Background: E. faecium infections are increasingly refractory to antibiotics, encouraging vaccine approaches.
Results: Structures of novel protective surface polysaccharides and a teichoic acid were identified from a multidrug-resistant strain.
Conclusion: Detection of these antigens on diverse clinical strains predicts broad coverage for a potential multivalent vaccine.
Significance: Results suggest the feasibility of a glycoconjugate vaccine to prevent E. faecium disease.
Keywords: antigen, carbohydrate structure, Enterococcus, teichoic acid, vaccine
Abstract
The incidence of multidrug-resistant Enterococcus faecium hospital infections has been steadily increasing. With the goal of discovering new vaccine antigens, we systematically fractionated and purified four distinct surface carbohydrates from E. faecium endocarditis isolate Tx16, shown previously to be resistant to phagocytosis in the presence of human serum. The two most abundant polysaccharides consist of novel branched heteroglycan repeating units that include signature sugars altruronic acid and legionaminic acid, respectively. A minor high molecular weight polysaccharide component was recognized as the fructose homopolymer levan, and a glucosylated lipoteichoic acid (LTA) was identified in a micellar fraction. The polysaccharides were conjugated to the CRM197 carrier protein, and the resulting glycoconjugates were used to immunize rabbits. Rabbit immune sera were evaluated for their ability to kill Tx16 in opsonophagocytic assays and in a mouse passive protection infection model. Although antibodies raised against levan failed to mediate opsonophagocytic killing, the other glycoconjugates induced effective opsonic antibodies, with the altruronic acid-containing polysaccharide antisera showing the greatest opsonophagocytic assay activity. Antibodies directed against either novel heteroglycan or the LTA reduced bacterial load in mouse liver or kidney tissue. To assess antigen prevalence, we screened a diverse collection of blood isolates (n = 101) with antibodies to the polysaccharides. LTA was detected on the surface of 80% of the strains, and antigens recognized by antibodies to the two major heteroglycans were co-expressed on 63% of these clinical isolates. Collectively, these results represent the first steps toward identifying components of a glycoconjugate vaccine to prevent E. faecium infection.
Introduction
Enterococcus species are a leading cause of nosocomial infections in hospital intensive care units where they affect patients debilitated by underlying co-morbidities such as malignancy, renal insufficiency, organ transplantation, poorly controlled diabetes, or recent surgery (1–3). In hospitals in the United States, they are the second most common organism associated with bloodstream infections and rank fourth for incidence of surgical site infections (4, 5). Enterococcus faecium is the most therapeutically challenging of the Enterococcus species due to a combination of intrinsic antibiotic resistance and the acquisition through horizontal gene transfer of additional drug resistance and virulence determinants that facilitate persistence in the hospital environment (6, 7). Of particular concern is the rapid rise of vancomycin-resistant Enterococcus (VRE)2 infections, which in 2009–2010 composed 83 and 62% of E. faecium bloodstream infections and surgical site infections in the United States, respectively (4). VRE strains are also associated with severalfold higher mortality rates than susceptible strains (8, 9). Combating multidrug-resistant E. faecium infections with newer antibiotics such as linezolid and daptomycin involves prolonged treatments during which resistance may arise, leaving physicians with limited treatment options (10–14). As a result, there is an urgent need to explore alternative vaccine-based approaches to treat E. faecium infections.
Capsular polysaccharide (CPS) conjugates of the Gram-positive Streptococcus pneumoniae pathogen have proven effective at controlling pneumococcal disease (15). There are also exploratory CPS conjugate vaccines currently in clinical trials to prevent Streptococcus agalactiae and Staphylococcus aureus disease (ClinicalTrials.gov registry numbers NCT01364571, NCT02046148 (16, 17)). Although putative E. faecium CPS biosynthetic genes have been predicted from comparative genomic analysis (18), definitive structures of high molecular weight polysaccharides have not been previously described. To date, structural characterization has been limited to the identification of E. faecium teichoic acids, including lipoteichoic acid and a wall teichoic acid (19, 20). E. faecium Tx16 was isolated from an endocarditis patient (21). This strain was shown to be refractory to phagocytosis by neutrophils and is representative of a nosocomial lineage responsible for the majority of multidrug-resistant E. faecium infections (21–24). The strain is resistant to and harbors genetic resistance determinants for chloramphenicol, macrolide, aminoglycoside, β-lactam, and tetracycline antibiotics (22). The presence of a protective surface polysaccharide antigen on this strain was implicated by the observation that exposure to periodate treatment rendered it susceptible to phagocytic killing, and crude carbohydrate extracted from this strain selectively blocked opsonophagocytic killing activity of whole cell immune serum (21, 25). Here, we describe the characterization of four distinct surface polysaccharides from this strain that may account for its resistance to killing by neutrophils. As neutrophils are essential for rapid clearance of E. faecium in mice (26), we used opsonophagocytic killing assays (OPA) to evaluate the functional activity of antibodies elicited by each polysaccharide and/or their carrier protein conjugates. To assess coverage of these antigens for a potential vaccine, we used flow cytometry to measure the presence of structurally related polysaccharides in a collection of clinically relevant E. faecium strains.
Experimental Procedures
Bacterial Strains and Culture Conditions
E. faecium strains were routinely cultured without aeration at 37 °C in Columbia broth supplemented with 2% glucose. For fermentations, a 500-ml seed culture was added to 7.5 liters of the same media in an 8-liter stirred tank reactor under pH control and was grown for 6 or 24 h. Polysaccharide extraction was based on an E. faecalis procedure (27). After killing by heat treatment (1 h, 65 °C), cells were harvested by centrifugation, resuspended in 150 ml of Tris/sucrose buffer, and treated overnight with 1 mg/ml lysozyme and 10 units/ml mutanolysin at 37 °C. After centrifugation (10,000 rpm for 20 min), the supernatant was treated with 100 μg/ml RNase and 10 units/ml DNase for 8 h at 37 °C and then Pronase (50 μg/ml) overnight at 42 °C. Ethanol was added to 25% (v/v), and the precipitate was discarded after centrifugation. The supernatant was adjusted to 75% (v/v) ethanol at room temperature, and the resulting precipitate was retained. After washing twice with 75% room temperature ethanol, the pellet was dried with a stream of nitrogen and resuspended in 20 ml of 50 mm Tris, pH 7.5, and 0.05% NaN3. In this way, 2–3 g of crude carbohydrate was obtained from 50 to 100 g of wet cells.
Polysaccharide Purification
Crude polysaccharides were initially separated on a size exclusion chromatography (SEC) Sephacryl S-400 column (16/60 and 26/100 columns in series) equilibrated with 50 mm Tris, pH 7.5, 100 mm NaCl with 0.5 ml/min flow rate. Fractions were monitored by UV absorption at 214, 254, and 280 nm, by native PAGE with Stains-All detection reagent and with colorimetric assays to detect the presence of carbohydrates (anthrone (28) and deoxy-sugar (29)). 2-Fold diluted individual SEC pools were subjected to anion exchange column chromatography (AEC) with two 5-ml HiTrap Q HP in series equilibrated with 25 mm Tris, 50 mm NaCl. Carbohydrates were eluted with a 1 m NaCl gradient spanning 10–15 column volumes. Fractions with carbohydrate activity were pooled, concentrated with 7-kDa MWCO spin filters (PALL Corp.), dialyzed against water, and freeze-dried prior to biochemical analysis.
Carrier Protein Conjugation
CRM197 carrier protein conjugates of purified polysaccharides were prepared via the activation of carbohydrate hydroxyl groups using the cyanylating reagent 1-cyano-4-dimethylamino-pyridinium tetrafluoroborate (30). 1-Cyano-4-dimethylamino-pyridinium tetrafluoroborate (50 μl at 100 mg/ml in acetonitrile) was added to a solution of the antigen (500 μl of 10 mg/ml polysaccharide in water), and the resulting solution was mixed by vortexing for 30 s. Subsequently, 50 μl of a 0.2 m triethylamine solution was added, and reaction mixture was mixed gently for 2 min. CRM197 (1 ml of 5 mg/ml in 200 mm HEPES buffer, pH 8.8) was introduced, and the mixture was stirred for 16 h at room temperature with a magnetic stir bar. The reaction mixture was then transferred to a centrifuge filter with 100-kDa MWCO (Millipore) and spun at 2000 rpm for 1–2 h to remove low molecular weight species. A 30-kDa MWCO was used for conjugations with nonlipidated teichoic acid to avoid loss of conjugate. The retentate was washed three times with 4 ml of sterile PBS. The final retentate was further chromatographed by size exclusion chromatography (as above) to remove free carrier protein and unconjugated polysaccharide from conjugate. Fractions were monitored by absorption at 280 nm. Fractions corresponding to the conjugate (positive for the presence of both protein and polysaccharide) were pooled, and the conjugate was characterized by the determination of the carbohydrate and protein content using anthrone (28) and Lowry (31) colorimetric assays, respectively.
Carbohydrate Structural Analysis
Structural determination of carbohydrate samples involved 1H NMR and two-dimensional NMR analysis (DQCOSY, TOCSY, NOESY/ROESY, and 1H/13C HSQC) and GC-MS. The molecular weight of each purified polysaccharide was determined by SEC-MALLS. NMR experiments were carried out on a Varian INOVA 500 MHz (1H) spectrometer with a 3-mm gradient probe at 25 °C or higher temperature as indicated in the text. Polysaccharide samples were dissolved in 99.8% D2O, and chemical shifts were referenced to an internal acetone reference (2.23 ppm for 1H and 31.45 ppm for 13C). All two-dimensional experiments were performed using standard pulse sequences, DQCOSY, TOCSY (mixing time = 120 ms), ROESY (mixing time = 500 ms), and 1H-13C HSQC/HMBC (long range relay transfer delay = 100 ms). Monosaccharide composition was determined by GC-MS of alditol acetate derivatives (32). Samples were methylated by the Ciucanu and Kerek procedure (33). After hydrolysis, the partially methylated monosaccharide derivatives were converted to the corresponding alditol acetates and analyzed by GC-MS with ion-trap MS detector (Varian Saturn 2000). To prepare acetylated 2-butyl glycosides, samples were treated with 1 m HCl in (R)-2-BuOH (90 °C, 2 h), dried with air stream, and acetylated. For the identification of the absolute configuration of altruronic acid, OS1 was treated with 1 m HCl/MeOH (90 °C, 2 h) to obtain methyl ester and reduced with NaBH4 in water (2 h, 30 °C), where the excess NaBH4 was quenched with 4 m HCl. The boric acid by-product was evaporated with methanol twice, and the residue was treated with (R)-2-BuOH-AcCl (10:1) for 3 h at 90 °C, dried, acetylated, and analyzed by GC-MS (Varian Saturn 2000 ion-trap instrument, capillary column DB-17, 160–260 °C by 4 °/min). Weight-average molecular weight (¯Mw) of the polysaccharide samples were determined by in-line SEC-RI-UV-MALLS analysis. The polysaccharide samples were separated on a TOSOH TSKgel GMPWxl mixed-bed analytical size exclusion column (7.8 × 300 mm) and eluted with a 150 mm NaCl, 100 mm sodium phosphate, 0.1 mm EDTA, pH 6.7, mobile phase using an Agilent 1200 HPLC with an isocratic pump at a flow rate of 0.8 ml/min. The eluent was detected with in-line UV light (280 nm), MALLS (Wyatt HELEOS 18-angle MALLS detector with laser wavelength at 658 nm), and RI (Wyatt OptiLab Rex) detectors. Data analysis was performed using the Wyatt ASTRA version 5.1 software, and a dn/dc value of 0.154 ml/g was used for all polysaccharide samples.
