Background: In the alternative NF-κB signaling pathway, p100 undergoes partial degradation to generate p52.
Results: Depletion of the p97-Npl4-Ufd1 complex or enzymatic inhibition of p97 significantly decreases the processing of p100 into p52.
Conclusion: The p97-Npl4-Ufd1 complex positively regulates the alternative NF-κB signaling pathway.
Significance: This work suggests that p97 is a potential target for diseases with dysregulation of the alternative NF-κB pathway.
Keywords: cell signaling, lymphoma, NF-κB (NF-κB), protein degradation, ubiquitylation (ubiquitination), p97-Npl4-Ufd1 complex
Abstract
Partial degradation of the p100 subunit to generate p52 subunit is a hallmark of the alternative NF-κB pathway, which has been implicated in cancer. Here, we uncovered a role of the p97-Npl4-Ufd1 complex in mediating p100-to-p52 processing and therefore positively regulating the alternative NF-κB pathway. We observed an elevation of p97 mRNA levels in lymphoma patients, which positively correlates with NFKB2 expression, a downstream target gene of the alternative NF-κB pathway. Moreover, NFKB2 mRNA levels were aberrantly down-regulated in patients with inclusion body myopathy associated with Paget's disease of the bone and frontotemporal dementia (IBMPFD), a disease caused by mutation of p97. Inactivation of p97 or depletion of the p97-Npl4-Ufd1 complex inhibits the processing of p100 into p52, decreasing transcription of the downstream target genes. Further analyses reveal that the p97-Npl4-Ufd1 complex interacts with F-box and WD repeats protein SCFβTrCP complex to regulate the partial degradation of p100, a process involving K48- and K11-linked ubiquitination. In line with this, in LPS-induced lung damage mice model, generation of p52 is significantly decreased in p97-KD mice compared with mock mice. Finally, abrogation of p97 ATPase activity by its specific inhibitor DBeQ, efficiently decreased proliferation of lymphoma cells. Collectively, our study revealed a regulatory role of the p97-Npl4-Ufd1 complex in regulating p100 partial degradation, highlighting the potential of p97 as a drug target for cancers with aberrant activation of the alternative NF-κB pathway.
Introduction
The mammalian transitional endoplasmic reticulum ATPase p97, also known as valosin-containing protein (VCP)4 in metazoans and cell division control protein 48 (CDC48) in yeast, is an evolutionarily conserved ATPase that participates in diverse cellular processes including endoplasmic reticulum-associated degradation (ERAD), ubiquitin-proteasome-dependent protein degradation, nuclear envelope reconstruction, transcriptional control, cell cycle regulation, and DNA damage response (1–5). p97 has been implicated in multiple pathological processes. For example, mutations of p97 can cause the syndrome of inclusion body myopathy associated with Paget's disease of the bone and frontotemporal dementia (IBMPFD) (6–9). p97 colocalizes with protein aggregates in Huntington, Machado-Joseph, and Parkinson diseases (10). In addition, elevated expression of p97 and/or its cofactors is associated with poor prognosis of cancers including non-small-cell lung carcinoma, breast cancer, prostate cancer, pancreatic cancer, and leukemia (11–15).
The diverse functions of p97 are directed by various cofactors including Npl4, Ufd1, p47, UBXD7, and FAF1 (16–22). p97 in complex with specific cofactors is differentially involved in the degradation of multiple non-ERAD proteins. For example, the p97-Npl4-Ufd1 complex regulates the turnover of HIF1α, Cdt1, and CD4 (18, 23). Recently, p97 was also shown to regulate IκBα degradation by promoting SCFβTrCP ubiquitin ligase-mediated K48 polyubiquitination, leading to the activation of canonical NF-κB signaling pathway (24). However, it remains unknown whether p97 is involved in the regulation of the alternative NF-κB signaling pathway.
The NF-κB subunit p100 is a major inhibitor of the alternative NF-κB signaling pathway (25, 26). In response to various developmental signals such as lymphotoxin (LTα1β2), B-cell activating factor (BAFF) and CD40 ligand, p100 is processed into p52 through a mechanism that requires IκBα kinase (IKKα)-dependent phosphorylation of p100 on Ser-866 and Ser-870, and the activity of SCFβTrCP ubiquitin ligase (27–32). Subsequent nuclear translocation of p52 activates the alternative NF-κB pathway, leading to the transcription of downstream target gene like NFKB2.
Tight control of p100 processing into p52 is important for proper function of NF-κB in cell growth and survival. Dysregulation of p100-p52 processing is associated with T-cell transformation by the human T-cell leukemia virus type 1, lymphoma, and many other human cancers (33–36). It is known that the SCFβTrCP ubiquitin ligase ubiquitinates p100 by K48-linked ubiquitination upon stimulation. However, the detailed mechanism of p100-p52 processing, especially the post-ubiquitinational events is not well defined. In this work, we found that p97 in complex with its cofactor Npl4 and Ufd1 positively regulates the alternative NF-κB pathway by promoting partial degradation of p100 into p52. This process depends on the ATPase activity of p97. Mutations of p97 characterized in IBMPFD impaired such regulatory function; while enzymatic inhibition of p97 blocked the alternative NF-κB signaling in cells and in mice.
