Background: AKAPs integrate intracellular signals by sequestering PKA with other kinases.
Results: Phosphorylation of Thr-1132 on AKAP220 initiates GSK3β recruitment, and PKA activity drives the release of GSK3β from the complex.
Conclusion: Cross-talk between PKA and GSK3β is optimized in the context of AKAP220 multienzyme complexes.
Significance: Signal responsive assembly of enzyme complexes may represent a general mechanism to diversify transduction through AKAPs.
Keywords: A-kinase anchoring protein (AKAP), β-catenin (B-catenin), cyclic AMP (cAMP), glycogen synthase kinase 3 (GSK-3), protein complex, protein kinase, protein kinase A (PKA), protein phosphorylation
Abstract
The proximity of an enzyme to its substrate can influence rate and magnitude of catalysis. A-kinase anchoring protein 220 (AKAP220) is a multivalent anchoring protein that can sequester a variety of signal transduction enzymes. These include protein kinase A (PKA) and glycogen synthase kinase 3β (GSK3β). Using a combination of molecular and cellular approaches we show that GSK3β phosphorylation of Thr-1132 on AKAP220 initiates recruitment of this kinase into the enzyme scaffold. We also find that AKAP220 anchors GSK3β and its substrate β-catenin in membrane ruffles. Interestingly, GSK3β can be released from the multienzyme complex in response to PKA phosphorylation on serine 9, which suppresses GSK3β activity. The signaling scaffold may enhance this regulatory mechanism, as AKAP220 has the capacity to anchor two PKA holoenzymes. Site 1 on AKAP220 (residues 610–623) preferentially interacts with RII, whereas site 2 (residues 1633–1646) exhibits a dual specificity for RI and RII. In vitro affinity measurements revealed that site 2 on AKAP220 binds RII with ∼10-fold higher affinity than site 1. Occupancy of both R subunit binding sites on AKAP220 could provide a mechanism to amplify local cAMP responses and enable cross-talk between PKA and GSK3β.
Introduction
Protein kinase A anchoring proteins (AKAPs)2 are a structurally diverse family of proteins united by the ability to bind protein kinase A (PKA) (1). Their principle function is to process a variety of intracellular signals by sequestering PKA with other enzymes such as kinases, phosphodiesterases, and protein phosphatases (2, 3). These macromolecular complexes limit the scope, duration, and flow of second messenger responsive signal transduction events (4). The physiological significance of this mechanism has been validated in several cellular and in vivo contexts (5–10).
The tetrameric PKA holoenzyme is composed of two regulatory (R) subunits and two catalytic (C) subunits (11–13). High resolution structural studies have revealed that the type I (RI) or type II (RII) subunits homodimerize and create a binding groove within a four-helix bundle (14, 15). An amphipathic helix within the AKAP binds to this groove with high affinity (16, 17). Electron microscopy of intact type II PKA-AKAP18γ complexes has shown that intrinsically disordered regions within each PKA regulatory subunit impart flexibility within the holoenzyme complex (18). As a result these macromolecular assemblies can adopt a range of conformations that may help to more efficiently position the PKA catalytic subunits in proximity to target substrates (18). By anchoring PKA at defined subcellular locations, AKAPs manage the phosphorylation-dependent modulation of downstream targets (19).
AKAP220 is a ubiquitously expressed 220-kDa protein that is encoded by the Akap11 gene (20). Its known enzymatic binding partners include PKA, glycogen synthase kinase 3β (GSK3β), and protein phosphatase 1 (PP1) (20–24). The potential for co-distribution of GSK3 with basophilic kinases such as PKA is interesting as phosphorylation of serine 21 on GSK3α or serine 9 on GSK3β attenuates its catalytic activity (25). GSK3 is a proline-directed kinase that often shows preference for a priming phosphate in the n+4 position. Substrate priming guides the phospho acceptor group on the substrate toward the catalytic site of the kinase to enhance catalytic activity (26, 27). However, substrate priming is not always a prerequisite, as this kinase can efficiently phosphorylate non-primed substrates such as β-catenin when both components are present in the same macromolecular complex (28, 29).
Our recent work has shown that AKAP220 regulates cytoskeletal dynamics through an interaction with IQ motif containing GTPase activating proteins (IQGAPs) (30). Signaling through anchored PKA and GSK3β regulates both actin polymerization and microtubule organization at the cell cortex (31). Gene silencing of AKAP220 decreases membrane ruffling, alters microtubule dynamics, and slows cell migration (31). Although AKAP220-associated PKA and GSK3β have been implicated in control of these latter cellular processes, it is imperative that we obtain a more mechanistic understanding of molecular interplay between these anchored enzymes. In this report we discover that active GSK3β phosphorylates AKAP220 to create and maintain its binding site and that AKAP220 localizes GSK3β with its substrate β-catenin in membrane ruffles. We also identify and characterize two separate PKA binding sites on AKAP220 and show that associated PKA regulates recruitment of GSK3β in a multistep and phosphorylation-dependent manner. This work enhances our understanding of kinase anchoring on AKAP220 and highlights the importance of molecular cross-talk within this macromolecular signaling complex.
Experimental Procedures
Cell Culture, Transfection, and Lysis
HEK293 cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin, and mouse inner medullary-collecting duct (mIMCD3) cells were maintained in DMEM:F-12 1:1 media supplemented with 10% FBS and penicillin/streptomycin. Cells were transfected with plasmids using either Mirus TransIT-LT1 or Lipofectamine 2000 transfection reagents and incubated for 48–72 h before fixation or lysis.
Co-immunoprecipitations
For pulldown assays investigating the interaction with R subunit, cells were lysed in ice-cold buffer (20 mm HEPES, 150 mm NaCl, 5 mm EDTA, 1% Triton, Pierce protease inhibitor mixture). For pulldown assays investigating the interaction with GSK3β and/or β-catenin, cells were washed in cold PBS and lysed in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Igepal, 5% glycerol, 25 mm β-glycerophosphate, 5 mm sodium orthovanadate, 5 mm NaF, 10 nm okadaic acid, and Pierce protease inhibitor mixture). Cleared lysates were combined with primary antibody or FLAG-agarose beads and rocked at 4 °C for 1–4 h. Beads were washed 3× at 4 °C with lysis buffer and resuspended in sample buffer for SDS-PAGE and Western blot analysis.
Peptide Array
A membrane-bound array of overlapping 20-mer peptides spanning AKAP220 residues 1009–1305 was synthesized with an Intavis MultiPep array as described previously (32). Membranes were overlaid with purified GSK3β, RI, or RII at 4 °C overnight. GSK3β phosphorylation of candidate AKAP220 peptides was detected by in vitro kinase assay. Increasing concentrations of purified GSK3β (7.5–75 ng/μl) and a buffer containing [γ-32P]ATP were incubated with the membrane-bound array of peptides, and γ-32P incorporation was detected by autoradiography.
CRISPR-Cas9 Genome Editing
The CRISPR/Cas9 expression vector pSpCas9(BB)-2A-GFP was obtained from the Zhang laboratory via Addgene (Addgene plasmid #48138) (33). A 20-nucleotide sequence followed by a protospacer adjacent motif from Streptococcus pyogenes (nucleotide sequence NGG) targeting the second exon of mouse Akap11 was selected using the Broad Institute online synthetic guide RNA designer described by Doench et al. (34). The guide sequence was selected for the least number of potential off-target sites. The guide (sequence-protospacer adjacent motif) was as follows (with a g in front necessary for the U6 promoter): 5′-GTCTTTAGGTGACTACTCCTGAGG-3′. The guide sequence without protospacer adjacent motif was cloned into px458 as previously described (33).The plasmid was transfected into mIMDC3 cells by Lipofectamine 2000. Two days after transfection, single cells were sorted into 96-well plates by FACS. Two weeks after sorting confluent clones were analyzed by genomic PCR using the primers 5′-AAGATTTTGCAGCTAAGGTGGC-3′ and 5′-ACTACTTTCATACACGTTCGCCT-3′. PCR products were sequenced to identify changes in the genomic sequence. Loss of AKAP220 protein expression was further verified by Western blot.
Immunostaining
Cells were seeded and transfected on 12-mm poly-d-lysine- and laminin-coated coverslips (Fisher catalog #08-774-385). To stimulate membrane ruffle formation, mIMCD3 cells were grown overnight on coverslips and treated with PDGF (50 ng/ml) for 30 min at 37 °C before fixation.
Cells were fixed with 4% paraformaldehyde in PBS at room temperature for 10 min. Coverslips were blocked in PBS with 10% donkey serum for 2 h at room temperature before primary antibody staining. Samples were washed 3× and incubated with donkey anti-mouse Alexafluor-488 and donkey anti-rabbit Alexafluor-555 secondary antibodies for 1 h at room temperature. Three final washes were performed before mounting in ProLong Gold anti-fade reagent with DAPI (Invitrogen catalog #P36935). Images were acquired by confocal microscopy with a 63× oil immersion objective.
