Background: The liver stage is an obligatory step in the Plasmodium life cycle.
Results: Liver stage tryptophan-rich protein is exported to host cells and plays a role in the development of the exoerythrocytic form. Immunization with this protein protects mice against sporozoite challenge, primarily via T-cells.
Conclusion: Protein immunization provides T-cell-based protection.
Significance: Tryptophan-rich proteins have vaccine potential.
Keywords: liver, malaria, parasitology, plasmodium, vaccine
Abstract
The liver stages of the malaria parasite are clinically silent and constitute ideal targets for causal prophylactic drugs and vaccines. Cellular and molecular events responsible for liver stage development are poorly characterized. Here, we show that sporozoite, liver stage tryptophan-rich protein (SLTRiP) forms large multimers. Mice immunized with a purified recombinant SLTRiP protein gave high antibody titers in both inbred and outbred mice. Immunized mice showed highly significant levels of protection upon challenge with sporozoites and exhibited 10,000-fold fewer parasite 18S-rRNA copy numbers in their livers. The protection offered by immunization with SLTRiP came mainly from T-cells, and antibodies had little role to play despite their high titers. Immunofluorescence assays showed that SLTRiP is expressed in the sporozoite and early to late liver stages of malaria parasites. SLTRiP protein is exported to the cytosol of infected host cells during the early hours of parasite infection. Parasites deficient in SLTRiP were moderately defective in liver stage parasite development. A transcriptome profile of SLTRiP-deficient parasite-infected hepatocytes highlighted that SLTRiP interferes with multiple pathways in the host cell. We have demonstrated a role for SLTRiP in sporozoites and the liver stage of malaria parasites.
Introduction
Malaria infection starts when an infected mosquito feeds on and inoculates sporozoites under the host skin (1). The sporozoites make their way into the bloodstream and reach the liver where they invade the hepatocytes. After productive invasion, a sporozoite develops into an exo-erythrocytic form (EEF),3 also known as the liver stage (LS). Inside the host cell, parasites reside within a parsitophorous vacuole that physically separates them from the host cell cytoplasm. EEFs are among the fastest growing eukaryotic cells. An EEF differentiates into thousands of merozoites, which are released in the form of merosomes (2, 3) and subsequently invade RBCs to commence the erythrocytic cycle.
Plasmodium sporozoites are transcriptionally programmed to express a unique set of virulent proteins required for productive hepatocyte invasion. Several genes that are expressed in sporozoites and LS of malaria parasites have been characterized, such as CSP, TRAP, SPECT1, SPECT2, CelTOS, UIS4, EXP1, and PPLP1 (4), some of which, like CSP and TRAP, are already in advanced stages of vaccine development (5, 6). CSP, which is expressed in sporozoites, LS, and exported to the hepatocyte cytoplasm, has previously been shown to modulate over 1000 genes of the host (7).
Sterile protective immunity against Plasmodium infection was achieved in human and mouse models by immunization with live radiation-attenuated sporozoites (8–10). Therefore, the liver stage biology of the malaria parasite is essential and represents a vital target for vaccine and drug development. Today, the only effective vaccine (in phase III clinical trials) against the disease is RTS,S, but this vaccine gives only partial protection of 30–50% (11–13). RTS,S is capable of providing protection for up to 45 months but at low efficacy, as demonstrated in a trial in Mozambique children in the 1–4-year-old age bracket (14). The RTS,S vaccine shows encouraging results, but there is a need to improve the vaccine's efficacy, either by modifying adjuvants or combining it with new antigens. Recently, a new approach of whole organism vaccine strategy was tested (15). Parasites that are able to invade liver cells but cannot complete the liver stage phase can be used as a live attenuated vaccine. These forms of the parasite are achieved either by targeted deletion of genes that are essential for liver stage development to generate genetically attenuated sporozoites or by chemical methods to produce chemically attenuated sporozoites. Immunization studies using genetically manipulated malaria parasites that do not allow complete development inside liver cells conferred sterile protection against the disease (16, 17). The protective immunity induced by genetically attenuated sporozoites and radiation-attenuated sporozoites is because of the CD8+ T-cell response against infected hepatocytes; however, the exact mechanism of protection is unknown (18, 19). Earlier, it was reported that the tryptophan-rich proteins found in viruses, Plasmodium, and other parasites are involved in host cell invasion (20), protein-protein interaction (21), and several signaling events (22, 23). Pv-trag, a tryptophan-rich protein in blood stages of Plasmodium vivax, has been characterized and shown to have vaccine potential (21).
A previous study showed that the growth of the liver stage of malaria parasites affects several pathways in infected host cells (24) related to immunity, metabolism, gap junctions, the cell cycle, p53 signaling, and cell transporters. In Toxoplasma gondii infection, the affected pathways in the host cell were related to metabolism, the cell cycle, the cytoskeleton, protein modification, DNA replication, transcription/translation, signaling, and cellular transport (25). Viruses also cause similar changes in infected host cells. Influenza viral proteins NS1 and NS2 interact with and affect several components of the host cell required for their successful infection. These affected host cell components are related to RNA transport and processing, nuclear transport, the cell cycle, the immune response, ribosomes and translation, signaling, and cytoskeleton (26). These studies suggest that there are some common themes in infected host cell modulation during infection with various pathogens. In this study, we explore the function of SLTRiP in liver stage parasite growth, its role in host modulation, and its vaccine potential.
Experimental Procedures
Experimental Animals and Parasites
We ensured that all animal work passed an ethical review process and was approved by NII's Institutional Animal Ethics Committee. Animals were maintained, and the work was carried out in accordance with the Committee for the Purpose of Control and Supervision on Experiments on Animals guidelines (Government of India) for the protection of animals used for experimental purposes. The Institutional Animal Ethics Committee approval number for the project is NII-217/09. Six- to 8-week-old male/female BALB/c, C57BL/6 mice, or 60–120-g SD1 rats obtained from the animal facility of the Institute were used for growing parasites. Plasmodium berghei ANKA or SLTRiP-KO parasites were cycled between C57BL/6 mice and Anopheles stephensi mosquitoes. Infected mosquitoes were fed on cotton pads soaked in 20% sucrose solution and maintained at 22 °C and 70–80% relative humidity. Sporozoites were extracted from dissected salivary glands of infected mosquitoes 18 days after the blood meal containing gametocytes. Infected mosquitoes were first rinsed with 50% ethanol, then washed, and dissected in RPMI 1640 media containing 10% FBS. To obtain sporozoites, salivary glands were ground gently and centrifuged at 800 rpm for 3 min to remove mosquito tissue. The number of sporozoites present per μl was then determined by counting in a hemocytometer.
Bioinformatics Analysis
Multiple sequence alignment was performed with the ClustalW program with default settings (27). B-cell epitope prediction was done with a program developed by Jens et al. (28) (available at the Immune Epitope Database analysis resource, accessed May, 2009), and the method used was the Kolaskar and Tongaonkar antigenicity scale and other options available within the program. For T-cell prediction, the Immune Epitope Database programs NETCHOP and NETCTL (in both cases the cutoff was set at +0.9, very high stringency) and IMMUNOGENICITY (for MHC-I, cutoff set at +0.1) were used with default settings.
For protein purification, the SLTRiP gene was codon-optimized (amino acids 67–419 of PBANKA_070100, previously denoted as PB000871.00.0) for Escherichia coli expression and chemically synthesized along with BamHI and XhoI sites (Entelechon, Germany). The codon-optimized SLTRiP gene was cloned at the BamHI and XhoI sites in the pGEX-6P1 vector (GE Healthcare) containing an N-terminal glutathione S-transferase (GST) tag. The plasmid pGEX-6P1-SLTRiP was transformed into E. coli strain BL21-CodonPlus®-RIL (Stratagene). The transformed cells were grown in LB medium containing 100 μg/ml ampicillin at 37 °C. The protein (GST-SLTRIP) was induced by adding isopropyl 1-thio-β-d-galactopyranoside (BioSynth Inc.) to a final concentration of 1 mm when the cell culture reached an A600 between 0.6 and 0.7, followed by an additional incubation of 16 h at 37 °C. The cells were harvested at 10,000 rpm for 10 min at 4 °C and suspended in buffer A (100 mm Tris, 250 mm NaCl, 10% glycerol, 0.5 mm EDTA, 0.05% Triton X-100, pH 8.8) with 0.02 mg/ml lysozyme (Bio Basic Inc.), 1× Protease Inhibitor mixture (Sigma, India) (Complete Lysis buffer) and sonicated at 4 °C (ice-cold H2O) for 40 min with 1 s on and 2 s off with an amplitude of 50. The lysed sample was cleared by centrifugation at 12,000 rpm for 30 min at 4 °C in an R247 rotor (REMI-CPR30 centrifuge). The supernatant was further cleared by filtration through a 0.45-μm glass fiber filter. The clear supernatant was loaded onto a pre-packed 5-ml GST-FF column and washed with 10 column volumes of buffer A. The protein was eluted with buffer A containing 13% v/v of buffer B (50 mm reduced glutathione (Amresco Inc.) is 100% of buffer B) using ÄKTA explorer. The purity was found to be >95%. Yields were typically in the range of 3–4 mg of purified protein/liter of bacterial culture. To obtain tag-free protein, the fusion protein (1 mg) was cleaved with 10 units of PreScission protease (GE Healthcare) at 4 °C for 16 h. The cleaved protein was passed through a GST column. The unbound fraction that contained tag-free SLTRiP protein was collected.
SLTRiP Biophysical Characterization
Circular Dichroism Spectroscopy
A secondary structural analysis of purified SLTRiP (3 μg/μl) in 10 mm Tris, 200 mm NaCl, pH 8.8, was performed on a CD spectrometer (JASCO model J815). The concentration of SLTRiP for the CD spectroscopy was calculated using the BCA method for protein estimation. The CD spectra were collected with a 1 nm bandwidth, cell length of 0.1 cm, and a scanning speed of 100 nm/min. PBS was used as the solvent, and CD spectra were collected at 25 °C. The effect of temperature on the secondary structure of SLTRiP was analyzed from 25 to 95 °C with intervals of 10 °C. CD data were analyzed by K2D2 and SOM CD software.