Experimental Procedures
Cloning, Reagents, and Antibodies
Mammalian expression vectors for Flag-βTrCP, Flag-Cul1, V5-p97, and its AA mutant were described previously (37). V5-NIK plasmid was kindly provided by Dr. Chen Wang. HA-p100 was bought from Open Biosystem. The p97 (3A) mutant was generated by substitution of amino acids Phe-52, Asp-55, and Tyr-110 with alanine in V5 vector. Recombinant human soluble CD40 ligand (rh-CD40L) was obtained from R&D systems (Minneapolis, MN). Antibodies specific for LTβR, Flag, Myc, and β-actin were purchased from Sigma. Antibodies specific for p100, p97, K11 Ub, and K48 Ub were from Millipore (Billerica, MA); those for V5, HA, Ufd1, and ubiquitin were from Santa Cruz Biotechnology (Santa Cruz, CA); and those for Npl4 and p52 were from Abcam (Cambridge, UK).
siRNA and shRNA
Duplexes of siRNA targeting p97 and negative control were synthesized by Genepharma (Shanghai, China). The siRNA sequences are as following. For human sip97 (targeting its CDS sequence), the forward oligo used was 5′-GAAUGCUCCUGCCAUCAUCTT-3′, and the reverse oligo was 5′-GAUGAUGGCAGGAGCAUUCTT-3′. For human sip97 (targeting its UTR sequence), the forward oligo used was 5′-CGCCACAUGUUCCCUAUAATT-3′, and the reverse oligo was 5′-UUAUAGGGAACAUGUGGCGTT-3′. For negative control, the forward oligo used was 5′-UUCUCCGAACGUGUCACGUTT-3′, the reverse oligo was 5′-ACGUGACACGUUCGGAGAATT-3′. For lentiviral transduction, Npl4 shRNA, p97 shRNA, Ufd1 shRNA, p47 shRNA, and control shRNA (scramble) were designed and constructed in the pLKO.1-puro backbone as previously described (38, 39). For human p97 shRNA, the forward oligo used was 5′-CCGGGGGCAC ATGTGATTGTTATCTCGAGATAACAATCACATGTGCCCTTTTTG-3′ and the reverse oligo was 5′-AATTCAAAAAGG GCACATGTGATTGTTATCTCGAGATAACAATCACATGTGCCC-3′. For human shNpl4, the forward oligo used was 5′-CCGGGTCCGCTACAGTCGAAATACTCGAGTATTTCGACTGTAGCGGACTTTTTG-3′ and the reverse oligo was 5′-AATTCAAAAAGTCCGCTACAGTCGAAATACTCGAGTATTTCGACTGTAGCGGAC-3′. For human Ufd1 shRNA, the forward oligo used was 5′-CCGGGCTGTT CAAACTGACCAATCTCGAGATTGGTCAGTTTGAACAGCTTTTTG-3′ and the reverse oligo was 5′-AATTCAAAAAGCT GTTCAAACTGACCAATCTCGAGATTGGTCAGTTTGAACAGC-3′. For human p47 shRNA, the forward oligo used was 5′-CCGGGGTGGATGATCTCTTTAAACTCGAGTTTAAAGAGATCATCCACCTTTTTG-3′ and the reverse oligo was 5′-AATTCAAAAAGGTGGATGATCTCTTTAAACTCGAGTTTAAAGAGATCATCCACC-3′.
Cell Culture and Transfection
HEK293T, MEF, and Raji lymphoma cells were purchased from Cell Resource Center (Institute of Life Science, Chinese Academy of Science). HEK293T and MEF cells were cultured in Dulbecco's modified Eagle's medium (DMEM), and Raji cells were maintained in RMPI 1640 medium. All culture was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a 37 °C humidified incubator and an atmosphere of 5% CO2 in air. Cells were transfected with siRNA or plasmids using the liposome transfection reagent Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or Lipofectamin RNA iMax (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation and Immunoblotting
For immunoprecipitation experiments, whole cell extracts were prepared after transfection or stimulation, and incubated overnight with indicated antibodies together with protein A/G beads (Santa Cruz Biotechnology). Beads were then washed three times with lysis buffer, and immunoprecipitates were eluted with SDS loading buffer and resolved in SDS-PAGE gels. The proteins were transferred to PVDF membrane (Bio-Rad) and further incubated with the indicated antibodies.
Immunofluorescence Imaging
MEF cells were plated on coverslips in 6-well plates and transfected with the indicated plasmids. Twenty-four hours later, cells were treated with or without anti-LTβR (100 ng/ml) for the indicated time points. Coverslips with the cells were washed once with PBS and fixed in 3.7% formaldehyde in PBS for 15 min. After permeabilization with Triton X-100 (0.25%) in PBS for 15 min, cells were blocked with PBS containing BSA (5%) for 1 h and then incubated with anti-p100/p52 antibodies for 1 h. After three separate washes, cells were incubated with secondary antibody for another hour and then stained with DAPI for 2 min. The coverslips were washed extensively and fixed on slides. Images were captured using a Leica laser scanning confocal microscope (Leica TCS SP2 AOBS).
Electrophoretic Mobility Shift Assays
The Raji cells transducing scramble or shp97 were stimulated with anti-LTβR, and then lysed in whole cell extraction buffer containing 20 mm HEPES pH 7.9, 350 mm NaCl, 20% glycerol, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm EGTA, 1% Nonidet P-40, 1 mm DTT, and protease inhibitors. For antibody supershift analysis, the binding reaction was performed in the absence of the probe, the appropriate antibody was added, and the mixture was incubated for 16 h at 4 °C. The probe was then added, the reaction mixture was incubated an additional 30 min at 25 °C, and the complexes were resolved by gel electrophoresis.