Line Plot Analysis
Membrane ruffles were identified by morphology and positive f-actin staining at the plasma membrane (phalloidin). Line plots were drawn from outside the cell, perpendicular to the ruffle edge, through a region with actin signal. The pixel intensity across the line was quantified in ImageJ for >18 cells per experiment, n = 3 experiments. Maximum projection images obtained with a 63× oil immersion objective.
Antibodies
Immunofluorescence and proximity ligation staining was done with mouse monoclonal anti-FLAG (Sigma catalog #F3165, FITC-conjugated catalog #F4049), rabbit polyclonal anti-RII (Santa Cruz sc-909), and polyclonal anti-RI (Santa Cruz sc-28893) antibodies. Co-immunoprecipitations were achieved with FLAG-agarose beads (Sigma catalog #:2220), cAMP-agarose beads (BioLog catalog #A020-06), mouse monoclonal anti-V5 (Life Technologies catalog #R96025), and rabbit anti-GFP (Life Technologies catalog #11122). Immunoblots were probed with mouse monoclonal anti-RII (BD Biosciences catalog #610626), mouse monoclonal anti-GSK3β (BD Biosciences catalog #610201), mouse monoclonal anti-β-catenin (BD Biosciences catalog #610154), or rabbit monoclonal anti-phospho-GSK3β(Ser-9) (Millipore catalog #04-1075) and rabbit polyclonal anti-AKAP220 produced by our laboratory. Membrane ruffling experiments were performed using Molecular Probes Actin-488 (Invitrogen), mouse monoclonal anti-β-catenin (BD Biosciences, and rabbit polyclonal anti-GSK3β (Cell Signaling catalog #12456P). Peptide array and purified protein overlays were probed with purified biotin-RII or biotin-RI and detected with Neutravidin-HRP (Life Technologies catalog #31030).
Fluorescence Polarization
FITC-labeled peptides were synthesized to >95% purity by BioMatik and resuspended to 100 μm stocks in assay buffer (20 mm HEPES, pH 7.5, 200 mm NaCl, and 0.1% Triton X-100). Assay were performed in black, round-bottomed 96-well plates (Corning catalog #3792). FITC-peptides were diluted to 5 nm final concentration in assay buffer and combined with varying concentrations of purified His-RII at a total volume of 100 μl per well. Fluorescence polarization measurements were made using a POLARstar Omega plate reader (BMG Labtech) and converted to anisotropy values. Binding data were fit using nonlinear regression to a one-site binding model in GraphPad Prism 6.0 (Fluorescence polarization = Bmax × [RIIα]/(Kd + RIIα) + NS × RIIα + background). In this equation Bmax is the maximum specific binding, Kd is the equilibrium constant, NS is the slope of nonspecific binding, and background is the amount of fluorescence polarization of each FITC-peptide with no RII added.
Duolink Proximity Ligation Assays
Cells were seeded onto acid-washed glass coverslips and transfected. They were fixed in 4% paraformaldehyde and stained according to the Duolink proximity ligation assay protocol. Before mounting, the cells were counterstained with a FITC-anti-FLAG antibody (Sigma catalog #F4049) to identify transfected cells. Images were obtained by confocal microscopy with a 63× oil immersion objective. For quantification of puncta, ImageJ was used to outline FITC-positive cells. The number of puncta in this region of interest was counted with the “find maxima” algorithm (35). To subtract background, this exact region of interest was moved to a region containing FITC-negative cells, and the number of puncta were counted.
Results
AKAP220 can sequester a variety of signal transduction enzymes. GSK3β was previously discovered as an AKAP220 binding partner by yeast two-hybrid screen, and biochemical studies mapped kinase binding to a central portion of the anchoring protein between residues 1017 and 1315 (24). Cell-based validation of this interaction was demonstrated by co-immunoprecipitation of endogenous GSK3β with AKAP220 from HEK293 cell lysates (Fig. 1A). The presence of GSK3β and the catalytic C-subunit of PKA was confirmed by immunoblot (Fig. 1A). None of these proteins was detected in IgG control immune complexes. These data confirm that AKAP220 has the capacity to anchor PKA and GSK3β.
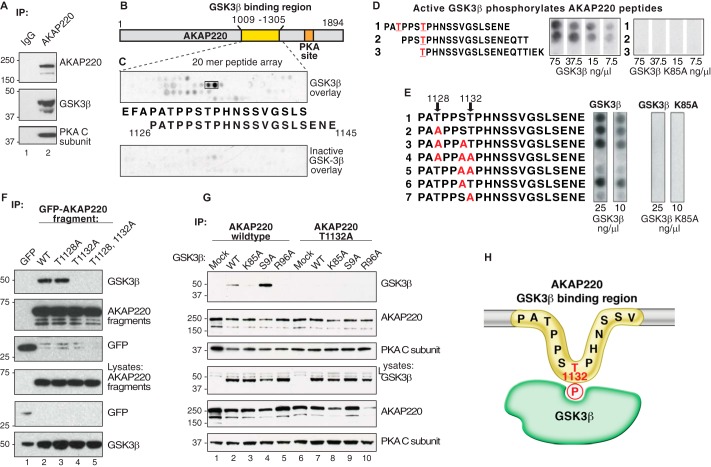
FIGURE 1.
The GSK3β phospho binding site on AKAP220 is Thr-1132. A, HEK293 cell lysates were immunoprecipitated (IP) with anti-AKAP220 antibody, and isolated complexes were analyzed by Western blot. B and C, peptide mapping of the GSK3β binding region, residues 1009–1305 of the mouse AKAP220 protein. Overlapping peptides (20-mers displaced by 3 amino acids sequentially) were synthesized onto a cellulose membrane and overlaid with purified GSK3β. D, Thr-1128 and Thr-1132 were predicted GSK3β phosphosites by Scansite analysis (red). Peptides containing these sites were synthesized by peptide array, and membranes were incubated with [γ32P]ATP and increasing concentrations of purified GSK3β or GSK3β K85A (kinase-dead). GSK3β phosphorylation was detected by autoradiography. E, AKAP220 1126–1145 peptides with candidate serine and threonine residues substituted with alanine were tested for GSK3β phosphorylation to locate the phosphosite. F, lysates from cells expressing either the GFP-AKAP220 1009–1305 fragment or one of the T1128A, T1132A, or the T1128A/T1132A mutant fragments were immunoprecipitated with anti-GFP antibody. Co-precipitation of endogenous GSK3β was assessed by immunoblot with an anti-GFP antibody. G, full-length V5-AKAP220 wild type or V5-AKAP220-T1132A were co-expressed in HEK293 cells with HA-GSK3β wild type, HA-GSK3β-K85A, HA-GSK3β-S9A, or HA-GSK3β-R96A. Immunocomplexes were isolated with a V5-antibody and probed for HA-GSK3β by Western blot. H, schematic representation of the AKAP220 Thr-1132 phosphosite binding to active GSK3β.
Mapping the GSK3β Binding Site
Detailed mapping of the GSK3β binding segment on AKAP220 was conducted by peptide array analyses (Fig. 1, B and C). A library of overlapping 20-mer peptides (displaced by 3 amino acids) spanning residues 1009–1305 of AKAP220 was spotted onto a peptide array membrane. Peptides were screened for direct interaction with purified GSK3β in a far Western overlay assay, and bound GSK3β was detected with an anti-GSK3β antibody. Two GSK3-binding peptides were detected, suggesting the interaction occurs within a minimum region of 17 amino acids encompassed by residues 1126–1142 (Fig. 1C). Control arrays covering the same region on AKAP220 gave negative results when probed with catalytically inactive GSK3β-K85A (25) (Fig. 1C, bottom panel).
A distinguishing feature of this sequence is a prevalence of proline, serine, and threonine residues. Scansite analysis of AKAP220 residues 1126–1142 predicted two consensus phosphorylation sites for GSK3 (Thr-1128 and Thr-1132) (36). To test whether purified GSK3β could phosphorylate one or both of these sites, overlapping AKAP220 peptides were generated by spot array and incubated with [γ-32P]ATP and increasing amounts of purified kinase (from 7.5–75 ng/μl; Fig. 1D). Radiolabeled phosphate incorporation into the immobilized peptides was assessed by autoradiography. Peptides 1 and 2 were phosphorylated in a dose-dependent manner, but peptide 3 lacking the upstream proline-rich segment was not (Fig. 1D, left array). Control experiments performed with a kinase-dead mutant (GSK3β-K85A) showed no phosphorylation of any of the peptides (Fig. 1D, right array).
To determine which site was phosphorylated, an additional array of AKAP220 peptides was generated in which alanine was substituted for candidate serine and threonine residues between 1128 and 1132 of AKAP220 (Fig. 1E). Phosphorylation was reduced in all peptides in which Thr-1132 was replaced with alanine (Fig. 1E, left arrays, peptides 4, 5, and 7). In contrast, single substitutions at Thr-1128 or Ser-1131 did not affect phosphorylation by GSK3β (Fig. 1E, left arrays, peptides 2, 3, and 6). Control experiments with the GSK3β-K85A mutant confirmed that this catalytically inactive form of the enzyme was unable to phosphorylate the AKAP220 peptides. Taken together these results suggest that Thr-1132 is a GSK3β phosphosite on AKAP220.