Peptidoglycan Quantitation
The peptidoglycan levels in purified polysaccharides were determined by HPAEC-PAD analysis using the E. coli peptidoglycan (InvivoGen, San Diego) as the standard, the concentration of which was determined by HPAEC-PAD (using commercially available muramic acid as reference) and SEC-RI analysis. The percent of peptidoglycan in polysaccharide (w/w) was calculated. The analysis was performed as follows: E. faecium polysaccharide samples (100 μg) and reference standard in the linear molar range 0.2 to 1.4 nmol of peptidoglycan after injection were hydrolyzed in 400 μl of 4 n HCl, for 24 h at 100 °C. Hydrolyzed samples and standards were evaporated to dryness using Speedvac (Thermo Scientific) and reconstituted in 250 μl of high purity water. Chromatography of the samples (25 μl injection) was performed using a Dionex ICS3000 system with automatic autosampler and a CarboPac PA 1 (4 × 250 mm) analytical column and guard column (4 × 50 mm) (Sunnyvale, CA). Analyses were carried out as described previously (34).
IgG ELISA
Indirect ELISAs were used to measure serum IgG levels in response to vaccination. For Pf1, Pf2, and Pf4 antigens, 5 μg/ml unconjugated polysaccharide was applied in pH 10.0 bicarbonate coating buffer to high binding microtiter plates (Maxisorp, NUNC). For ELISAs measuring Pf3 LTA serum titers, 1 μg/ml antigen was combined with 1 μg/ml methylated human serum albumin for microplate coating to improve binding. After blocking with 1% BSA in PBS, microplates were incubated for 1 h room temperature with serially diluted sera from animals sampled at day 0 and at post-vaccination test bleed time points. Plates were subsequently processed according to a commercial peroxidase-based ELISA kit (KPL, Gaithersburg, MD).
Polyclonal Antibodies
Rabbits were prescreened prior to vaccination to exclude those showing detectable anti-LTA ELISA titers at serum dilutions of 1:100 or greater in assays using microplates coated with 1 μg/ml commercial E. faecalis LTA (Sigma) and 1 μg/ml methylated human serum albumin.
Polysaccharide Conjugate-specific Rabbit Antibodies
Groups of 3–4 rabbits were vaccinated i.m. with 25 μg of conjugated polysaccharide (based on polysaccharide) and 75 μg (75 ISCO units) of Iscomatrix adjuvant (CSL Biotherapies Inc.) at weeks 0, 6, and 8. Sera were sampled from each rabbit prior to vaccination (week 0) and at the post-dose three time point (week 10). The CRM197 glycoconjugates elicited robust ELISA responses with IgG end point titers for all vaccinated rabbit sera exceeding 10,000 at week 10. ELISA responses of the matched preimmune week 0 sera were negligible.
Nonconjugated Polysaccharide Antibodies
The vaccination schedule for the Pf3 LTA antigen, adapted from a previously described procedure (20), involved subcutaneous injections of 100 μg with 100 μg of Iscomatrix adjuvant at weeks 0 and 1, followed by three weekly i.v. injections of 10 μg (minus adjuvant) during weeks 2–4. Sera were sampled from each rabbit before vaccination (week 0) and after the final vaccination (week 5). ELISA IgG end point titer responses for all four vaccinated rabbits at week 5 were ∼10,000. Studies were carried out in accordance with established guidelines and protocols and were reviewed and approved by Pfizer Institutional Animal Care and Use Committee.
Mouse Passive Protection Model
The animal model was performed as described previously (35). In brief, 6–8-week-old female BALB/c mice (Charles River Laboratories Germany GmbH) were immunized intraperitoneally with either 200 μl of normal rabbit serum (NRS) or test sera. Immunizations were done 48 and 24 h before the bacterial challenge. Mice were infected intravenously with 7.0 × 106 CFUs of E. faecium Tx16 via the tail vein and sacrificed 24 h after challenge; colony counts in liver and kidney were determined. Statistical significance was determined by nonparametric Kruskal-Wallis test with Dunn's post-test using Graphpad Prism (version 5.0); p values ≤0.05 were considered statistically significant. Experiments were performed in compliance with the German animal protection law (TierSchG). The mice were housed and handled in accordance with good animal practice as defined by FELASA and the national animal welfare body GV-SOLAS. The animal welfare committees of the University of Freiburg (Regierungspräsidium Freiburg Az 35/9185.81/G-07/15) approved the animal experiments.
Immunofluorescence and Flow Cytometry
Polysaccharide-specific immune rabbit antisera and matched pre-bleed control sera were used as primary antibodies to assess surface expression of polysaccharides by flow cytometry. Overnight bacterial cultures were washed in 1× PBS (Life Technologies, Inc.) and killed by heating (1 h 60 °C). Cells were then blocked with 1% BSA in PBS (1 h room temperature) and incubated with primary rabbit antibody (1 h room temperature). After washing, this was followed by incubation with phycoerythrin-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) for fluorescence detection. The bacterial cells were fixed with 1% paraformaldehyde and analyzed with an Accuri C6 flow cytometer (BD Biosciences). The mean fluorescence intensity (MFI) of the phycoerythrin channel was determined for each sample (counting 20,000 events). E. faecium strains were tested in batches of ∼15× strains each, with Tx16 included as an internal control.
Opsonophagocytic Assays
Pre-frozen bacterial stocks of Tx16 were grown in the same fermentation media as used for polysaccharide purification (Columbia broth supplemented with 2% dextrose, cultured without aeration at 37 °C). Cells were pelleted and suspended to a concentration of 1 A600 unit/ml in PBS supplemented with 20% glycerol and frozen. Pretitered thawed bacteria were diluted to 1 × 105 CFU/ml in OPA buffer (Hanks' balanced salt solution (Life Technologies, Inc.), 0.1% gelatin, 1 mm MgCl2, 2.5 mm CaCl2) and 10 μl (103 CFU) opsonized with 10 μl of serially diluted sera for 1 h at 4 °C in a U-bottomed tissue culture microplate. Subsequently, 10 μl of 10% complement (baby rabbit serum, Pel-Freez 31061-3) and 20 μl of HL-60 cells (0.5 × 107/ml) were added to each well, and the mixture was shaken at 350 rpm (ThermoFisher MaxQ2000) for 1 h at 37 °C in a 5% CO2 incubator. 10 μl of each 50-μl reaction was transferred into the corresponding wells of a prewetted Millipore MultiScreenHTS HV filter plate containing 200 μl of water. After vacuum filtering the liquid, 150 μl of Columbia broth (with 2% glucose) was applied and filtered and plate incubated overnight at 37 °C. The next day, the colonies were enumerated after staining with Coomassie dye using an ImmunoSpot® analyzer and ImmunoCapture software. To establish the specificity of OPA activity, immune sera were preincubated with 20 μg/ml purified antigen prior to the opsonization step. The OPA includes control reactions without HL60 cells or complement, to demonstrate dependence of any observed killing on these components.
Bioinformatic Analysis
Strain E0155 was sequenced using both Ion Torrent (Life Technologies, Inc.) and MiSeq benchtop sequencer (Illumina). FASTQ sequence read files were imported into CLC Genomic Workbench to run de novo assembly workflow. The assembled contigs were then exported as FASTA format. 17 E. faecium genome sequences (1 complete, 16 draft) were collected from the NCBI GenBank database. Tx16 strain orthologues of Campylobacter jejuni legionaminic acid biosynthetic genes (36) were identified using BLAST searches (with e-values <10−30). The presence of this configuration of genes in other E. faecium strains was queried by loading the genomes in FASTA format into BIGSdb (Bacterial Isolate Genome Sequence Database (37)) and running a BLAST scan for all genes located between the M24 aminopeptidase and galE anchor genes that flank this 35-kb region of the Tx16 genome (22). After extracting related gene sequences, any structural (gene content) change was visualized by pairwise alignment against the TX16 genome using MultiPipMaker (38).
Results
Purification of Polysaccharides
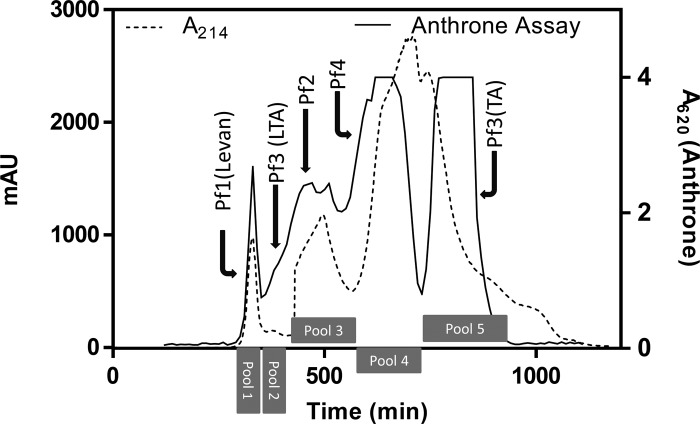
Extracellular polysaccharides were isolated from E. faecium Tx16 bacterial cells as described under “Experimental Procedures.” Cells were digested with mutanolysin and lysozyme to release cell wall-associated polysaccharides, and the soluble fraction was treated with nucleases and protease to degrade nucleic acids and proteins. Crude carbohydrate was concentrated by ethanol precipitation prior to fractionation by SEC. SEC fractions were consolidated into five major pools according to the major peaks of anthrone carbohydrate activity (Fig. 1). Four distinct polysaccharides designated Pf1 to Pf4 were subsequently identified and purified from these pools by AEC.
FIGURE 1.
Size exclusion chromatography profile of E. faecium Tx16 extracellular polysaccharides. Pools corresponding to the major anthrone activity peaks from which individual Pf1 to Pf4 polysaccharides were purified are indicated. mAU, one-thousandth of an absorbance unit.