Real-time PCR Analysis
Real-time PCR was performed on Applied Biosystems Step Two Real-Time PCR System (Applied Biosystems) using the comparative Ct quantization method as described previously. Real-time PCR Master Mix (Toyobo, JP) was used to detect and quantify the expression level of the target gene. GAPDH was used as internal control. The primers are as the following: hNFKB2: 5′-ATGGAGAGTTGCTACAACCCA-3′ (F), 5′-CTGTTCCACGATCACCAGGTA-3′ (R); mNFKB2: 5′-TGGCATCCCCGAATATGATGA-3′ (F), 5′-TGACAGTAGGATAGGTCTTCCG-3′ (R); hMCP1: 5′-CAGCCAGATGCAATCAATGCC-3′ (F), 5′-TGGAATCCTGAACCCACTTCT-3′ (R); mMCP1: 5′-TAAAAACCTGGATCGGAACCAAA-3′ (F), 5′-GCATTAGCTTCAGATTTACGGGT-3′ (R); hRANTES: 5′-CCAGCAGTCGTCTTTGTCAC-3′ (F), 5′-CCAGCAGTCGTCTTTGTCAC-3′ (R); hBAFF: 5′-GGGAGCAGTCACGCCTTAC-3′ (F), 5′-GATCGGACAGAGGGGCTTT-3′ (R); mVCAM-1: 5′-TTGGGA GCCTCAACGGTACT-3′ (F), 5′-GCAATCGTTTTGTATTCAGGGGA-3′ (R); mp97: 5′-TGGCTCATCTGTGCGAAGAG-3′ (F), 5′-ATTTGCCACCAGTAGACCTAGA-3′ (R); hIL6: 5′-ACTCACCTCTTCAGAACG AATTG-3′ (F), 5′-CCATCTTTGGAAGGTTCAGGTTG-3′ (R); hGAPDH: 5′-TGTGGGCATCAATGGATTTGG-3′ (F), 5′-AC ACCATGTATTCCGGGTCAAT-3′ (R); mGAPDH: 5′-TTGTCATGGGAGTGAACGAGA-3′ (F), 5′-CAGGCAGTTGGTGGTACAGG-3′ (R).
Ubiquitination Assay
Cells were harvested and lysed in a buffer containing 8 m urea, 50 mm Tris, pH 7.5, 150 mm NaCl, and 1× proteases inhibitor mixture. Before immunoprecipitation, the urea was diluted to a final concentration of 0.4 m with 50 mm Tris/HCl, pH 7.5. The mixture was then incubated with anti-HA-conjugated Sepharose beads overnight at 4 °C. Immobilized proteins were washed five times with 1× PBS buffer before being resolved by SDS-PAGE and immunoblotted with indicated antibodies. HA-K48Ub and HA-K11Ub are HA-tagged K48 only and K11 only ubiquitin mutants, respectively.
Cell Proliferation Assay
Cells were transfected with the indicated plasmids. ATP cell viability assay was used for detecting cell proliferation. ATP content was measured in accordance with the protocol of the CellTiter-GloTM luminescent cell viability assay kit (Promega, Madison, WI). Briefly, 100 μl of assay reagent was added to the wells and mixed for 2 min at room temperature. After 10 min, intracellular ATP content was measured using a luminescence multilabel counter (Envision, Perkin Elmer). Cell proliferation was calculated using the following equation: Cell proliferation (%) = [value (test) − value (blank)]/[value (control) − value (blank)] × 100.
Mice
Chow-fed, 6-week-old male C57BL/6 mice (Slac) were housed under a reverse light-dark cycle. shp97 (or control shRNA, 500 nm) in 200 μl of 5% glucose, including transfection reagent jetPEI (Polyplus, France, Illkirchcedex), was intravenously injected into mice once every day for 7 days. Subsequently, animals were injected intraperitoneally with LPS (20 mg/kg) for 6 h. All procedures for animal experimentation were performed in accordance with the Institutional Animal Care and Use Committee guidelines of the Animal Core Facility of the Institutes of Biochemistry and Cell Biology (SIBCB). The approval ID for using the animals was No. 081 by the Animal Core Facility of SIBCB.
Statistics
The data are presented as means ± S.D. One-way analysis of variance (ANOVA) and Student's t test were used for continuous variables. Pearson coefficient test were used for correlation analysis. p values of <0.05 were considered significant.
Results
p97 Positively Regulates p100 Processing in a Manner Dependent on Its ATPase Activity
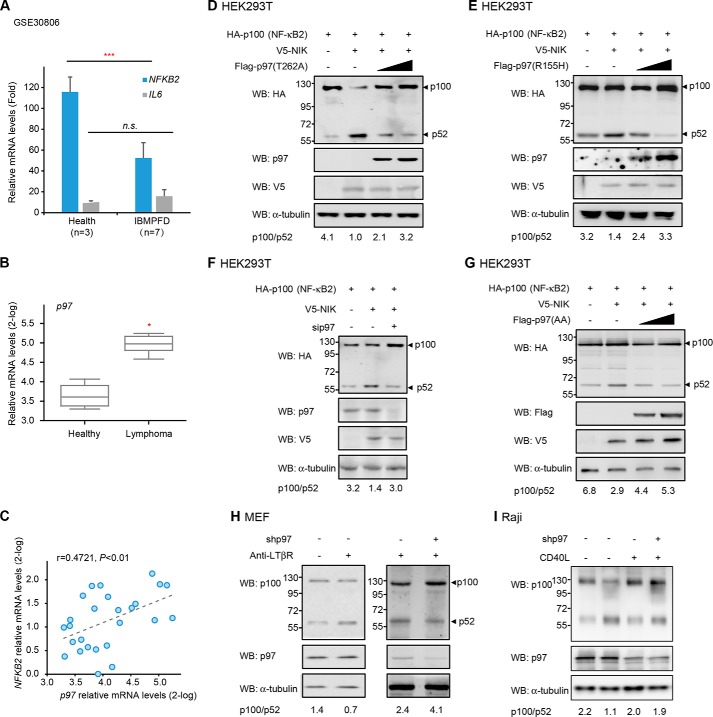
Previously, we reported that Ter94, a Drosophila homolog of p97, regulates Hedgehog signaling pathway through partial degradation of the full-length transcriptional factor Ci155 into its active form Ci75, a process involving K11-linked ubiquitination of Ci by the Cul1-Slimb-based E3 ubiquitin ligase (37). Inspired by the similarity between Ci155-Ci75 processing and p100-p52 maturation, we hypothesized that p97 might also target p100 for partial degradation in the alternative NF-κB signaling pathway. To test this possibility, we first examined the potential correlation of p97 with the alternative NF-κB pathway by analyzing clinical data from the Gene Expression Omnibus (GEO) Database. We observed a significant down-regulation of NFKB2 mRNA levels in the quadriceps muscle samples of Paget disease of the bone and frontotemporal dementia (IBMPFD) patients with p97 mutation (datasets GSE30806) (p = 0.0005), when compared with those of healthy people (Fig. 1A). On the other hand, no significant alteration was observed in IBMPFD samples for mRNA levels of IL6, a downstream target gene of the classical NF-κB signaling (p = 0.1964) (Fig. 1A). Since a previous study has implicated the alternative NF-κB pathway in lymphoma (33), we also examined the potential correlation of p97 with lymphoma by analyzing clinical data from the Oncomine Database (40). An up-regulation of p97 mRNA levels was observed in the tissue samples of lymphoma patients (p < 0.05) (Fig. 1B). Moreover, p97 mRNA levels were positively correlated with those of NFKB2 in lymphoma (p < 0.01) (Fig. 1C). Together, these observations suggest that p97 is involved in the alternative NF-κB signaling pathway.