Cellular validation of this GSK3β binding event in HEK293 cells was confirmed by transfection of a GFP-tagged fragment of the anchoring protein (residues 1009–1305). Lysates from cells expressing wild type, T1128A, T1132A, or T1128A/T1132A AKAP220 fragments were prepared, and signaling complexes were isolated by immunoprecipitation with anti-GFP antibody. Immune complexes were probed for co-precipitation of endogenous GSK3β by immunoblot (Fig. 1F, top panel). These experiments revealed that mutation of Thr-1132 to an alanine abolished GSK3β binding (Fig. 1F, top panel, lanes 4 and 5). However, mutation of Thr-1128 to alanine did not effect GSK3β interaction when compared with wild type (Fig. 1F, top panel, lanes 2 and 3). Control immunoblots verified the enrichment of AKAP220 fragments and determined the expression levels of the protein components. Thus, Thr-1132 is required for AKAP220-GSKβ interaction inside cells.
Kinase Activity Drives GSK3β Interaction with AKAP220
We next investigated the GSK3β interaction in the context of full-length AKAP220. HEK293 cells were co-transfected with plasmids encoding V5-tagged AKAP220 and either wild type GSK3β, a kinase-dead mutant (K85A), a constitutively active mutant (S9A), or a phosphate binding site mutant (R96A) (25). The R96A mutation specifically prevents GSK3β from phosphorylating primed substrates but has no effect on non-primed substrates such as β-catenin and Axin (37). Immunoblot analysis revealed that AKAP220 had increased binding with the constitutively active GSK3β-S9A mutant compared with wild type GSK3β (Fig. 1G, lanes 2 and 4). However, neither the catalytically inactive GSK3β-K85A nor the phosphate binding site mutant GSK3β-R96A co-fractionated with AKAP220 (Fig. 1G, lanes 3 and 5). In addition, the AKAP220-T1132A mutant did not interact with wild type or any of the mutant GSK3β forms (Fig. 1G, lanes 6–10). Together these data indicate the catalytic activity of GSK3β is required for binding and that Thr-1132 on AKAP220 is a GSK3β substrate required for the interaction.
In light of these results we propose a mechanism of interaction between GSK3β and AKAP220 that is depicted in Fig. 1H. The active kinase phosphorylates Thr-1132 on AKAP220 to initiate complex formation. Mutating this residue to alanine abolishes GSK3β binding, indicating that Thr-1132 is required and that AKAP220 interacts with a single molecule of GSK3β. Catalytic activity is involved in formation of the AKAP220-GSK3β complex, as kinase-dead mutants did not associate with full-length AKAP220 in cells. Additional validation of this hypothesis comes from our evidence of enhanced AKAP220 binding to the constitutively active GSK3β-S9A mutant.
AKAP220-GSK3β Complexes Are Present in Membrane Ruffles
The subcellular location of AKAP220-GSK3β complexes was observed by immunofluorescence microscopy. HEK293 cells were transfected with full-length YFP-AKAP220 and stained with antibodies against YFP (AKAP220) and endogenous GSK3β (Fig. 2A). Both proteins were enriched at membrane ruffles (identified by phalloidin f-actin staining) and were readily detected in the cytosol (Fig. 2A). These results are consistent with previous reports of AKAP220 localization in HT1080 cells where it binds IQGAP1 and promotes actin polymerization in membrane ruffles (38).
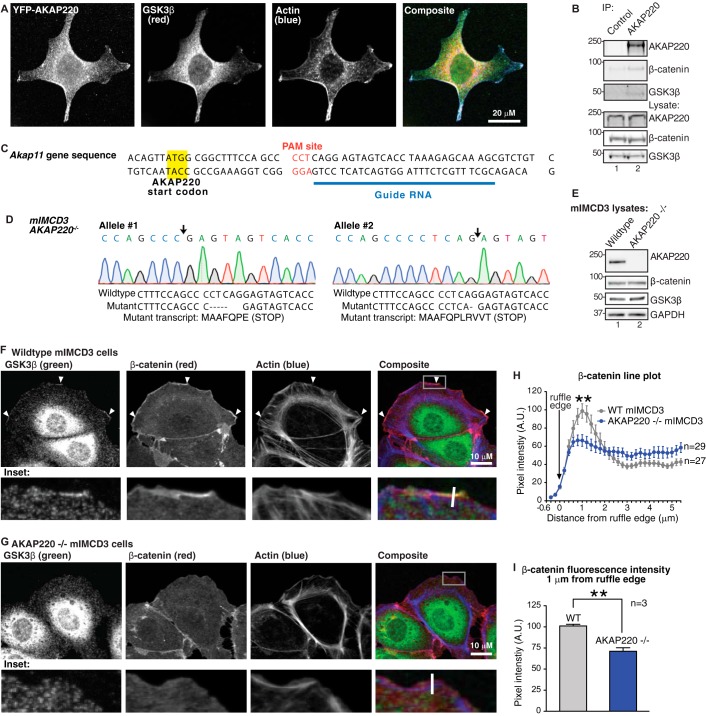
FIGURE 2.
AKAP220 localizes GSK3β and β-catenin to membrane ruffles. A, HEK293 cell expressing YFP-AKAP220 were stained for YFP (AKAP220, green) endogenous GSK3β (red), and actin (phalloidin, blue). AKAP220 and GSK3β co-distribute in the cytosol and are both enriched in membrane ruffles. Scale bar, 20 μm. B, co-immunoprecipitation (IP) of endogenous AKAP220 from wild type mIMCD3 cells detected an interaction with GSK3β and β-catenin. C, CRISPR-Cas9 gene editing was used to disrupt the Akap11 gene using the indicated synthetic guide RNA to the mouse Akap11 gene that encodes AKAP220. D, sequencing analysis of the AKAP220−/− clone demonstrates deletions on both Akap11 alleles (site of deletion indicated by black arrows). The wild type and mutant sequences are listed, with the resulting mutant transcripts from each allele are indicated. Both mutations create early in-frame stop codons. E, complete loss of AKAP220 protein expression was confirmed by Western blot. F, wild type and AKAP220−/− mIMCD3 cells were stained for endogenous GSK3β (green), β-catenin (red), and f-actin (phalloidin, blue). Membrane ruffles were identified as regions with positive f-actin stain. The location of zoomed insets is indicated (white box). In wild type cells, both GSK3β and β-catenin were enriched in ruffles (arrowheads). G, no GSK3β or β-catenin enrichment was observed in membrane ruffles of AKAP220−/− cells. Average peak intensity occured at 1 μm from the ruffle edge in both wild-type and AKAP220−/− cells. This peak intensity was significantly higher in the wild-type cells compared to AKAP220−/− cells (p < 0.01). H, representative quantification of β-catenin distribution at the membrane from one experiment. Fluorescence intensity was measured by line plot analysis, with lines drawn perpendicular to the ruffle edge (representative white lines are indicated on insets). A.U., arbitrary units. I, average β-catenin fluorescence intensity 1 μm from the ruffle edge was significantly reduced in AKAP220−/− cells (p < 0.01; error bars represent S.E.). In wild-type cells, the average intensity 1 μm from the ruffle edge was 101.1 ± 1.9 A.U. In AKAP220−/− cells, the average intensity at the ruffle edge was 71.12 ± 4.1 A.U. (average of n = 3 experiments, >18 cells per condition per experiment).
Based on this information, we next investigated whether AKAP220 may serve to anchor GSK3β near substrates in membrane ruffles. For this purpose we selected an immortalized epithelial line produced from mIMCD3. These cells were chosen for their high endogenous expression of AKAP220 and amenability to genome-editing techniques. Using co-immunoprecipitation, we first confirmed that endogenous AKAP220 and GSK3β interact in the wild type mIMCD3 cell line (Fig. 2B). Interestingly, we also detected endogenous β-catenin in AKAP220 immunocomplexes, a well known downstream target of GSK3β (Fig. 2B). Both GSK3β and β-catenin signals were absent in control IgG pulldown assays.
Based on this information, we next decided to produce an AKAP220-null cell line to gain further understanding of its function in membrane ruffles. Using CRISPR-Cas9 technology, the AKAP220−/− cell line was created using a synthetic guide RNA that recognized the first coding exon of AKAP220 (Fig. 2C). Sequencing analysis of several positive clones revealed one clone with frameshift mutations and early stop-codons on both alleles of the Akap11 gene (Fig. 2D). Complete loss of AKAP220 protein expression in the AKAP220−/− line was verified by Western blot analysis (Fig. 2E).