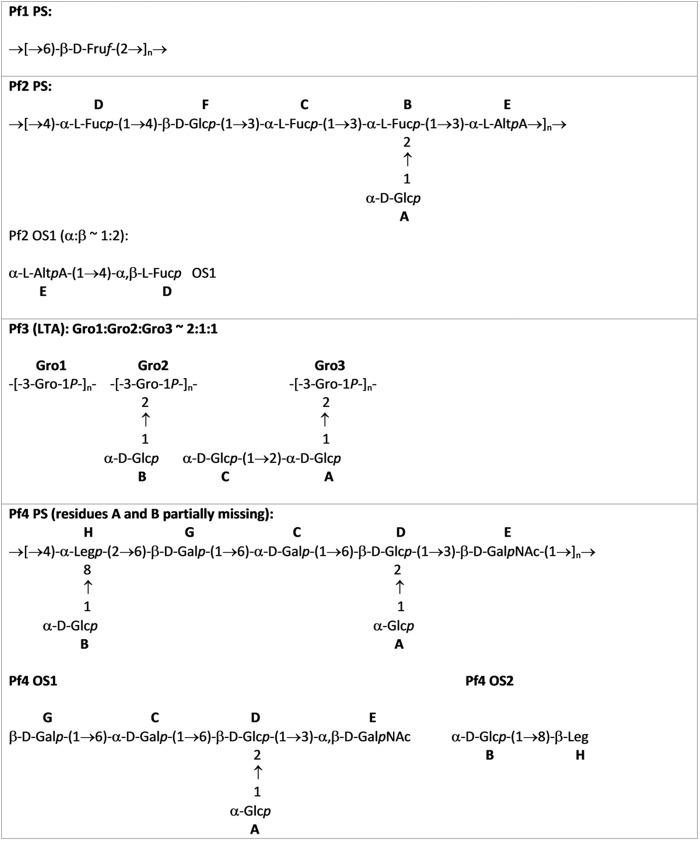
Pf1 Polysaccharide (a Fructose Homopolymer)
Pf1 had the highest molecular mass of the polysaccharides characterized and eluted as a single peak near the SEC void volume (pool 1, Fig. 1). 1H NMR spectrum contained no anomeric signals but showed multiple signals between 3.5 and 4.3 ppm. Analysis of the two-dimensional NMR data (Table 1 and supplemental Fig. S1) indicated regular structure of a levan-like polymer with the −6-β-d-Fruf-2-repeating unit. The linkage type was determined by comparison of the NMR spectra with published data for various fructose polymers of Bacillus species isolated from cloud water (39). In addition to the major series, the spectra contained minor signals of fructose of undefined origin, possibly ends of chain or different substitution type. GC analysis of the alditol acetates was consistent with the levan determination, as glucose and mannose derivatives were observed in equal amounts (the reduction of the C2 carbonyl group of fructose after hydrolysis produces mannitol and glucitol).
TABLE 1.
Pf1 (levan) 1H and 13C assignments (ppm) at 25 °C (600 MHz)
The acetone reference is 2.23/31.45 ppm.
| Residue | H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 |
|---|---|---|---|---|---|---|
| Fru | 3.67; 3.77/61.2 | /105.5 | 4.19/77.5 | 4.10/76.4 | 3.95/81.5 | 3.56; 3.90/64.6 |
Pf2 Polysaccharide (an Altruronic Acid-containing Heteroglycan)
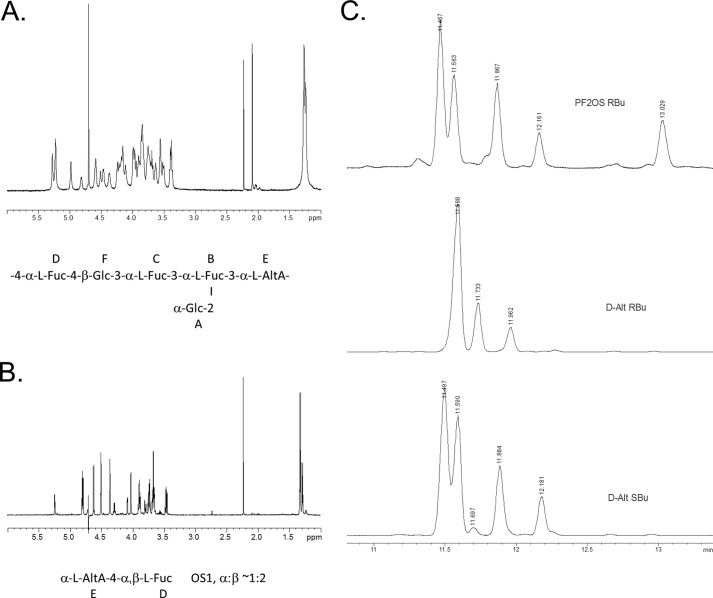
The Pf2 polysaccharide was purified from SEC pool 3 (Fig. 1) by AEC. The Pf2-containing anthrone positive peak eluted from the AEC column with 0.22 m NaCl (data not shown). The sample was concentrated and desalted on Sephadex G-15 prior to NMR analysis. GC analysis of the alditol acetates showed the presence of fucose and glucose in the ratio of 3:2. A set of two-dimensional NMR spectra of the phosphatidylserine (DQCOSY, TOCSY, NOESY/ROESY, and 1H/13C HSQC) was recorded and assigned (Table 2; Fig. 2A, and supplemental Fig. S2). Spectra contained spin systems of six monosaccharides. Three fucose and two glucose residues and a monosaccharide with TOCSY signal pattern and vicinal H-H coupling constants typical for β-galacturonic acid in pyranose form were identified. However, an attempt to determine absolute configuration by GC-MS of acetylated glycosides/esters with optically pure 2-butanol yielded no derivatives of galacturonic acid. In addition, 13C NMR data of this monosaccharide were not consistent with the expected values of β-galacturonic acid. Also, no NOE correlation between H-1 and H-5, which is typical for β-Galp, was observed. This situation pointed to the presence of α-l-altruronic acid in 1C4 conformation, which has H-1,2,3,4 orientation as in β-Gal and equatorial H-5, resulting in the absence of NOE between H-1 and H-5. To further confirm the tentative identification of α-l-altruronic acid, a partial hydrolysis of Pf2 was performed (0.5 m TFA, 90 °C, 2 h) to yield an acidic disaccharide (OS1), which was isolated by anion exchange chromatography and gel chromatography on Sephadex G-15. The purified OS1 generated a clean, completely interpretable 1H NMR spectrum (Fig. 2B and Table 2). Full assignment of one- and two-dimensional NMR spectra confirmed that the uronic acid had α-altro-configuration, being in good agreement with published data (40).
TABLE 2.
Pf2 polysaccharide (PS) and oligosaccharide (OS) 1H and 13C assignments (ppm) at 32 °C (D2O, 600 MHz)
NA indicates not applicable; ND indicates not detected.
| Residue | H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6a;b |
|---|---|---|---|---|---|---|
| A α-Glc | 5.27/100.3 | 3.51/72.8 | 3.69/74.4 | 3.38/70.7 | 3.76/73.5 | 3.75; 3.85/61.8 |
| B α-Fuc | 5.23/100.8 | 4.21/71.4 | 4.24/73.4 | 4.15/67.8 | 4.18/68.0 | 1.27/16.5 |
| C α-Fuc | 5.22/93.8 | 3.98/67.3 | 4.17/77.8 | 4.00/70.4 | 4.37/68.1 | 1.24/16.5 |
| D α-Fuc PS | 4.98/100.9 | 3.84/69.9 | 3.86/69.9 | 4.11/81.1 | 4.46/68.5 | 1.28/16.5 |
| D α-Fuc OS1 | 5.24/93.3 | 3.79/69.7 | 3.88/69.5 | 4.08/81.0 | 4.29/67.6 | 1.29/16.6 |
| D β-Fuc OS1 | 4.62/97.2 | 3.46/73.2 | 3.67/73.1 | 4.02/80.1 | 3.89/71.9 | 1.32/16.7 |
| E α-l-AltA PS | 4.82/102.6 | 3.90/71.5 | 3.73/77.3 | 4.51/70.7 | 4.57/78.3 | NA/ND |
| E α-l-AltA OS1 | 4.79/102.3 | 3.73/71.5 | 3.66/71.1 | 4.36/70.6 | 4.50/77.8 | NA/ND |
| J, Hz | J1,2 7 | J3,4 3 | J4,5 3 | |||
| F β-Glc | 4.60/100.1 | 3.40/74.5 | 3.63/75.5 | 3.56/78.3 | 3.56/76.7 | 3.83;3.95/61.2 |
FIGURE 2.
Structural analysis of altruronic acid heteroglycan (Pf2). A,1H NMR spectrum peaks around 2 ppm are acetone (internal standard) and acetic acid (from column buffer) B,1H NMR spectrum of the OS1 derivative. C, GC traces of acetylated 2-butyl glycosides derived from OS1 and d-altrose standard establishing l-configuration of altruronic acid.
Connections between monosaccharides in Pf2 were identified on the basis of NOE correlations (A1:B1,2; B1:E3; C1:B3; D1:F4; and F1:C3) and 13C chemical shifts. Absolute configurations of l-fucose and d-glucose were determined by GC analysis of the acetylated 2-butyl glycosides prepared with optically pure isomers of 2-butanol. For the determination of the absolute configuration of altruronic acid, OS1 was treated with HCl/MeOH, and the methyl ester was reduced with NaBH4. Resulting altrose was treated with (R)-2-BuOH and acetylated (see under “Experimental Procedures”). GC analysis of the product showed that it was identical to the standard prepared from d-altrose and (S)-2-BuOH and differed from the derivative obtained with (R)-2-BuOH, indicating l-configuration of the altruronic acid (Fig. 2C).
Methylation analysis of the Pf2 (GC of partially methylated alditol acetates) (33) showed the presence of 3-, 4-, and 2,3-substituted fucose residues, terminal and 4-substituted glucose residues, in agreement with NMR data. High orifice voltage ESI mass spectra of the polysaccharide confirmed its structure, containing a prominent peak at 937.7 atomic mass units (negative mode) or 939.7 atomic mass units (positive mode), which corresponded to the proposed repeating unit minus one H2O (calculated exact mass 938.3 atomic mass units) (Fig. 3). Other peaks corresponded to addition or loss of hexose (162 Da) or 6-deoxyhexose (146 Da) residues. The level of residual peptidoglycan in the Pf2 sample, resulting presumably from incomplete enzyme cleavage, was determined by HPAEC-PAD analysis of muramic acid to be ∼4% by mass.
FIGURE 3.
Structures of Pf1 to Pf4 polysaccharides (PS) and derived oligosaccharides (OS).
