Abstract
G protein-coupled receptors (GPCRs) are allosteric proteins, because their signal transduction relies on interactions between topographically distinct, yet conformationally linked, domains. Much of the focus on GPCR allostery in the new millennium, however, has been on modes of targeting GPCR allosteric sites with chemical probes due to the potential for novel therapeutics. It is now apparent that some GPCRs possess more than one targetable allosteric site, in addition to a growing list of putative endogenous modulators. Advances in structural biology are also shedding new insights into mechanisms of allostery, although the complexities of candidate allosteric drugs necessitate rigorous biological characterization.
Keywords: allosteric regulation, chemical biology, drug discovery, G protein, G protein-coupled receptor (GPCR), structural biology, biased agonism, bitopic ligand
Introduction
G protein-coupled receptors (GPCRs),4 also known as 7-transmembrane (7TM) receptors, are the largest superfamily of cell surface receptor proteins encoded by the human genome (1). These integral membrane proteins are highly dynamic and exist in an equilibrium between various functionally distinct conformational states (2). GPCRs fulfil the vital biological function of transducing a wide range of extracellular signals (e.g. photons, lipids, neurotransmitters, hormones, peptides, enzymes, ions, odorants) across the cell membrane into the cytosolic space. Physiologically, the process begins when an endogenous extracellular signal interacts with the primary (“orthosteric”) binding site of GPCR, resulting in a conformational rearrangement that conveys the signal through the plasma membrane-spanning 7TM region and subsequently triggering intracellular signaling cascades via heterotrimeric G proteins and other accessory proteins (3). Because GPCR-mediated signaling systems are involved in regulating a multitude of physiological and pathophysiological processes, it is not surprising that the GPCR superfamily encompasses the targets of more actual and potential drugs than any other family of proteins (4, 5).
To date, the majority of probe compounds and marketed drugs that target GPCRs are small molecules, but it is noteworthy that there is a growing interest in utilizing biologics and antibodies to target these receptors as well (6). In addition, although the mode of action of the bulk of GPCR-targeting agents remains orthosteric, the turn of the millennium has witnessed substantial efforts in alternative methods of modulating GPCR activity, specifically by targeting topographically distinct allosteric sites. This minireview discusses some of the key characteristics associated with GPCR allostery and ongoing challenges and opportunities in understanding and exploiting the phenomenon.
Characteristics of GPCR Allostery
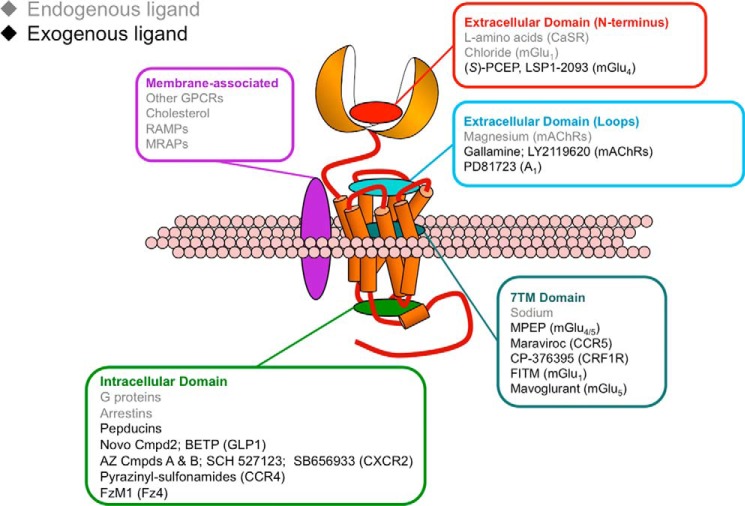
Allostery is a widespread biological phenomenon that describes the ability of interactions occurring at one site of a macromolecule to modulate interactions at a spatially distinct binding site on the same macromolecule in a reciprocal manner. Since allosteric effects were first described in archetypal examples, such as the heme-heme interactions of hemoglobin, allostery has been acknowledged as a means by which proteins and other molecules (e.g. DNA) may amplify, attenuate, bias, and otherwise fine-tune their physiological functions (2, 7–9). Initial observations of allosteric phenomena in enzymes were mechanistically summarized first in the Monod-Wyman-Changeux (MWC) and subsequently in the Koshland-Nemethy-Filmer (KNF) models (10, 11). Although the MWC model depicts allostery as a concerted process (i.e. conformational selection), and the KNF model describes it as a sequential process (i.e. conformational induction), each model reflects valid key aspects of the nature of allostery, which involves ligand-mediated shifts in the population of pre-existing macromolecular conformational ensembles and resulting changes in the interactive properties of the new ensembles (12). In addition to enzymes, it became apparent that other protein classes, including GPCRs, possess many of the characteristics associated with allosteric proteins (13). GPCRs are conformationally dynamic proteins that act as conduits for the transfer of energy over a distance. Indeed, GPCR signal transduction is intrinsically allosteric as it involves the binding of an extracellular stimulus and subsequent propagation of the signal through the protein to a topographically distinct (e.g. ∼50 Å) intracellular site recognized by G proteins, β-arrestins, and others. Moreover, because of the broad diversity of endogenous activators of GPCRs, an orthosteric region on one type of receptor (e.g. class A biogenic amine receptor) may represent an allosteric domain in another type of receptor (e.g. class B secretin family or class C glutamate family receptors) (14) (Fig. 1).
FIGURE 1.
Potential allosteric ligand-binding regions on GPCRs; some representative allosteric modulators recognizing each region are also listed. RAMP, receptor activity-modifying protein; MRAP, melanocortin receptor accessory protein; Cmpd, compound; AZ, AstraZeneca; Fz4, frizzled4 receptor; CaSR, calcium-sensing receptor; PCEP, 3-amino-3-carboxypropyl-2′-carboxyethyl phosphinic acid.
Perhaps not surprisingly, therefore, the attractiveness and tractability of GPCRs as drug targets, coupled with advances in drug screening at these receptors, have uncovered different types of allosteric GPCR modulators (Table 1) that are generally classified operationally based on their modes of pharmacology. An allosteric ligand that potentiates an agonist-mediated receptor response is referred to as a “positive allosteric modulator” (PAM), whereas one that attenuates activity is known as a “negative allosteric modulator” (NAM) (15). Mechanistically, these effects can be achieved through modulation of the binding affinity of orthosteric ligands and/or through changes in the ability of the orthosteric ligand-occupied receptor complex to interact with intracellular transducer proteins. In contrast a ligand that binds at the allosteric site without affecting receptor or orthosteric ligand activity (at equilibrium) is classed as “neutral allosteric ligand” (NAL). “Allosteric agonists” are ligands that are capable of directly activating the receptor from an allosteric site even in the absence of an orthosteric agonist (15), whereas “PAM agonists” and “NAM agonists” display mixed modes of modulation and direct GPCR activation depending on the cellular context (15). More recently, “bitopic ligands” have also been described, which are defined as hybrid molecules possessing separate orthosteric and allosteric pharmacophores that concomitantly engage with their respective sites on a single GPCR to mediate novel pharmacology; several molecules initially classified as allosteric agonists in the literature have since been reclassified as bitopic ligands (15–18).
TABLE 1.