Size Exclusion Chromatography
Gel filtration analysis of purified SLTRiP (10 mg/ml) in Tris 100 mm, NaCl 200 mm, 0.01% Triton X-100, 10% glycerol, pH 8.8, was performed on a Sephadex 200PG 16/60 column (GE Healthcare) and a Sephacryl S-500 HR 16/60 column (GE Healthcare, catalog no. 28-9356-06) using ÄKTA explorer (GE Healthcare). The molecular weight of SLTRiP was calculated from the standard curve obtained using molecular weight standards (GE Healthcare, catalog no. 28-4038-42).
Native PAGE Analysis of Purified SLTRiP
Native PAGE analysis of purified SLTRiP was performed to check the multimeric nature of SLTRiP. Samples were prepared in native PAGE loading buffer. A 6% native PAGE was run at 150 V for 8 h in 1× native PAGE running buffer (29).
Immunization with SLTRiP-purified Protein
Two groups of BALB/c mice (experimental and control, 10 mice/group) aged 6–8 weeks were immunized either with purified SLTRiP or PBS only. Priming was done with 50 μg of protein in complete Freund's adjuvant (Sigma, India) per mouse. In the three subsequent boosters the amount of protein used was 25 μg per mouse mixed with incomplete Freund's adjuvant (Sigma, India). Boosts were given on days 15, 21, and 28 post-priming. The control group was immunized in an identical manner except there was no SLTRiP protein. RAG-1null mice (30), five per group, were immunized in the same way as above.
ELISA
ELISA was used to determine the titer of the anti-SLTRiP antibody. In the ELISA, 3,3′,5,5′-tetramethylbenzidine was used as a substrate for detection of the HRP-linked secondary antibody (Santa Cruz Biotechnology). Purified SLTRiP protein (0.1 μg/well) in bicarbonate coating buffer was coated overnight in the ELISA plate at 4 °C. The ELISA plate was then washed with washing buffer and blocked with 3% BSA for 1 h at 37 °C. The ELISA plate was then incubated with anti-SLTRiP polyclonal sera in triplicate wells at different dilutions for 1 h at room temperature. The ELISA plate was washed three times with washing buffer, and the plate was further incubated with HRP-conjugated secondary antibody at a dilution of 1:5000 for 45 min at room temperature. After washing the ELISA plate three times, 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added. Once optimal color development was achieved, the reaction was stopped by adding the stopping reagent (HCl). The absorbance of the ELISA plate was read in a spectrophotometer (TECAN, Infinite M200) at 450 nm.
Challenge with P. berghei ANKA Sporozoites
Two weeks after the final boost, both the control and experimental groups were challenged with 1 × 104 P. berghei ANKA sporozoites by the intravenous route. Five mice (from each group) were sacrificed after 48 h of sporozoite infection, to check the liver parasite burden by quantitative RT-PCR using parasite 18S-rRNA primers. The remaining five mice in each group were used for checking the emergence and growth of blood stage parasites by scoring Giemsa-stained thin blood smears prepared on a daily basis from day 3 to post-sporozoite challenge. RAG-1null-immunized and sporozoite-challenged mice were followed by checking parasitemia in thin blood smears.
Sporozoite Neutralization Assay (SNA) for Testing Antibody Protective Efficacy
SNA was performed as per Kumar et al. (31) except that wild type P. berghei sporozoites were used instead of a transgenic strain. Briefly, sporozoites were incubated with dilutions of anti-SLTRiP polyclonal sera at 4 °C for 30 min followed by HepG2 cell infection. Liver stage parasite growth at 48 h post-sporozoite infection was measured by real time PCR.
Immunofluorescence Assay (IFA) Salivary Gland Sporozoites
WT sporozoites were isolated from the salivary glands of infected mosquitoes and coated overnight at 4 °C on 12-chambered Teflon-coated slides (TEKDON). The sporozoites were fixed with 4% formaldehyde, permeabilized with 0.2% saponin, and blocked with 3% BSA (Sigma, India) for 1 h at room temperature. The slides were first incubated with anti-SLTRiP mouse polyclonal sera (1:1000 dilution) for 2.0 h at room temperature followed by three washes with PBS. The slides were further incubated with goat anti-mouse Alexa-488-conjugated secondary antibody (dilution 1:2500) and washed as before. For the IFA of SLTRiP-KO sporozoites, anti-circumsporozoite biotin-coupled antibody (1°Ab) and streptavidin-rhodamine (2°Ab) were used. Nuclei were stained with DAPI (Sigma, India). Slides were observed under a fluorescence microscope to visualize the SLTRiP in sporozoites.
IFA to Check Expression in EEFs and Export of SLTRiP to Hepatocyte Cytoplasm
The SLTRiP expression profile during the liver stage was determined by IFA using mouse anti-SLTRiP antibody. HepG2 cells were grown on circular 15-mm coverslips in a 24-well plate containing media (DMEM with 10% FBS) and maintained at 37 °C and 5% CO2. Subconfluent HepG2 cells in each well were infected with 10,000 P. berghei ANKA sporozoites isolated from the salivary glands of infected mosquitoes (extracted in RPMI 1640 medium with 20% FBS). Infected HepG2 cells were harvested at 6, 12, 24, 36, 42, and 48 h post-infection, fixed with 4% formaldehyde (EMS, Hatfield, PA) for 10 min, and permeabilized with 0.2% saponin in PBS for 15 min. The cells were blocked with 3% BSA in PBS (Sigma, India) for 1 h at room temperature followed by incubation with mouse anti-SLTRiP antibody (1:1000) overnight at 4 °C. Then the cells were washed three times with PBS and incubated with goat anti-mouse Alexa-488 secondary antibody (1:2000) for 1 h at room temperature followed by three washes with PBS. Stained cells were mounted on glass slides along with anti-fade reagent (Bio-Rad) and imaged with a fluorescence microscope (Zeiss, Axio Imager M2).
Blood Stage IFA
On a glass slide, a thin smear of blood with 3–4% parasitemia was prepared, fixed with methanol, and permeabilized with 0.2% saponin for 10 min at room temperature. Permeabilized cells were blocked with 1% BSA (Sigma, India) for 30 min at room temperature and incubated with mouse anti-SLTRiP antibody (1:1000) for 2 h, also at room temperature. The cells were washed three times with PBS and further incubated with goat anti-mouse IgG-secondary antibody (1:2000) conjugated to Alexa-488 for 1 h at room temperature. Nuclei were stained with DAPI (Sigma, India) for 10 min at room temperature.
Parasite Transfection for Generating SLTRiP-deficient Parasites
SLTRiP knock-out (SLTRiP-KO) parasites were generated by a double homologous recombination method. P. berghei ANKA wild type parasites were used to generate SLTRiP-KO (Pb Δ SLTRiP::hDHFR&GFP). For this, first we prepared a targeting construct by cloning two PCR products, the 5′ UTR (711 bp) and 3′ UTR (832 bp) of the SLTRiP gene (UTR nomenclature according to PB000871.00.0 orientation in the contig RP0739, which matches in reverse orientation to the current chromosome 7 sequence). The targeting construct contained the hDHFR gene for drug selection and GFP for microscopic analysis of transfected parasites. The SLTRiP 5′ UTR was amplified by PCR using the following primers: DKJ1-FP having an XhoI restriction site and DKJ2-RP having a ClaI restriction site. The SLTRiP 3′ UTR was amplified by PCR using primers DKJ3-FP containing a NotI restriction site and DKJ4-RP containing a SacII restriction site. The targeting construct DNA was purified using an Endo-free plasmid midi prep kit (Qiagen, Germany). The targeting construct was linearized with ScaI restriction enzyme, and the linearized plasmid was electroporated into P. berghei ANKA schizonts using an Amaxa nucleofector device (LONZA, India). The electroporated parasites were intravenously injected into C57BL/6 mice and kept under pyrimethamine selection for 4–5 days to obtain a mixed selected population of transformed parasites. Genomic DNA from the mixed population was analyzed by diagnostic PCR to identify the integration event. The mixed selected population was used for dilution cloning following the limiting dilution method to obtain a clonal population of a single transformed parasite.
Diagnostic PCR and Southern Hybridization to Confirm Gene Deletion in SLTRiP-KO
Genotyping of transformed parasites was done by diagnostic PCR and Southern blot analysis. Genomic DNA from transformed parasites was extracted from erythrocyte stage parasites and analyzed by diagnostic PCR using the following pair of primers DKJ5-FP (hDHFR-3′ UTR) and DKJ6-RP, (SLTRiP-3′ UTR) to check for correct integration. Transformed parasites analyzed using the SLTRiP gene-specific primers DKJ7-FP and DKJ8-RP also confirmed loss of the SLTRiP gene. For Southern blot analysis, genomic DNA from transformed parasites was digested with AccI and ScaI for 3 h. The digested DNA was run on a 1% agarose gel at 25 V for 14 h and transferred overnight by the capillary method to a positively charged nylon membrane (Roche Applied Science). The transferred DNA was UV cross-linked using a charge of 120 mJ to the nylon membrane. The SLTRiP-3′ UTR (832 bp) digoxigenin-labeled PCR product was used as a probe. The probe was prepared according to the protocol provided by the kit's manufacturer (Roche Applied Science).
Infectivity Assay for SLTRiP-KO
Five C57BL/6 mice were intravenously injected with 6000 sporozoites of WT or SLTRiP-KO parasites. The parasite liver load was measured as described previously (7). In another experiment, five BALB/c mice per group were intravenously injected with 4000 sporozoites of WT or SLTRiP-KO parasites. The emergence of blood stage parasites was followed in Giemsa-stained blood smears on a daily basis. For in vitro analysis, we used the human hepatoma cell line HepG2. Infection (in triplicate) was initiated with 2 × 104 sporozoites per well in a 24-well tissue culture plate. Total RNA was extracted using the TRIzolTM method. Parasite load was measured as described previously (7).
SLTRiP-KO Phenotypic Characterization in the Mosquito and Blood Stages of Malaria Parasites
A. stephensi mosquitoes (3–5 days old, female) were infected with SLTRiP-KO or WT parasites by blood feeding for 10 min on parasite-infected mice with a 0.5 ± 0.1% gametocyte count. The infected mosquitoes were maintained at 22 °C, 80% humidity and under a cyclic condition of 12 h light/12 h dark. Infected mosquitoes were examined on day 14 for oocyst numbers and on day 18 for numbers of salivary gland sporozoites.