Pf3 Polysaccharide (a Glucosylated Lipoteichoic Acid)
The Pf3 polysaccharide was identified in SEC pools 1 and 2 of the fractionated crude polysaccharide extract from strain Tx16 (Fig. 1). The Pf3 eluted from the AEC column at 0.58 m NaCl and fractions showed very weak reactivities in the anthrone carbohydrate detection assay. Because the 1H NMR spectra of polysaccharides eluted from AEC with 0.58 m NaCl of SEC pool 1 and 2 were identical, these were combined, desalted on Sephadex G-15, and called Pf3. A set of two-dimensional NMR spectra (DQCOSY, TOCSY, ROESY, 1H/31P HMQC, and 1H/13C HSQC) was recorded and assigned (Table 3 and supplemental Fig. S3). Spectra contained spin systems of three monosaccharides, all α-Glcp, and phosphorylated glycerol. Structures of the four discrete teichoic acid forms representing the glycerol phosphate backbone with or without mono- and disaccharide substitutions and terminal Gro-1P units are shown in Fig. 3. Lipid signals were present (broad peak around 1 ppm in 1H spectrum), suggesting that the carbohydrate is a cell membrane-anchored LTA. The lipid part was not analyzed further. The nonglycosylated form is prominent (Gro1) and present in approximately equal proportions to the glycosylated forms (Gro2 + Gro3). Mono- and disaccharide-modified forms were present in equal proportions (Gro2 = Gro3). Connections between monosaccharides were identified on the basis of NOE correlations (A1:Gro3–2; B1:Gro2–2; and C1:A1,2) and 13C chemical shifts. 1H-31P HMQC showed correlations between glycerol H-1 and H-3 with 31P at 0.4 ppm (data not shown). The overall structure of the glucosylated LTA matches that of de-alanylated LTA purified from E. faecalis (41). The presence of this LTA in high molecular weight SEC fractions presumably reflects its micellar nature. SDS-PAGE analysis confirmed that Pf3 migrates as a low molecular weight compound in the presence of SDS detergent. Compounds of identical structure lacking lipid tail were purified by AEC from SEC pools 4 and 5. As they lack detectable amino sugars associated with wall teichoic acids (42), we speculate that these may have arisen through lipase cleavage during processing of the bacteria. The lipidated and nonlipidated teichoic acids were designated Pf3(LTA) and Pf3(TA), respectively.
TABLE 3.
Pf3 (LTA) 1H and 13C assignments (D2O, 28 °C, 600 MHz)
Data are collected from de-alanylated LTA (no d-alanine substitution at C2 of glycerol residues) as shown in the structures in Fig. 3.
| Residue | H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6a;b |
|---|---|---|---|---|---|---|
| A α-Glc | 5.46/95.9 | 3.69/75.4 | 3.89/72.4 | 3.48/70.6 | 3.96/73.0 | 3.78–3.89/61.7 |
| B α-Glc | 5.19/99.0 | 3.54/72.7 | 3.77/74.2 | 3.42/70.8 | 3.94/73.0 | 3.78–3.89/61.7 |
| C α-Glc | 5.18/96.6 | 3.59/72.4 | 3.82/73.7 | 3.46/70.6 | 3.92/73.0 | 3.78–3.89/61.7 |
| Gro1 | 3.93/67.6 | 4.07/70.5 | 4.00/67.6 | |||
| Gro2 | 4.05/65.9a | 4.14/76.5 | 4.05/66.4a | |||
| Gro3 | 4.05/65.9a | 4.22/76.1 | 4.05/66.4a | |||
| Gro4 (terminal Gro-1P) | 3.89;3.94/67.6 | 3.92/71.9 | 3.62;3.68/63.2 |
a These signals can be interchanged.
Pf4 Polysaccharide (a Legionaminic Acid-containing Heteroglycan)
A second novel polysaccharide designated Pf4 was recovered from Tx16 SEC pool 4 (Fig. 1), eluting from AEC with 0.2 m NaCl. Samples were heterogeneous, containing a predominant heteroglycan with small amounts of rhamnan and peptide contaminants. As it was not possible to purify them further by anion exchanger (retained) and Sephadex G-50 (eluted with void volume), these contaminants were most likely covalently linked to the primary polysaccharide. HPAEC-PAD analysis determined the molar ratio of Rha (from minor polymer contaminant) to Gal (from Pf4) to be ∼1:3. Absolute configurations of Glc, Gal, and GalN were determined by GC of acetylated 2-butanol derivatives and found to be all d (data not shown).
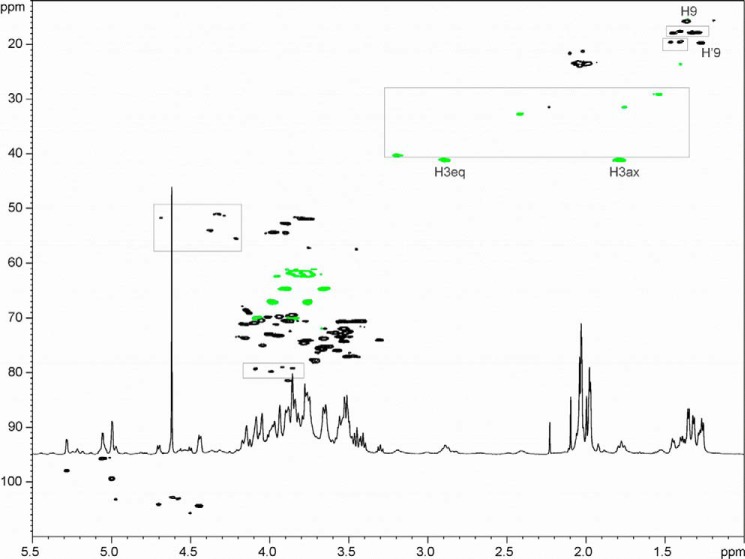
NMR analysis of the sample and derived fragments led to a heteroheptameric branched structure, where the side chain GlcA and -B were linked to the residues H and D (Table 4 and Figs. 3 and 4). Side chain glucose residues A and B were not present on all repeating units (both residues A and B are present in about 50% of the repeats, A:B ∼1:1); thus spectra also contained signals of monosubstituted residues H′ and D′. Residue H was identified as di-N-acetyl-legionaminic acid (Leg, 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid) based on NMR data. It had nine carbon chains with deoxy carbons at C-3 and C-9 and amino groups at C-5 and C-7 (identified by chemical shift of C-5 and C-7 around 54 ppm), typical for bacterial nonulosonic acids. It had the configuration of Leg (same as sialic acid) with H-3a,4,5,6 in an axial orientation, as deduced from large coupling constants, ∼10 Hz. Configurations of C-7 and C-8 were inferred from 1H and 13C NMR shifts of H/C 7–9, which are similar to the values described for the d-glycero-d-galacto-isomer (43). Amino groups were acetylated as indicated by the presence of the NMR signals of two N-acetyl groups.
TABLE 4.
Pf4 polysaccharide 1H and 13C assignments (D2O, 40 °C, 600 MHz)
Pf4 is heterogeneous and can be partially nonglucosylated at residue H (α-Leg) or residue D (β-Glc). NA indicates not applicable; ND indicates not determined.
| Residue | H/C-1 | H/C-2 | H/C-3(a,e) | H/C-4 | H/C-5 | H/C-6a;b | H/C-7 | H/C-8 | H/C-9 |
|---|---|---|---|---|---|---|---|---|---|
| A α-Glc | 5.28/97.9 | 3.52/72.8 | 3.76/74.1 | 3.45/70.5 | 4.00/72.9 | 3.77;3.83/61.7 | NA | NA | NA |
| B α-Glc | 5.06/95.7 | 3.50/72.5 | 3.55/73.4 | 3.41/70.5 | 3.52/74.3 | 3.76;3.84/61.7 | NA | NA | NA |
| C α-Gal | 5.00/99.4 | 3.85/69.4 | 3.89/70.5 | 4.05/70.4 | 4.09/70.9 | 3.84;4.07/70.0 | NA | NA | NA |
| D β-Glc | 4.70/104.1 | 3.51/77.0 | 3.56/75.9 | 3.54/70.6 | 3.64/75.2 | 3.76;3.98/67.1 | NA | NA | NA |
| D′ β-Glc | 4.50/105.7 | 3.30/74.0 | 3.46/76.9 | 3.53/70.5 | 3.61/75.2 | 3.75;3.99/67.0 | NA | NA | NA |
| E β-GalNAc | 4.58;4.61/103.0;102.8 | 3.89;3.88/52.7 | 3.88/81.4 | 4.13/69.1 | 3.67/75.6 | 3.76;3.82/62.1 | NA | NA | NA |
| G β-Gal | 4.44/104.4 | 3.52/72.0 | 3.65/73.7 | 3.94/69.7 | 3.78/74.5 | 3.65;3.90/64.7 | NA | NA | NA |
| H α-Leg | NA/ND | NA/ND | 1.79;2.89/41.1 | 3.72/77.7 | 3.79/51.8 | 41.5/73.6 | 3.97/54.3 | 3.94/73.2 | 1.35/15.7 |
| H′ α-Leg | NA/ND | NA/ND | 1.79;2.89/41.1 | 3.72/77.7 | 3.75/51.9 | 4.04/75.0 | 3.90/54.4 | 4.01/69.8 | 1.26/19.7 |
| H β-Leg OS2 | NA/ND | NA /96.7 | 1.87;2.30/40.6 | 3.94/68.3 | 3.75/54.0 | 4.14/71.5 | 4.03/53.8 | 3.86/73.0 | 1.20/15.6 |
| B α-Glc OS2 | 5.02/95.7 | 3.49/72.4 | 3.54/73.3 | 3.40/70.5 | 3.51/74.2 | 3.75;3.83/61.7 | NA | NA | NA |
FIGURE 4.
1H-13C HSQC spectrum of the legionaminic acid heteroglycan (PF4). Some major signals not related to the described polysaccharide structure are marked by boxes.
Partial acid hydrolysis of Pf4 with 0.5 m TFA (90 °C, 1.5 h) produced a mixture of the oligosaccharides resulting from the complete cleavage of Leg (H) and β-GlcNAc (E) glycosidic linkages and partial cleavage of the Gal linkage. A higher molecular mass peak in the gel chromatographic separation of hydrolysis products was attributed to peptidoglycan fragments. The level of peptidoglycan contamination in the Pf4 sample was determined independently by HPAEC-PAD analysis of muramic acid to be ∼9% by mass. The rhamnan was not recovered and was probably completely depolymerized under these conditions. Oligosaccharides were separated by anion exchange chromatography to give a neutral mixture of oligosaccharides (OS1) and acidic disaccharides (OS2). OS1 was heterogeneous due to partial substitution with Glc (A) and Gal (G) (Gal (G) was partially lost due to hydrolysis). NMR spectra of the OS1 were in agreement with the major product expected from partial acid hydrolysis (data not shown). ESI MS (negative mode) of OS1 showed peaks of Hex2HexNAc1 (m/z 544.6), Hex3HexNAc1 (m/z 706.7), and Hex4HexNAc1 (m/z 868.8). NMR data for Pf4 OS2 are included in Table 4. Negative mode ESI MS of OS2 contained main peak at m/z 495.4 (calculated molecular mass 496.19).
The sequence of monosaccharides in the repeating unit was established from NOE and HMBC data as follows: correlations A1:D1,2; B1:H8, C1:D6; D1:E3; E1:H4 were observed. Linkage of the Leg (H) residue to Gly-6 was based on the downfield shift of C-6 of G (64.7 ppm), which returned to a nonsubstituted position in OS1 (62 ppm). Structures of the repeat unit and OS1/OS2 oligosaccharides are shown in Fig. 3.