Representative allosteric modulators of G protein-coupled receptors
AZ, AstraZeneca; BCQA, benzyl quinolone carboxylic acid; BMS, Bristol-Myers Squibb; Cmpd, compound; CGRP, calcitonin gene-related peptide; FSHR, follicle-stimulating hormone receptor; GH, growth hormone; GLP1R, glucagon-like peptide 1 receptor; GnRH, gonadotropin-releasing hormone; HMA, 5-(N,N-hexamethylene)amiloride; P2Y12, purine P2Y12; RXFP, relaxin family peptide.
| Receptor | Exogenous modulator(s) | Endogenous modulator(s)a |
|---|---|---|
| Class A | ||
| 5-HT1A | Anandamide; cholesterol | |
| 5-HT1B | 5-HT-moduline; sodium | |
| 5-HT1D | 5-HT-moduline | |
| 5-HT2A | Anandamide; oleamide; sodium | |
| 5-HT2C | PNU-69176E | Anandamide; oleamide |
| 5-HT4 | IgG | |
| 5-HT7 | Anandamide | |
| Adenosine A1 | LUF5484; PD 71,605; PD 81,723; PD 117,975 | Sodium |
| Adenosine A2A | Amilorides | Sodium |
| Adenosine A3 | DU124183; VUF5455; VUF8504 | 2-arachidonylglycerol; sodium |
| Adrenergic α1 | Amilorides; benzodiazepines; conopeptide; ρ-TIA | IgG; sodium |
| Adrenergic α2A | Amilorides | Cholesterol; sodium |
| Adrenergic α2B | Amilorides | Sodium |
| Adrenergic α2D | Agmatine; sodium | |
| Adrenergic β1 | Magnesium; manganese; IgG | |
| Adrenergic β2 | Cholesterol; IgG; zinc | |
| Angiotensin AT1 | IgG; sodium | |
| Cannabinoid CB1 | Org 27569; Org 27759; Org 29647; PSNCBAM-1; RTI-371 | Lipoxin A4; Pepcan-12; pregnenolone; sodium |
| Chemokine CCK1 | Benzodiazepines | |
| Chemokine CXCR1 | AZ Cmpds. A & B; Reparixin; SCH 527123 | |
| Chemokine CXCR2 | AZ Cmpds. A & B; Reparixin; SCH 527123; SB 656933 | |
| Chemokine CXCR3 | IP-10; I-TAC | |
| Chemokine CXCR4 | ASLW; RSVM; trichosanthin; plerixafor | |
| Chemokine CXCR7 | AMD3100; GSLW | |
| Chemokine CCR1 | BX-471; CP-481715; UCB35625 | |
| Chemokine CCR2 | CCR2-RA-[R]; JNJ-27141491; S.D.-24 | |
| Chemokine CCR3 | TAK779; UCB35625 | |
| Chemokine CCR4 | Pyrazinyl sulfonamides | |
| Chemokine CCR5 | AK602; AK530; ancriviroc; aplaviroc; maraviroc SCH 351125; TAKK220; TAK779; Trichosanthin; vicriviroc; | |
| Cholecystokinin CCK1 | Devazepide; T-0632; GI181771X; Bdz-1 | |
| Cholecystokinin CCK2 | GI181771X; Bdz-2 | |
| Dopamine D1 | Zinc | |
| Dopamine D2 | Amilorides; l-prolyl-l-leucyl-glycinamide; PAOPA SB269652; | Homocysteine; IgG; melanotropin release inhibiting factor 1; zinc |
| Endothelin ETA | Acetylsalicylic acid; sodium salicylate | IgG, sodium |
| Free fatty acid FFAR1 | AM-1638; TAK-875 | Docosahexaenoic acid |
| Free fatty acid FFAR2 | Phenylacetamides 1 & 2 | |
| Free fatty acid FFAR3 | hexahydroquinolone-3-carboxamide Cmpd 1 | |
| FSHR | BMS cmpds. 2–7 | |
| GnRH receptor | Furan-1; FD-1; HMA | |
| GH secretagogue | l-692,429; GH-releasing peptide 6 | |
| Luteinizing hormone | Org 41841; [3H]-Org 43553 | |
| M1 mAChR | Brucine; BQCA; MK7622; ML169; MT3; MT7; staurosporine; tacrine VU0029627; VU0119498 | Arachidonic acid; IgG; sodium |
| M2 mAChR | Alcuronium; C7/3-phth; DUO3; gallamine; LY2033298; LY2119620; tacrine; W84 | Arachidonic acid; dynorphin-A; IgG; myelin basic protein; major basic protein; protamine; sodium |
| M3 mAChR | Brucine; N-chloromethyl; WIN62577; VU0119498 | Arachidonic acid; IgG; sodium |
| M4 mAChR | Alcuronium; LY2033298; LY2119620; MT3; Thiochrome; VU0010010; VU0152099; VU0152100 | Arachidonic acid; sodium |
| M5 mAChR | ML326; ML375; ML380; ML381; VU0119498; VU0238429 | Arachidonic acid |
| Neurokinin 1 | Heparin | |
| Neurokinin 2 | [N-Benzyl, N- (2-naphthylmethyl)-amino]-acetonitrile | |
| μ-opioid | BMS-986121; BMS-986122; Cannabidiol | Magnesium; manganese; sodium |
| δ-opioid | BMS-986187; BMS-986188; Cannabidiol | Manganese; sodium |
| κ-opioid | Magnesium; manganese; sodium | |
| Oxytocin | Cholesterol; Progesterone (rat); 5-dihydroprogesterone (human) | |
| Purine P2Y12 | 2,2′-Pyridylsatogen tosylate | |
| Rhodopsin | Cholesterol | |
| RXFP1 | ML290 | |
| RXFP3 | 135PAM1 | |
| S1PR2 | CYM-5520 | |
| S1PR3 | CYM-5541 | |
| Thyrotropin receptor | IgG | |
| Class B | ||
| Calcitonin receptor | Pyrazolopyrimidines 2d, 2e, 2f, 2g | |
| CGRP receptor | BIBN4096BS | |
| CRF1 receptor | Antalarmin; CP-376395; NBI 35965; DMP696; NBI 27914 | |
| Glucagon | Bay27–9955; l-168,049 | |
| GLP1R | Novo Nordisk Compounds 1–6; BETP | |
| Class C | ||
| Calcium-sensing receptor | Fendeline; cinacalcet; NPS 467; NPS 568; NPS 2143; Calhex 231 | l-Phe; l-Trp; l-Tyr; Glutathione; IgG |
| GABAB | CGP7930; CGP13501; GS39783 | l-Leu; l-Ile; l-Phe; IgG |
| mGlu1 | (−)-CPCCOEt; [3H]R214127; BAY36–7620; cis-64a; EM-TBPC; JNJ16259685; NPS2390; PHCCC; Ro01–6128; Ro67–7476; Ro0711401; YM-298198 | IgG |
| mGlu2 | BINA; CBiPES; JNJ-40068782; LY181837; LY2607540; LY487379; MNI-137; RO4491533; RO4988546; RO5488608 | |
| mGlu3 | AZD8529; MNI-137; RO4491533; RO4988546; RO5488608 | |
| mGlu4 | SIB-1893; MPEP; PHCCC; VU0080421; VU0155041; VU0155094; VU0422288 | |
| mGlu5 | 5-MPEP; 5PAM523; ADX-47273; CDPPB; CPPHA; DFB; DMeOB; DCB; DPFE; Fenobam; Mavoglurant; M-5MPEP; MPEP; MTEP; VU-29; VU0357121; VU0360172 | IgG |
| mGlu6 | VU0155041; VU0422288; PHCCC | |
| mGlu7 | ADX71743; AMN082; MMPIP; VU0155041; VU0155094; VU0422288 | |
| mGlu8 | AZ12216052; VU0155041; VU0155094; VU0422288 | |
| Taste T1R1 | S807; IMP | |
| Taste T1R2 | S819; S.E.-2; S.E.-3; Senomyx | |
| Taste T1R3 | Cyclamate, lactisole | |
| Class F | ||
| Smoothened | Sant1; Sant2; nat-20(S)-OHC | Oxysterols |
| Frizzled4 | FzM1 | |
a Note that some of these examples are more appropriately considered putative endogenous allosteric modulators.
There are a number of general characteristics associated with allosteric GPCR modulators that present both unique advantages over orthosteric ligands as well as challenges to successful detection or validation of allosteric compounds. The first characteristic is the potential for allosteric ligands to exhibit greater receptor subtype selectivity. This property has two potential origins: i) a decreased evolutionary pressure for sequence conservation within allosteric sites relative to the orthosteric site between GPCR subtypes (assuming there is no endogenous allosteric ligand for such a site, but see the next section) and/or ii) selective cooperativity with an orthosteric site at one receptor subtype while exhibiting neutral cooperativity at other subtypes of that receptor family (19). The nature of the cooperativity between orthosteric and allosteric sites on a GPCR also represents a second important characteristic of allostery that has practical and therapeutic implications. If the modulator displays minimal direct allosteric agonism in its own right, then it will act as a PAM or NAM only when and where the endogenous ligand is released, thus maintaining the natural spatiotemporal “rhythms” of the endogenous orthosteric ligand. Furthermore, very subtle degrees of positive or negative cooperativity (which may be all that is necessary for certain GPCRs and disease states) result in an allosteric “effect ceiling” that increases the likelihood of on-target safety in overdose situations, although this also poses a challenge for the screening of modulators with low degrees of cooperativity (20). The ability to achieve unprecedented modes of on-target selectivity and/or fine-tune endogenous responses as a consequence of pure PAM or NAM activity may prove particularly important in diseases where tight physiological regulation is vital, such as neurodegeneration, schizophrenia, diabetes, and endocrine disorders.
