesang M., Aktories K. (2003) Rho-specific Bacillus cereus ADP-ribosyltransferase C3cer cloning and characterization. Biochemistry 42, 9694–9702 [DOI] [PubMed] [Google Scholar]
- 29. Sehr P., Joseph G., Genth H., Just I., Pick E., Aktories K. (1998) Glucosylation and ADP ribosylation of Rho proteins: effects on nucleotide binding, GTPase activity, and effector coupling. Biochemistry 37, 5296–5304 [DOI] [PubMed] [Google Scholar]
- 30. Genth H., Gerhard R., Maeda A., Amano M., Kaibuchi K., Aktories K., Just I. (2003) Entrapment of Rho ADP-ribosylated by Clostridium botulinum C3 exoenzyme in the Rho-guanine nucleotide dissociation inhibitor-1 complex. J. Biol. Chem. 278, 28523–28527 [DOI] [PubMed] [Google Scholar]
- 31. Aktories K., Wilde C., Vogelsgesang M. (2004) Rho-modifying C3-like ADP-ribosyltransferases. Rev. Physiol. Biochem. Pharmacol. 152, 1–22 [DOI] [PubMed] [Google Scholar]
- 32. Han S., Tainer J. A. (2002) The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol. 291, 523–529 [DOI] [PubMed] [Google Scholar]
- 33. Ménétrey J., Flatau G., Stura E. A., Charbonnier J. B., Gas F., Teulon J. M., Le Du M. H., Boquet P., Menez A. (2002) NAD binding induces conformational changes in Rho ADP-ribosylating Clostridium botulinum C3 exoenzyme. J. Biol. Chem. 277, 30950–30957 [DOI] [PubMed] [Google Scholar]
- 34. Evans H. R., Sutton J. M., Holloway D. E., Ayriss J., Shone C. C., Acharya K. R. (2003) The crystal structure of C3stau2 from Staphylococcus aureus and its complex with NAD. J. Biol. Chem. 278, 45924–45930 [DOI] [PubMed] [Google Scholar]
- 35. Wilde C., Genth H., Aktories K., Just I. (2000) Recognition of RhoA by Clostridium botulinum C3 exoenzyme. J. Biol. Chem. 275, 16478–16483 [DOI] [PubMed] [Google Scholar]
- 36. Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. (2004) The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure 12, 1955–1965 [DOI] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogelsgesang M., Stieglitz B., Herrmann C., Pautsch A., Aktories K. (2008) Crystal structure of the Clostridium limosum C3 exoenzyme. FEBS Lett. 582, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 40. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3, Schrodinger, LLC, New York [Google Scholar]
- 44. Joosten R. P., Joosten K., Cohen S. X., Vriend G., Perrakis A. (2011) Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank. Bioinformatics 27, 3392–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsuge H., Nagahama M., Oda M., Iwamoto S., Utsunomiya H., Marquez V. E., Katunuma N., Nishizawa M., Sakurai J. (2008) Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc. Natl. Acad. Sci. U.S.A. 105, 7399–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsurumura T., Tsumori Y., Qiu H., Oda M., Sakurai J., Nagahama M., Tsuge H. (2013) Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex. Proc. Natl. Acad. Sci. U.S.A. 110, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsuge H., Tsurumura T. (2015) Reaction Mechanism of Mono-ADP-Ribosyltransferase Based on Structures of the Complex of Enzyme and Substrate Protein. Curr. Top. Microbiol. Immunol. 384, 69–87 [DOI] [PubMed] [Google Scholar]
- 48. O'Neal C. J., Jobling M. G., Holmes R. K., Hol W. G. (2005) Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 309, 1093–1096 [DOI] [PubMed] [Google Scholar]
- 49. Aktories K. (2011) Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 9, 487–498 [DOI] [PubMed] [Google Scholar]
- 50. Xiao J., Worby C. A., Mattoo S., Sankaran B., Dixon J. E. (2010) Structural basis of Fic-mediated adenylylation. Nat. Struct. Mol. Biol. 17, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilde C., Barth H., Sehr P., Han L., Schmidt M., Just I., Aktories K. (2002) Interaction of the Rho-ADP-ribosylating C3 exoenzyme with RalA. J. Biol. Chem. 277, 14771–14776 [DOI] [PubMed] [Google Scholar]
- 52. Holbourn K. P., Sutton J. M., Evans H. R., Shone C. C., Acharya K. R. (2005) Molecular recognition of an ADP-ribosylating Clostridium botulinum C3 exoenzyme by RalA GTPase. Proc. Natl. Acad. Sci. U.S.A. 102, 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pautsch A., Vogelsgesang M., Tränkle J., Herrmann C., Aktories K. (2005) Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme. EMBO J. 24, 3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karplus P. A., Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]