AKAP220 Anchors Both GSK3β and β-Catenin in Membrane Ruffles
To visualize protein localization, mIMCD3 cells were grown on glass coverslips and stimulated with PDGF (50 ng/ml for 30 min) to induce membrane ruffling. The cells were fixed and stained with antibodies against endogenous GSK3β and β-catenin, and membrane ruffles were identified by phalloidin staining for f-actin. In wild type mIMCD3 cells, both GSK3β and β-catenin were enriched in membrane ruffles (Fig. 2F, arrowheads). Regions of distinct co-distribution were evident in these actin-rich structures (Fig. 2F, insets). As expected, loss of AKAP220 caused a redistribution of GSK3β away from membrane ruffles (Fig. 2G). Interestingly, β-catenin was also lost from ruffles and showed significantly reduced enrichment at the ruffle edge in AKAP220−/− cells (Fig. 2G, insets). The distribution of β-catenin at the ruffle edge was quantified by line plot analysis of fluorescence intensity. In wild type cells, the β-catenin signal increased adjacent to the ruffle edge (Fig. 2H, gray line). In AKAP220−/− cells, however, the peak fluorescence intensity at the ruffle edge was significantly reduced (Fig. 2H, blue line). Comparison of β-catenin fluorescence intensity 1 μm from the plasma membrane confirmed a significant reduction in AKAP220−/− cells (average of n = 3 experiments, ≥18 cells per experiment) (Fig. 2I). Collectively, these functional studies in AKAP220−/− cells confirm that AKAP220 localizes GSK3β with its substrate β-catenin and recruits both proteins to membrane ruffles.
A logical next question was to establish if other enzymes in the AKAP220 scaffold could also participate in regulating the location and activity of GSK3β. Because GSK3β activity can be inhibited by PKA (25), we hypothesized that sequestering both kinases in proximity to each other AKAP220 could provide a mechanism for local molecular cross-talk.
Identification of a Second PKA Anchoring Site on AKAP220
The initial discovery and characterization of AKAP220 mapped an R subunit binding site to residues 1633–1646 in the mouse protein (20). However, subsequent experiments revealed that deletion of this 14-amino acid segment does not eliminate all PKA association with AKAP220 (Fig. 3A). HEK293 cells were transfected with wild type AKAP220 or a form of the anchoring protein lacking the originally defined PKA binding site (AKAP220 Δ1633–1646). As expected, the C subunit of PKA co-fractionated with wild type AKAP220 immune complexes as assessed by immunoblot (Fig. 3A). Likewise, PKA also co-fractionated with the mutant form lacking the previously identified PKA binding site (Fig. 3A). Control immunoblots confirmed that equivalent levels of each protein were expressed and enriched in the co-immunoprecipitates. Therefore we concluded that AKAP220 contains multiple PKA anchoring sites.
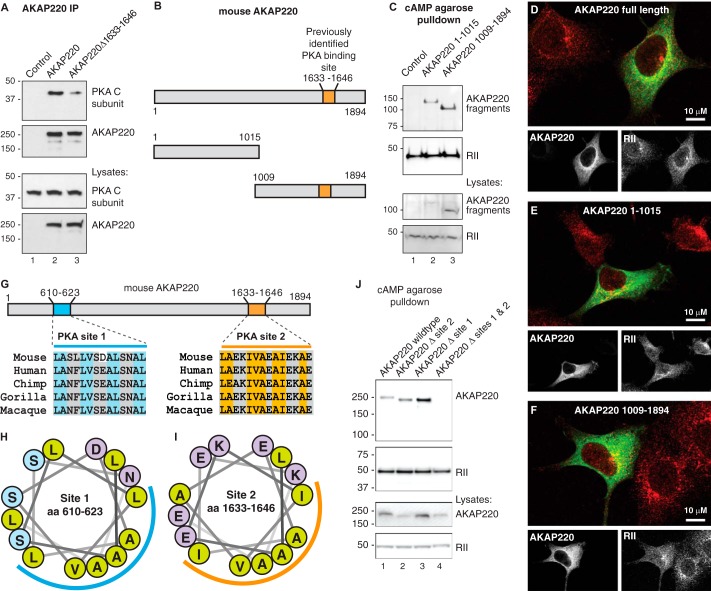
FIGURE 3.
There are two PKA binding sites on AKAP220. A, full-length wild type AKAP220 or a mutant form of AKAP220 lacking residues 1633–1646 were immunoprecipitated (IP) from HEK293 cell lysates. Co-purification of the PKA catalytic (C) subunit confirms PKA binding to the indicated form of the anchoring protein. B, schematic diagram of full-length mouse AKAP220 and two fragments that were used to assess the presence of a second PKA binding site: C-terminal (residues 1–1015) and N-terminal (residues 1009–1894) halves of AKAP220. The previously identified PKA binding region is indicated: residues 1633–1646. C, HEK293 cells expressing each construct were lysed, and then RII and its associated proteins were isolated by cAMP-agarose pulldown assays. Immunoblots detected co-purification of the FLAG-AKAP220 fragments using an anti-FLAG antibody. D–F, co-distribution of RII (red) with AKAP220 full-length (D), the 1–1015 fragment (E), or the 1009–1984 fragment (F) (green). Single focal plane images collected by confocal microscopy are shown; scale bars, 10 μm. G, amino acid sequence analysis of mouse AKAP220 revealed a predicted amphipathic helix at residues 610–623 (site 1) that is highly conserved. The previously identified PKA binding site sequence (site 2) also forms a helix and is highly conserved. H and I, the helical wheel prediction for the candidate PKA binding site at residues 610–623 presents a hydrophobic face (blue curved line) suitable for R subunit binding similar to that of the known PKA site at residues 1633–1646 (orange curved line). The hydrophobic residues are marked as light green circles. J, AKAP220 mutants in which the RII binding amphipathic helices were deleted were expressed in HEK293 cells. RII and any associated proteins were isolated by cAMP-agarose pulldown. Single deletion of AKAP220 site 1 or site 2 did not prevent the interaction with the PKA regulatory subunit, but double deletion of both sites was sufficient to abolish the interaction.
To map the location of the other PKA binding site, we used a complementary pulldown approach. HEK293 cells were transfected with FLAG-tagged constructs encoding the N-terminal (1–1015) or C-terminal (1009–1894) halves of the anchoring protein (Fig. 3B). AKAP220 complexes were co-purified by virtue of the R subunit affinity for cAMP-agarose. As expected, the C-terminal half of AKAP220 containing the known PKA site was readily detected by immunoblot (Fig. 3C, lane 3). However, the N-terminal half of the anchoring protein was also detected by cAMP-agarose pulldown and must, therefore, contain a binding site for the PKA R subunit (Fig. 3C, lane 2). Control immunoblots confirmed enrichment of RII on the cAMP affinity resin and the expression levels of each protein in cell lysates.
Next, immunofluorescence microscopy was used to visualize the subcellular distribution of AKAP220 and fragments with RII inside cells. Transfected HEK293 cells fixed in 4% paraformaldehyde were stained for AKAP220 and fragments (anti-FLAG; green) and endogenous RII subunit of PKA (anti-RII; red). Each AKAP220 form co-distributed with RII (Figs. 3, D–F). Taken together these biochemical and cell-based experiments infer that at least one additional PKA binding site resides within the first 1015 residues of AKAP220.
Structural approaches have shown that the hydrophobic face of an amphipathic helix on AKAPs interacts with the docking and dimerization domain on the R subunit (39, 40). Algorithms have been developed to screen for prospective R subunit anchoring helices (41, 42). Using these approaches, residues 610–623 of AKAP220 were predicted to be a putative PKA binding helix (Fig. 3, G and H; blue line). This region is almost invariant in mammalian orthologs of AKAP220 and resembles the previously defined PKA binding helix on AKAP220 and other AKAPs (Figs. 3, G, orange, and 2I).
To test the PKA binding capacity of this newly identified region, cAMP agarose pulldown assays were conducted with wild type and helix deletion mutants of AKAP220 (Fig. 3J). These constructs were full-length AKAP220, AKAP220 Δ site 1 (Δ610–623), AKAP220 Δ site 2 (Δ1633–1646), and AKAP220 double delete (Δ610–623, Δ1633–1646; Fig. 3J). Each construct encoded a V5-epitope tag for probing by Western blot. Full-length AKAP220 and each single-deletion mutant were pulled down by cAMP-agarose affinity resin (Fig. 3J, lanes 1–3). This demonstrates that each binding helix on AKAP220 is sufficient for interaction with the PKA R subunit. In contrast, the double-deletion mutant was not detected by cAMP pulldown, indicating that removal of both helical regions abolished interaction with R subunits (Fig. 3J, lane 4). Control immunoblots confirmed the enrichment of RII on the affinity resin and expression levels of each protein in cell lysates. These experiments establish the presence of two PKA binding sites on AKAP220 that are capable of independent interaction with the PKA holoenzyme.
PKA Binding Sites on AKAP220 Have Differential Isoform Selectivity
Although the majority of anchoring proteins preferentially interact with type II PKA, a few type I PKA-selective AKAPs have also been reported (2, 43–45). Another subset, designated dual function AKAPs, can interact with either RI or RII (46, 47). Therefore, a logical next step was to evaluate the R subunit binding preferences of each anchoring helix on AKAP220. His-tagged MBP-fusion proteins encompassing each PKA binding site were expressed in Escherichia coli and purified on a nickel-nitrilotriacetic acid column. Protein purity and concentration was confirmed by stain-free gel (Fig. 4A, top panel). Both AKAP220 fragments bound RII as detected by biotin-RII overlay (Fig. 4A, middle panel). However, only site 2 of AKAP220 was detected by biotin-RI overlay (Fig. 4A, bottom panel, lane 2). These initial findings infer that site 2 on AKAP220 is a dual function PKA anchoring helix, whereas site 1 is RII-selective.