A particularly interesting phenomenon associated with allostery that has been most noted at GPCRs is the property of “probe dependence,” wherein the magnitude and direction of an allosteric effect can change depending on the nature of the interacting ligands (15, 21). Probe dependence has substantial implications for GPCR drug discovery and GPCR biology. For instance, many GPCRs have more than one endogenous orthosteric agonist, and totally different effects can be observed (or missed altogether) depending on which agonist is used to activate the receptor in the presence of a given allosteric modulator (22). In addition, the allosteric nature of GPCR signal transduction means that the ability of a ligand-bound receptor to recognize cellular effector molecules is also subject to probe dependence, this time directed intracellularly rather than between two ligands. Specifically, the ability of orthosteric or allosteric GPCR ligands to stabilize different functionally relevant conformations of the same GPCR can give rise to the phenomenon of “biased agonism,” whereby only a subset of the possible signaling repertoire of the receptor is recruited at the relative expense of other pathways (21, 23, 24).
Although beyond the scope of the current minireview, the allosteric pharmacology of bitopic ligands also presents a unique set of characteristics. Because of their dual pharmacophore nature, bitopic ligands can provide both greater selectivity through interaction with an allosteric site and higher affinity through concomitant engagement of the orthosteric site. A judicious choice of orthosteric and allosteric building blocks can also yield bitopic ligands that display novel biased agonism (25). Although the spatiotemporal control of endogenous signaling afforded by pure allosteric modulator ligands is lost with bitopic ligands, the latter may prove particularly useful in situations where endogenous agonist tone is progressively lost (e.g. neurodegenerative disorders) (18).
Endogenous Allosteric Modulators
Although most studies of GPCR allostery have traditionally focused on the actions of exogenous allosteric modulators because of the implications for novel drug discovery (see below), these receptors can also be modulated by a variety of endogenous substances (26). As mentioned above, the best characterized allosteric interaction at GPCRs is the positive cooperativity exhibited between the intracellular G protein-binding site with the orthosteric site (3). The GPCR intracellular face can also interact with β-arrestins: endogenous GPCR accessory proteins that were originally characterized as scaffolding proteins involved in the termination of GPCR signaling, internalization, and recycling of receptors. It is now recognized that β-arrestins can be involved in G protein-independent signal transduction (27–30). These unique signaling properties emerge as a result of specific receptor conformations that have been shown to interact more readily with β-arrestins, leading to increased β-arrestin binding and biased signaling through non-canonical signaling pathways (31–33). Although beyond the scope of the current review, it is also widely acknowledged that different types of GPCRs have the potential to associate with other proteins, such as receptor activity-modifying proteins (RAMPs) or melanocortin receptor accessory proteins (MRAPs), or with each other in the form of homo- or hetero-dimers (or higher order oligomers) with novel pharmacological properties (26). In a number of such instances, cooperative interactions have been noted between orthosteric ligands within such complexes (34).
In addition to the ubiquitous allosteric sites utilized by G proteins and β-arrestins, individual GPCR classes or subclasses possess more specific allosteric sites that may be targeted by endogenous allosteric modulators. For example, pharmacological and crystallographic evidence has shown that sodium is vital for stabilizing the inactive state of many class A GPCRs via an allosteric site centered on a highly conserved aspartate residue, Asp2.50 (26). Additionally, aromatic amino acids (e.g. l-phenylalanine, l-tryptophan, and l-tyrosine) bind at a site near to, but spatially distinct from, the orthosteric site within the “Venus flytrap” (VFT) N-terminal domain of the calcium-sensing class C GPCR to potentiate the actions of extracellular calcium at a number of intracellular signaling pathways (35–37). A similar allosteric site has been shown to be located in the Venus flytrap of the metabotropic glutamate (mGlu) class C GPCRs. This site was initially reported as an allosteric chloride-binding site in the mGlu1 subtype, but some synthetic small molecule agonists have since been reported to bind this site in mGlu4 (38–40).
As summarized in Table 1, a growing number of substances encompassing not only amino acids and ions, but also lipids, peptides, and proteins, have been proposed to act as putative endogenous allosteric modulators of different types of GPCRs (26). It should be noted that in many of these instances, conclusive validation of an allosteric mechanism remains to be established, but the study of endogenous GPCR modulators may prove to be a fertile ground for uncovering novel biology in health and disease. For instance, the nature and composition of endogenous lipidic substances and numerous types of peptides and proteins can vary dramatically in inflammation; if it can be shown that some of these substances are bona fide allosteric modulators of specific GPCRs, then this may represent a novel avenue for understanding and targeting GPCR functionality in inflammatory disease (26). In addition, GPCR-directed autoantibodies have been identified as potential endogenous allosteric ligands related to a number of chronic disease states. In particular, autoantibodies have been identified in patients with a variety of cardiovascular diseases, including autoantibodies against the AT1 receptor, the β1-adrenergic receptor, the M2 muscarinic acetylcholine receptor (mAChR), the α1-adrenergic receptor, the ETA receptor, and the 5-HT4 receptor (26). Additionally, the central nervous system can be affected by autoantibody activity. For instance, autoantibodies against GABAB have been found in the cerebrospinal fluid of limbic encephalitis patients, whereas patients with basal ganglia encephalitis with dominant movement and psychiatric disease present with autoantibodies against the dopamine D2 receptor (26).
Exogenous Allosteric Modulators
Numerous synthetic small molecules with an allosteric mode of action have been reported for GPCRs (Table 1). The mAChR family is arguably the most well studied class A GPCR system in this regard (41). Indeed, the first examples of GPCR allosteric modulators, the alkane bis-ammonium family of ligands, were identified at the mAChRs (42). Since that time, allosteric modulators of nearly every mode of action have been found to target the mAChRs, including PAMs, NAMs, PAM and NAM agonists, and bitopic ligands (17, 43–48). Interestingly, most of these ligands bind to the mAChRs at a shared site (albeit with different affinities depending on the subtype), often referred to as the “common” allosteric site and highlighting how the targeting of a common allosteric domain can yield markedly different biological behaviors (49, 50). However, evidence has been provided for the existence of a second allosteric site on the mAChRs, recognized by indolocarbazole analogues of staurosporine (e.g. KT5720 and KT5823) and the benzimidazole derivatives WIN63577 and WIN51708. As noted with the “common site” modulators, the second-site compounds also demonstrated positive, negative, or neutral interactions with acetylcholine depending on the mAChR subtype, but showed largely neutral interactions with the binding of common site modulators, further supporting the presence of at least two allosteric sites on a single GPCR (51–54).
The notion that a single GPCR may possess more than one allosteric site for exogenous small molecules is likely more widespread. For instance, a number of allosteric modulators have been described for the gonadotropin-releasing hormone receptor, including Furan-1 and its derivative, FD-1 (55), as well as a series of amiloride derivatives (56). Interestingly, interaction studies between the two different classes of allosteric ligand revealed neutral cooperativity, despite each class individually modulating orthosteric ligands (56), again suggestive of more than one allosteric site on this receptor family. The intracellular face of GPCRs may also harbor allosteric sites for selective small molecules. For instance, binding sites for allosteric antagonists of the CXCR2, CCR4, and CCR5 chemokine receptors have been identified among the intracellular loops (57–59). Intracellular allosteric sites are also utilized by pepducins: cell-penetrating, lipidated peptides that target the intracellular loops. By tailoring the peptide's design to the intracellular domain of a particular GPCR, researchers have been successful in producing pepducins selectively targeting a number of GPCRs involved in inflammatory diseases, including protease-activated receptors (PAR1, -2, -4) and chemokine receptors (CXCR1, -2, -4) (60, 61).