FIGURE 1.
p97 regulates the processing of p100 to p52 in a manner dependent on its ATPase activity. A, bar plot for the mRNA levels of NFKB2 and IL6 in the IBMPFD datasets (***, p < 0.001, n.s., no significance, same below). B, box plot for p97 mRNA levels in the lymphoma datasets from the Oncomine database (*, p < 0.05, same below). C, scatter plots between p97 and NFKB2 in the lymphoma datasets. D–E, immunoblotting for p100/p52 in cells overexpressing HA-p100 and V5-NIK with or without IBMPFD-related p97 mutants T262A (D) and R155H (E). F, immunoblotting for p100/p52 in cells overexpressing HA-p100 and V5-NIK with or without sip97 (p97 siRNA, same below). G, immunoblotting for p100/p52 in cells overexpressing HA-p100 and V5-NIK with or without p97 (AA, K251AK524A, same below) mutant. H, immunoblotting for p100/p52 in the lysates of control (scramble, same below) and p97-knockdown (shp97, p97 shRNA, same below) MEF cells after treatment with anti-LTβR for 4 h. I, immunoblotting for p100/p52 in the lysates of control and p97-knockdown Raji cells after stimulation with CD40L for 6 h.
To further explore the potential correlation between the IBMPFD-related mutations of p97 and the aberrant activation of the alternative NF-κB pathway, we then constructed two p97 mutants commonly found in IBMPFD, namely, R155H and T262A, which have been reported to induce conformational defects of p97 in cells (41). The processing of p100 into p52 was then induced by overexpression of NF-κB inducing kinase (NIK), an upstream kinase that strongly activates the alternative NF-κB signaling pathway. As shown in Fig. 1, D and E, overexpression of the p97 mutant R155H or T262A dose-dependently elevated the ratio of p100/p52, and inhibited the generation of p52, suggesting a regulatory role of p97 in the alternative NF-κB signaling pathway. To further validate such potential function of p97, we transfected p97 specific siRNA into HEK293T cells. Knockdown of p97 significantly elevated the ratio of p100/p52, and inhibited the generation of p52 (Fig. 1F). Meanwhile, p97 (AA), a mutant version of p97 that lost ATPase activity, also led to a marked increase in the ratio of p100/p52 (Fig. 1G). Together, these data indicate that p97 indeed is involved in the processing of p100 to p52 and that such regulatory role of p97 is dependent on its ATPase activity.
Previous studies reveal that crosslinking of the lymphotoxin β receptor (LTβR) with an agonistic antibody is able to trigger p52 generation and DNA binding through inducing the processing of p100 (28). Consistently, we found that anti-LTβR strongly induced the processing of p100 to p52 in mouse embryonic fibroblasts (MEFs) (Fig. 1H). Knockdown of p97 by its specific shRNA increased p100/p52 ratio, and resulted in a decrease in the processing of p100 after stimulation with anti-LTβR (Fig. 1H). Moreover, ligation of CD40 can activate the alternative NF-κB signaling in lymphocytes (32), we then examined the inhibitory effect of p97 on CD40L-induced p100 processing. Consistent with our observations in MEF cells, p97 depletion in Raji cells also potently blocked CD40L-induced p52 generation (Fig. 1I). Together, these data further indicate that p97 plays a positive role in the processing of p100 to p52.
p97 Facilitates the Activation of the Alternative NF-κB Signaling
Activation of the alternative NF-κB signaling is a multistep process that includes at least p100 processing, p52 nuclear translocation, DNA binding and transcription of downstream target genes. To further corroborate the regulatory role of p97 in the alternative NF-κB signaling, we first evaluated whether IBMPFD-related mutations of p97 affect the transcription of downstream target genes of the alternative NF-κB pathway. Consistent with our observations for p100-p52 processing (Fig. 1, D and E), real-time PCR analysis in anti-LTβR-treated MEF cells showed that overexpression of p97 mutant T262A or R155H substantially reduced the mRNA levels of the alternative NF-κB pathway target genes NFKB2 and MCP1 (Fig. 2, A and B). Next, we examined the effect of p97 on the nuclear translocation of p52 in MEF cells after incubation with anti-LTβR. Immunofluorescence results demonstrated that p97 knockdown decreased the nuclear translocation of p52, when compared with scramble group (Fig. 2C). Subsequently, the effect of p97 on p52 binding to κB promoter was evaluated by using a 22-bp probe containing κB promoter sequence. As shown in Fig. 2D, p52 binding to the wild-type κB probe (supershift) was reduced ∼50% by silencing p97 in Raji cells. Finally, these results were further confirmed by analyzing the effect of p97 knockdown on the transcription of the alternative NF-κB pathway downstream target genes. Compared with scramble groups, the mRNA levels of target genes NFKB2, MCP1, VCAM-1, and RANTES were significantly decreased by depletion of p97 in either anti-LTβR-treated MEF or CD40L-treated Raji cells (Fig. 2, E and F). Taken together, these results clearly demonstrate that p97 facilitates the activation of the alternative NF-κB signaling pathway.