Evaluation of Tx16 Polysaccharides as Vaccine Antigens
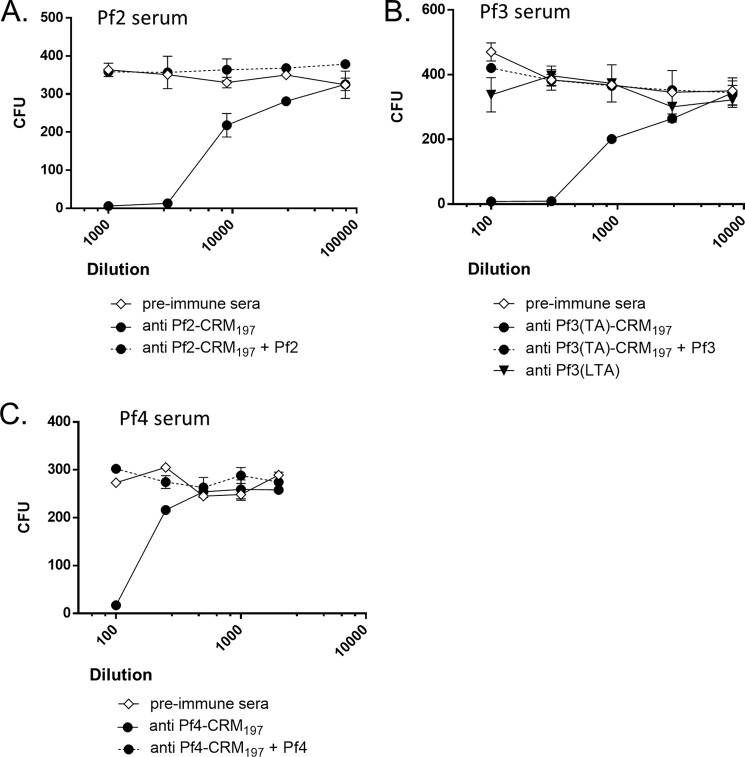
Rabbits were vaccinated with CRM197 carrier protein conjugates of the Pf1, Pf2, Pf3(TA), and Pf4 polysaccharides, as well as unconjugated Pf3(LTA), to determine whether these antigens could induce a protective immune response. Sera from rabbits with the highest polysaccharide antigen-specific ELISA antibody titers were tested in OPAs with HL60 (neutrophil-like promyelocyte) cells and baby rabbit serum as source of complement (Fig. 5). E. faecium Tx16 was highly susceptible to killing in OPA by the Pf2-CRM197 antisera (Fig. 5A). The killing was antigen-specific as opsonic activity was not observed with preimmune sera from the same rabbit, and the OPA activity could be completely blocked by preincubating the immune sera with purified unconjugated polysaccharide. The Pf3(TA)-CRM197 and Pf4-CRM197 antisera also killed this strain in an antigen-specific manner but were less potent than the Pf2-CRM197 antisera (Fig. 5, B and C). In contrast, the Pf1-CRM197 immune antisera (data not shown) and Pf3(LTA) antisera (Fig. 5B) were unable to kill Tx16 in the OPA.
FIGURE 5.
Susceptibility of E. faecium Tx16 to opsonophagocytic killing by antibodies to polysaccharide antigens Pf2 to Pf4. Preincubation of antisera with 20 μg/ml unconjugated antigen was used to test specificity. A, Tx16 and Pf2-CRM197 antisera. B, Tx16 and Pf3(TA)-CRM197 or Pf3(LTA) antisera. C, Tx16 and Pf4-CRM197 antisera.
The ability of a subset of the antisera to protect against E. faecium in vivo was tested in a mouse passive protection model. Mice were injected with rabbit sera prior to challenge with Tx16 bacteria. Mice were sacrificed 24 h later, and cfu counts were determined in homogenized kidney or liver tissue. Compared with unvaccinated normal rabbit sera (NRS), immune sera elicited by Pf2-CRM197, Pf3(LTA), or Pf4-CRM197 antigens significantly reduced cfus in disseminated Tx16 infections by ≥100-fold in kidneys and ≥50-fold in liver tissue (Fig. 6).
FIGURE 6.
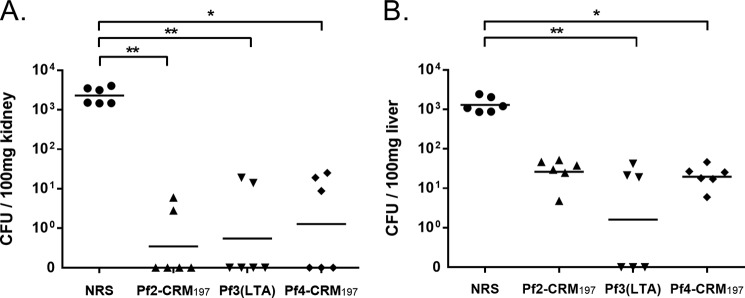
Protective activity of Pf2 to Pf4 immune sera in passive protection infection model. Passive immunization with anti Pf2-CRM197, Pf3(LTA), and Pf4-CRM197 sera promotes clearance of E. faecium Tx16 in kidneys (A) and livers (B) of mice in comparison with normal rabbit serum (NRS). 24 h after challenge, mice were sacrificed, and organs were aseptically removed to assess viable counts. Statistical analysis was done by Kruskal Wallis test and Dunn's post-test; horizontal bars represent geometric means, and statistical significance is denoted with asterisk (*, p < 0.1; **, p < 0.01).
Immunodetection of Antigens in Invasive Disease Isolates
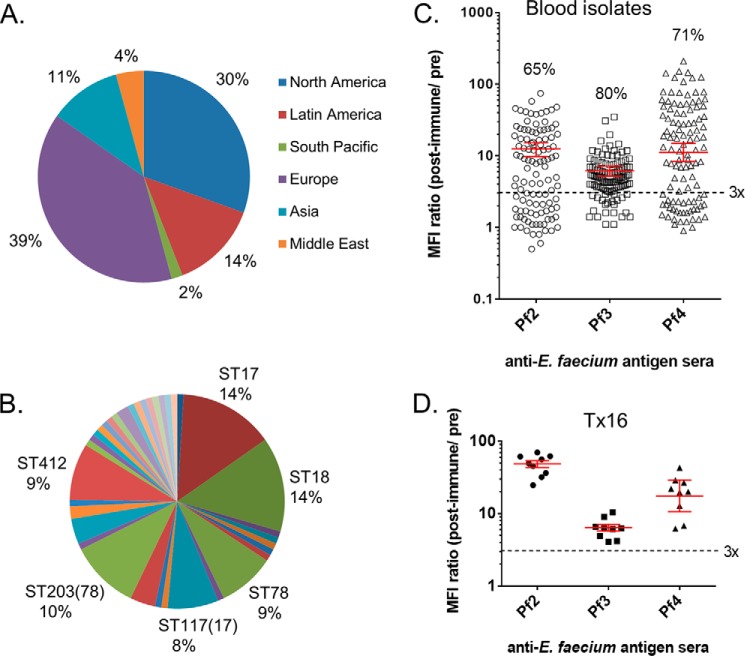
For comparative analysis of antigen presentation on E. faecium clinical strains, we used flow cytometry with Pf1-Pf4 immune sera to measure the binding of specific antibodies to whole bacteria. A collection of 18 E. faecium reference strains (including E. faecium Tx16) that had previously been characterized by genome sequencing were evaluated first (Table 5) (44, 45). These strains include isolates from a variety of clinical sources as well as carriage strains from healthy donors. The antigen expression level was reported as the ratio of MFI obtained with postimmune serum to that of matched preimmune control serum at 1:500 serum dilution. MFI values of greater than 3-fold above preimmune serum background were considered positive. The most commonly expressed antigen detected on 17/18 (95%) of the E. faecium strains was Pf3(LTA). As expected from its common structure (41), a cross-reacting LTA antigen was also detected on E. faecalis control strain FA2-2 (46), which exhibited a 6-fold higher MFI signal with postimmune sera than with preimmune control sera (Table 5). Pf2 sera reacted with 7 of the 18 E. faecium (42%) and Pf4 sera with 8 of the 18 (44) strains, but neither sera bound to the E. faecalis strain. In contrast, Pf1 antiserum failed to show binding activity above baseline levels with any of these strains.
TABLE 5.
E. faecium reference strains and Tx16 antigen antisera reactivity
| Strain | Source | Date | MLST STa | VRE? | Ref. | MFI ratio (immune/preimmune sera) |
Leg IGCF orthologues? | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Pf1 | Pf2 | Pf3 | Pf4 | |||||||
| E0155 | Fecal (hospital) | 1995 | 17 (17) | Yes | 26 | 2 | 30 | 6 | 106 | Yes |
| E0980 | Fecal (community) | 1998 | 94 | 45 | 3 | 25 | 8 | 100 | Yes | |
| E1039 | Fecal (community) | 1998 | 42 | 45 | 1 | 1 | 11 | 2 | ||
| E1071 | Fecal (hospital) | 2000 | 32 | Yes | 45 | 2 | 2 | 16 | 5 | |
| E1162 | Clinical (blood) | 1997 | 17 (17) | 45 | 2 | 1 | 7 | 3 | ||
| E1636 | Clinical (blood) | 1961 | 106 | 45 | 1 | 1 | 6 | 1 | ||
| E1679 | Clinical (catheter) | 1998 | 114 | Yes | 45 | 2 | 1 | 13 | 1 | |
| U0317 | Clinical (urine) | 2005 | 78 (78) | 45 | 2 | 6 | 11 | 5 | Yes | |
| 1,231,502 | Clinical (blood) | 2005 | 203 (78) | Yes | 44 | 3 | 14 | 21 | 45 | Yes |
| 1,231,410 | Clinical (SSTI) | 2005 | 17 (17) | Yes | 44 | 3 | 8 | 21 | 11 | Yes |
| 1,230,933 | Clinical (blood) | 2005 | 18 (18) | Yes | 44 | 3 | 5 | 21 | 11 | Yes |
| 1,231,501 | Clinical (blood) | 2005 | 52 | 44 | 2 | 1 | 8 | 1 | ||
| 1,231,408 | Clinical (blood) | 2005 | 582 | 44 | 3 | 1 | 26 | 1 | ||
| 1,141,733 | Clinical (wound) | 2005 | 52 | 44 | 2 | 1 | 2 | 1 | ||
| Com12 | Fecal (community) | 2006 | 107 | 44 | 2 | 1 | 6 | 1 | ||
| Com15 | Fecal (community) | 2006 | 583 | 44 | 2 | 1 | 3 | 1 | ||
| Tx16 | Clinical (blood) | 1992 | 18 (18) | 22 | 3 | 19 | 11 | 13 | Yes | |
| Tx1330 | Fecal (community) | 1994 | 107 | 22 | 2 | 1 | 6 | 1 | ||
| E. faecalis FA2-2 | JH2 (plasmid-cured) | ≤1973 | 8 | 46 | 1 | 1 | 6 | 1 | ||
a Hospital-associated lineage is indicated in parentheses (24).