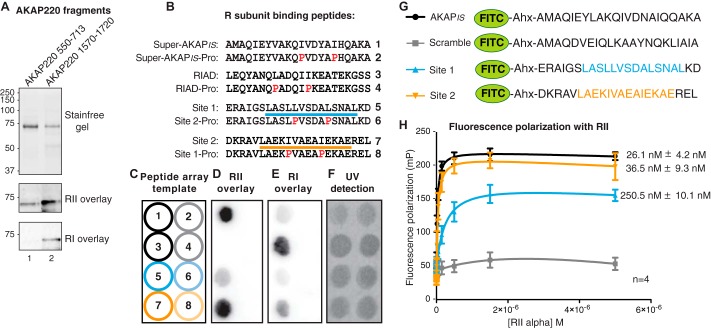
FIGURE 4.
Characterization of isoform-specific PKA binding to AKAP220. A, AKAP220 fragments encompassing site 1 (550–713) and site 2 (1570–1720) were expressed as His-MBP fusion proteins E. coli. Nickel-nitrilotriacetic acid affinity purification and equal loading of protein was verified by UV exposure of stain-free gel. R-subunit overlay detected isoform-specific binding characteristics of each fragment. B, the amino acid sequences of peptides synthesized in peptide arrays (C–F) are shown for PKA binding site 1 and site 2 along with sequences for Super-AKAPis and the RI anchoring disruptor peptide (RIAD), the positive controls for RII and RI binding, respectively. Negative binding controls incorporated helix-disrupting proline substitutions (red, peptides designated as -Pro). Amphipathic helices are indicated by a blue line for site 1 (peptide 5) and an orange line for site 2 (peptide 7). C, each numbered peptide in B was synthesized onto cellulose as illustrated in panel C and then submitted to a far Western with purified R-subunit. D, biotin-RII overlay detected binding to Super-AKAPis and AKAP220 site 2 (peptides 1 and 7). A weak signal was detected for AKAP220 site 1 (peptide 5). No signal was detected for RIAD or any of the proline mutant peptides (peptides 2–4, 6, and 8). E, overlay with biotin-RI detected RIAD and AKAP220 site 2 (peptides 3 and 7). No signal was detected for AKAP220 site 1, Super-AKAPis, or any of the proline mutant peptides (peptides 1, 2, 4–6, and 8). F, peptide array synthesis and spotting efficiency was detected by exposure to UV light. G, FITC-conjugated peptides for AKAPis, site 1, site 2, and a scrambled AKAPis sequence that were used to determine RII binding affinity measurements by fluorescence polarization are shown. Identified binding sites are indicated in blue (site 1) and orange (Site 2). H, fluorescence polarization was used to assess RII binding affinities of the peptides shown in G. Saturation binding experiments determined the binding affinity of each peptide for RII. Increasing concentrations of purified His-RIIα were added to 5 nm FITC-peptide, and fluorescence polarization was measured with a plate reader. Polarization (P) was calculated using the emitted fluorescence intensity (I) both parallel and perpendicular to the excitation source using the following equation: P = (I parallel − I perpendicular)/(I parallel + I perpendicular). Values were converted to millipolarization (mP) units for analysis. Kd values were calculated by non-linear regression to a one-site binding model in GraphPad Prism 6.0.
To consolidate this finding, AKAP220 peptides encompassing each binding site (site 1, residues 604–625; site 2, residues 1628–1649) were synthesized onto a cellulose membrane by peptide array (Fig. 4, B and C). Super-AKAPis and RIAD peptides were used as positive binding sequences for RII and RI, respectively (48, 49). Helix-disrupting proline mutations were introduced into each peptide sequence as negative binding controls. As anticipated, both the AKAP220 site 1 and site 2 peptides were detected by biotin-RII overlay. This signal was completely abolished in proline mutant forms, indicating the formation of an amphipathic helix was required for RII binding (Fig. 4D). Only the AKAP220 site 2 peptide was detected by biotin-RI overlay, indicating this site has dual-specific PKA binding capacity, whereas site 1 binds only to RII (Fig. 4E). Peptide spotting efficiency was confirmed by illumination with UV light (Fig. 4F).
Next, fluorescence polarization experiments were performed to measure the apparent binding affinities of AKAP peptides for RII. FITC-conjugated peptides (22-mers) encompassing site 1 or site 2 of AKAP220 were synthesized (Fig. 4G, bottom two peptides). FITC-conjugated AKAPis served as a positive control, whereas a scrambled peptide of identical amino acid composition served as a negative binding control (Fig. 4G, top two peptides). FITC-labeled peptides (5 nm) were combined with increasing concentrations of purified RIIα (50 pm to 5 μm). After incubation at room temperature for 30 min to reach binding equilibrium, fluorescence polarization was measured with a plate reader.
Using this approach, the dissociation constant (Kd) of AKAP220 site 2 peptide binding to RIIα was 36.5 ± 9.3 nm (n = 4; Fig. 4H). This value was comparable with AKAPis (26.1 ± 4.2 nm n = 4; Fig. 4H). In contrast, the Kd of the AKAP220 site 1 peptide for RIIα was almost 10-fold lower (250.5 ± 10.1 nm, n = 4; Fig. 4H). The scrambled peptide did not bind RII. Unfortunately, it was not possible to calculate RI binding affinities, as saturation was not reached with this assay. These measurements support and explain the differences in signal strength seen in Fig. 4D. Together, the data in Fig. 4 determine that site 1 (residues 610–623) of AKAP220 is an RII-selective site, whereas site 2 (residues 1633–1646) is a dual-specificity PKA anchoring site.
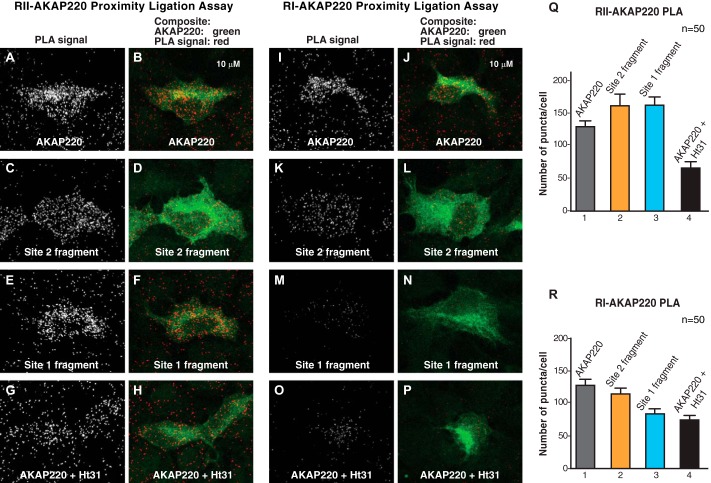
AKAP220 Interacts with Type I and Type II PKA in Situ
To examine AKAP220 interaction with type I and type II PKA in cells, we used a sensitive proximity ligation assay (PLA). This in situ approach uses two secondary antibody probes conjugated to DNA oligonucleotides (35). Enzymatic ligation and rolling circle DNA amplification occurs when both probes bind their targets within a radius of 40–60 nm. Subsequent in situ hybridization of fluorescently tagged complementary oligonucleotides generates PLA puncta that are visible with a fluorescence microscope. Detection of fluorescent PLA puncta denoted the location and abundance of individual AKAP220-R subunit pairs.
HEK293 cells were transfected with the full-length AKAP220 or fragments encompassing residues 1–1015 (site 1) or 1009–1894 (site 2) of the anchoring protein. As a control, cells expressing full-length AKAP220 were treated with the anchoring inhibitor peptide Ht31 to universally disrupt R subunit-AKAP interactions (16). Cells were fixed and incubated with a mixture of antibodies that recognized AKAP220 (anti-FLAG) and antibodies against endogenous RI or RII followed by a FITC-anti-FLAG counterstain to identify transfected cells. Scanning laser confocal microscopy was used to generate maximum projection images, and PLA puncta were quantified by semi-automatic analysis as described previously (35). PLA puncta were defined as local intensity maxima and measured with the Find Maxima algorithm in ImageJ. For quantification, FITC-positive cells were delineated by hand (50 cells per experimental condition), and numbers of puncta were counted. Background was defined as the number of PLA puncta in adjacent untransfected (FITC-negative) cells.
As expected, PLA signal was detected between full-length AKAP220 and endogenous RII (Fig. 5, A and Q, column 1). A comparable number of PLA puncta were counted for both the site 1 and site 2 AKAP220 fragments, verifying that each fragment effectively binds RII inside cells (Fig. 5, C, E, and Q, columns 2 and 3). Treatment with Ht31 reduced the number of puncta, confirming the fidelity of the assay (Fig. 5, G and Q, column 4).