FIGURE 2.
p97 regulates the alternative NF-κB signaling. A–B, transcription of NFKB2 and MCP1, in anti-LTβR-treated MEF cells after transfection with p97 mutant T262A (A) or R155H (B) (**, p < 0.01, same below). C, immunofluorescent staining of p100 and p52 in MEF cells transfected with n.c. (negative control) or sip97 after incubation with anti-LTβR for 12 h. Cells were labeled with DAPI (blue) to visualize the nuclei (left), fluorescent intensities of p100/p52 in cytosol and nuclear were calculated (n = 100, right). D, EMSA assay for detecting the p52 binding to κB promoter by using a 22-bp probe containing κB promoter sequence. E-F, mRNA levels of NFKB2, MCP1, and VCAM-1 in control and p97-knockdown MEF cells after stimulation with anti-LTβR (E), and those in Raji cells after treatment with CD40L (F).
p97 in Complex with Npl4-Ufd1 as a Regulator of the Alternative NF-κB Pathway
The diverse functions of p97 are directed by various adaptor proteins, such as Npl4 and Ufd1. We previously showed that the Drosophila homolog of p97 acts together with Npl4 and Ufd1 to regulate the partial degradation of the transcription factor Ci in the Hedgehog signaling pathway (37). Here we also examined the functional relevance of the p97-Npl4-Ufd1 complex in the alternative NF-κB signaling pathway. Similar to the effects of p97, knockdown of either Npl4 or Ufd1 significantly impaired NIK- or anti-LTβR- induced p100-p52 processing (Fig. 3, A and B). Consistently, depletion of Npl4/Ufd1 markedly reduced CD40L-induced transcription of downstream target genes including NFKB2, RANTES, MCP1, and BAFF, as did p97 knockdown (Fig. 3C). Together, these observations indicate that Npl4 and Ufd1 are the cofactors required for p97 during its regulation of the alternative NF-κB pathway.
FIGURE 3.
The p97-Npl4-Ufd1 complex regulates the p100 processing and the alternative NF-κB signaling. A–B, immunoblotting for p100/p52 in NIK-transfected HEK293T cells (A) or anti-LTβR-incubated MEF cells (B) after transfection with indicated shRNAs. C, transcription of NFKB2, MCP1, RANTES, and BAFF in CD40L-stimulated Raji cells after transfection with indicated shRNAs. D, immunofluorescent staining of p100 and p52 in MEF cells transfected with vector or p97 (3A, F52AD55AY110A) mutant after treatment with CD40L. Cells were labeled with DAPI (blue) to visualize the nuclei (left), fluorescent intensities of p100/p52 in cytosol and nuclear were calculated (n = 100, right). E, mRNA levels of NFKB2, MCP1, RANTES, and BAFF in control and p97 (3A) Raji cells after treatment with CD40L.
To further assess the importance of the p97-Npl4-Ufd1 complex assembly on the regulation of the alternative NF-κB signaling pathway, we took advantage of the available structural information and created a p97 mutant (with Phe-52, Asp-55, Tyr-110 all mutated to alanines, termed 3A) that disabled its interaction with Npl4 (42). Notably, overexpressing the mutant p97 (3A) decreased the nuclear translocation of p52 (Fig. 3D), as well as the mRNA levels of downstream target genes (Fig. 3E), suggesting a dominant negative effect of such mutation on the processing of p100 to p52. Together, these results indicate that p97 in complex with Npl4-Ufd1 is essential to regulate the alternative NF-κB signaling pathway.
p97 Regulation of p100 Partial Degradation Involves SCFβTrCP-mediated Both K48 and K11-linked Ubiquitination
Our previous study indicates that K11-linked ubiquitination is involved in the partial degradation of the transcriptional factor Ci in the Hedgehog signaling pathway. To test whether K11-linked ubiquitination is also involved in the processing of p100 to p52 in the alternative NF-κB signaling pathway, we co-transfected HEK293T cells with HA-p100 and V5-NIK. As shown in Fig. 4A, overexpression of NIK led to significant accumulation of K48- and K11-linked ubiquitination of p100; while knockdown of p97 further enhanced such effect. Consistent with this observation, transfection of the p97 (AA) mutant also enhanced NIK-induced total and K11-linked ubiquitination of p100 (Fig. 4B). Furthermore, we co-transfected Flag-p100, V5-NIK, and different shRNA with HA-K48Ub or HA-K11Ub into HEK293T cells and then evaluated p100 ubiquitination by anti-HA immunoprecipitate and subsequent anti-Flag immunoblotting. Depletion of either Npl4 or Ufd1 further increased NIK-induced accumulation of K48- and K11-linked ubiquitination of p100, as did p97 knockdown (Fig. 4C). Taken together, these results indicate that regulation of the p100 processing by the p97-Npl4-Ufd1 complex involves both K48- and K11-linked ubiquitination.