Class B GPCRs, such as the secretin, glucagon, and glucagon-like peptide 1 receptors, have proven notoriously intractable to small molecule discovery and have remained an area of intense research with regard to allosteric drugs. One common observation is the discovery of direct-acting allosteric agonists, in addition to PAM agonists or NAMs (62). Pure class B GPCR PAMs have thus far remained relatively more elusive, but whether this reflects a fundamental property of this class of GPCR or the relative immaturity of detailed studies of Class B allostery remains to be determined. In common with the preceding examples, however, there is clear evidence suggesting the presence of more than one allosteric site on Class B GPCRs (63, 64).
Within the class C GPCRs, the mGlu receptors have a very rich allosteric pharmacology. In particular, allosteric modulation of the mGlu5 receptor has been of interest as a target for the treatment of schizophrenia and Alzheimer disease (65, 66). As such, multiple small molecule PAMs have been developed for mGlu5 (67–72), with the bulk acting at a common site in the upper region of the 7TM domain often termed the “MPEP-binding site” after the prototypical mGlu5 NAM, MPEP (65, 66). However, several ligands have been identified that are non-competitive with the MPEP site, including the PAMs VU357121 and CPPHA, indicating the presence of at least two distinct allosteric sites (67, 69, 73, 74). Further investigations have shown that PAMs acting on each of these sites may have varied effects on different signaling pathways, indicating that allosteric modulation of discrete allosteric sites may have significant effects on the response of the GPCR (75, 76).
Finally, the Class F GPCR family, which includes the Smoothened and Frizzled receptors, has also been considered difficult to target with small molecules, but recent breakthroughs have revealed that these receptors possess allosteric binding sites for exogenous ligands. For example, a series of small molecules that were originally designed to act as pharmacological chaperones for a misfolded mutant of Frizzled4 were subsequently identified as novel allosteric modulators of the wild-type form of Frizzled4; the binding site for these compounds was proposed to be located in the vicinity of intracellular loop 3 of the receptor (77). The recent crystal structure of the Smoothened receptor bound to the allosteric modulator, Sant1, also revealed an allosteric pocket that is located deep within the transmembrane-spanning cavity of the receptor, toward the cytosolic end; this is in contrast to the binding site that is closer to the extracellular entrance and utilized by canonical ligands of this receptor (78).
It is important to note that although small molecules have made an undeniable impact on the study of allostery, there is growing interest in utilizing biologics to target allosteric sites. Thus far, allosteric antibodies have shown the most promise in targeting receptor tyrosine kinases, such as the insulin receptor, where the monoclonal antibody XMetA allosterically binds and activates the receptor (79, 80). A subcategory of allosteric antibodies known as “allosteric ligand-modifying antibodies” (ALMA) acts by binding the endogenous ligand prior to interacting with the receptor (15). One example is gevokizumab, an anti-interleukin-1β antibody that binds interleukin and alters the conformation of the endogenous ligand. Subsequently, the gevokizumab-interleukin-1β complex binds in a characteristic ternary complex with the receptor (81, 82). Although examples of rationally designed allosteric antibodies targeting GPCRs have yet to be described, the novel mechanisms of allosteric antibodies and allosteric ligand-modifying antibodies offer a valuable new strategy for the study of GPCR allostery in the future.
Chemical Biology Challenges in Designing Allosteric Modulators of GPCRs
The production of highly site-specific allosteric probe molecules is a crucial factor in the study of allosteric interactions. Structure-based drug design is still a relatively nascent field with respect to GPCRs, although substantial progress has occurred in recent years (see below). Within both industry and academia, a large number of resources have been dedicated to the generation, collection, and curation of large libraries of natural and synthetic small molecules from which to screen for and optimize novel allosteric ligands, and this remains the major source of such compounds (83, 84). Given this significant investment, the methodologies with which these novel ligands are interrogated are central to any research campaign. The field of chemical biology has provided a number of tools to probe allosteric mechanisms of GPCR structure and function. These techniques may provide the biochemical and biophysical information necessary to garner a clear understanding of the structural dynamics and signaling behavior of allosteric interactions on a chemical level. Techniques such as bioluminescent and Förster resonance energy transfer as well as single-molecule detection fluorescence have gained popularity for the study of GPCRs and validated binding partners. However, there are limitations for using these techniques in the process of screening for novel ligands, not least of which is the challenge of applying these methods to a high throughput screening format (85, 86). Measurement of second messengers of G protein signaling (e.g. calcium and cAMP) or β-arrestin recruitment has proven more successful in this respect; nevertheless, it remains challenging to develop selective small molecule allosteric modulators.
For example, allosteric ligands often possess delicate structure-activity relationships. That is, in a given chemical series, a seemingly minor modification to a molecule's steric or electronic properties often leads to the complete ablation of its activity. Similarly, certain chemical series demonstrate “mode switching” whereby small modifications to the structure can result in dramatically changed pharmacological profiles (87). These concerns are particularly relevant when further modifications are applied to the probes, for instance in the generation of irreversible or photoactivatable allosteric molecules (88, 89), or through the use of these chemical probes in vivo, where biochemical transformations undertaken by metabolic processes may alter a modulator's potency, cooperativity, receptor selectivity, or mode of action.
Novel chemotypes of allosteric modulators also pose challenges to pharmacological characterization. For instance, many allosteric modulators display phenomena such as probe dependence, and can impose biased signaling on the actions of orthosteric ligands (Table 2). These may go unnoticed with the use of a single screening methodology. Only through the measurement of an allosteric ligand's effect on multiple downstream pathways in the presence of all relevant endogenous orthosteric ligands can a compound's properties be fully elucidated, and this must be factored into all allosteric discovery programs (90). Further complexities in characterizing a novel allosteric ligand may be caused by differences between species' receptor isoforms, differential effects on orthosteric ligand affinity and efficacy, varied kinetics between different signaling pathways, and the possibility of undetected endogenous allosteric interactions. Therefore efforts must be made to fully characterize and validate novel chemical probes in as many experimental paradigms as possible to compose the clearest picture of the compound's properties.
TABLE 2.
Representative examples of biased allosteric modulation
CaSR, calcium-sensing receptor; Oxo, oxotremorine (1-(4-pyrrolidin-1-ylbut-2-yn-1-yl)pyrrolidin-2-one); Oxo-M, oxotremorine methiodide (N,N,N-trimethyl-4-(2-oxo-1-pyrolidinyl)-2-butyn-1-ammonium iodide); PLD, phospholipase D; TMA, tetramethylammonium.
| Ligand | Receptor | Bias profile (and orthosteric agonist probe) |
|---|---|---|
| Cinacalcet | CaSR | PAM for intracellular Ca2+ mobilization (Ca2++) |
| NAL for ERK1/2 phosphorylation (Ca2+) | ||
| LPI805 | Neurokinin 2 | NAM for cAMP production (Neurokinin A) |
| NAL for intracellular Ca2+ mobilization (Neurokinin A) | ||
| 2-amino-3-benzothiophene derivative (MIPS#?) | Adenosine A1 | Allosteric agonist for cAMP production |
| VU0029767 | M1 | PAM for intracellular Ca2+ mobilization (ACh) |
| NAL for PLD activation (ACh) | ||
| Weak PAM for PI hydrolysis (ACh) | ||
| LY203398 | M2 | PAM for ERK1/2 phosphorylation (ACh, Oxo, Oxo-M, TMA, or McN-343) |
| NAM for ERK1/2 phosphorylation (Pilocarpine or Xanomeline) | ||
| Brucine (mutant) | M3K7.32E | Allosteric PAM agonist for Gαq (CCh) |
| PAM for Gα12 (CCh) | ||
| NAL for Gαi (CCh) | ||
| 1-(4-Ethoxyphenyl)-5-methoxy-2-methylindole-3-carboxylic acid and Nα-tosyltryptophan | CTRH2 | NAMs for arrestin recruitment (prostaglandin D2) |
| NALs for G protein coupling | ||
| CPPHA | mGlu5 | PAM for intracellular Ca2+ mobilization (Glutamate or DHPG) |
| NAM for ERK1/2 phosphorylation (Glutamate or DHPG, low concentrations) | ||
| NAM for ERK1/2 phosphorylation (Glutamate or DHPG, high concentrations) | ||
| M-5MPEP | mGlu5 | NAM for intracellular Ca2+ mobilization (quisqualate or DHPG) |
| Weak NAM for PI hydrolysis (quisqualate or DHPG) | ||
| PDC113.824 | PGF2α | NAM for Gα12 (PGF2α) |
| PAM for Gαq (PGF2α) | ||
| ORG27569 | CB1 | NAM for Gαi (CP55940) |
| Allosteric agonist for ERK1/2 phosphorylation |
What Can Structural Biology Reveal About Allosteric Mechanisms?