FIGURE 5.
AKAP220 interactions with type I and type II PKA in cells. A–H, a PLA was used to detect protein-protein interactions between AKAP220 and RII inside intact HEK293 cells. PLA signal between RII and the wild type, site 1, and site 2 fragments of AKAP220 is seen as puncta (grayscale; A, C, and E). Treatment with the R subunit anchoring inhibitor peptide Ht31 was used to disrupt the interaction (grayscale, G). Transfected cells were identified by FITC anti-FLAG counterstain (green; B, D, F, and H). I–P, the PLA assay was repeated to detect type I regulatory subunit interactions with AKAP220. PLA signal was detected between RI and the wild type and site 2 fragments of AKAP220 but was reduced with the site 1 fragment (grayscale; I, K, and M). The interaction with RI was disrupted when Ht31 was present (grayscale; O). Transfected cells were identified by FITC anti-FLAG counterstain (green; J, L, N, and P). Q, quantification of AKAP220-RII interaction as evidenced by PLA puncta per cell in each condition (n = 50 cells per condition). R, data for RI interaction with AKAP220 as measured by PLA puncta per cell are presented (n = 50 cells per condition).
Parallel experiments also detected an interaction between full-length AKAP220 and RI in cells (Fig. 5, I and R, column 1). The number of puncta for the AKAP220 site 2 fragment was comparable to wild type, confirming this site has dual-specific R subunit binding capacity (Fig. 5, K and R, column 2). However, the AKAP220 site 1 fragment had reduced PLA signal comparable with that seen in Ht31-treated cells (Fig. 5, M, O, and R, columns 3 and 4). These results further support that AKAP220 site 2 (residues 1633–1646) can bind RI or RII, whereas site 1 (residues 610–623) is RII-selective.
Anchored PKA Modulates GSK3β Binding to AKAP220
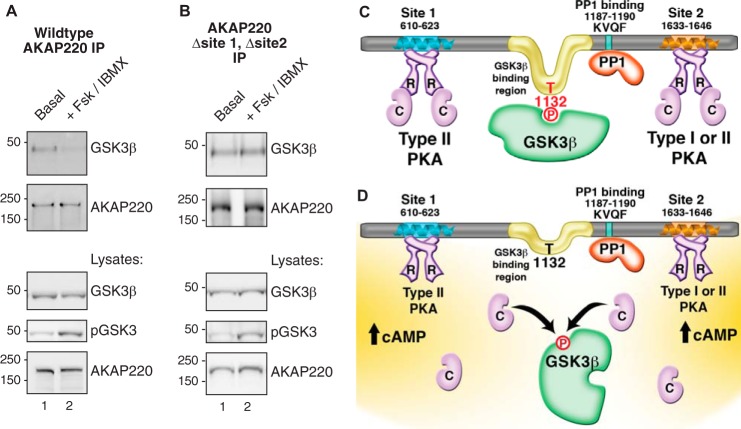
PKA phosphorylation of GSK3β on serine 9 suppresses enzyme activity (50). Therefore, we assessed whether cross-talk between both kinases could influence complex assembly with AKAP220. To test this hypothesis we transfected HEK293 cells with wild type AKAP220 and stimulated them with 10 μm FSK and 75 μm IBMX to increase intracellular cAMP. AKAP220 immune complexes were isolated. Elevation of intracellular cAMP led to a decrease in GSK3β co-fractionation with AKAP220 compared with basal conditions (Fig. 6A). Thus, GSK3β dissociates from the anchoring protein in response to mobilization of the cAMP cascade. To determine whether this event was mediated by PKA sequestered within the same complex, we repeated the experiment with a mutant form of AKAP220 that is unable to bind PKA (AKAP220-Δsite1-Δsite2). GSK3β dissociation from AKAP220 in response to FSK/IBMX treatment was blunted in this context (Fig. 6B). Therefore, an anchored pool of PKA primarily controls this event. Control blots of cell lysates in both experiments confirmed that phospho-GSK3β levels increased after FSK/IBMX stimulation (Fig. 6, A and B, bottom panels, lane 2). Taken together these experiments show that AKAP220 complexes are dynamic, and the reorganization of binding partners occurs in response to cellular signals.
FIGURE 6.
GSK3β dissociates from AKAP220 complexes in response to cAMP. A, HEK293 cells were stimulated with 10 μm FSK and 75 μm IBMX for 10 min at 37 °C to increase intracellular cAMP and activate PKA. AKAP220 immunocomplexes were isolated from cell lysates and probed by immunoblot. GSK3β binding was decreased in stimulated cells. Control blots confirmed equal expression and pulldown of AKAP220. An antibody specific for phospho-GSK3β(Ser-9) detected a higher signal in the treated lysates, confirming stimulation. No phospho-GSK3β(Ser-9) was detected in the pulldown assays. IP, immunoprecipitation. B, the experiment was repeated with a mutant form of AKAP220 that does not bind PKA. Dissociation of GSK3β was blunted in these conditions. C, schematic representation of enzyme binding sites on AKAP220. PKA can bind to two sites. Site 1 is RII-selective, and site 2 has dual-specificity for RI or RII. One copy of active GSK3β interacts with AKAP220 between residues 1126 and 1145, a region containing the GSK3β phosphosite at Thr-1132 that is required for GSK3β binding. Additionally, PP1 binds to the KVQF motif at residues 1187–1190. D, proposed model of cAMP responsive GSK3β dissociation from AKAP220. In response to cAMP, anchored PKA is activated, and the C subunits are released. Anchored GSK3β is phosphorylated, preventing it from phosphorylating Thr-1132 causing its release from the complex. Anchored PP1 may relieve PKA-mediated inhibition of GSK3β by dephosphorylating Ser-9, providing bi-directional control of AKAP220 complex formation in response to cAMP.
Discussion
A-kinase anchoring proteins utilize various mechanisms to exert dynamic and local control of signal transduction events. For example, sequestering kinases and phosphatases with their substrates allows for efficient and bi-directional modulation of phospho-signals (51). The processive relay of information through anchored enzyme cascades can precisely direct messages to downstream targets (52, 53). Allosteric modulation of catalytic activity through association with anchoring or targeting subunits also fine-tunes the substrate specificity of broad spectrum signaling enzymes (54, 55). The predominant mechanistic theme emerging from this study is that cross-talk between PKA and GSK3β is augmented by AKAP220. Specifically, PKA negatively regulates local GSK3β activity and binding to the signaling complex in response to cAMP.
The biochemical and cell-based studies presented in Figs. 3 and 4 demonstrate that AKAP220 is one of a select group of AKAPs with the capacity to sequester multiple PKA holoenzymes. For example, the multivalent scaffolding protein yotiao/AKAP450 contains two PKA binding sites and constrains a range of protein kinases and phosphatases at the Golgi and centrosomes (56, 57). Likewise, the sphingosine kinase interacting protein (SKIP) is an RI-selective anchoring protein that tethers a pair of type I PKA holoenzymes with preferred substrates at the inner face of the mitochondrial matrix (43, 44). Dimerization of anchoring proteins is another means to increase the local concentration of PKA as exemplified by membrane-associated AKAP79/150 (58). Thus, various modes of interaction can be used to locally concentrate PKA in the vicinity of AKAPs.
Our work on AKAP220 illustrates an interesting variation on this theme whereby a single AKAP protomer contains two anchoring sites with differential PKA isoform selectivity. Site 2 on AKAP220 (residues 1633–1646) exhibits a dual specificity for RI and RII, whereas the newly identified site 1 (residues 610–623) preferentially interacts with RII. Accordingly, each site could anchor distinct PKA isoforms that act on diverse downstream targets. This is particularly relevant because RI and RII not only display different sensitivities to cAMP but also exhibit tissue-specific and developmentally regulated patterns of expression. It is also important to note that although site 2 has a higher apparent affinity for RII than RI, we observed RI binding inside cells by proximity ligation assay. These findings indicate that RI can successfully compete for binding to AKAP220 in a cellular context. Therefore, we propose that AKAP220 constrains a range of macromolecular complexes that include either type I or type II PKA.
Mechanistically, this model could impact the duration of second messenger responses, particularly because type I PKA is more sensitive to cAMP than type II PKA (59, 60). Additionally, such signaling responses may be more robust if two PKA holoenzymes are simultaneously anchored by AKAP220. An illustrative example of this concept comes from evidence that SKIP complexes exist in different states of RI occupancy. Sophisticated biophysical analyses show that up to 59% of SKIP sequesters a single molecule of RI, whereas 41% of the anchoring protein sequesters two molecules of RI (44). Thus, future experiments will investigate the stoichiometry and range of AKAP220 PKA complexes that exist in side cells.