FIGURE 4.
p97 regulation of the p100 processing involves both K48- and K11-linked ubiquitination. A, ubiquitination assay for p100 in p97-knockdown cells with its specific siRNA. B, ubiquitination assay for p100 in HEK293T cells after transfection with p97 (AA) mutant. C, ubiquitination assay for p100 in HEK293T cells transfected with indicated plasmids. Flag-p100, V5-NIK, and different shRNA were transfected together with HA-K48 ubiquitin (HA-K48 Ub, left) or HA-K11 ubiquitin (HA-K11 Ub, right) into HEK293T cells. Anti-HA immunoprecipitates (IP) were analyzed by immunoblotting (IB) with anti-Flag antibodies to detect ubiquitination of p100. D, co-IP assay for detection of interactions among p97, SCFβTrCP, and p100. E, ubiquitination assay for p100 in cells overexpressing βTrCP.
p97 binds to many ubiquitin ligases, including SCFβTrCP-related E3s (17). It is known that the SCFβTrCP ubiquitin ligase ubiquitinates p100 by K48-linked ubiquitination upon stimulation. Thus we hypothesized that p97 and the SCFβTrCP complex may function together to mediate the partial degradation of p100. To test this possibility, we examined interactions among p97, SCFβTrCP, and p100 by co-IP assay. Consistent with previous study (30), our results showed that p97 can interact with SCF components βTrCP and Cul1 (Fig. 4D). Moreover, both βTrCP and p97 interacted with p100 (Fig. 4D), suggesting that p97, together with the SCFβTrCP complex physically associates with p100 to regulate its partial degradation into p52. Furthermore, overexpression of βTrCP also increased NIK-induced K48- and K11-linked ubiquitination of p100 (Fig. 4E). Taken together, these results indicate that p97 regulates p100-p52 processing by targeting p100 for partial degradation, and that this process involves SCFβTrCP-mediated both K48- and K11-linked ubiquitination of p100.
p97 as a Potential Therapeutic Target for Cancers with Aberrant Activation of the Alternative NF-κB Pathway
To further corroborate our mechanistic study of p97 regulation of the p100 processing, we developed LPS-induced lung pathology model in mock mice and p97-KD mice by intraperitoneal injection of LPS (20 mg/kg) every day (Fig. 5A). Consistent with previous reports, LPS challenge triggered the processing of p100 into p52; whereas p97 knockdown decreased the generation of p52 when compared with the mock mice (Fig. 5B). Moreover, silencing p97 impaired LPS-induced transcription of NFKB2 and MCP1 in the lung tissues of mice (Fig. 5C). These in vivo studies confirm that p97 positively regulates the processing of p100 into p52 during activation of the alternative NF-κB signaling pathway.
FIGURE 5.
In vivo evaluation of the regulatory effect of p97 on the alternative NF-κB signaling. A, expression of p97 in the lung tissue from mock mice and p97-KD mice. B, immunoblotting for p100/p52 in the lung tissues of mock and p97-KD mice after challenge with LPS. C, transcription of NFKB2 and MCP1 in the lung tissues of mock and p97-KD mice after stimulation with LPS. D, immunoblotting for p100/p52 in the lysates of HEK293T cells overexpressing HA-p100 and V5-NIK after treatment with p97 inhibitor DBeQ for 4 h. E, cell proliferation and NFKB2 mRNA levels in Raji cells after stimulation with DBeQ. F, model of p97-regulated partial degradation of p100 in the alternative NF-κB pathway.
To evaluate the potential of p97 as a therapeutic target against cancers with aberrant activation of the alternative NF-κB pathway, we took advantage of p97 inhibitor compound DBeQ, which specifically turns off p97 ATPase activity. As shown in Fig. 5D, DBeQ treatment substantially blocked the processing of p100 into p52, again supporting the notion that p97 facilitates the partial degradation of p100. Moreover, treatment of the lymphoma cell line Raji with DBeQ efficiently decreased its proliferation (Fig. 5E). Consistently, such catalytic inhibition of p97 dramatically down-regulated the transcription of NFKB2 (Fig. 5E).
Discussion
Multiple functions of p97 have been studied with a common theme of “extraction and segregation of ubiquitinated substrates.” Recently, our study has shown that Ter94, the Drosophila homolog of p97, regulates the Hedgehog signaling pathway by targeting the transcription factor Ci for partial degradation. In this work, we showed that p97 in complex with its cofactor Npl4 and Ufd1, regulates the alternative NF-κB signaling pathway by facilitating the partial degradation of p100 into p52 in a manner dependent on ATPase activity of p97. Moreover, p97 regulation of the p100 processing involves SCFβTrCP-mediated both K48- and K11-linked ubiquitination. These studies suggest that p97-mediated partial degradation may function as a general regulatory mechanism for the processing and maturation of certain important proteins especially transcription factors.
The NF-κB subunit p100 has been long realized as a repressor of the alternative NF-κB signaling pathway by inhibiting the nuclear translocation of RelB (43, 44). Upon stimulation by lymphotoxin, B-cell-activating factor, or CD40 ligand, NIK and IKKα induce the phosphorylation of p100, which then interacts with the SCFβTrCP complex for subsequent generation of p52 (30, 45, 46). Our data indicate that the p97-Npl4-Ufd1 complex promotes the processing of p100 into p52 by directing p100 for SCFβTrCP-mediated partial degradation. In keeping with this notion, depletion of p97, Npl4, or Ufd1, disruption of the p97-Npl4 interaction, or catalytic inhibition of p97, all resulted in impaired p100-p52 processing, as well as reduced transcription of target genes. Thus an intact p97-Npl4-Ufd1 complex with ATPase activity is required for its regulatory function in the alternative NF-κB signaling. Furthermore, silencing of p97, Npl4, or Ufd1, or enzymatic inactivation of p97 by abrogation of its ATP binding ability, or overexpression of βTrCP, all caused accumulation of K48- and K11-linked ubiquitination of p100, suggesting a functional involvement of both K48- and K11-linked ubiquitination in p97-mediated partial degradation of p100. Actually, we also observed alterations of both K48- and K11-linked ubiquitination in p97-mediated processing of Ci. One possibility is that branched ubiquitin chains containing both K48- and K11-linked blocks play a role in p97-mediated protein partial degradation. In this regard, a recent paper reported that brunched ubiquitin chains containing multiple K11-linked blocks are important to provide an improved signal for proteasomal degradation (47).