Immunocytochemistry of SLTRiP-KO Liver Stages
WT or SLTRiP-KO sporozoites (1 × 106) were injected (i.v.) separately into two C57BL/6 mice. The livers of infected mice were extracted 48 hpi and cut into 3–5-μm thick sections using a manual rotatory microtome (Leica Biosystems). The liver sections were permeabilized with 0.2% saponin and blocked with 5% milk in PBS for 1 h at room temperature. The liver sections were then incubated with monoclonal parasite anti-HSP70 antibody (1:1000) overnight at 4 °C followed by three washes with PBST. Then they were incubated with the secondary antibody (goat anti-mouse Alexa 594) for 90 min at room temperature followed by three washes with PBST.
Transcriptome Analysis
Cell Infection, Cell Sorting, and RNA Isolation for RNA Sequencing
In vitro infection of HepG2 cells was carried out in 6-well plates by adding either WT-GFP or SLTRiP-KO or unrelated KO control sporozoites. Sporozoites (2 × 106) were added to the monolayer of HepG2 cells (1 × 106 cells/well). The plates were incubated at 37 °C and 5% CO2. After 22 h of sporozoite infection, the cells were washed with 1× PBS and trypsinized with 250 μl of trypsin/EDTA (Gibco) per well of the 6-well plate. Trypsinized cells were centrifuged at 800 rpm for 5 min to remove the trypsin and washed twice with 1× PBS, 10% FBS. The HepG2 cells were suspended in 1× PBS, 2 mm EDTA for fluorescence-activated cell sorting (BD FACS AriaTM). The FACS-sorted cells were analyzed by fluorescence microscopy for parasitized GFP-positive cells. More than 90% of the cells in all three samples were GFP-positive. Total RNA was isolated from sorted cells using the TRIzol method, and the concentration of RNA was determined by a Qubit fluorimeter (Invitrogen) using the QubitTM RNA assay kit. The total RNA for each sample was analyzed on an Agilent bioanalyzer using the Agilent RNA 6000 Pico kit for quality control purposes. Forty thousand sorted cells gave ∼100 ng of RNA, which was used for NGS analysis after selecting the mRNA population.
RNA Sequencing and Data Analysis
RNA sequencing was performed as recommended by the stranded mRNA sample preparation kit from IlluminaTruseq. Briefly, the poly(A) containing mRNA molecules were purified from total RNA using poly(T) oligo-attached magnetic beads. The mRNA was fragmented into small pieces using divalent cations under elevated temperature and converted into first strand cDNA using reverse transcriptase and random hexamer primers. The second strand of cDNA was synthesized using DNA polymerase I and RNase-H enzymes. The synthesized cDNAs have an additional “A” base, which is required for adapter ligation. The products were purified and enriched with PCR to create the final cDNA library. The cDNA library for each sample was analyzed on an Agilent 2100 Bioanalyzer using the Agilent DNA 1000 kit. Deep sequencing of cDNA libraries was performed with the help of Illumina Genome analyzer II for 78 cycles, including six additional cycles for an index read. Sequence reads were independently aligned using CLC Genomic Workbench 6.0 with default settings against the human genome assembly (GRCH 37. P13). Differential expression, p values, and fold changes for each gene were determined using CLC Genomic Workbench 6.0 with default parameters.
Validation of Target Genes in NGS by Real Time PCR
To validate the next generation sequencing data, we performed quantitative real time PCR for selected genes on cDNA made from independent biological replicates using SYBR Green PCR master mix (Life Technologies, Inc.) on the RealPlex detection system (Eppendorf, Germany). For real time PCR, the primers for selected genes were designed using primer blast software from NCBI. The sequence of each primer is provided in Table 4. HSP90AB and ACTB (β-actin) were used as housekeeping gene controls for all the samples. The real time PCR conditions were one cycle at 95 °C for 2 min and then 40 cycles of 95 °C for 30 s, 55 °C for 20 s, and 68 °C for 30 s followed by a melting curve. For each sample, the real time PCR was performed in triplicate in a 20-μl reaction volume. The relative expression or fold change in each selected gene between WT and SLTRiP KO was determined using the 2−ΔΔCt method.
TABLE 4.
List of primers used in this study
List showing the primers along with their sequences and ID numbers used in this study.

*, 9–54 primers were used for NGS transcriptomic data validation by real time PCR.
Host Cell Gene Network Analysis
Differentially expressed host genes showing greater than ±3-fold differences and with p values <0.05 were selected for pathway analysis using GeneCodis3 software (32). The GO analysis of selected genes in each pathway was done with the help of the Bingo 2.3 plugin in Cytoscape 2.8.3.
Statistical Analysis
All the data were analyzed, except where indicated otherwise, using SigmaPlot version 11.0 (Systat Software, Inc., San Jose, CA). p values were calculated using the t test method. p values less than 0.05 were considered to indicate significant differences.
Results
SLTRiP Is a Conserved Tryptophan-rich Protein with Potential B- and T-cell Epitopes
Plasmodium LS-exported proteins could be excellent vaccine candidates due to their potential processing by hepatocytes and presentation on their surfaces. We endeavored to identify a Plasmodium protein that specifically expresses in sporozoites and the liver stage of malaria parasites and is likely exported to the hepatocyte cytoplasm. Previously, it was determined that PF08_0003 encodes a protein that expresses in the sporozoite stage of P. falciparum (33). Therefore, we searched for orthologs of PF08_0003 and found them in various Plasmodium species (PBANKA_070100 in P. berghei ANKA (previous ID, PB000871.00.0, containing amino acids 67–419 of PBANKA_070100), PCAS_093700 in Plasmodium chabaudi chabaudi, PY17X_0701300 (or PY04984) in Plasmodium yoelii 17XNL, PVX_088850 in P. vivax Sal-1 and PKH_050090 in Plasmodium knowlesi H). Rodent malaria genes PBANKA_070100, PCAS_093700, and PY17X_0701300 are syntenic and show a two-exon and one-intron genomic structure, whereas PVX_088850, PKH_050090, and PF8_003 are nonsyntenic. PBANKA_070100 encodes a 419-amino acid protein with a calculated mass of 51,092 Da. PBANKA_070100 (SLTRiP) shows significant sequence identity to orthologous proteins in other rodent Plasmodium species (Fig. 1A). Multiple sequence alignment shows that a segment of SLTRiP is conserved in Plasmodium species and contains a large number of position-constrained conserved tryptophan residues, which are depicted in bold and underlined letters in Fig. 1B. SLTRiP also contains a few cysteines (three in P. berghei), but these are not conserved among other species, indicating they are not crucial for protein structure/function. Apparently, in the absence of disulfide bond formation in SLTRiP, conserved tryptophans may play a key role in the structural stability of the protein.
FIGURE 1.
Purification and biophysical characterization of SLTRiP. A, SLTRiP encodes a conserved tryptophan-rich protein. Sequence identity percentage among orthologous SLTRiPs from different Plasmodium species is shown. B, multiple sequence alignment (MSA) of Pb-SLTRiP with orthologous proteins of Plasmodium species. The GenBankTM accession numbers for proteins in the respective Plasmodium species are indicated next to the protein sequences. Conserved tryptophan residues are shown in boldface and underlined letters. C, purification of SLTRiP by affinity chromatography. Lane 1, induced lysate; lane 2, flow-through; lane 3, wash; lane 4, protein size markers; lanes 5–10, elution fractions of SLTRiP. D, SLTRiP cleavage by PreScission protease. Lane 1, GST-SLTRiP-purified recombinant protein; lanes 2 and 10, showing the cleavage of purified GST-SLTRiP; lanes 3–5, and lane 7, purified SLTRiP protein without the GST tag; lane 6, protein size markers. E, gel filtration analysis (GFC) of purified SLTRiP showing different molecular mass complexes of SLTRiP. F, SDS-PAGE analysis of eluted sample from GFC as shown in E. mAU, milli-absorbance unit. G, native PAGE analysis of purified SLTRiP confirming that SLTRiP forms aggregates of high molecular weight. * indicates the SLTRiP protein stuck in the well, and the arrow represents the SLTRiP protein that comes out of the well. A single x indicates the ferritin monomer, and xx represents the ferritin dimer. H, secondary structure analysis of SLTRiP by CD spectroscopy and effect of temperature on its secondary structure. The helical content was calculated from CD data using K2D2 and SOM CD programs.
P. vivax tryptophan-rich proteins expressed in blood stage parasites have been shown to induce humoral immune responses in humans (34) who are naturally exposed to malaria. Bioinformatics analysis of the SLTRiP amino acid sequence using a program developed by Jens et al. (28) (available at the Immune Epitope Database) showed the presence of multiple strong B (ten) and T-cell (four) epitopes. Based on the predicted peptide sequences, two of the B-cell epitopes (SNKSPKNVSVN and GSKVKSSERS) when conjugated to keyhole limpet hemocyanin gave strong antibody responses in mice (data not shown), an indication of the immunogenic potential of SLTRiP.
Cloning, Expression, Purification, and Biophysical Characterization of SLTRiP
Because expression of the Plasmodium-derived SLTRiP gene in E. coli failed, we synthesized the SLTRiP gene according to E. coli codon usage. The codon-optimized and chemically synthesized SLTRiP gene was then cloned into pGEX-6P-1, an E. coli expression vector, and expressed as a GST-tagged recombinant protein. SLTRiP was purified by glutathione-Sepharose chromatography to a purity of >95% (Fig. 1C, lanes 5–10). Plasmid pGEX-6p-1 has a thrombin cleavage site, which allows the GST tag to be removed from the SLTRiP-GST fusion protein after cleavage with PreScission protease (Fig. 1D, lanes 3–5 and 7 (cleaved purified) and lanes 2 and 10 (cleaved unpurified)). Purified SLTRiP was obtained after passing the cleaved SLTRiP-GST through a glutathione-Sepharose column, where the cleaved GST bound to the column and free SLTRiP passed through in the flow-through (Fig. 1D, lanes 3–5). To check the size and molecular association, gel filtration chromatography (GFC) was performed using Sephacryl S200HP and S500HR. Using the S200HP column, we found that SLTRiP appeared in the void volume, indicating that SLTRiP forms a large complex of high molecular weight. Next we ran the protein on the S500HR, where it eluted over an extended time in three peaks having overlapping boundaries (Fig. 1E). The GFC results showed that SLTRiP forms multimers with different molecular weights such as dodecamers, octamers, and tetramers. SDS-PAGE analysis of eluted samples from GFC confirmed that the SLTRiP protein was present in these eluted samples (Fig. 1F). The native PAGE analysis of purified SLTRiP supported the GFC conclusion as to its multimeric nature (Fig. 1G). SLTRiP multimerization was also confirmed through the dynamic light scattering method (data not shown). The multimeric nature of SLTRiP might be advantageous with respect to its antigenic properties due to its slow release in immunized mice.