One biological role for AKAP220-associated pools of PKA is to ensure the compartmentalized regulation of GSK3β. Unlike most serine/threonine protein kinases, GSK3β is active under basal conditions (50). As previously stated, suppression of GSK3β activity occurs upon phosphorylation of serine 9 by upstream kinases including PKA and Akt. In this report we show that AKAP220-associated PKA contributes to the phosphorylation-dependent modulation of GSK3β, in part because both enzymes reside within the same macromolecular complex. Our work further indicates that AKAP220 preferentially sequesters catalytically active GSK3β. We show that the constitutively active GSK3β-S9A mutant has enhanced binding to AKAP220, whereas neither the kinase-dead GSK3β-K85A nor the phosphate binding site mutant GSK3-R96A was able to bind. A previous study demonstrated binding between a C-terminal AKAP220 fragment (residues 1011–1901) and two catalytically inactive forms of GSK3β, H85M, and Y216F (24). The conflicting results in these studies may be due to a varied degree of kinase inhibition or conformational differences between the GSK3β-H85M and Y216F mutants used in the previous work and the K85A and R96A mutant forms used here. Additionally, our studies utilized full-length AKAP220 rather than a truncated fragment of the anchoring protein. Thus, secondary contact points on AKAP220 may be involved in modulating the interaction with catalytically active GSK3β. Future structural analyses of the binding interface on GSK3β may ultimately provide details to resolve these outstanding questions.
The preferential association of catalytically active GSK3β with AKAP220 is further underscored by our identification of Thr-1132 as a phosphosite that is required for recruitment of this kinase. Substitution with an alanine at this position abolishes GSK3β binding, emphasizing a role for the phosphate moiety as a molecular determinant of GSK3β anchoring. Our findings can be consolidated into a mechanism presented in Fig. 5C, whereby catalytically active GSK3β is required to both establish and maintain the interaction with AKAP220 by phosphorylating Thr-1132 on the anchoring protein.
It has long been recognized that GSK3β often has a preference for “primed” or pre-phosphorylated substrates, although under certain conditions it is not required (50). This is consistent with our peptide array data showing that Thr-1132 on AKAP220 is phosphorylated by GSK3β irrespective of a priming phosphorylation event. However, we also note Ser-1136 on the anchoring protein is optimally positioned to serve as a priming site and may be a substrate for a different kinase in a cellular context. Another interesting possibility is that phosphorylation of Thr-1132 may initiate the processive phosphorylation of an additional consensus GSK3β site located four residues upstream at Thr-1128. Indirect support for this concept comes from our peptide array data in Fig. 1E. We noted that a single alanine substitution at Thr-1132 reduced but did not entirely eliminate peptide phosphorylation. Hence, GSK3β-mediated priming of Thr-1132 may serve as a prerequisite for the efficient phosphorylation of Thr-1128 in vivo. Taken together, these findings underscore the importance of phospho-Thr-1132 as a primary determinant for recruitment of active GSK3β to the AKAP220 signaling scaffold.
The significance of GSK3β anchoring is emphasized by the discovery that AKAP220 interacts with β-catenin, a well known GSK3β substrate. In fact, AKAP220 is involved in localizing both GSK3β and β-catenin to membrane ruffles, as our quantitative image analysis shows deficient enrichment of GSK3β or β-catenin at the ruffle edge in AKAP220−/− cells. These results suggest that AKAP220 may facilitate signaling between these two proteins during lamellipodia and ruffle formation. These new observations are consistent with work by our laboratory demonstrating that siRNA-mediated knockdown of AKAP220 reduced membrane ruffling and slowed cell migration. These phenotypes were linked to the interaction between AKAP220 and IQGAP1, a known β-catenin binding partner. This large macromolecular complex, therefore, contains several signaling components implicated in ruffling and migration that are recruited to the appropriate cellular location by AKAP220.
Another important facet of this study is the discovery that anchored PKA modulates the interaction between GSK3β and AKAP220. Our cell-based studies infer that cAMP stimulation promotes GSK3β dissociation from AKAP220 complexes. Importantly this effect is not observed with an AKAP220 mutant that is unable to anchor PKA. This finding raises interesting questions about cross-talk between these and other enzyme binding partners. This is particularly relevant as AKAP220 also targets the catalytic subunit of protein phosphatase PP1, an enzyme that may allow for bidirectional regulation of phosphorylation events (22). For example, inhibition of GSK3β via PKA-mediated phosphorylation of Ser-9 can be relieved by PP1 dephosphorylation of this site (61). It is also possible that Thr-1132 on AKAP220 is a prospective PP1 substrate. Because dephosphorylation of this residue could enhance GSK3β dissociation from the complex, PP1 may act cooperatively with PKA to strengthen the cAMP-mediated response. These ideas have been consolidated into a schematic that emphasize the major findings of this study, presented in Fig. 6, C and D.
In conclusion we show that sequestering of GSK3β by AKAP220 is regulated at multiple levels. First, GSK3β activity is required to phosphorylate Thr-1132 on AKAP220. This phosphorylation event both establishes and sustains the GSK3β-AKAP220 subcomplex. Second, activation of AKAP220-associated PKA promotes GSK3β dissociation, likely through phosphorylation-dependent inhibition of GSK3β activity. Finally, tethering of either type I or type II PKA holoenzymes may alter the magnitude and duration of the local cAMP response. The presence of two independent R subunit-anchoring helices on AKAP220 may represent a mechanism to amplify local cAMP responses, especially if two PKA holoenzymes can be simultaneously anchored. Our findings highlight the diverse and dynamic assembly of AKAP220 signaling complexes and the utility of enzyme cross-talk as a mechanism to integrate and disseminate cellular information.
Author Contributions
J. L. W. and B. J. T. designed, performed, and analyzed pulldown, overlay, and peptide array experiments. J. L. W. conducted all imaging and PLA studies. O.-M. S. designed and verified cell lines produced by CRISPR-Cas9 genome editing. P. J. N. purified protein and performed fluorescence polarization experiments. J. L. W., L. K. L., and J. D. S. prepared figures and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK105542 and DK054441.
- AKAP
- PKA anchoring protein
- GSK3β
- glycogen synthase kinase 3β
- PP1
- protein phosphatase 1
- IQGAP
- IQ motif containing GTPase activating protein
- mIMCD3
- mouse inner medullary-collecting duct
- PLA
- proximity ligation assay
- R
- subunit
- C
- catalytic subunit
- SKIP
- sphingosine kinase interacting protein
- IBMX
- isobutylmethylxanthine
- FSK
- forskolin.
REFERENCES
- 1. Welch E. J., Jones B. W., Scott J. D. (2010) Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol. Interv. 10, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scott J. D., Dessauer C. W., Taskén K. (2013) Creating order from chaos: cellular regulation by kinase anchoring. Annu. Rev. Pharmacol. Toxicol. 53, 187–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taskén K., Aandahl E. M. (2004) Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 84, 137–167 [DOI] [PubMed] [Google Scholar]
- 4. Langeberg L. K., Scott J. D. (2015) Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16, 232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bauman A. L., Soughayer J., Nguyen B. T., Willoughby D., Carnegie G. K., Wong W., Hoshi N., Langeberg L. K., Cooper D. M., Dessauer C. W., Scott J. D. (2006) Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol. Cell 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hinke S. A., Navedo M. F., Ulman A., Whiting J. L., Nygren P. J., Tian G., Jimenez-Caliani A. J., Langeberg L. K., Cirulli V., Tengholm A., Dell'Acqua M. L., Santana L. F., Scott J. D. (2012) Anchored phosphatases modulate glucose homeostasis. EMBO J. 31, 3991–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Navedo M. F., Nieves-Cintrón M., Amberg G. C., Yuan C., Votaw V. S., Lederer W. J., McKnight G. S., Santana L. F. (2008) AKAP150 is required for stuttering persistent Ca2+ sparklets and angiotensin II-induced hypertension. Circ. Res. 102, e1–e11 [DOI] [PubMed] [Google Scholar]
- 8. Tunquist B. J., Hoshi N., Guire E. S., Zhang F., Mullendorff K., Langeberg L. K., Raber J., Scott J. D. (2008) Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 12557–12562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith F. D., Langeberg L. K., Cellurale C., Pawson T., Morrison D. K., Davis R. J., Scott J. D. (2010) AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat. Cell Biol. 12, 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kritzer M. D., Li J., Passariello C. L., Gayanilo M., Thakur H., Dayan J., Dodge-Kafka K., Kapiloff M. S. (2014) The scaffold protein muscle A-kinase anchoring protein β orchestrates cardiac myocyte hypertrophic signaling required for the development of heart failure. Circ. Heart Fail. 7, 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taylor S. S., Ilouz R., Zhang P., Kornev A. P. (2012) Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 13, 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N.-H., Taylor S. S., Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 13. Su Y., Dostmann W. R., Herberg F. W., Durick K., Xuong N.-H., Ten Eyck L., Taylor S. S., Varughese K. I. (1995) Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding proteins. Science 269, 807–813 [DOI] [PubMed] [Google Scholar]