Based on previous studies and our findings, we propose a mechanistic model for p97 regulation of the alternative NF-κB signaling pathway (Fig. 5F). Upon activation by receptor/NIK/IKKα signaling, p100 undergoes phosphorylation and recruits the SCFβTrCP complex and p97-Npl4-Ufd1 complex for subsequent ubiquitination and partial degradation to generate p52, which then enters the nucleus in association with RelB to activate target gene transcription. In this process, both K48- and K11-linked ubiquitin chains are added to p100 by the SCFβTrCP complex; while the p97-Npl4-Ufd1 complex may selectively direct p100 modified with both K48- and K11-linked ubiquitination to the proteasome for partial degradation.
Consistent with our mechanistic study of p97 in the regulation of p100-p52 processing, dysregulation of the alternative NF-κB pathway was observed in IBMPFD patients with p97 mutations. We showed that IBMPFD mutation of p97 impaired its regulatory function in the alternative NF-κB signaling pathway. Moreover, we found that p97 is up-regulated in lymphoma patients, and that p97 mRNA levels are positively correlated with those of NFKB2 in patients with lymphoma, further indicating the pathological relevance of p97 in the alternative NF-κB signaling pathway.
It has been well established that the classical NF-κB pathway has a tumor-promoting effect. Meanwhile, the alternative NF-κB pathway has been implicated in the initiation of a subset of tumors including lymphoma, multiple myeloma, and melanoma (31, 36, 48). Regardless of their specific roles, these two pathways are usually activated concurrently in tumorigenesis. Our current work, together with previous reports now indicates that p97 can positively regulate both classical and alternative NF-κB signaling. During our preparation of this manuscript, a independent research group reported the involvement of p97 in the alternative NF-κB signaling pathway (49), supporting our conclusion. More importantly, we showed that the p97 specific inhibitor DBeQ can potently inhibit the viability of lymphoma and melanoma cells by down-regulating the alternative NF-κB pathway, highlighting the therapeutic potential of p97 as an anticancer target, especially for patients with aberrant activation of the alternative NF-κB signaling pathway.
Author Contributions
Z. Z. and Y. W. performed experiments and analyzed data. Z. S. did the structural modeling. C. L., H. Q., and W. W. contributed to data analysis. Y. Z. contributed to experimental design. Z. C. Z., Y. W,, and S. J. wrote the manuscript. Z. C. Z. and S. J. supervised the project.
Acknowledgments
We thank Drs. Aning Lin and Shaocong Sun for generous support.
This work was supported by the 973 program of the Ministry of Science and Technology of China (2012CB910204), the National Natural Science Foundation of China (31270808, 31300734, 31470736, 31470868, 91442125), the Science and Technology Commission of Shanghai Municipality (13ZR1446400), the “Cross and Cooperation in Science and Technology Innovation Team” project of the Chinese Academy of Sciences, the Youth Innovation Promotion Association of Chinese Academy of Sciences, and the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (2014KIP202). The authors declare that they have no conflicts of interest with the contents of this article.
- VCP
- valosin-containing protein
- ERAD
- endoplasmic reticulum-associated degradation
- NIK
- NF-κB inducing kinase
- IKKα
- IκBα kinase.
REFERENCES
- 1. Erzurumlu Y., Kose F. A., Gozen O., Gozuacik D., Toth E. A., Ballar P. (2013) A unique IBMPFD-related P97/VCP mutation with differential binding pattern and subcellular localization. Int. J. Biochem. Cell Biol. 45, 773–782 [DOI] [PubMed] [Google Scholar]
- 2. Meyer H. (2012) p97 complexes as signal integration hubs. BMC Biol. 10, 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lilley B. N., Ploegh H. L. (2005) Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodman P. G. (2003) p97, a protein coping with multiple identities. J. Cell Sci. 116, 4283–4290 [DOI] [PubMed] [Google Scholar]
- 5. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) AAA-ATPase p97/Cdc48p, a Cytosolic Chaperone Required for Endoplasmic Reticulum-Associated Protein Degradation. Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar K. R., Needham M., Mina K., Davis M., Brewer J., Staples C., Ng K., Sue C. M., Mastaglia F. L. (2010) Two Australian families with inclusion-body myopathy, Paget's disease of bone and frontotemporal dementia: novel clinical and genetic findings. Neuromuscul. Disord. 20, 330–334 [DOI] [PubMed] [Google Scholar]
- 7. Ju J. S., Weihl C. C. (2010) Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy. Hum. Mol. Genet. 19, R38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stojkovic T., Hammouda el H., Richard P., López de Munain A., Ruiz-Martinez J., Camaño P., Gonzalez P. C., Laforêt P., Pénisson-Besnier I., Ferrer X., Lacour A., Lacomblez L., Claeys K. G., Maurage C. A., Fardeau M., Eymard B. (2009) Clinical outcome in 19 French and Spanish patients with valosin-containing protein myopathy associated with Paget's disease of bone and frontotemporal dementia. Neuromuscul. Disord. 19, 316–323 [DOI] [PubMed] [Google Scholar]
- 9. Watts G. D., Wymer J., Kovach M. J., Mehta S. G., Mumm S., Darvish D., Pestronk A., Whyte M. P., Kimonis V. E. (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381 [DOI] [PubMed] [Google Scholar]
- 10. Mizuno Y., Hori S., Kakizuka A., Okamoto K. (2003) Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci. Lett. 343, 77–80 [DOI] [PubMed] [Google Scholar]
- 11. Yamamoto S., Tomita Y., Hoshida Y., Iizuka N., Kidogami S., Miyata H., Takiguchi S., Fujiwara Y., Yasuda T., Yano M., Nakamori S., Sakon M., Monden M., Aozasa K. (2004) Expression level of valosin-containing protein (p97) is associated with prognosis of esophageal carcinoma. Clin. Cancer Res. 10, 5558–5565 [DOI] [PubMed] [Google Scholar]