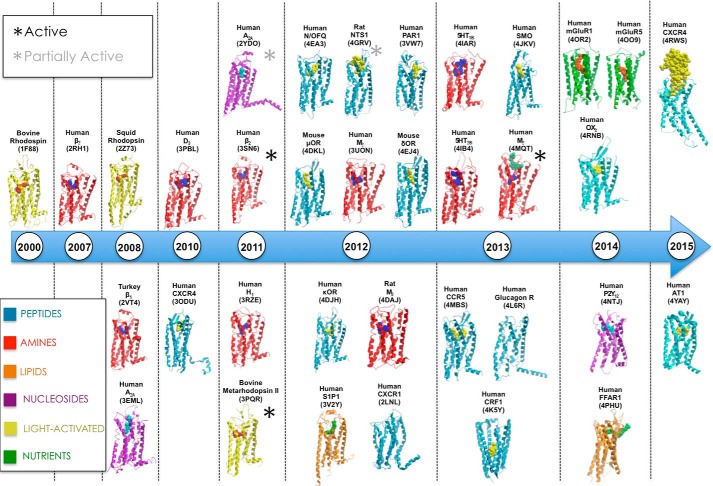
Although the use of selective allosteric probe molecules and functional assays can reveal much about allosteric pharmacology, the information provided by crystal structures and other high resolution approaches is invaluable for more direct, molecular level insights into GPCR allostery. Since the turn of the millennium, when the first high resolution GPCR crystal structure was solved for bovine rhodopsin (91), there has been a seemingly exponential growth in the number of crystal structures solved for a range of class A, B, and C GPCRs (Fig. 2), although the majority of these rely on some form of protein engineering to improve stability and crystal formation. This has allowed the observation of conformational snapshots adopted by structures co-bound to ligands and/or interacting proteins, offering an unprecedented view into the fundamental structural basis for receptor function.
FIGURE 2.
Timeline of representative GPCR crystal structures, 2000–2015. N/OFQ, nociception/orphanin FQ receptor; SMO, Smoothened receptor; OR, opioid receptor; H1, histamine H1 receptor; S1P1, sphingosine-1-phosphate 1 receptor.
Of note, there have been a number of inactive state crystal structures of GPCRs in binary complexes with small molecule NAMs. These include the chemokine receptor CCR5 bound to maraviroc, the corticotropin-releasing factor receptor CRF1 bound to CP-376395, the mGlu1 bound to FITM, the mGlu5 bound to mavoglurant, and the aforementioned Smoothened receptor bound to Sant1 (78, 92–95). Despite providing new insights into the binding behavior of these NAMs, the inactive and binary complex natures of these structures do not capture the complete structural mechanisms that underlie cooperativity between the allosteric and orthosteric sites. Indeed, the CRF1 and mGlu structures lack the key N-terminal domains that constitute much (or all) of the orthosteric binding site.
A major breakthrough in recent years has been the crystal structure depicting the classic ternary complex of an orthosteric agonist binding the β2-adrenergic receptor coupled to a Gs protein (96). This structure offered the first observation of how a GPCR orthosteric site is allosterically coupled to G protein activation. In terms of small molecule allostery, the recent structure of an active M2 mAChR in complex with the high efficacy orthosteric agonist, iperoxo, and the PAM, LY02119620 (97), also represented a major structural biology advance in understanding allostery at a GPCR. For example, the structure is consistent with the predictions of the MWC model of allostery in that the PAM preferentially recognizes and stabilizes a preformed active state of the receptor. Nonetheless, these structures still represent first steps in our molecular level understanding of mechanisms underlying GPCR allostery. Phenomena such as biased allosteric modulation, probe dependence, and the actual mechanisms underlying transmission of cooperativity remain challenging as they require the ability to capture multiple states in the absence and presence of multiple ligands. In the meantime, additional insights into the structural and dynamic mechanisms of allostery are being obtained via other methods. For example, the inactive M2 mAChR structure has been subjected to long timescale molecular dynamics simulations with a diverse range of small molecule allosteric modulators, in the absence or presence of an orthosteric ligand, to identify binding poses and mechanisms underlying cooperativity for a broad set of NAMs (98). The use of NMR has also allowed researchers to study the conformational flexibility of the receptor as a whole, providing more information on receptor dynamics than can be obtained through static crystal structures, although no study has directly applied this approach to GPCR allosteric modulators to date (99, 100).
Conclusions
Allosteric modulation of GPCRs is now a widely accepted phenomenon with substantial implications for novel drug discovery, yet many fundamental issues remain to be addressed. For example, it is now clear that a single GPCR can possess more than one allosteric site, but whether the targeting of such sites can lead to differential behaviors remains unknown. The prevalence of endogenous allosteric modulators remains to be determined, but they may prove to be a mechanism of tissue-specific regulation in normal physiology or disease that may be amenable to chemical manipulation. The ascendance of biologics as therapeutics opens new vistas for targeting of GPCRs with a greater degree of specificity than previously possible, yet the extent with which such substances can interact allosterically with GPCRs is largely unexplored. As more detailed GPCR structural information becomes available, it too will profoundly affect our understanding of allostery in GPCRs as well as the manner by which allosteric molecules are designed. In preparing for such an eventuality, the rigorous biological characterization of each new allosteric site and allosteric ligand discovered remains paramount.
Author Contributions
All authors wrote the manuscript.
This work was supported by Program Grant APP1055134 of the National Health and Medical Research Council (NHMRC) of Australia. This is the second article in the Thematic Minireview series “New Directions in G Protein-coupled Receptor Pharmacology.” The authors declare that they have no conflicts of interest with the contents of this article.
- GPCR
- G protein-coupled receptor
- 7TM
- 7-transmembrane
- 5-HT
- 5-hydroxytryptamine
- CCh
- carbachol (2-[(aminocarbonyl)oxy]-N,N,N-trimethylethanaminium chloride)
- CCR
- chemokine CC motif receptor
- CRF1
- corticotropin-releasing factor 1
- CXCR
- CXC motif chemokine receptor
- DHPG
- dihydroxyphenylglycine
- FFAR
- free fatty acid receptor
- GnRH
- gonadotropin-releasing hormone
- mAChR
- muscarinic acetylcholine receptor
- mGlu
- metabotropic glutamate receptor
- PGF2α
- prostaglandin F2α
- MWC
- Monod-Wyman-Changeux
- KNF
- Koshland-Nemethy-Filmer
- PAM
- positive allosteric modulator
- NAL
- neutral allosteric ligand
- NAM
- negative allosteric modulator
- MPEP
- 2-methyl-6-(phenylethynyl)pyridine.