Next, we evaluated the reactivity of Pf2, Pf3, and Pf4 antisera against a globally sourced collection of 101 E. faecium clinical blood isolates from 2009 to 2010, obtained from the multicenter Tigecycline Evaluation and Surveillance Trial (T.E.S.T.) (Fig. 7A, supplemental Table S1). The Pf1 antisera were not investigated further due to the low level of reactivity observed with the reference strains and its lack of OPA activity against Tx16. MLST analysis of the T.E.S.T. isolates showed they are genotypically diverse and include 12 new sequence types (Fig. 7B and supplemental Table S1). Seventy five percent (18/24 total) of the United States strains in this collection were resistant to vancomycin, which approximates the E. faecium VRE prevalence of 83% reported for all bloodstream infections in United States hospitals during 2009–2010 (4). The binding of the Pf2, Pf3, and Pf4 antisera to these isolates measured by flow cytometry (with Tx16 as control) is shown in Fig. 7, C and D. As observed with the reference isolates, the Pf3(LTA) antisera bound to the highest number of strains (80%). The proportion of the T.E.S.T. strains expressing antigens reacting with the Pf2 and Pf4 glycoconjugate sera was substantially higher than observed with the genome-sequenced reference strains as follows: 65% of strains reacted with antisera to Pf2, 71% for Pf4 sera, and 63% of strains reacted with both Pf2 and Pf4 antisera.
FIGURE 7.
Antigen detection in E. faecium blood isolates. A, geographic, and B, MLST distribution of E. faecium clinical strains (n = 101). C, bacterial surface antigen expression in this set of strains analyzed separately by flow cytometry with matched pre- and postimmune antisera to Pf2-CRM197, Pf3(LTA), and Pf4-CRM197. Percentages of strains binding antibodies to individual antigens (above three times immune to preimmune serum activity threshold) are indicated. D, variability of independent preparations of Tx16 used as internal control for each batch of strains analyzed. Corresponding geometric mean titers and 95% confidence intervals (error bars) are shown in red.
Identification of Legionaminic Acid Biosynthetic Genes
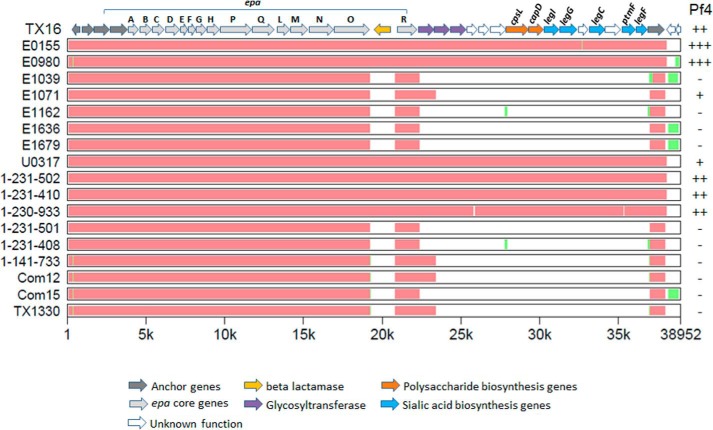
Seven of the eight genome-sequenced E. faecium reference strains that reacted with Pf4 antisera harbor orthologues of genes legIGCF responsible for legionaminic acid biosynthesis in C. jejuni (36), consistent with the presence of this rare monosaccharide in the Pf4 heteroglycan (Table 5 and Fig. 8). In contrast, all strains not reacting with Pf4 antisera did not contain these biosynthetic genes. These genes lie at the distal end of a megacluster of putative carbohydrate biosynthetic genes, the proximal half of which is related to the E. faecalis epa operon responsible for the biosynthesis of the E. faecalis rhamnose-containing heteroglycan (18, 22, 47). This 3′-cluster of 17 genes is characterized by a flanking β-lactamase gene insertion that separates the epaR gene from the core epa region and reconfigures it as the most proximal member of a potential operon that includes multiple carbohydrate and sialic acid biosynthesis genes (Fig. 8). Legionaminic acid has the same stereochemistry as the sialic acid 5-N-acetylneuraminic acid (Neu5NAc) but contains instead 9-deoxy and 7-amino groups. Although previously annotated as sialic acid synthesis genes (18), our BLAST analysis reveals that the predicted proteins are more closely related to C. jejuni legionaminic acid biosynthetic enzymes than to the NeuBCDA proteins responsible for sialic acid biosynthesis in S. agalactiae CPS (supplemental Table S2). In addition, clear orthologues of the C. jejuni LegC transaminase and PtmF isomerase (which co-catalyzes the first step in legionaminic biosynthesis (36)) are present in the E. faecium cluster but are absent from the genome of S. agalactiae.
FIGURE 8.
Conservation of Tx16 legionaminic acid biosynthesis gene cluster (legIGCF) among E. faecium strains. Orthologous sequences of E. faecalis epa, polysaccharide biosynthesis, and sialic acid biosynthesis genes (including anchor genes at two ends) were selected from each E. faecium genome and aligned by MultiPipMaker using default settings (38). As the reference, Tx16 genes are shown at top, with individual genes represented by arrows. Genes with homology to S. agalactiae and S. aureus CPS biosynthesis genes are labeled cpsL and capD, respectively. Orthologues of C. jejuni legionaminic acid biosynthesis genes are labeled accordingly (supplemental Table S2) (36). The x axis denotes the location of evolutionarily conserved regions in relation to the Tx16 sequence (22). Red regions represent areas of high sequence conservation (at least 100 bp with no gaps and >70% identity), and green represents regions where an alignment could be made but with lower sequence conservation. White regions denote the absence of gene sequences present in Tx16. The reactivity of strains with antisera to legionaminic acid heteroglycan Pf4 is indicated (see Table 5 for values and supplemental Table S1 for key). GenBankTM accession numbers for genome sequences are as follows: Tx16 (NC_017960); E0155 (AUWX00000000); E0980 (ABQA01000000); E1039(ACOS01000000); E1071 (ABQI01000000); E1162 (ABQJ01000000); E1636 (ABRY01000000); E1679 (ABSC01000000); U0317 (ABSW01000000); 1-231-502 (ACAX01000000); 1-231-410 (ACBA01000000); 1-230-933 (ACAS01000000); 1-231-501 (ACAY01000000); 1-231-408 (ACBB01000000); 1-141-733 (ACAZ01000000); Com12 (ACBC01000000); Com15 (ACBD01000000); and TX1330 (ACHL01000000).
Discussion
E. faecium is a problem pathogen that is becoming increasingly hard to treat with available antibiotics. The identification of protective antigens is needed to facilitate vaccine-based prophylactic approaches. This study provides the first structural identification of surface polysaccharides expressed by E. faecium other than teichoic acids (19, 20). A polyglucan and heteroglycan isolated from a VRE clinical isolate were previously described without elucidating their structures (48). Structures, yields, and sizes of the polysaccharides purified from the Tx16 strain are summarized in Fig. 3 and Table 6. The highest molecular mass polysaccharide purified from Tx16 (Pf1) is a fructose homopolymer known as levan with molecular mass of ∼30 MDa. A glycoconjugate of this polymer failed to elicit antibodies that were able to kill this strain in OPA. Pf1 was recovered in relatively low yield from in vitro grown Tx16 and was only detected at low levels on the bacterial surface (Tables 5 and 6). Although levan is not commonly associated with pathogenic bacteria, it contributes to biofilm production in the plant pathogen Erwinia amylovora (49) and was detected as an exopolysaccharide component of the opportunistic pathogen Burkholderia cepacia (50).
TABLE 6.
Yields and identities of polysaccharides purified from E. faecium Tx16
Quantities recovered are milligrams of pure compound/g of crude polysaccharide starting material.
| Tx16 polysaccharides | |||
|---|---|---|---|
| Antigen | Identity | Mass (kDa) | Yield (mg/g) |
| Pf1 | Levan | 31,920a | 2 |
| Pf2 | Altruronic acid heteroglycan | 300a | 20 |
| Pf3(LTA) | Glucosylated lipoteichoic acid | 15b | 3 |
| Pf3(TA) | Glucosylated teichoic acid | 15b | 2 |
| Pf4 | Legionaminic acid heteroglycan | 63a | 74 |
a Data were determined by SEC-MALLS.
b Data were estimated by SDS-PAGE.
Discovery of Novel Protective Polysaccharides of E. faecium
Although intrinsically resistant to neutrophil-mediated phagocytosis, the Tx16 strain can be killed by antibodies directed against bacterial surface carbohydrate in the presence of neutrophils and complement (25). We identified two novel predominant surface polysaccharides from E. faecium Tx16 (Table 6) that are the likely targets of these functional antibodies. The 300-kDa altruronic acid-containing heteroglycan (Pf2) is within the size range of native capsular polysaccharides purified from Streptococcus pneumoniae (100 to >900 kDa) (51), and Pf2-specific polyclonal antibodies are highly potent in OPAs with the Tx16 strain. Antibodies specific for the 63-kDa legionaminic containing heteroglycan (Pf4) were also functional in this OPA but appeared less potent than Pf2 antibodies. Antibodies to Pf2 and Pf4 heteroglycans were also capable of reducing Tx16 infection in a mouse passive protection model (Fig. 6). Surface heteroglycans have also been described in E. faecalis species and consist of a 130-kDa capsular diheteroglycan and smaller 50-kDa rhamnose-containing polymer of incompletely defined structure (27, 52, 53).
The altruronic acid and legionaminic acid monosaccharides in the Pf2 and Pf4 heteroglycans are rare in the context of polysaccharides described for other Gram-positive bacteria. Altruronic acid was previously identified as a component of surface lipopolysaccharide (LPS) O-antigens of the Gram-negative bacteria Plesiomonas shigelloides (Shigella) and Proteus mirabilis (40, 54) and in a capsular polysaccharide of Aerococcus viridans var. homari, a Gram-positive pathogen of marine lobsters (55). Legionaminic acid was first discovered as the principal component of serotype 1 O-chain LPS from Legionella pneumophila (56, 57), the cause of Legionnaires' disease. Aside from its association with the LPS of pathogenic Gram-negative bacteria, it is also a component of glycosylated flagella proteins of Campylobacter, Helicobacter, and Clostridium (58). Legionaminic acid may have analogous biological functions as the structurally similar, more common sialic acid Neu5NAc. In the context of group B Streptococcus (S. agalactiae) capsular polysaccharides, terminal Neu5NAc residues of the branched repeat units mediate binding to host leukocyte lectins in a manner that is thought to mimic structurally analogous host cell glycans (59). Another example of molecular mimicry is the sialic acid capsular polysaccharides of Neisseria meningitidis serogroup B and Escherichia coli K1, which are thought to subvert host cellular immune responses due to structural identity with polysialic acid chains expressed on the neural cell adhesion molecule (60). The immunological impact of legionaminic acid in the context of the branched heteropentameric repeat of Pf4 is unknown, although a branched pentameric repeat of similar composition (Gal, Glc, GalNAc, and Leg) forms the core repeat of the LPS O-antigen of Cronobacter tuicensis (61).