Circular dichroism is a valuable tool to study protein secondary structure. CD spectroscopy results showed that SLTRiP-GST is mostly (86%) α-helices when measured at 25°C, which is reduced to 5% under heat-denatured conditions at 95 °C. The results also show that the protein is stable from 25 to 55 °C (no major change in helical content), and it starts to denature above 55 °C (Fig. 1H). SLTRiP without its fusion partner (GST cleaved) showed stability from 25 to 55 °C and thereafter a sharp decline in helical content (data not shown).
Immunization Provides Protection against Sporozoite Challenge, Generating Specific and High Titer Antibodies
Mice (BALB/c) immunized with purified SLTRiP protein yielded a very high antibody titer of 5 × 106 after the second booster immunization (Fig. 2A). The specificity of the mouse anti-SLTRiP antibodies was determined by Western blot (Fig. 2B) and IFA (Fig. 3A). Mouse anti-SLTRiP antibodies specifically recognized SLTRiP in P. berghei wild type sporozoite lysate but not in SLTRiP-KO sporozoite or P. berghei (wild type) blood stage parasite lysate (Fig. 2B), showing a very high degree of specificity. The absence of SLTRiP antibody cross-reactivity with blood stage parasites highlights that SLTRiP is different from other tryptophan-rich proteins expressed during the blood stages (for example P. vivax TRAg and P. yoelii pAg1 and pAg3).
FIGURE 2.
SLTRiP immunization, antibody specificity, and sporozoite challenge assay. A, antibody titers of mouse anti-SLTRiP polyclonal sera determined by the ELISA method. Error bar represents the standard deviation. Cont, control. B, mouse polyclonal sera generated against purified SLTRiP specifically recognizes SLTRiP in sporozoite lysate. Lane 1, purified recombinant GST-SLTRIP (72.5 kDa, E. coli expressed); lane 2, SLTRiP-KO sporozoite lysate; lane 3, WT blood stage lysate; lane 4, WT sporozoite lysate; and lane 5, protein molecular mass markers. Western blotting with anti-CSP antibody serves as control for KO and WT sporozoite samples. β-Actin was used as loading control. C, SLTRiP immunized mice show a 4-day delay in the first emergence of blood stage infection as compared with control group. Each data point represents the average parasitemia from five mice. D, parasite load in the livers of SLTRiP immunized mice as measured by qPCR for parasite 18S-rRNA copy numbers. The parasite load in the livers of SLTRiP immunized mice is 4 log scale (10,000 times) lower as compared with the control group of mice. Each bar represents the average 18S-rRNA copy number from five mice. Error bars represent the standard deviation in each group. *, p < 0.005. E, SNA with SLTRiP-Ab compared with the control sera. Infection was determined by measuring parasite 18S-rRNA copy numbers in samples using real time PCR. Error bars represent the standard deviation in each group. ns, not significant. p > 0.1.
FIGURE 3.
SLTRiP is expressed in the sporozoite and liver stages. A, SLTRiP is expressed in sporozoites. Fluorescent images (green) show the expression and localization of SLTRiP in P. berghei sporozoites. Sporozoite nuclei are visualized by staining with DAPI (blue), and sporozoite morphology is shown in a differential interference contrast (DIC) image. Merged refers to a blend of the green and blue images. Anti-circumsporozoite staining (red) was used as positive control for SLTRiP-KO parasites. SLTRiP (green fluorescence) was absent in the SLTRiP-KO parasites. B, SLTRiP is expressed in the liver stage. Fluorescent images of P. berghei liver stages in HepG2 cells, stained with mouse anti-SLTRiP antibody (green), at different hours post-infection. SLTRiP expression is abrogated (not detectable in IFA) in 48-h liver stage parasites. C, average pixel intensity of SLTRiP (in IFA images) at various time points in liver stage parasite growth. D, expression of SLTRiP during the blood stage of P. berghei parasites. The nucleus of the parasite is stained with DAPI (blue), and the merged image shows an overlay of all three images.
The only promising vaccine against malaria in its pre-erythrocytic stage is RTS,S, but it gives only partial protection of 30–50%. RTS,S immunization is capable of providing protection for up to 45 months, albeit at low efficacy, as was demonstrated in a trial in children in the 1–4-year-old age bracket (32). In the future, the efficacy of the RTS,S vaccine may be increased by combining it with new antigens that have high vaccine potential. To test the antigenic potential of SLTRiP protein, BALB/c mice were immunized with purified SLTRiP protein and challenged with 1 × 104 wild type sporozoites. In the control and SLTRiP immunized groups, blood stage parasites were detected on the 3rd and 7th days, respectively (Fig. 2C), highlighting that SLTRiP gives strong (4-day delay in the pre-patent period, see Table 1) but partial protection against sporozoite challenge. The above result was further verified by checking the parasite burden in the livers of immunized and control mice 48 h post-challenge with sporozoites. The results showed that the parasite load in liver of SLTRiP immunized mice was four log scale (10,000 times) less as compared with the control group of mice (Fig. 2D). These results emphasize that SLTRiP has high vaccine potential. The efficacy of SLTRiP protein as a vaccine can be further increased by modifying either the properties of the adjuvant or the delivery method.
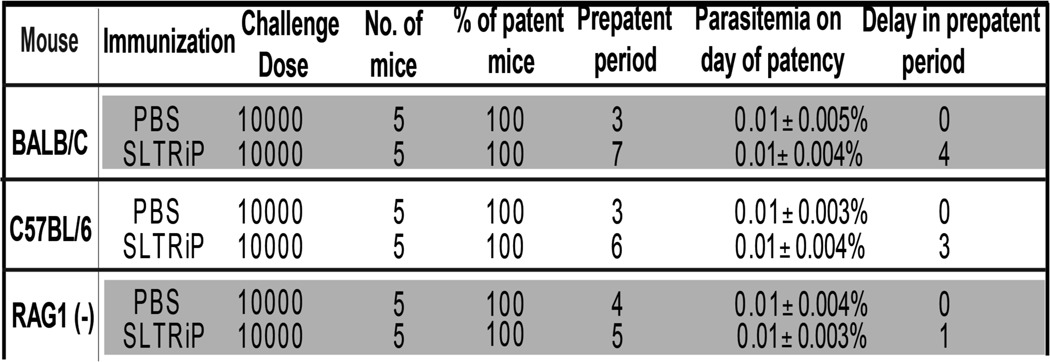
TABLE 1.
Delay in pre-patent period in the SLTRiP immunized mice that were challenged with sporozoites
Protection after immunization with recombinant SLTRiP is shown. The table summarizes the delay in the pre-patent period of SLTRiP immunized versus control group mice when challenged with WT sporozoites. RAG1null mice immunized with SLTRIP and subsequently challenged with sporozoites show only a 1-day delay in the pre-patent period as against a 3-day delay in genotype-matched control immunized mice, thus showing a major loss of protection in B- and T-cell-deficient mice (RAG1null). Because the antibodies against SLTRiP show insignificant protection (Fig. 2E), the above loss of protection in RAG1null mice implies that T cells are responsible for a majority of the protection observed in wild type mice. The dose represents the number of sporozoites injected intravenously per mouse.
Protection against Sporozoite Challenge in SLTRiP Immunized Mice Is T-cell-dependent
SLTRiP immunization generated very high antibody titers, and one would naturally expect these antibodies to play a role in protection against sporozoite challenge. Using a sporozoite antibody neutralization assay, we tested the contribution of anti-SLTRiP antibodies for protection. To our surprise, at a 1:5 dilution we observed only 2-fold less infection compared with control (Fig. 2E), indicating a minimal role for the antibodies in conferring protection. To ascertain the contribution of cell-mediated immunity, we immunized RAG-1null mice with SLTRiP and measured the loss of protection compared with genotype-matched controls (C57BL/6). RAG-1null mice, which lack both B- and T-cells, showed a 100-fold loss in protection (out of a total of 1000-fold enhanced protection in matched wild type controls (Table 1)), indicating T-cells play a major role in protection, because antibodies give only minimal protection (Fig. 2E). In RAG-1null mice 10-fold protection was observed (1 day delay in the pre-patent period, Table 1), which likely derives from innate immune responses.
SLTRiP Is Expressed in Sporozoites, EEFs, and Exported to Hepatocyte Cytosol
To determine the stage-specific expression, we checked SLTRiP protein expression in sporozoites, liver stages, and blood stages. IFA using anti-SLTRiP polyclonal antibody confirmed the SLTRiP protein has cytosolic expression in sporozoites (Fig. 3A). Mouse anti-SLTRiP antibody specifically recognized SLTRiP in wild type sporozoites but not in SLTRiP-KO sporozoites. Treatment of sporozoites with anti-circumsporozoite antibody coupled with biotin was used as a positive control for IFA of SLTRiP-KO sporozoites. The IFA results also showed that this protein was expressed between 12 and 42 h of LS growing in vitro (HepG2 cells). The expression of SLTRiP protein was maintained until 42 h and was completely abrogated in 48-h-old EEFs (Fig. 3B). SLTRiP levels in EEFs were more or less same from 12 to 26 h but reduced by nearly 20% at 36 h and ∼40% at 42 h (Fig. 3C). Thus SLTRiP protein expression initiated in salivary gland sporozoites was maintained from the early to mid-liver stages and paused at the post-48-h liver stage. Finally, we checked expression during the blood stage of malaria parasites but could not detect any SLTRiP expression (Fig. 3D).