REFERENCES
- 1. Fredriksson R., Lagerström M. C., Lundin L.-G., Schiöth H. B. (2003) The G-protein-coupled receptors in the human genome form five main families: phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 2. Kenakin T., Miller L. J. (2010) Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol. Rev. 62, 265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oldham W. M., Hamm H. E. (2006) Structural basis of function in heterotrimeric G proteins. Q. Rev. Biophys. 39, 117–166 [DOI] [PubMed] [Google Scholar]
- 4. Rask-Andersen M., Almén M. S., Schiöth H. B. (2011) Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 10, 579–590 [DOI] [PubMed] [Google Scholar]
- 5. Conn P. J., Lindsley C. W., Meiler J., Niswender C. M. (2014) Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat. Rev. Drug Discov. 13, 692–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 7. Monod J., Changeux J.-P., Jacob F. (1963) Allosteric proteins and cellular control systems. J. Mol. Biol. 6, 306–329 [DOI] [PubMed] [Google Scholar]
- 8. Christopoulos A., Kenakin T. (2002) G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54, 323–374 [DOI] [PubMed] [Google Scholar]
- 9. Fenton A. W. (2008) Allostery: an illustrated definition for the “second secret of life.” Trends Biochem. Sci. 33, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monod J., Wyman J., Changeux J.-P. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 11. Koshland D. E., Jr., Némethy G., Filmer D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 [DOI] [PubMed] [Google Scholar]
- 12. Motlagh H. N., Wrabl J. O., Li J., Hilser V. J. (2014) The ensemble nature of allostery. Nature 508, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canals M., Sexton P. M., Christopoulos A. (2011) Allostery in GPCRs: “MWC” revisited. Trends Biochem. Sci. 36, 663–672 [DOI] [PubMed] [Google Scholar]
- 14. Conn P. J., Christopoulos A., Lindsley C. W. (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 8, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christopoulos A., Changeux J.-P., Catterall W. A., Fabbro D., Burris T. P., Cidlowski J. A., Olsen R. W., Peters J. A., Neubig R. R., Pin J.-P., Sexton P. M., Kenakin T. P., Ehlert F. J., Spedding M., Langmead C. J. (2014) International union of basic and clinical pharmacology. XC. Multisite pharmacology: recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol. Rev. 66, 918–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohr K., Tränkle C., Kostenis E., Barocelli E., De Amici M., Holzgrabe U. (2010) Rational design of dualsteric GPCR ligands: quests and promise. Br. J. Pharmacol. 159, 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valant C., Gregory K. J., Hall N. E., Scammells P. J., Lew M. J., Sexton P. M., Christopoulos A. (2008) A novel mechanism of G protein-coupled receptor functional selectivity. Muscarinic partial agonist McN-A-343 as a bitopic orthosteric/allosteric ligand. J. Biol. Chem. 283, 29312–29321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valant C., Robert Lane J., Sexton P. M., Christopoulos A. (2012) The best of both worlds? Bitopic orthosteric/allosteric ligands of G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 52, 153–178 [DOI] [PubMed] [Google Scholar]
- 19. Lazareno S., Dolezal V., Popham A., Birdsall N. J. M. (2004) Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: receptor subtype selectivity via cooperativity rather than affinity. Mol. Pharmacol. 65, 257–266 [DOI] [PubMed] [Google Scholar]
- 20. Christopoulos A. (2002) Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat. Rev. Drug Discov. 1, 198–210 [DOI] [PubMed] [Google Scholar]
- 21. Kenakin T. (2005) New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat. Rev. Drug Discov. 4, 919–927 [DOI] [PubMed] [Google Scholar]
- 22. Wootten D., Christopoulos A., Sexton P. M. (2013) Emerging paradigms in GPCR allostery: implications for drug discovery. Nat. Rev. Drug. Discov. 12, 630–644 [DOI] [PubMed] [Google Scholar]
- 23. Leach K., Sexton P. M., Christopoulos A. (2007) Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol. Sci. 28, 382–389 [DOI] [PubMed] [Google Scholar]
- 24. Kenakin T., Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 12, 205–216 [DOI] [PubMed] [Google Scholar]
- 25. Valant C., May L. T., Aurelio L., Chuo C. H., White P. J., Baltos J.-A., Sexton P. M., Scammells P. J., Christopoulos A. (2014) Separation of on-target efficacy from adverse effects through rational design of a bitopic adenosine receptor agonist. Proc. Natl. Acad. Sci. U.S.A. 111, 4614–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Westhuizen E. T., Valant C., Sexton P. M., Christopoulos A. (2015) Endogenous allosteric modulators of G protein-coupled receptors. J. Pharmacol. Exp. Ther. 353, 246–260 [DOI] [PubMed] [Google Scholar]
- 27. Luttrell L. M., Ferguson S. S., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F., Kawakatsu H., Owada K., Luttrell D. K., Caron M. G., Lefkowitz R. J. (1999) β-Arrestin-dependent formation of β2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661 [DOI] [PubMed] [Google Scholar]
- 28. Lefkowitz R. J., Shenoy S. K. (2005) Transduction of receptor signals by β-arrestins. Science 308, 512–517 [DOI] [PubMed] [Google Scholar]
- 29. Luttrell L. M., Gesty-Palmer D. (2010) Beyond desensitization: physiological relevance of arrestin-dependent signaling. Pharmacol. Rev. 62, 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reiter E., Ahn S., Shukla A. K., Lefkowitz R. J. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gurevich V. V., Pals-Rylaarsdam R., Benovic J. L., Hosey M. M., Onorato J. J. (1997) Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J. Biol. Chem. 272, 28849–28852 [DOI] [PubMed] [Google Scholar]
- 32. Galandrin S., Oligny-Longpré G., Bonin H., Ogawa K., Galés C., Bouvier M. (2008) Conformational rearrangements and signaling cascades involved in ligand-biased mitogen-activated protein kinase signaling through the β1-adrenergic receptor. Mol. Pharmacol. 74, 162–172 [DOI] [PubMed] [Google Scholar]
- 33. Tang W., Strachan R. T., Lefkowitz R. J., Rockman H. A. (2014) Allosteric modulation of β-arrestin-biased angiotensin II type 1 receptor signaling by membrane stretch. J. Biol. Chem. 289, 28271–28283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Milligan G. (2009) G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br. J. Pharmacol. 158, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conigrave A. D., Quinn S. J., Brown E. M. (2000) l-Amino acid sensing by the extracellular Ca2+-sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 4814–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Qiu W., Quinn S. J., Conigrave A. D., Brown E. M., Bai M. (2002) Three adjacent serines in the extracellular domains of the CaR are required for l-amino acid-mediated potentiation of receptor function. J. Biol. Chem. 277, 33727–33735 [DOI] [PubMed] [Google Scholar]
- 37. Mun H. C., Culverston E. L., Franks A. H., Collyer C. A., Clifton-Bligh R. J., Conigrave A. D. (2005) A double mutation in the extracellular Ca2+-sensing receptor's Venus flytrap domain that selectively disables l-amino acid sensing. J. Biol. Chem. 280, 29067–29072 [DOI] [PubMed] [Google Scholar]
- 38. Ogawa H., Qiu Y., Philo J. S., Arakawa T., Ogata C. M., Misono K. S. (2010) Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: possible allosteric regulation and a conserved structural motif for the chloride-binding site. Protein Sci. 19, 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Selvam C., Oueslati N., Lemasson I. A., Brabet I., Rigault D., Courtiol T., Cesarini S., Triballeau N., Bertrand H. O., Goudet C., Pin J.-P., Acher F. C. (2010) A virtual screening hit reveals new possibilities for developing group III metabotropic glutamate receptor agonists. J. Med. Chem. 53, 2797–2813 [DOI] [PubMed] [Google Scholar]
- 40. Acher F. C., Selvam C., Pin J.-P., Goudet C., Bertrand H. O. (2011) A critical pocket close to the glutamate binding site of mGlu receptors opens new possibilities for agonist design. Neuropharmacology 60, 102–107 [DOI] [PubMed] [Google Scholar]