Novel E. faecium Heteroglycans as Potential Virulence Factors
The detection of Pf2 and Pf4 antigens in invasive blood isolates at a higher rate than that observed in a collection of genome-sequenced strains (which includes commensals) raises the question of whether these antigens contribute to bacterial virulence or hospital adaptation. Consistent with this possibility, six of the seven strains in the reference set of 18 that co-express surface antigens reacting with Pf2 and Pf4 sera are multidrug-resistant and belong to three hospital lineages originating from sequence types 17, 18, and 78 that are associated with nosocomial infections (Table 5) (24). These strains include VRE strains (E0155, E1071, 1,231502, 1,231,410, and 1,230,933) and non-VRE strains resistant to aminoglycosides, ampicillin, and tetracycline or ciprofloxacin (Tx16, U0317). The association of VRE strains 1,231502, 1,231,410, and 1,230,933 with the presence of the unique legIGCF-containing gene cluster was previously noted (18). Commensal strain E0980, the sole exception among the reference strains, belongs neither to these hospital-associated groups nor is it multidrug-resistant (18, 22), yet it contains the legIGCF genes and binds to antibodies elicited by both Pf2 and Pf4 heteroglycans.
The correlation of Pf4 antisera reactivity with the presence of the legIGCF gene cluster begs the question of whether this region might be responsible for the biosynthesis of the legionaminic acid-containing heteroglycan Pf4. Also present in this cluster are orthologues of genes from other bacterial pathogens that are associated with polysaccharide chain initiation and repeat unit formation (Fig. 8 and supplemental Table S2). The epaR gene encodes a predicted sugar transferase that adds galactose to the C55 lipid carrier. When used as TBLASTN query sequences, the Tx16 open reading frames identified orthologues of glycosyltransferases and orthologues of CPS biosynthetic genes from S. aureus (Cap5D) and S. agalactiae (CpsL). Therefore, it is tempting to speculate that the E. faecium leg cluster may be involved in both legionaminic acid production and Pf4 biosynthesis. If so, the association of this locus primarily with hospital-adapted strains may be a reflection of enhanced virulence properties conferred by this surface polysaccharide.
Putative genetic determinants responsible for Pf2 antigen biosynthesis have been harder to locate. A second predicted E. faecium CPS biosynthetic locus was previously identified that contains conserved orthologues of S. pneumoniae cpsABCD (18), a CPS phosphoregulatory system. In S. agalactiae, orthologous cpsABCD genes reside at the 5′ end of the CPS operon and regulate CPS polymer length and cell wall attachment (62). However, the downstream region in E. faecium is highly variable, and no obvious correlations between the presence of plausible genes and Pf2 antigen expression are evident based on available genome sequence information. Sequencing of additional strains may improve the ability to identify candidates. Alternatively, differential gene regulation or dispersal of genes across multiple loci may confound attempts to link phenotype with genotype. Ultimately, mutational analysis will be required to unambiguously identify pathways for Pf2 and Pf4 antigen biosynthesis.
E. faecium LTA Is a Structurally Conserved, Protective Antigen
E. faecium LTA (Pf3) was recovered from the second SEC pool as a micellar aggregate and purified by elution from AEC using 0.6 m NaCl. The glucosylated structure of the Tx16 Pf3 (LTA) is identical to that previously reported for the LTA of E. faecalis strain 12030 but without the d-Ala modification (41). Tx16 has the potential for LTA d-Ala modification as its genome harbors a dltABCD operon that is responsible for d-alanylation of teichoic acids in Gram-positive bacteria, including orthologues of d-alanine-d-alanyl carrier ligase (DltA) and d-alanine transferase (DltB) (22). Loss of labile d-Ala groups from native Tx16 LTAs occurred presumably due to the basic buffer conditions used for polysaccharide purification, as we were able to purify d-alanine-modified LTAs from E. faecalis and other E. faecium strains using conventional (63) butanol extraction and acidic buffer (data not shown). Antibodies against de-alanylated E. faecalis LTA of the same structure were previously shown to induce functional antibodies that protected against E. faecalis and E. faecium infections after active and passive immunization in mice (35, 41). Antibodies to Tx16 LTA were also able to protect against Tx16 infection in mice (Fig. 6). Our results predict that this conserved enterococcal polysaccharide has the potential for broad protective coverage against E. faecium, given that 80% of bacteremia strains express this antigen. We also showed that OPA titers can be significantly enhanced through conjugation of the glucosylated (Pf3) teichoic acid to the CRM197 carrier protein (Fig. 5B). The feasibility of making a Pf3 polysaccharide conjugate has implications for vaccine design, as polysaccharide conjugates induce B-cell memory in contrast to unconjugated polysaccharides (64).
Antigen Coverage and Composition of a Multivalent Vaccine
The goal of any vaccine-based prophylaxis targeting E. faecium is to prevent invasive disease. We evaluated a global collection of clinical blood isolates to assess prevalence and potential coverage of antigens reacting with antibodies specific for E. faecium LTA and novel heteroglycan antigens. A trivalent glycoconjugate prophylactic vaccine against E. faecium composed of enterococcal glucosylated teichoic acid (Pf3(TA)), altruronic acid heteroglycan (Pf2), and legionaminic acid heteroglycan (Pf4) could potentially cover at least 90% of the blood isolates, considering at least one of these three antigens is expressed on the bacterial surface (Fig. 7C and supplemental Table S1). Alternatively, for therapeutic intervention to protect patients during a period of risk in an intensive care unit setting, a combination of prophylactic monoclonal antibodies targeting the same three antigens might be considered.
Author Contributions
S. K. carried out all of the antigen purifications, carrier protein conjugations, and flow cytometry analysis. E. V. determined polysaccharide structures. H. J. contributed clinical strains and ELISA data. F. L. and N. K. provided carbohydrate analytical support. L. H. and R. D. performed bioinformatic analysis. D. L. provided OPA support and mouse model data analysis. R. D. generated Tx16 OPA data and wrote the manuscript. R. D., V. P., J. H., A. S. A., and K. U. J. contributed to experimental strategies and provided additional oversight. All authors contributed to the final manuscript.
Supplementary Material
Acknowledgments
Baseclear (Leiden, Netherlands) generated MLST data for the E. faecium blood isolates. We acknowledge the use of the E. faecium MLST database, which is located at Imperial College London and funded by The Wellcome Trust. We thank Michael Gilmore, Barbara Murray, and Rob Willems for strains and Philip Lee (Pfizer) and Lorna Nunez (Pfizer) for help with genome sequencing. We thank Ellen Murphy for help with bioinformatic analysis of putative biosynthetic genes. Dominique Wobser contributed expert technical support for the mouse model. We also thank Dee Illenburger, Jennifer Obregon, and Kathryn Mason for generating rabbit antibodies.
This work was supported by Pfizer Inc. E. V. is an employee of the National Research Council (Canada) whose services were contracted by Pfizer. J. H. and D. L. received financial support from Pfizer in connection with research conducted and reported in this work. The remaining contributors are Pfizer employees and as such were paid by Pfizer and may own Pfizer stock.

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- VRE
- vancomycin-resistant Enterococcus
- OPA
- opsonophagocytic assay
- LTA
- lipoteichoic acid
- SEC
- size exclusion chromatography
- MFI
- mean fluorescence intensity
- CPS
- capsular polysaccharide
- Leg
- 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid
- MWCO
- molecular weight cutoff
- AEC
- anion exchange column chromatography
- NRS
- normal rabbit serum
- T.E.S.T.
- Tigecycline Evaluation and Surveillance Trial
- Neu5NAc
- 5-N-acetylneuraminic acid
- MALLS
- multiangle laser light scattering
- RI
- refractive index
- HPAEC-PAD
- high performance anion-exchange liquid chromatography with pulsed amperometric detection.
REFERENCES
- 1. Theilacker C., Jonas D., Huebner J., Bertz H., Kern W. V. (2009) Outcomes of invasive infection due to vancomycin-resistant Enterococcus faecium during a recent outbreak. Infection 37, 540–543 [DOI] [PubMed] [Google Scholar]
- 2. Fernández Guerrero M. L., Goyenechea A., Verdejo C., Roblas R. F., de Górgolas M. (2007) Enterococcal endocarditis on native and prosthetic valves: a review of clinical and prognostic factors with emphasis on hospital-acquired infections as a major determinant of outcome. Medicine 86, 363–377 [DOI] [PubMed] [Google Scholar]
- 3. Joels C. S., Matthews B. D., Sigmon L. B., Hasan R., Lohr C. E., Kercher K. W., Norton J., Sing R. F., Heniford B. T. (2003) Clinical characteristics and outcomes of surgical patients with vancomycin-resistant enterococcal infections. Am. Surg. 69, 514–519 [PubMed] [Google Scholar]
- 4. Sievert D. M., Ricks P., Edwards J. R., Schneider A., Patel J., Srinivasan A., Kallen A., Limbago B., Fridkin S., National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. (2013) Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 34, 1–14 [DOI] [PubMed] [Google Scholar]
- 5. Hidron A. I., Edwards J. R., Patel J., Horan T. C., Sievert D. M., Pollock D. A., Fridkin S. K., National Healthcare Safety Network Team, Participating National Healthcare Safety Network Facilities. (2008) NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29, 996–1011; Correction (2009) Infect. Control Hosp. Epidemiol. 30, 107 [DOI] [PubMed] [Google Scholar]
- 6. Arias C. A., Murray B. E. (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Willems R. J., van Schaik W. (2009) Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 4, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 8. Edmond M. B., Ober J. F., Dawson J. D., Weinbaum D. L., Wenzel R. P. (1996) Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin. Infect. Dis. 23, 1234–1239 [DOI] [PubMed] [Google Scholar]
- 9. Vergis E. N., Hayden M. K., Chow J. W., Snydman D. R., Zervos M. J., Linden P. K., Wagener M. M., Schmitt B., Muder R. R. (2001) Determinants of vancomycin resistance and mortality rates in enterococcal bacteremia. A prospective multicenter study. Ann. Int. Med. 135, 484–492 [DOI] [PubMed] [Google Scholar]
- 10. Schulte B., Heininger A., Autenrieth I. B., Wolz C. (2008) Emergence of increasing linezolid-resistance in enterococci in a post-outbreak situation with vancomycin-resistant Enterococcus faecium. Epidemiol. Infect. 136, 1131–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allen G. P., Bierman B. C. (2009) In vitro analysis of resistance selection by linezolid in vancomycin-susceptible and -resistant Enterococcus faecalis and Enterococcus faecium. Int. J. Antimicrob. Agents 34, 21–24 [DOI] [PubMed] [Google Scholar]