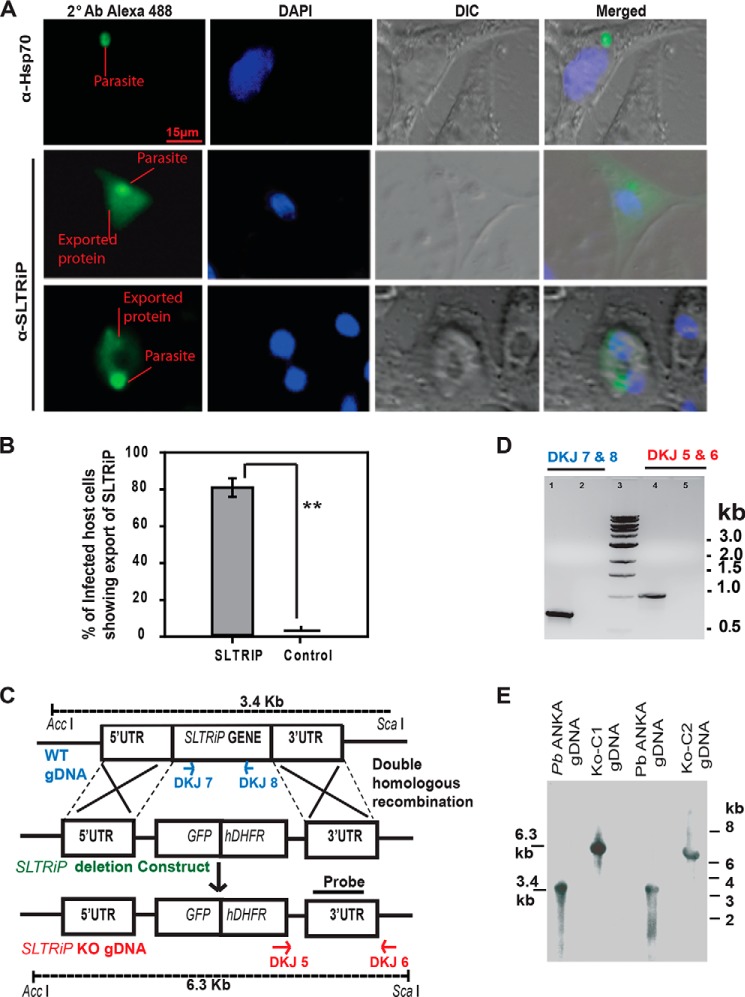
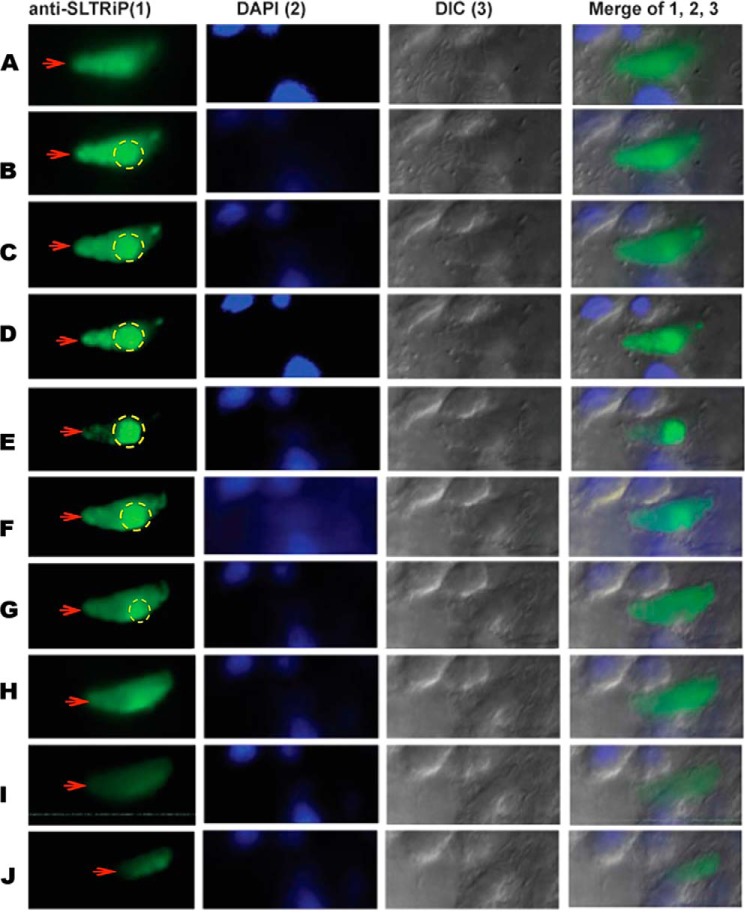
IFA with EEFs also confirmed that this protein is exported to the cytosol of infected HepG2 cells (Figs. 4A and 5). Export of SLTRiP into the cytosol of infected host cells is more clearly visible in (Fig. 5), which represents the z stacks of an IFA image of a 12-h EEF. When the parasite was in focus, the exported protein was faintly visible (stack E). Conversely, the exported protein is clearly visible in some stacks, but the parasite is not visible (stack A and I). We calculated the percentage of infected host cells showing export of SLTRiP and found that 80% of WT-infected host cells showed export of SLTRiP at 12 h post-sporozoite infection (Fig. 4B).
FIGURE 4.
SLTRiP is exported to the cytosol of infected hepatocytes. A, IFA image of infected HepG2 cells 12 hpi showing SLTRiP translocation (export across the parasitophorous vacuole membrane) in the cytosol of host cells. Infected cells were stained with mouse anti-SLTRiP polyclonal sera, and in control (export negative) experiments cells were stained with anti-parasite HSP70 antibody. In these images, the emphasis was on keeping the exported protein in focus and not the parasite. DIC, differential interference contrast. B, analysis of infected host cells for export (% showing export of SLTRiP) as compared with control where the parasites were stained with HSP-70 antibody. **, p < 0.0001. The error bar represents the standard deviation. C, strategy of targeted gene disruption: A schematic representation of the replacement strategy to generate the SLTRiP-KO parasite. The SLTRiP gene locus was targeted with a ScaI linearized replacement plasmid containing the 5′ and 3′ UTR regions of the SLTRiP locus. The replacement plasmid also contained GFP and a human dihydrofolate reductase (hDHFR) cassette for positive selection of transfected parasites. After a double crossover event, the SLTRiP gene was replaced by the GFP/human dihydrofolate reductase cassette, resulting in the SLTRiP-KO parasite (P. berghei Δ SLTRiP::hDHFR&GFP)]. D, genotyping of knock-out parasites. PCR analysis using genomic DNA from P. berghei-WT (lane 1) or SLTRiP-KO (lane 2) with SLTRiP gene-specific primers (DKJ-7 and DKJ-8) is shown, which should give a 640-bp fragment, confirming the SLTRiP gene deletion (no PCR product in lane 2). Lane 3 has a 1-kb DNA ladder. Diagnostic PCR (using primers DKJ-5 and DKJ-6) performed on SLTRiP-KO genomic DNA gave a desired size product (lane 4, expected size 990 bp) that was absent (lane 5) when WT-P. berghei ANKA genomic DNA was used as template. E, genotyping by Southern blot hybridization. The SLTRiP-KO and WT-P. berghei ANKA genomic DNA were digested with AccI and ScaI and probed with a 3′ UTR probe (as shown in C). The restriction fragments recognized by the 3′ UTR probe should be 3.4 kb for WT and 6.3 kb for SLTRiP-KO. The expected size fragments are detected in the blots.
FIGURE 5.
z stacks of a 12-h-old EEF-infected host cell. The infected cell was stained with anti-SLTRiP antibody, and fluorescent images were captured on an Axio-Imager M2 (Zeiss) fitted with ApoTome. The figure shows the export of SLTRiP into the cytosol of the infected host cell (red arrows). Parasite-associated fluorescence is marked with a dotted circle; anything outside the circle is due to exported SLTRiP. In this image, when the parasite is in focus, the exported protein is faintly visible (stack E). Conversely, the exported protein is clearly visible in some stacks, but the parasite is not visible (stacks A and I). A total of 20 z stacks were captured per event, and each stack was 0.2 μm thick. Figure shows only the images from the alternate z stacks. DIC, differential interference contrast.
Role of SLTRiP in Sporozoite and Liver Stage Parasites
SLTRiP Plays No Role in the Blood and Mosquito Stages
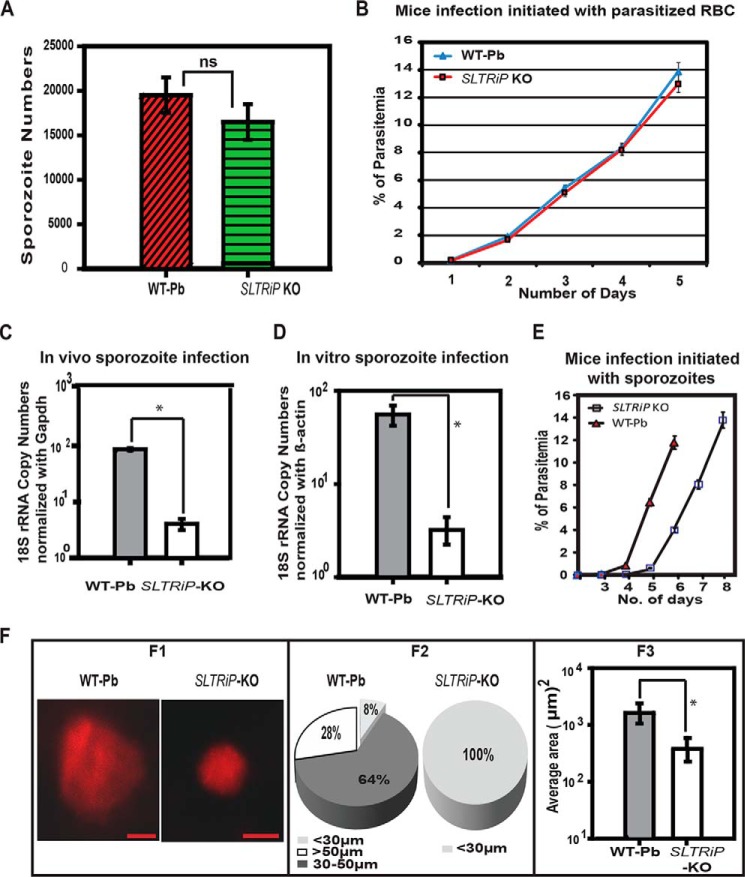
To investigate the function of SLTRiP during the Plasmodium life cycle, we generated P. berghei parasites deficient in the SLTRiP gene using a replacement strategy (Fig. 4C). Knock-out of the SLTRiP gene was confirmed by diagnostic PCR (Fig. 4D) and Southern blot analysis (Fig. 4E). Using SLTRiP-KO parasites, we found there were no significant differences in the development of the oocyst or in the number of sporozoites in oocysts or salivary glands between SLTRiP-KO and WT-infected mosquitoes. In the case of SLTRiP-KO, the average number of sporozoites in mosquito salivary glands was 14,000 ± 2000 compared with 16,000 ± 2000 in WT parasites, which is not a significant difference (Fig. 6A, p = 0.14). This finding also indicates that earlier mosquito stages were not affected in SLTRiP-KO.