- 41. Gregory K. J., Sexton P. M., Christopoulos A. (2007) Allosteric modulation of muscarinic acetylcholine receptors. Curr. Neuropharmacol. 5, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lüllmann H., Ohnesorge F. K., Schauwecker G. C., Wassermann O. (1969) Inhibition of the actions of carbachol and DFP on guinea pig isolated atria by alkane-bis-ammonium compounds. Eur. J. Pharmacol. 6, 241–247 [DOI] [PubMed] [Google Scholar]
- 43. Clark A. L., Mitchelson F. (1976) The inhibitory effect of gallamine on muscarinic receptors. Br. J. Pharmacol. 58, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stockton J. M., Birdsall N. J., Burgen A. S., Hulme E. C. (1983) Modification of the binding properties of muscarinic receptors by gallamine. Mol. Pharmacol. 23, 551–557 [PubMed] [Google Scholar]
- 45. Proska J., Tucek S. (1994) Mechanisms of steric and cooperative actions of alcuronium on cardiac muscarinic acetylcholine receptors. Mol. Pharmacol. 45, 709–717 [PubMed] [Google Scholar]
- 46. Chan W. Y., McKinzie D. L., Bose S., Mitchell S. N., Witkin J. M., Thompson R. C., Christopoulos A., Lazareno S., Birdsall N. J. M., Bymaster F. P., Felder C. C. (2008) Allosteric modulation of the muscarinic M4 receptor as an approach to treating schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 105, 10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leach K., Loiacono R. E., Felder C. C., McKinzie D. L., Mogg A., Shaw D. B., Sexton P. M., Christopoulos A. (2010) Molecular mechanisms of action and in vivo validation of an M4 muscarinic acetylcholine receptor allosteric modulator with potential antipsychotic properties. Neuropsychopharmacology 35, 855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steinfeld T., Mammen M., Smith J. A. M., Wilson R. D., Jasper J. R. (2007) A novel multivalent ligand that bridges the allosteric and orthosteric binding sites of the M2 muscarinic receptor. Mol. Pharmacol. 72, 291–302 [DOI] [PubMed] [Google Scholar]
- 49. Christopoulos A., Lanzafame A., Mitchelson F. (1998) Allosteric interactions at muscarinic cholinoceptors. Clin. Exp. Pharmacol. Physiol. 25, 185–194 [DOI] [PubMed] [Google Scholar]
- 50. Ellis J., Seidenberg M. (2000) Interactions of alcuronium, TMB-8, and other allosteric ligands with muscarinic acetylcholine receptors: studies with chimeric receptors. Mol. Pharmacol. 58, 1451–1460 [DOI] [PubMed] [Google Scholar]
- 51. Lazareno S., Gharagozloo P., Kuonen D., Popham A., Birdsall N. J. (1998) Subtype-selective positive cooperative interactions between brucine analogues and acetylcholine at muscarinic receptors: radioligand binding studies. Mol. Pharmacol. 53, 573–589 [DOI] [PubMed] [Google Scholar]
- 52. Lazareno S., Popham A., Birdsall N. J. (2000) Allosteric interactions of staurosporine and other indolocarbazoles with N-[methyl-3H]scopolamine and acetylcholine at muscarinic receptor subtypes: identification of a second allosteric site. Mol. Pharmacol. 58, 194–207 [DOI] [PubMed] [Google Scholar]
- 53. Lazareno S., Popham A., Birdsall N. J. M. (2002) Analogs of WIN 62,577 define a second allosteric site on muscarinic receptors. Mol. Pharmacol. 62, 1492–1505 [DOI] [PubMed] [Google Scholar]
- 54. Lanzafame A. A., Sexton P. M., Christopoulos A. (2006) Interaction studies of multiple binding sites on M4 muscarinic acetylcholine receptors. Mol. Pharmacol. 70, 736–746 [DOI] [PubMed] [Google Scholar]
- 55. Sullivan S. K., Brown M. S., Gao Y., Loweth C. J., Lio F. M., Crowe P. D., Struthers R. S., Betz S. F. (2006) Allosteric and orthosteric binding modes of two nonpeptide human gonadotropin-releasing hormone receptor antagonists. Biochemistry 45, 15327–15337 [DOI] [PubMed] [Google Scholar]
- 56. Heitman L. H., Ye K., Oosterom J., Ijzerman A. P. (2008) Amiloride derivatives and a nonpeptidic antagonist bind at two distinct allosteric sites in the human gonadotropin-releasing hormone receptor. Mol. Pharmacol. 73, 1808–1815 [DOI] [PubMed] [Google Scholar]
- 57. Nicholls D. J., Tomkinson N. P., Wiley K. E., Brammall A., Bowers L., Grahames C., Gaw A., Meghani P., Shelton P., Wright T. J., Mallinder P. R. (2008) Identification of a putative intracellular allosteric antagonist binding-site in the CXC chemokine receptors 1 and 2. Mol. Pharmacol. 74, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 58. Andrews G., Jones C., Wreggett K. A. (2008) An intracellular allosteric site for a specific class of antagonists of the CC chemokine G protein-coupled receptors CCR4 and CCR5. Mol. Pharmacol. 73, 855–867 [DOI] [PubMed] [Google Scholar]
- 59. Salchow K., Bond M. E., Evans S. C., Press N. J., Charlton S. J., Hunt P. A., Bradley M. E. (2010) A common intracellular allosteric binding site for antagonists of the CXCR2 receptor. Br. J. Pharmacol. 159, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hollenberg M. D., Saifeddine M., Sandhu S., Houle S., Vergnolle N. (2004) Proteinase-activated receptor-4: evaluation of tethered ligand-derived peptides as probes for receptor function and as inflammatory agonists in vivo. Br. J. Pharmacol. 143, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaneider N. C., Agarwal A., Leger A. J., Kuliopulos A. (2005) Reversing systemic inflammatory response syndrome with chemokine receptor pepducins. Nat. Med. 11, 661–665 [DOI] [PubMed] [Google Scholar]
- 62. Bortolato A., Doré A. S., Hollenstein K., Tehan B. G., Mason J. S., Marshall F. H. (2014) Structure of Class B GPCRs: new horizons for drug discovery. Br. J. Pharmacol. 171, 3132–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen D., Liao J., Li N., Zhou C., Liu Q., Wang G., Zhang R., Zhang S., Lin L., Chen K., Xie X., Nan F., Young A. A, Wang M.-W. (2007) A nonpeptidic agonist of glucagon-like peptide 1 receptors with efficacy in diabetic db/db mice. Proc. Natl. Acad. Sci. U.S.A. 104, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nolte W. M., Fortin J.-P., Stevens B. D., Aspnes G. E., Griffith D. A., Hoth L. R., Ruggeri R. B., Mathiowetz A. M., Limberakis C., Hepworth D., Carpino P. A. (2014) A potentiator of orthosteric ligand activity at GLP-1R acts via covalent modification. Nat. Chem. Biol. 10, 629–631 [DOI] [PubMed] [Google Scholar]
- 65. Gregory K. J., Dong E. N., Meiler J., Conn P. J. (2011) Allosteric modulation of metabotropic glutamate receptors: structural insights and therapeutic potential. Neuropharmacology 60, 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gregory K. J., Conn P. J. (2015) Molecular insights into metabotropic glutamate receptor allosteric modulation. Mol. Pharmacol. 88, 188–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Brien J. A., Lemaire W., Wittmann M., Jacobson M. A., Ha S. N., Wisnoski D. D., Lindsley C. W., Schaffhauser H. J., Rowe B., Sur C., Duggan M. E., Pettibone D. J., Conn P. J., Williams D. L. (2004) A novel selective allosteric modulator potentiates the activity of native metabotropic glutamate receptor subtype 5 in rat forebrain. J. Pharmacol. Exp. Ther. 309, 568–577 [DOI] [PubMed] [Google Scholar]
- 68. Chen Y., Nong Y., Goudet C., Hemstapat K., de Paulis T., Pin J.-P., Conn P. J. (2007) Interaction of novel positive allosteric modulators of metabotropic glutamate receptor 5 with the negative allosteric antagonist site is required for potentiation of receptor responses. Mol. Pharmacol. 71, 1389–1398 [DOI] [PubMed] [Google Scholar]