- 14. Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 15. Kinderman F. S., Kim C., von Daake S., Ma Y., Pham B. Q., Spraggon G., Xuong N. H., Jennings P. A., Taylor S. S. (2006) A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carr D. W., Stofko-Hahn R. E., Fraser I. D., Bishop S. M., Acott T. S., Brennan R. G., Scott J. D. (1991) Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266, 14188–14192 [PubMed] [Google Scholar]
- 17. Newlon M. G., Roy M., Morikis D., Carr D. W., Westphal R., Scott J. D., Jennings P. A. (2001) A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 20, 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith F. D., Reichow S. L., Esseltine J. L., Shi D., Langeberg L. K., Scott J. D., Gonen T. (2013) Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. eLife 2, e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dessauer C. W. (2009) Adenylyl cyclase-A-kinase anchoring protein complexes: the next dimension in cAMP signaling. Mol. Pharmacol. 76, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lester L. B., Coghlan V. M., Nauert B., Scott J. D. (1996) Cloning and characterization of a novel A-kinase anchoring protein: AKAP 220, association with testicular peroxisomes. J. Biol. Chem. 271, 9460–9465 [DOI] [PubMed] [Google Scholar]
- 21. Day M. E., Gaietta G. M., Sastri M., Koller A., Mackey M. R., Scott J. D., Perkins G. A., Ellisman M. H., Taylor S. S. (2011) Isoform-specific targeting of PKA to multivesicular bodies. J. Cell Biol. 193, 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schillace R. V., Scott J. D. (1999) Association of the type 1 protein phosphatase PP1 with the A-kinase anchoring protein AKAP220. Curr. Biol. 9, 321–324 [DOI] [PubMed] [Google Scholar]
- 23. Schillace R. V., Voltz J. W., Sim A. T., Shenolikar S., Scott J. D. (2001) Multiple interactions within the AKAP220 signaling complex contribute to protein phosphatase 1 regulation. J. Biol. Chem. 276, 12128–12134 [DOI] [PubMed] [Google Scholar]
- 24. Tanji C., Yamamoto H., Yorioka N., Kohno N., Kikuchi K., Kikuchi A. (2002) A-kinase anchoring protein AKAP220 binds to glycogen synthase kinase-3β (GSK-3β) and mediates protein kinase A-dependent inhibition of GSK-3β. J. Biol. Chem. 277, 36955–36961 [DOI] [PubMed] [Google Scholar]
- 25. Fang X., Yu S. X., Lu Y., Bast R. C., Jr., Woodgett J. R., Mills G. B. (2000) Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. U.S.A. 97, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doble B. W., Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. ter Haar E., Coll J. T., Austen D. A., Hsiao H. M., Swenson L., Jain J. (2001) Structure of GSK3β reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 8, 593–596 [DOI] [PubMed] [Google Scholar]
- 28. Scales T. M., Lin S., Kraus M., Goold R. G., Gordon-Weeks P. R. (2009) Nonprimed and DYRK1A-primed GSK3 β-phosphorylation sites on MAP1B regulate microtubule dynamics in growing axons. J. Cell Sci. 122, 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weston C. R., Davis R. J. (2001) Signal transduction: signaling specificity: a complex affair. Science 292, 2439–2440 [DOI] [PubMed] [Google Scholar]
- 30. Logue J. S., Whiting J. L., Scott J. D. (2011) Sequestering Rac with PKA confers cAMP control of cytoskeletal remodeling. Small GTPases 2, 173–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Logue J. S., Whiting J. L., Tunquist B., Sacks D. B., Langeberg L. K., Wordeman L., Scott J. D. (2011) AKAP220 protein organizes signaling elements that impact cell migration. J. Biol. Chem. 286, 39269–39281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith F. D., Samelson B. K., Scott J. D. (2011) Discovery of cellular substrates for protein kinase A using a peptide array screening protocol. Biochem. J. 438, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B. L., Xavier R. J., Root D. E. (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gajadhar A., Guha A. (2010) A proximity ligation assay using transiently transfected, epitope-tagged proteins: application for in situ detection of dimerized receptor tyrosine kinases. Biotechniques 48, 145–152 [DOI] [PubMed] [Google Scholar]
- 36. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frame S., Cohen P., Biondi R. M. (2001) A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 38. Logue J. S., Whiting J. L., Tunquist B., Langeberg L. K., Scott J. D. (2011) Anchored protein kinase A recruitment of active Rac GTPase. J. Biol. Chem. 286, 22113–22121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burns-Hamuro L. L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G. L., Cantor C. R., Braun A., Taylor S. S. (2003) Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. U.S.A. 100, 4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gold M. G., Fowler D. M., Means C. K., Pawson C. T., Stephany J. J., Langeberg L. K., Fields S., Scott J. D. (2013) Engineering A-kinase anchoring protein (AKAP)-selective regulatory subunits of protein kinase A (PKA) through structure-based phage selection. J. Biol. Chem. 288, 17111–17121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Asirvatham A. L., Galligan S. G., Schillace R. V., Davey M. P., Vasta V., Beavo J. A., Carr D. W. (2004) A-kinase anchoring proteins interact with phosphodiesterases in T lymphocyte cell lines. J. Immunol. 173, 4806–4814 [DOI] [PubMed] [Google Scholar]
- 42. Burgers P. P., van der Heyden M. A., Kok B., Heck A. J., Scholten A. (2015) A systematic evaluation of protein kinase A-A-kinase anchoring protein interaction motifs. Biochemistry 54, 11–21 [DOI] [PubMed] [Google Scholar]
- 43. Kovanich D., van der Heyden M. A., Aye T. T., van Veen T. A., Heck A. J., Scholten A. (2010) Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. Chembiochem 11, 963–971 [DOI] [PubMed] [Google Scholar]
- 44. Means C. K., Lygren B., Langeberg L. K., Jain A., Dixon R. E., Vega A. L., Gold M. G., Petrosyan S., Taylor S. S., Murphy A. N., Ha T., Santana L. F., Tasken K., Scott J. D. (2011) An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, E1227–E1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burgers P. P., Ma Y., Margarucci L., Mackey M., van der Heyden M. A., Ellisman M., Scholten A., Taylor S. S., Heck A. J. (2012) A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J. Biol. Chem. 287, 43789–43797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang L. J., Wang L., Ma Y., Durick K., Perkins G., Deerinck T. J., Ellisman M. H., Taylor S. S. (1999) NH2-terminal targeting motifs direct dual specificity A-kinase- anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J. Cell Biol. 145, 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang L. J., Durick K., Weiner J. A., Chun J., Taylor S. S. (1997) D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. U.S.A. 94, 11184–11189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alto N. M., Soderling S. H., Hoshi N., Langeberg L. K., Fayos R., Jennings P. A., Scott J. D. (2003) Bioinformatic design of A-kinase anchoring protein-in silico: A potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. U.S.A. 100, 4445–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carlson C. R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J. D. (2006) Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281, 21535–21545 [DOI] [PubMed] [Google Scholar]
- 50. Cohen P., Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 51. Carnegie G. K., Means C. K., Scott J. D. (2009) A-kinase anchoring proteins: from protein complexes to physiology and disease. IUBMB Life 61, 394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wong W., Goehring A. S., Kapiloff M. S., Langeberg L. K., Scott J. D. (2008) mAKAP compartmentalizes oxygen-dependent control of HIF-1α. Sci. Signal. 1, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carnegie G. K., Soughayer J., Smith F. D., Pedroja B. S., Zhang F., Diviani D., Bristow M. R., Kunkel M. T., Newton A. C., Langeberg L. K., Scott J. D. (2008) AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol Cell 32, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoshi N., Langeberg L. K., Gould C. M., Newton A. C., Scott J. D. (2010) Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol. Cell 37, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prince J. T., Ahn N. G. (2010) The case of the disappearing drug target. Mol. Cell 37, 455–456 [DOI] [PubMed] [Google Scholar]
- 56. Westphal R. S., Tavalin S. J., Lin J. W., Alto N. M., Fraser I. D., Langeberg L. K., Sheng M., Scott J. D. (1999) Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 285, 93–96 [DOI] [PubMed] [Google Scholar]
- 57. Witczak O., Skålhegg B. S., Keryer G., Bornens M., Taskén K., Jahnsen T., Orstavik S. (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 18, 1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gold M. G., Stengel F., Nygren P. J., Weisbrod C. R., Bruce J. E., Robinson C. V., Barford D., Scott J. D. (2011) Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc. Natl. Acad. Sci. U.S.A. 108, 6426–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Amieux P. S., McKnight G. S. (2002) The essential role of RI α in the maintenance of regulated PKA activity. Ann. N.Y. Acad. Sci. 968, 75–95 [DOI] [PubMed] [Google Scholar]
- 60. Brandon E. P., Idzerda R. L., McKnight G. S. (1997) PKA isoforms, neural pathways, and behaviour: making the connection. Curr. Opin. Neurobiol. 7, 397–403 [DOI] [PubMed] [Google Scholar]
- 61. Hernández F., Langa E., Cuadros R., Avila J., Villanueva N. (2010) Regulation of GSK3 isoforms by phosphatases PP1 and PP2A. Mol. Cell Biochem. 344, 211–215 [DOI] [PubMed] [Google Scholar]