FIGURE 6.
Characterization of SLTRiP-deficient parasites and SLTRiP's role in liver stage parasite development. A, number of sporozoites in the salivary glands of infected mosquitoes. Bars depict the average number of sporozoites in salivary glands of 50 mosquitoes. ns, not significant (n = 3, p = 0.14). Error bars represent the standard deviation in the average number of sporozoites from 50 mosquitoes. B, growth during the blood stage. There was no significant difference in the growth of SLTRiP-KO and WT parasites during the blood stage. Each data point shows, on that day, the average parasitemia from five mice along with the standard deviation (n = 3). C, in vivo growth assay. Parasite 18S-rRNA copy numbers in the livers of SLTRiP-KO or WT parasite infected mice were measured using quantitative RT-PCR to check the parasite load at 46 hpi. Mouse Gapdh was used as a housekeeping gene for normalization of the 18S-rRNA copy number quantification. Each bar represents the average of five mice. Error bars represent the standard deviation in each group (n = 3). *, p < 0.05. D, in vitro growth assay. SLTRiP-KO or WT parasite 18S-rRNA copy numbers in HepG2 cells at 46 hpi were quantified by qPCR. Human β-actin was used as a housekeeping gene control for normalization of parasite 18S-rRNA copy numbers. Each bar represents the average of two independent invasion experiments. Error bars represent the S.E. of each group. *, p < 0.05. E, delay in growth during the liver stage. Each data point shows the average parasitemia of six mice (n = 2), and error bars represent the standard deviation in each group. F, microscopic analyses of growing parasites in vivo. Panel F1, liver stage parasite images, captured on an Axio Imager M2 microscope (Zeiss) attached to a CCD camera. Panel F2, pie chart showing the percent distribution of liver stage parasite diameters. The smallest group has ∼8% parasites in the case of wild type and 100% in the case of SLTRiP-KO. Panel F3, average area measured from images of liver stage parasites in the livers of C57BL/6 mice infected with SLTRiP-KO or WT sporozoites. Each bar represents the average of 50 parasites, and the error bar represents the standard deviation in each group. *, p < 0.05.
To check the effect of SLTRiP-KO parasites on blood stage growth, 100,000 SLTRiP-KO or WT blood stage parasites were intravenously injected. SLTRiP-KO parasites showed a normal phenotype during the erythrocytic cycle when compared with WT parasites (Fig. 6B). These findings demonstrate that SLTRiP plays no role in blood, gametocyte, or mosquito stage parasite development.
SLTRiP-KO Parasites Are Defective in Growth during the Liver Stage
To ascertain the role of SLTRiP in liver stage parasite development, we used quantitative real time PCR. The results revealed that there were 21-fold fewer 18S-rRNA copy numbers in SLTRiP-KO parasites during liver stage growth in vivo (Fig. 6C). SLTRiP-KO infection in HEPG2 cells also showed defects comparable with the in vivo findings (Fig. 6D). The LS growth defect in vivo was also confirmed, in a separate experiment, by checking the pre-patent period (Fig. 6E and Table 2). The results showed there was more than a 1-day delay in the pre-patent period of SLTRiP-KO as compared with WT LS parasites. These results indicate that SLTRiP plays an auxiliary role in liver stage parasite development.
TABLE 2.
Delay in pre-patent period (in mice) when SLTRiP-KO sporozoites were compared with WT sporozoites
Delay in the pre-patent period in mice infected with SLTRiP-KO was compared with WT sporozoites (n = 3). The dose represents the number of sporozoites injected intravenously per mouse.

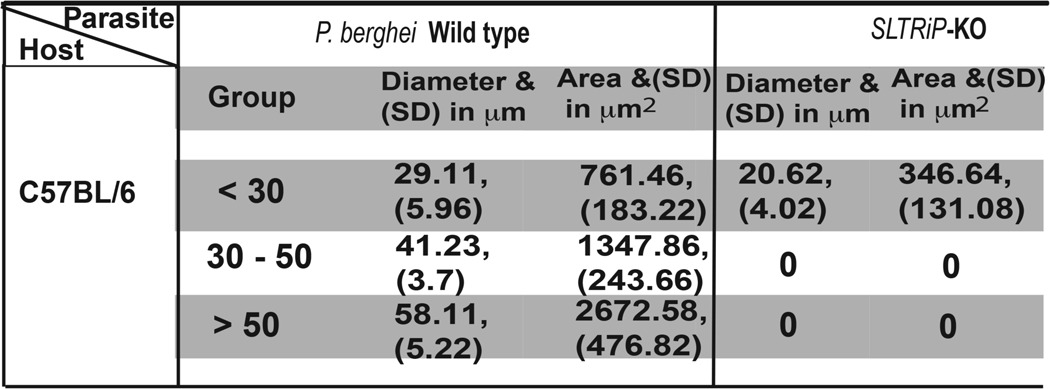
To understand whether SLTRiP affects the growth of liver trophozoites and schizonts, we injected 1 × 105 SLTRiP-KO or WT sporozoites intravenously into two mice, and 48 h EEF images were acquired from liver sections processed for IFA (Fig. 6F). Two-dimensional images were measured using software to calculate the area and diameter of each parasite. Based on their diameter, the parasites were divided into three groups (Fig. 6F2 and Table 3). Fig. 6F2 shows that SLTRiP-KO parasites were smaller in size as compared with WT parasites (p = 0.01). The average area of SLTRiP-KO parasites was 4.67-fold less compared with WT parasites (Fig. 6F3, p < 0.05). The above findings confirm the role of SLTRiP in liver trophozoite and schizont development.
TABLE 3.
SLTRiP-KO parasites are defective in growth during liver stage as compared with Pb-ANKA parasite
Table shows the average values for the area and diameter of SLTRiP-KO and WT parasites, measured in the livers of C57BL/6 mice after 48 h of infection with the respective parasites. On the basis of size, the SLTRiP-KO and WT parasite populations were divided into three size groups as depicted. For each group, the standard deviation in the area and diameter is given in parentheses. The diameter range for PBANKA and SLTRiP KO parasites in the liver of C57BL/6 mice is 15–65 and 13–27 μm, respectively. The area range for PBANKA and SLTRiP KO parasites in the liver of C57BL/6 mice is 272–3406 and 133–606 μm2, respectively. S.D. represents the standard deviation in respective average value for the diameter and area, and it is given in parentheses.
SLTRiP Plays a Role in Host Modulation
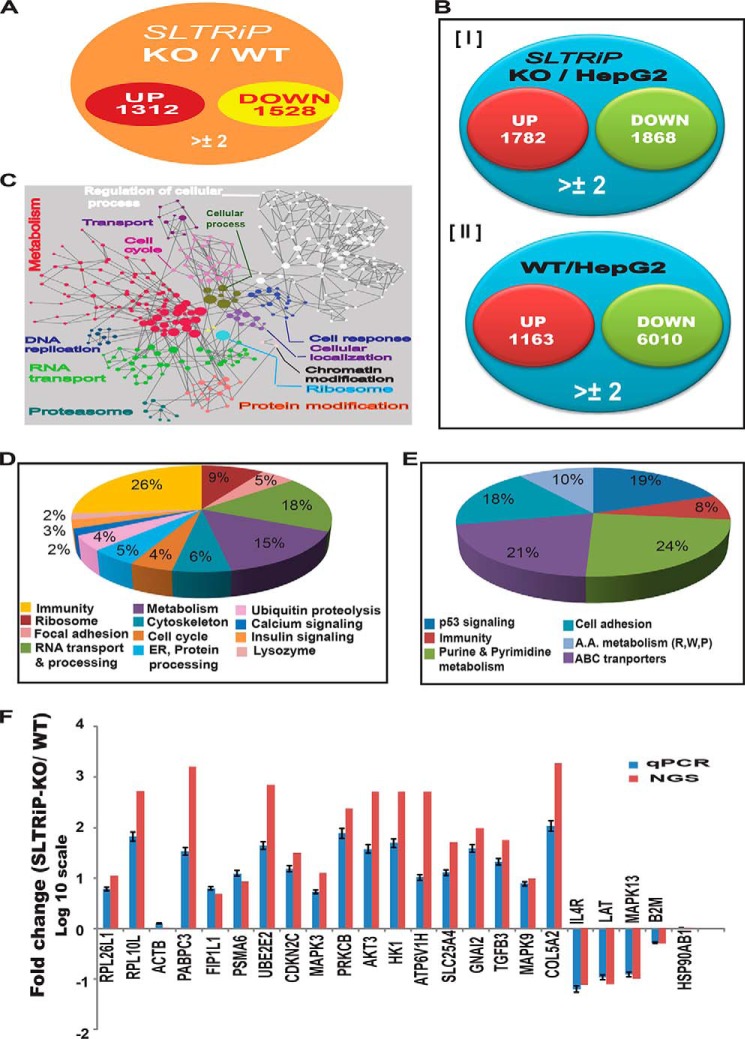
To determine the function of exported SLTRiP in host modulation, we performed whole transcriptome analyses of host transcripts at 22 h post-sporozoite infection of HepG2 cells. We chose this time point on the basis of a previous microarray-based study showing that the maximum number of gene changes occur at around 24 h (24) as well as knowing the maximum export of SLTRiP to hepatocyte cytoplasm occurs from 12 to 20 h post-infection. When compared with WT-infected cells, 2840 transcripts changed more than 2-fold (Fig. 7A and supplemental Table S1). When compared with uninfected HepG2 cells, SLTRiP-KO-infected cells showed the expression of 1782 genes was up-regulated and 1868 were down-regulated (Fig. 7B, panel I), although wild type-infected cells showed the expression of 1163 genes was up-regulated and that of 6010 was down-regulated (Fig. 7B, panel II). Functional cluster analysis of host genes that were showing ≥3-fold changes are depicted in Fig. 7C, and Fig. 7, D and E, illustrates the KEGG pathway analysis of affected host genes in the case of SLTRiP-KO compared with WT parasite infection. The pie charts in Fig. 7, D and E, show the major pathways affected were RNA transport/processing, metabolism, immunity, and ribosome structure/function. To validate the NGS data, we selected two or more genes from various pathways (both up-/down-regulated), and a total of 23 genes were tested by qPCR on independent biological repeats of the experiment (Fig. 7F), which showed the qPCR results coincided with the NGS data. The list of primers used for qPCR, along with their sequences, is shown in Table 4.