- 12. Tran T. T., Panesso D., Gao H., Roh J. H., Munita J. M., Reyes J., Diaz L., Lobos E. A., Shamoo Y., Mishra N. N., Bayer A. S., Murray B. E., Weinstock G. M., Arias C. A. (2013) Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arias C. A., Torres H. A., Singh K. V., Panesso D., Moore J., Wanger A., Murray B. E. (2007) Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 14. Arias C. A., Panesso D., McGrath D. M., Qin X., Mojica M. F., Miller C., Diaz L., Tran T. T., Rincon S., Barbu E. M., Reyes J., Roh J. H., Lobos E., Sodergren E., Pasqualini R., et al. (2011) Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Roux A., Schmöle-Thoma B., Schmöele-Thoma B., Siber G. R., Hackell J. G., Kuhnke A., Ahlers N., Baker S. A., Razmpour A., Emini E. A., Fernsten P. D., Gruber W. C., Lockhart S., Burkhardt O., Welte T., Lode H. M. (2008) Comparison of pneumococcal conjugate polysaccharide and free polysaccharide vaccines in elderly adults: conjugate vaccine elicits improved antibacterial immune responses and immunological memory. Clin. Infect. Dis. 46, 1015–1023 [DOI] [PubMed] [Google Scholar]
- 16. Anderson A. S., Miller A. A., Donald R. G., Scully I. L., Nanra J. S., Cooper D., Jansen K. U. (2012) Development of a multicomponent Staphylococcus aureus vaccine designed to counter multiple bacterial virulence factors. Hum. Vaccin. Immunother. 8, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paoletti L. C., Madoff L. C. (2002) Vaccines to prevent neonatal GBS infection. Semin. Neonatol. 7, 315–323 [DOI] [PubMed] [Google Scholar]
- 18. Palmer K. L., Godfrey P., Griggs A., Kos V. N., Zucker J., Desjardins C., Cerqueira G., Gevers D., Walker S., Wortman J., Feldgarden M., Haas B., Birren B., Gilmore M. S. (2012) Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3, e00318–00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bychowska A., Theilacker C., Czerwicka M., Marszewska K., Huebner J., Holst O., Stepnowski P., Kaczyński Z. (2011) Chemical structure of wall teichoic acid isolated from Enterococcus faecium strain U0317. Carbohydr. Res. 346, 2816–2819 [DOI] [PubMed] [Google Scholar]
- 20. Huebner J., Wang Y., Krueger W. A., Madoff L. C., Martirosian G., Boisot S., Goldmann D. A., Kasper D. L., Tzianabos A. O., Pier G. B. (1999) Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 67, 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arduino R. C., Jacques-Palaz K., Murray B. E., Rakita R. M. (1994) Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62, 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin X., Galloway-Peña J. R., Sillanpaa J., Roh J. H., Nallapareddy S. R., Chowdhury S., Bourgogne A., Choudhury T., Muzny D. M., Buhay C. J., Ding Y., Dugan-Rocha S., Liu W., Kovar C., Sodergren E., et al. (2012) Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 12, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lam M. M., Seemann T., Bulach D. M., Gladman S. L., Chen H., Haring V., Moore R. J., Ballard S., Grayson M. L., Johnson P. D., Howden B. P., Stinear T. P. (2012) Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194, 2334–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willems R. J., Top J., van Schaik W., Leavis H., Bonten M., Sirén J., Hanage W. P., Corander J. (2012) Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3, e00151–00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rakita R. M., Quan V. C., Jacques-Palaz K., Singh K. V., Arduino R. C., Mee M., Murray B. E. (2000) Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol. Med. Microbiol. 28, 291–299 [DOI] [PubMed] [Google Scholar]
- 26. Leendertse M., Willems R. J., Giebelen I. A., Roelofs J. J., Bonten M. J., van der Poll T. (2009) Neutrophils are essential for rapid clearance of Enterococcus faecium in mice. Infect. Immun. 77, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thurlow L. R., Thomas V. C., Hancock L. E. (2009) Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191, 6203–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trevelyan W. E., Forrest R. S., Harrison J. S. (1952) Determination of yeast carbohydrates with the anthrone reagent. Nature 170, 626–627 [DOI] [PubMed] [Google Scholar]
- 29. Dische Z., Shettles L. B. (1948) A specific color reaction of methylpentoses and a spectrophotometric micromethod for their determination. J. Biol. Chem. 175, 595–603 [PubMed] [Google Scholar]
- 30. Lees A., Nelson B. L., Mond J. J. (1996) Activation of soluble polysaccharides with 1-cyano-4-dimethylaminopyridinium tetrafluoroborate for use in protein-polysaccharide conjugate vaccines and immunological reagents. Vaccine 14, 190–198 [DOI] [PubMed] [Google Scholar]
- 31. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 32. Sawardeker J. S., Sloneker J. H., Jeanes A. (1965) Quantitative determination of monosaccharides as their alditol acetates by gas-liquid chromatography. Anal. Chem. 10.1021/ac60231a048 [DOI] [Google Scholar]
- 33. Ciucanu J., Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 34. Clarke A. J. (1993) Compositional analysis of peptidoglycan by high-performance anion-exchange chromatography. Anal. Biochem. 212, 344–350 [DOI] [PubMed] [Google Scholar]
- 35. Huebner J., Quaas A., Krueger W. A., Goldmann D. A., Pier G. B. (2000) Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect. Immun. 68, 4631–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schoenhofen I. C., Vinogradov E., Whitfield D. M., Brisson J.-R., Logan S. M. (2009) The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725 [DOI] [PubMed] [Google Scholar]
- 37. Jolley K. A., Maiden M. C. (2010) BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartz S., Zhang Z., Frazer K. A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. (2000) PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matulová M., Husárová S., Capek P., Sancelme M., Delort A.-M. (2011) NMR structural study of fructans produced by Bacillus sp. 3B6, bacterium isolated in cloud water. Carbohydr. Res. 346, 501–507 [DOI] [PubMed] [Google Scholar]
- 40. St Swierzko A., Shashkov A. S., Senchenkova S. N., Toukach F. V., Ziolkowski A., Cedzynski M., Paramonov N. A., Kaca W., Knirel Y. A. (1996) Structural and serological studies of the O-specific polysaccharide of the bacterium Proteus mirabilis O10 containing l-altruronic acid, a new component of O-antigens. FEBS Lett. 398, 297–302 [DOI] [PubMed] [Google Scholar]
- 41. Theilacker C., Kaczynski Z., Kropec A., Fabretti F., Sange T., Holst O., Huebner J. (2006) Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect. Immun. 74, 5703–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weidenmaier C., Peschel A. (2008) Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 [DOI] [PubMed] [Google Scholar]
- 43. Knirel Y. A., Shashkov A. S., Tsvetkov Y. E., Jansson P.-E., Zãhringer U. (2003) 5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 58, 371–417 [DOI] [PubMed] [Google Scholar]
- 44. Palmer K. L., Carniol K., Manson J. M., Heiman D., Shea T., Young S., Zeng Q., Gevers D., Feldgarden M., Birren B., Gilmore M. S. (2010) High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192, 2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Schaik W., Top J., Riley D. R., Boekhorst J., Vrijenhoek J. E., Schapendonk C. M., Hendrickx A. P., Nijman I. J., Bonten M. J., Tettelin H., Willems R. J. (2010) Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clewell D. B., Tomich P. K., Gawron-Burke M. C., Franke A. E., Yagi Y., An F. Y. (1982) Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152, 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teng F., Singh K. V., Bourgogne A., Zeng J., Murray B. E. (2009) Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect. Immun. 77, 3759–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsu C. T., Ganong A. L., Reinap B., Mourelatos Z., Huebner J., Wang J. Y. (2006) Immunochemical characterization of polysaccharide antigens from six clinical strains of Enterococci. BMC Microbiol. 6, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koczan J. M., McGrath M. J., Zhao Y., Sundin G. W. (2009) Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99, 1237–1244 [DOI] [PubMed] [Google Scholar]
- 50. Cescutti P., Impallomeni G., Garozzo D., Sturiale L., Herasimenka Y., Lagatolla C., Rizzo R. (2003) Exopolysaccharides produced by a clinical strain of Burkholderia cepacia isolated from a cystic fibrosis patient. Carbohydr. Res. 338, 2687–2695 [DOI] [PubMed] [Google Scholar]
- 51. Harding S. E., Abdelhameed A. S., Morris G. A., Adams G., Laloux O., Cerny L., Bonnier B., Duvivier P., Conrath K., Lenfant C. (2012) Solution properties of capsular polysaccharides from Streptococcus pneumoniae. Carbohydr. Polym. 90, 237–242 [DOI] [PubMed] [Google Scholar]
- 52. Theilacker C., Kaczyński Z., Kropec A., Sava I., Ye L., Bychowska A., Holst O., Huebner J. (2011) Serodiversity of opsonic antibodies against Enterococcus faecalis–glycans of the cell wall revisited. PLoS ONE 6, e17839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hancock L. E., Gilmore M. S. (2002) The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. U.S.A. 99, 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tóth A., Medgyes A., Bajza I., Lipták A., Batta G., Kontrohr T., Péterffy K., Pozsgay V. (2000) Synthesis of the repeating unit of the O-specific polysaccharide of Shigella sonnei and quantitation of its serologic activity. Bioorg. Med. Chem. Lett. 10, 19–21 [DOI] [PubMed] [Google Scholar]
- 55. Hermansson K., Kenne L., Lindberg B., Arie B., Brown R. G., Stewart J. E. (1990) Structural studies of the capsular polysaccharide from Aerococcus viridans var. homari. Carbohydr. Res. 208, 145–152 [DOI] [PubMed] [Google Scholar]
- 56. Knirel Y. A., Rietschel E. T., Marre R., Zähringer U. (1994) The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur. J. Biochem. 221, 239–245 [DOI] [PubMed] [Google Scholar]
- 57. Palusińska-Szysz M., Russa R. (2009) Chemical structure and biological significance of lipopolysaccharide from Legionella. Recent Pat. Antiinfect. Drug Discov. 4, 96–107 [DOI] [PubMed] [Google Scholar]
- 58. Zunk M., Kiefel M. J. (2014) The occurrence and biological significance of the [small α]-keto-sugars pseudaminic acid and legionaminic acid within pathogenic bacteria. RSC Adv. 4, 3413–3421 [Google Scholar]
- 59. Carlin A. F., Lewis A. L., Varki A., Nizet V. (2007) Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J. Bacteriol. 189, 1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vimr E., Lichtensteiger C. (2002) To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10, 254–257 [DOI] [PubMed] [Google Scholar]
- 61. MacLean L. L., Vinogradov E., Pagotto F., Perry M. B. (2011) Characterization of the lipopolysaccharide O-antigen of Cronobacter turicensis HPB3287 as a polysaccharide containing a 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-non-2-ulosonic acid (legionaminic acid) residue. Carbohydr. Res. 346, 2589–2594 [DOI] [PubMed] [Google Scholar]
- 62. Toniolo C., Balducci E., Romano M. R., Proietti D., Ferlenghi I., Grandi G., Berti F., Ros I. M., Janulczyk R. (2015) Streptococcus agalactiae capsule polymer length and attachment is determined by the proteins CpsABCD. J. Biol. Chem. 290, 9521–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morath S., Geyer A., Hartung T. (2001) Structure–function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193, 393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Clutterbuck E. A., Lazarus R., Yu L.-M., Bowman J., Bateman E. A., Diggle L., Angus B., Peto T. E., Beverley P. C., Mant D., Pollard A. J. (2012) Pneumococcal conjugate and plain polysaccharide vaccines have divergent effects on antigen-specific B cells. J. Infect. Dis. 205, 1408–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.