- 69. Hammond A. S., Rodriguez A. L., Townsend S. D., Niswender C. M., Gregory K. J., Lindsley C. W., Conn P. J. (2010) Discovery of a novel chemical class of mGlu5 allosteric ligands with distinct modes of pharmacology. ACS Chem. Neurosci. 1, 702–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rodriguez A. L., Grier M. D., Jones C. K., Herman E. J., Kane A. S., Smith R. L., Williams R., Zhou Y., Marlo J. E., Days E. L., Blatt T. N., Jadhav S., Menon U. N., Vinson P. N., Rook J. M., Stauffer S. R., Niswender C. M., Lindsley C. W., Weaver C. D., Conn P. J. (2010) Discovery of novel allosteric modulators of metabotropic glutamate receptor subtype 5 reveals chemical and functional diversity and in vivo activity in rat behavioral models of anxiolytic and antipsychotic activity. Mol. Pharmacol. 78, 1105–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lamb J. P., Engers D. W., Niswender C. M., Rodriguez A. L., Venable D. F., Conn P. J., Lindsley C. W. (2011) Discovery of molecular switches within the ADX-47273 mGlu5 PAM scaffold that modulate modes of pharmacology to afford potent mGlu5 NAMs, PAMs and partial antagonists. Bioorg. Med. Chem. Lett. 21, 2711–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Noetzel M. J., Rook J. M., Vinson P. N., Cho H. P., Days E., Zhou Y., Rodriguez A. L., Lavreysen H., Stauffer S. R., Niswender C. M., Xiang Z., Daniels J. S., Jones C. K., Lindsley C. W., Weaver C. D., Conn P. J. (2012) Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol. Pharmacol. 81, 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao Z., Wisnoski D. D., O'Brien J. A., Lemaire W., Williams D. L., Jr., Jacobson M. A., Wittman M., Ha S. N., Schaffhauser H., Sur C., Pettibone D. J., Duggan M. E., Conn P. J., Hartman G. D., Lindsley C. W. (2007) Challenges in the development of mGluR5 positive allosteric modulators: the discovery of CPPHA. Bioorg. Med. Chem. Lett. 17, 1386–1391 [DOI] [PubMed] [Google Scholar]
- 74. Chen Y., Goudet C., Pin J.-P., Conn P. J. (2008) N-{4-Chloro-2-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-hydroxybenzamide (CPPHA) acts through a novel site as a positive allosteric modulator of group 1 metabotropic glutamate receptors. Mol. Pharmacol. 73, 909–918 [DOI] [PubMed] [Google Scholar]
- 75. Zhang Y., Rodriguez A. L., Conn P. J. (2005) Allosteric potentiators of metabotropic glutamate receptor subtype 5 have differential effects on different signaling pathways in cortical astrocytes. J. Pharmacol. Exp. Ther. 315, 1212–1219 [DOI] [PubMed] [Google Scholar]
- 76. Noetzel M. J., Gregory K. J., Vinson P. N., Manka J. T., Stauffer S. R., Lindsley C. W., Niswender C. M., Xiang Z., Conn P. J. (2013) A novel metabotropic glutamate receptor 5 positive allosteric modulator acts at a unique site and confers stimulus bias to mGlu5 signaling. Mol. Pharmacol. 83, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Generoso S. F., Giustiniano M., La Regina G., Bottone S., Passacantilli S., Di Maro S., Cassese H., Bruno A., Mallardo M., Dentice M., Silvestri R., Marinelli L., Sarnataro D., Bonatti S., Novellino E., Stornaiuolo M. (2015) Pharmacological folding chaperones act as allosteric ligands of Frizzled4. Nat. Chem. Biol. 11, 280–286 [DOI] [PubMed] [Google Scholar]
- 78. Wang C., Wu H., Evron T., Vardy E., Han G. W., Huang X.-P., Hufeisen S. J., Mangano T. J., Urban D. J., Katritch V., Cherezov V., Caron M. G., Roth B. L., Stevens R. C. (2014) Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bhaskar V., Goldfine I. D., Bedinger D. H., Lau A., Kuan H. F., Gross L. M., Handa M., Maddux B. A., Watson S. R., Zhu S., Narasimha A. J., Levy R., Webster L., Wijesuriya S. D., Liu N., Wu X., Chemla-Vogel D., Tran C., Lee S. R., Wong S., Wilcock D., White M. L., Corbin J. A. (2012) A fully human, allosteric monoclonal antibody that activates the insulin receptor and improves glycemic control. Diabetes 61, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bhaskar V., Lau A., Goldfine I. D., Narasimha A. J., Gross L. M., Wong S., Cheung B., White M. L., Corbin J. A. (2013) XMetA, an allosteric monoclonal antibody to the insulin receptor, improves glycaemic control in mice with diet-induced obesity. Diabetes Obes. Metab. 15, 272–275 [DOI] [PubMed] [Google Scholar]
- 81. Roell M. K., Issafras H., Bauer R. J., Michelson K. S., Mendoza N., Vanegas S. I., Gross L. M., Larsen P. D., Bedinger D. H., Bohmann D. J., Nonet G. H., Liu N., Lee S. R., Handa M., Kantak S. S., Horwitz A. H., Hunter J. J., Owyang A. M., Mirza A. M., Corbin J. A., White M. L. (2010) Kinetic approach to pathway attenuation using XOMA 052, a regulatory therapeutic antibody that modulates interleukin-1β activity. J. Biol. Chem. 285, 20607–20614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Issafras H., Corbin J. A., Goldfine I. D., Roell M. K. (2014) Detailed mechanistic analysis of gevokizumab, an allosteric anti-IL-1β antibody with differential receptor-modulating properties. J. Pharmacol. Exp. Ther. 348, 202–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Schultz Kirkegaard H., Valentin F. (2014) Academic drug discovery centres: the economic and organisational sustainability of an emerging model. Drug Discov. Today 19, 1699–1710 [DOI] [PubMed] [Google Scholar]
- 84. Nicolaou K. C. (2014) Advancing the drug discovery and development process. Angew Chem. Int. Ed. Engl. 53, 9128–9140 [DOI] [PubMed] [Google Scholar]
- 85. Bouvier M., Heveker N., Jockers R., Marullo S., Milligan G. (2007) BRET analysis of GPCR oligomerization: newer does not mean better. Nat. Methods 4, 3–4; author reply 4, 10.1038/nmeth0107-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huber T., Sakmar T. P. (2014) Chemical Biology Methods for Investigating G Protein-Coupled Receptor Signaling. Chem. Biol. 21, 1224–1237 [DOI] [PubMed] [Google Scholar]
- 87. Wood M. R., Hopkins C. R., Brogan J. T., Conn P. J., Lindsley C. W. (2011) “Molecular switches” on mGluR allosteric ligands that modulate modes of pharmacology. Biochemistry 50, 2403–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Davie B. J., Valant C., White J. M., Sexton P. M., Capuano B., Christopoulos A., Scammells P. J. (2014) Synthesis and pharmacological evaluation of analogues of benzyl quinolone carboxylic acid (BQCA) designed to bind irreversibly to an allosteric site of the M1 muscarinic acetylcholine receptor. J. Med. Chem. 57, 5405–5418 [DOI] [PubMed] [Google Scholar]
- 89. Pittolo S., Gómez-Santacana X., Eckelt K., Rovira X., Dalton J., Goudet C., Pin J.-P., Llobet A., Giraldo J., Llebaria A., Gorostiza P. (2014) An allosteric modulator to control endogenous G protein-coupled receptors with light. Nat. Chem. Biol. 10, 813–815 [DOI] [PubMed] [Google Scholar]
- 90. Wootten D., Savage E. E., Willard F. S., Bueno A. B., Sloop K. W., Christopoulos A., Sexton P. M. (2013) Differential activation and modulation of the glucagon-like peptide-1 receptor by small molecule ligands. Mol. Pharmacol. 83, 822–834 [DOI] [PubMed] [Google Scholar]
- 91. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 92. Tan Q., Zhu Y., Li J., Chen Z., Han G. W., Kufareva I., Li T., Ma L., Fenalti G., Li J., Zhang W., Xie X., Yang H., Jiang H., Cherezov V., Liu H., Stevens R. C., Zhao Q., Wu B. (2013) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 341, 1387–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hollenstein K., Kean J., Bortolato A., Cheng R. K. Y., Doré A. S., Jazayeri A., Cooke R. M., Weir M., Marshall F. H. (2013) Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 [DOI] [PubMed] [Google Scholar]
- 94. Wu H., Wang C., Gregory K. J., Han G. W., Cho H. P., Xia Y., Niswender C. M., Katritch V., Meiler J., Cherezov V., Conn P. J., Stevens R. C. (2014) Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Doré A. S., Okrasa K., Patel J. C., Serrano-Vega M., Bennett K., Cooke R. M., Errey J. C., Jazayeri A., Khan S., Tehan B., Weir M., Wiggin G. R., Marshall F. H. (2014) Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 [DOI] [PubMed] [Google Scholar]
- 96. Rasmussen S. G. F., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T. A., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Crystal structure of the β2-adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kruse A. C., Ring A. M., Manglik A., Hu J., Hu K., Eitel K., Hübner H., Pardon E., Valant C., Sexton P. M., Christopoulos A., Felder C. C., Gmeiner P., Steyaert J., Weis W. I., Garcia K. C., Wess J., Kobilka B. K. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dror R. O., Green H. F., Valant C., Borhani D. W., Valcourt J. R., Pan A. C., Arlow D. H., Canals M., Lane J. R., Rahmani R., Baell J. B., Sexton P. M., Christopoulos A., Shaw D. E. (2013) Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature 503, 295–299 [DOI] [PubMed] [Google Scholar]
- 99. Didenko T., Liu J. J., Horst R., Stevens R. C., Wüthrich K. (2013) Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr. Opin. Struct. Biol. 23, 740–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu J. J., Horst R., Katritch V., Stevens R. C., Wüthrich K. (2012) Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]