FIGURE 7.
Host modulation by SLTRiP. A, Venn diagram for host genes affected during SLTRiP-KO infection compared with WT parasite infection. B, Venn diagram for host genes affected during SLTRiP-KO (panel I) or WT (panel II) parasite infection compared with uninfected cells. C, functional cluster analysis of host genes affected by SLTRiP-KO parasites. Nodes represented in a single color are involved in same biological process. D and E, pie chart showing pathways and % of total gene changed during SLTRiP-KO infection compared with WT parasite. D, up-regulated transcripts; E, down-regulated transcripts. F, validation of NGS data with qPCR. ACTB (β-actin) and HSP90AB1 were taken as housekeeping genes. Blue and red bars represent the fold changes (log10 scale) in host genes observed by qPCR and NGS, respectively. Error bars represent the standard deviation in the average value of the qPCR data.
Discussion
LS are clinically silent and constitute ideal targets for causal prophylactic drugs and vaccines, because parasite numbers are limited, and intervention at this stage prevents progression of the disease to the blood stage (35). Tryptophan-rich proteins in blood stages of P. vivax (PvTRAg39.8) have been shown to generate antibody responses in naturally exposed humans. PvTRAg39.8 also contains strong B- and T-cell epitopes (34). Recombinant SLTRiP forms multimers in solution and is stable over a wide range of temperatures (25–55 °C). The multimeric nature of SLTRiP might be the reason for its strong immunogenicity, as it allows a slow release of antigen in immunized mice. The liver stage vaccine candidate CSP (recombinant) also shows oligomerization (36) and is known to be a strong antigen. SLTRiP's multimeric nature and thermostability are advantageous from a vaccine point of view. Polyclonal antibodies against SLTRiP do not cross-react with any protein of blood stage parasites, an indication that SLTRiP is distinct from blood stage-specific tryptophan-rich proteins.
SLTRIP Immunization Protects against Sporozoite Challenge Primarily through the Cellular Arm of the Immune System
Mouse immunization with SLTRiP conferred highly significant protection against sporozoite challenge, indicating SLTRiP has vaccine potential. SLTRiP generated a strong antibody response in immunized animals. Previous work in P. yoelii with PypAg1 also showed a protective immune response against blood stage parasites (37). In contrast to this, anti-SLTRiP polyclonal antibodies showed minimal protection against sporozoite challenge. The above finding also implies that parasites might be using SLTRiP's highly antigenic exposed epitopes as a smoke screen to hide critical protective epitopes from the immune system.
SLTRIP immunization offers protection that mainly comes from the cellular arm (T-cells), although the humoral arm (antibodies) plays a minimal role. Innate immune responses are likely to play a small role in protection. Protective immunity during the liver stage of malaria parasite infection is mainly because of T-cell responses against sporozoite and liver stage antigens, with the CSP being the most abundant and prominent in protection (38). Recently, it has been shown via the use of CSP transgenic mice (CSP-tolerized) or CSP-swapped transgenic parasites that CSP is dispensable for protection, and other antigens may also provide strong protection in the absence of CSP (39, 40).
Role of SLTRiP in Parasite Development
Deletion of the SLTRiP gene resulted in defective LS growth. Parasites lacking SLTRiP were able to invade HepG2 cells with the same efficiency as WT, indicating that it was not essential for sporozoite invasion of the host cell. Previous studies showed that sporozoites migrate through several hepatocytes before invading one with the formation of a parsitophorous vacuole (41). Other parasite proteins like CSP, TRAP, SPECT1, and SPECT2 play roles in movement, receptor interaction, and induction of the protective immune response (42–44).
Parasites deficient in SLTRiP had an unchanged phenotype during the mosquito stage, indicating SLTRiP-KO parasites had normal development in the mosquito tissues. Because we could not detect expression of SLTRiP in blood stages, we therefore anticipated it had no role, and indeed, SLTRiP-deficient parasites had a normal phenotype during the blood stages. Previous studies have shown that proteins like UIS3, UIS4, SLARP, L-FABP, and others regulate liver stage development of parasites without affecting the blood or mosquito stage (45–47). These proteins perform liver stage-specific functions and are only essential during the liver stage. SLTRiP is also a parasite protein that performs stage-specific functions during the liver stage of malaria parasites.
SLTRiP is likely exported to the hepatocyte cytoplasm through the PEXEL-mediated export mechanism, as it contains putative PEXEL motifs. Previously, three liver-stage proteins have been shown to be exported to hepatocyte cytoplasm CSP (7, 48), IBIS1 (49), and LISP2 (50). In the case of CSP, ectopic overexpression of CSP in HeLa cells led to change in levels of over 1000 transcripts (7). This led us to ponder whether SLTRIP also has a similar effect on infected hepatocytes.
Role of SLTRiP in Host Modulation
To understand the function of SLTRiP with respect to the host modulation, we performed RNAseq-based transcriptome profiling of HepG2 cells infected with WT-GFP, SLTRiP-KO, unrelated transfection control parasites, and normal HepG2 cells. By comparison of infected host cell transcriptome profiles, it was observed that SLTRiP interferes with multiple pathways in the host cell, e.g. RNA processing and transport, structural components of the ribosome, immunity, focal adhesion, and the cytoskeleton. Several components of the proteasomal machinery, signaling, and solute transporters of host cells were also modified. Although deletion of the SLTRiP gene from the parasite caused a 1-day delay in liver stage growth, it led to the modulation of roughly 2800 host genes. At this juncture, it is not clear how these contrasting observations can best be reconciled. It is possible that deletion of SLTRiP may activate a back-up mechanism in the parasite.
The presence of 19 tryptophan residues in SLTRiP makes it a unique protein associated with protein-protein interaction and signaling events. We hypothesize that due to the presence of a large number of tryptophan residues, SLTRiP might be able to interact with several proteins in the host cell and thus regulate various pathways of the host cell. Alternatively, although we do not have any direct proof, it might be targeting host miRNA machinery to affect so many genes. There have been many reports demonstrating that a single protein (e.g. CSP, NS1, and ROP16) in diverse pathogens can regulate various pathways in the host cell (7, 26, 51).
In short, this study demonstrates that a tryptophan-rich protein has vaccine potential, and it regulates the development of the liver stage of the malaria parasite, where it plays a role in the growth of the parasite.
Supplementary Material
Acknowledgments
We thank Santosh Kumar Jena, Vivek Kumar Pandey, and Surender Singh Rawat for their technical assistance in mosquito breeding. We thank Dr. Arnab Mukhopadhyay and Dr. Awadhesh Pandit for their help in collecting the NGS data at the NII NGS core facility and Vikash Kumar at the NII Central FACS facility for cell sorting.
This work was supported in part by Ramalingaswami Fellowship Grant BT/HRD/35/02/13/2008 from the Department of Biotechnology, Government of India, conferred (to A. P. S.) and generous funding from an NII core grant. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- EEF
- exo-erythrocytic form
- IFA
- immunofluorescence assay
- LS
- liver stage
- hpi
- hour post-infection
- NGS
- next-generation sequencing
- SLTRiP
- sporozoite, liver stage tryptophan-rich protein
- SNA
- sporozoite neutralization assay
- CSP
- circumsporozoite protein
- qPCR
- quantitative PCR
- GFC
- gel filtration chromatography.
REFERENCES
- 1. Amino R., Thiberge S., Martin B., Celli S., Shorte S., Frischknecht F., Ménard R. (2006) Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat. Med. 12, 220–224 [DOI] [PubMed] [Google Scholar]
- 2. Prudêncio M., Rodriguez A., Mota M. M. (2006) The silent path to thousands of merozoites: the Plasmodium liver stage. Nat. Rev. Microbiol. 4, 849–856 [DOI] [PubMed] [Google Scholar]
- 3. Sturm A., Amino R., van de Sand C., Regen T., Retzlaff S., Rennenberg A., Krueger A., Pollok J. M., Menard R., Heussler V. T. (2006) Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 313, 1287–1290 [DOI] [PubMed] [Google Scholar]
- 4. Aly A. S., Vaughan A. M., Kappe S. H. (2009) Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 63, 195–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaughan A. M., Kappe S. H. (2012) Malaria vaccine development: persistent challenges. Curr. Opin. Immunol. 24, 324–331 [DOI] [PubMed] [Google Scholar]
- 6. Hill A. V., Reyes-Sandoval A., O'Hara G., Ewer K., Lawrie A., Goodman A., Nicosia A., Folgori A., Colloca S., Cortese R., Gilbert S. C., Draper S. J. (2010) Prime-boost vectored malaria vaccines: progress and prospects. Hum. Vaccin. 6, 78–83 [DOI] [PubMed] [Google Scholar]
- 7. Singh A. P., Buscaglia C. A., Wang Q., Levay A., Nussenzweig D. R., Walker J. R., Winzeler E. A., Fujii H., Fontoura B. M., Nussenzweig V. (2007) Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell 131, 492–504 [DOI] [PubMed] [Google Scholar]
- 8. Clyde D. F. (1975) Immunization of man against Falciparum and Vivax malaria by use of attenuated sporozoites. Am. J. Trop. Med. Hyg. 24, 397–401 [DOI] [PubMed] [Google Scholar]
- 9. Hoffman S. L., Goh L. M., Luke T. C., Schneider I., Le T. P., Doolan D. L., Sacci J., de la Vega P., Dowler M., Paul C., Gordon D. M., Stoute J. A., Church L. W., Sedegah M., Heppner D. G., et al. (2002) Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J. Infect. Dis. 185, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 10. Mellouk S., Lunel F., Sedegah M., Beaudoin R. L., Druilhe P. (1990) Protection against malaria induced by irradiated sporozoites. Lancet 335, 721. [DOI] [PubMed] [Google Scholar]
- 11. Agnandji S. T., Lell B., Soulanoudjingar S. S., Fernandes J. F., Abossolo B. P., Conzelmann C., Methogo B. G., Doucka Y., Flamen A., Mordmüller B., Issifou S., Kremsner P. G., Sacarlal J., Aide P., Lanaspa M., et al. (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365, 1863–1875 [DOI] [PubMed] [Google Scholar]
- 12. Bejon P., Lusingu J., Olotu A., Leach A., Lievens M., Vekemans J., Mshamu S., Lang T., Gould J., Dubois M. C., Demoitié M. A., Stallaert J. F., Vansadia P., Carter T., Njuguna P., et al. (2008) Efficacy of RTS,S/AS01E vaccine against malaria in children 5–17 months of age. N. Engl. J. Med. 359, 2521–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olotu A., Lusingu J., Leach A., Lievens M., Vekemans J., Msham S., Lang T., Gould J., Dubois M. C., Jongert E., Vansadia P., Carter T., Njuguna P., Awuondo K. O., Malabeja A., et al. (2011) Efficacy of RTS,S/AS01E malaria vaccine and exploratory analysis on anti-circumsporozoite antibody titers and protection in children aged 5–17 months in Kenya and Tanzania: a randomised controlled trial. Lancet Infect. Dis. 11, 102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sacarlal J., Aide P., Aponte J. J., Renom M., Leach A., Mandomando I., Lievens M., Bassat Q., Lafuente S., Macete E., Vekemans J., Guinovart C., Sigaúque B., Sillman M., Milman J., et al. (2009) Long-term safety and efficacy of the RTS,S/AS02A malaria vaccine in Mozambican children. J. Infect. Dis. 200, 329–336 [DOI] [PubMed] [Google Scholar]
- 15. Pinzon-Charry A., Good M. F. (2008) Malaria vaccines: the case for a whole-organism approach. Expert Opin. Biol. Ther. 8, 441–448 [DOI] [PubMed] [Google Scholar]
- 16. Kappe S. H., Mikolajczak S. A. (2011) Immunology. Another shot at a malaria vaccine. Science 334, 460–461 [DOI] [PubMed] [Google Scholar]
- 17. van Dijk M. R., Douradinha B., Franke-Fayard B., Heussler V., van Dooren M. W., van Schaijk B., van Gemert G. J., Sauerwein R. W., Mota M. M., Waters A. P., Janse C. J. (2005) Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc. Natl. Acad. Sci. U.S.A. 102, 12194–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jobe O., Lumsden J., Mueller A. K., Williams J., Silva-Rivera H., Kappe S. H., Schwenk R. J., Matuschewski K., Krzych U. (2007) Genetically attenuated Plasmodium berghei liver stages induce sterile protracted protection that is mediated by major histocompatibility complex class I-dependent interferon-γ-producing CD8+ T-cells. J. Infect. Dis. 196, 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weiss W. R., Sedegah M., Beaudoin R. L., Miller L. H., Good M. F. (1988) CD8+ T-cells (cytotoxic/suppressors) are required for protection in mice immunized with malaria sporozoites. Proc. Natl. Acad. Sci. U.S.A. 85, 573–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Broer R., Boson B., Spaan W., Cosset F. L., Corver J. (2006) Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 80, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jalah R., Sarin R., Sud N., Alam M. T., Parikh N., Das T. K., Sharma Y. D. (2005) Identification, expression, localization and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol.. Biochem. Parasitol. 142, 158–169 [DOI] [PubMed] [Google Scholar]
- 22. Kitagawa Y., Yamaguchi M., Zhou M., Komatsu T., Nishio M., Sugiyama T., Takeuchi K., Itoh M., Gotoh B. (2011) A tryptophan-rich motif in the human parainfluenza virus type 2 V protein is critical for the blockade of toll-like receptor 7 (TLR7)- and TLR9-dependent signaling. J. Virol. 85, 4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komla-Soukha I., Sureau C. (2006) A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 80, 4648–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albuquerque S. S., Carret C., Grosso A. R., Tarun A. S., Peng X., Kappe S. H., Prudêncio M., Mota M. M. (2009) Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events. BMC Genomics 10, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skariah S., Mordue D. G. (2012) Identification of Toxoplasma gondii genes responsive to the host immune response during in vivo infection. PLoS One 7, e46621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Chassey B., Aublin-Gex A., Ruggieri A., Meyniel-Schicklin L., Pradezynski F., Davoust N., Chantier T., Tafforeau L., Mangeot P. E., Ciancia C., Perrin-Cocon L., Bartenschlager R., André P., Lotteau V. (2013) The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 9, e1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X. Version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 28. Larsen J. E., Lund O., Nielsen M. (2006) Improved method for predicting linear B-cell epitopes. Immunome Res. 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arndt C., Koristka S., Bartsch H., Bachmann M. (2012) Native polyacrylamide gels. Methods Mol. Biol. 869, 49–53 [DOI] [PubMed] [Google Scholar]
- 30. Mombaerts P., Iacomini J., Johnson R. S., Herrup K., Tonegawa S., Papaioannou V. E. (1992) RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 [DOI] [PubMed] [Google Scholar]
- 31. Kumar K. A., Oliveira G. A., Edelman R., Nardin E., Nussenzweig V. (2004) Quantitative Plasmodium sporozoite neutralization assay (TSNA). J. Immunol. Methods 292, 157–164 [DOI] [PubMed] [Google Scholar]
- 32. Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. (2012) GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40, W478–W483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 [DOI] [PubMed] [Google Scholar]
- 34. Garg S., Chauhan S. S., Singh N., Sharma Y. D. (2008) Immunological responses to a 39.8 kDa Plasmodium vivax tryptophan-rich antigen (PvTRAg39.8) among humans. Microbes Infect. 10, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 35. Hoffman S. L., Doolan D. L. (2000) Malaria vaccines-targeting infected hepatocytes. Nat. Med. 6, 1218–1219 [DOI] [PubMed] [Google Scholar]
- 36. Plassmeyer M. L., Reiter K., Shimp R. L., Jr., Kotova S., Smith P. D., Hurt D. E., House B., Zou X., Zhang Y., Hickman M., Uchime O., Herrera R., Nguyen V., Glen J., Lebowitz J., et al. (2009) Structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J. Biol. Chem. 284, 26951–26963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burns J. M., Jr., Adeeku E. K., Dunn P. D. (1999) Protective immunization with a novel membrane protein of Plasmodium yoelii-infected erythrocytes. Infect. Immun. 67, 675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuji M., Zavala F. (2003) T-cells as mediators of protective immunity against liver stages of Plasmodium. Trends Parasitol. 19, 88–93 [DOI] [PubMed] [Google Scholar]
- 39. Grüner A. C., Mauduit M., Tewari R., Romero J. F., Depinay N., Kayibanda M., Lallemand E., Chavatte J. M., Crisanti A., Sinnis P., Mazier D., Corradin G., Snounou G., Rénia L. (2007) Sterile protection against malaria is independent of immune responses to the circumsporozoite protein. PLoS ONE 2, e1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar K. A., Sano G., Boscardin S., Nussenzweig R. S., Nussenzweig M. C., Zavala F., Nussenzweig V. (2006) The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature 444, 937–940 [DOI] [PubMed] [Google Scholar]
- 41. Mota M. M., Pradel G., Vanderberg J. P., Hafalla J. C., Frevert U., Nussenzweig R. S., Nussenzweig V., Rodríguez A. (2001) Migration of Plasmodium sporozoites through cells before infection. Science 291, 141–144 [DOI] [PubMed] [Google Scholar]
- 42. Frevert U., Engelmann S., Zougbédé S., Stange J., Ng B., Matuschewski K., Liebes L., Yee H. (2005) Intravital observation of Plasmodium berghei sporozoite infection of the liver. PLoS Biol. 3, e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishino T., Yano K., Chinzei Y., Yuda M. (2004) Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PLoS Biol. 2, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishino T., Chinzei Y., Yuda M. (2005) A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell. Microbiol. 7, 199–208 [DOI] [PubMed] [Google Scholar]
- 45. Mueller A. K., Camargo N., Kaiser K., Andorfer C., Frevert U., Matuschewski K., Kappe S. H. (2005) Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U.S.A. 102, 3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Silvie O., Goetz K., Matuschewski K. (2008) A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog 4, e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mikolajczak S. A., Jacobs-Lorena V., MacKellar D. C., Camargo N., Kappe S. H. (2007) L-FABP is a critical host factor for successful malaria liver stage development. Int. J. Parasitol. 37, 483–489 [DOI] [PubMed] [Google Scholar]
- 48. Khan Z. M., Ng C., Vanderberg J. P. (1992) Early hepatic stages of Plasmodium berghei: release o f circumsporozoite protein and host cellular inflammatory response. Infect. Immun. 60, 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ingmundson A., Nahar C., Brinkmann V., Lehmann M. J., Matuschewski K. (2012) The exported Plasmodium berghei protein IBIS1 delineates membranous structures in infected red blood cells. Mol. Microbiol. 83, 1229–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orito Y., Ishino T., Iwanaga S., Kaneko I., Kato T., Menard R., Chinzei Y., Yuda M. (2013) Liver-specific protein 2: a Plasmodium protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol. Microbiol. 87, 66–79 [DOI] [PubMed] [Google Scholar]
- 51. Jensen K. D., Hu K., Whitmarsh R. J., Hassan M. A., Julien L., Lu D., Chen L., Hunter C. A., Saeij J. P. (2013) Toxoplasma gondii rhoptry 16 kinase promotes host resistance to oral infection and intestinal inflammation only in the context of the dense granule protein GRA15. Infect. Immun. 81, 2156–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.