- 12. Asaka S., Fujimoto T., Akaishi J., Ogawa K., Onda M. (2006) Genetic prognostic index influences patient outcome for node-positive breast cancer. Surg. Today 36, 793–801 [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto S., Tomita Y., Hoshida Y., Iizuka N., Monden M., Yamamoto S., Iuchi K., Aozasa K. (2004) Expression level of valosin-containing protein (p97) is correlated with progression and prognosis of non-small-cell lung carcinoma. Ann. Surg. Oncol. 11, 697–704 [DOI] [PubMed] [Google Scholar]
- 14. Zhao C. H., Li Q. F. (2005) Altered profiles of nuclear matrix proteins during the differentiation of human gastric mucous adenocarcinoma MGc80–3 cells. World J. Gastroenterol. 11, 4628–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asai T., Tomita Y., Nakatsuka S., Hoshida Y., Myoui A., Yoshikawa H., Aozasa K. (2002) VCP (p97) regulates NFκB signaling pathway, which is important for metastasis of osteosarcoma cell line. Jpn J Cancer Res. 93, 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye Y. (2006) Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 156, 29–40 [DOI] [PubMed] [Google Scholar]
- 17. Wójcik C., Rowicka M., Kudlicki A., Nowis D., McConnell E., Kujawa M., DeMartino G. N. (2006) Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell 17, 4606–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Magadán J. G., Pérez-Victoria F. J., Sougrat R., Ye Y., Strebel K., Bonifacino J. S. (2010) Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 6, e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Acs K., Luijsterburg M. S., Ackermann L., Salomons F. A., Hoppe T., Dantuma N. P. (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 21. Franz A., Orth M., Pirson P. A., Sonneville R., Blow J. J., Gartner A., Stemmann O., Hoppe T. (2011) CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol. Cell 44, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011) Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kahn M., Sugawara H., McGowan P., Okuno K., Nagoya S., Hellström K. E., Hellström I., Greenberg P. (1991) CD4+ T cell clones specific for the human p97 melanoma-associated antigen can eradicate pulmonary metastases from a murine tumor expressing the p97 antigen. J. Immunol. 146, 3235–3241 [PubMed] [Google Scholar]
- 24. Li J. M., Wu H., Zhang W., Blackburn M. R., Jin J. (2014) The p97-UFD1L-NPL4 protein complex mediates cytokine-induced IκBα proteolysis. Mol. Cell. Biol. 34, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silverman N., Maniatis T. (2001) NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342 [DOI] [PubMed] [Google Scholar]
- 26. Fan C. M., Maniatis T. (1991) Generation of p50 subunit of NF-κB by processing of p105 through an ATP-dependent pathway. Nature 354, 395–398 [DOI] [PubMed] [Google Scholar]
- 27. Fukushima H., Matsumoto A., Inuzuka H., Zhai B., Lau A. W., Wan L., Gao D., Shaik S., Yuan M., Gygi S. P., Jimi E., Asara J. M., Nakayama K., Nakayama K. I., Wei W. (2012) SCF(Fbw7) modulates the NFκB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep 1, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z.-W., Karin M., Ware C. F., Green D. R. (2002) The Lymphotoxin-β Receptor Induces Different Patterns of Gene Expression via Two NF-κB Pathways. Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 29. Claudio E., Brown K., Park S., Wang H., Siebenlist U. (2002) BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat. Immunol. 3, 958–965 [DOI] [PubMed] [Google Scholar]
- 30. Liang C., Zhang M., Sun S. C. (2006) β-TrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell. Signal. 18, 1309–1317 [DOI] [PubMed] [Google Scholar]
- 31. Giardino Torchia M. L., Conze D. B., Jankovic D., Ashwell J. D. (2013) Balance between NF-κB p100 and p52 regulates T cell costimulation dependence. J. Immunol. 190, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coope H. J., Atkinson P. G., Huhse B., Belich M., Janzen J., Holman M. J., Klaus G. G., Johnston L. H., Ley S. C. (2002) CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 21, 5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thakur S., Lin H. C., Tseng W. T., Kumar S., Bravo R., Foss F., Gélinas C., Rabson A. B. (1994) Rearrangement and altered expression of the NFKB-2 gene in human cutaneous T-lymphoma cells. Oncogene 9, 2335–2344 [PubMed] [Google Scholar]
- 34. Fracchiolla N. S., Lombardi L., Salina M., Migliazza A., Baldini L., Berti E., Cro L., Polli E., Maiolo A. T., Neri A. (1993) Structural alterations of the NF-κB transcription factor lyt-10 in lymphoid malignancies. Oncogene 8, 2839–2845 [PubMed] [Google Scholar]
- 35. Cogswell P. C., Guttridge D. C., Funkhouser W. K., Baldwin A. S., Jr. (2000) Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 19, 1123–1131 [DOI] [PubMed] [Google Scholar]
- 36. Rayet B., Gélinas C. (1999) Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18, 6938–6947 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Z., Lv X., Yin W.-C., Zhang X., Feng J., Wu W., Hui C.-C., Zhang L., Zhao Y. (2013) Ter94 ATPase complex targets K11-linked ubiquitinated Ci to proteasomes for partial degradation. Dev. Cell 25, 636–644 [DOI] [PubMed] [Google Scholar]
- 38. Jiao S., Zhang Z., Li C., Huang M., Shi Z., Wang Y., Song X., Liu H., Li C., Chen M., Wang W., Zhao Y., Jiang Z., Wang H., Wong C. C., Wang C., Zhou Z. (2015) The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat. Immunol. 16, 246–257 [DOI] [PubMed] [Google Scholar]
- 39. Chen C., Shi Z., Zhang W., Chen M., He F., Zhang Z., Wang Y., Feng M., Wang W., Zhao Y., Brown J. H., Jiao S., Zhou Z. (2014) Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex. J. Biol. Chem. 289, 9651–9661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piccaluga P. P., Agostinelli C., Califano A., Rossi M., Basso K., Zupo S., Went P., Klein U., Zinzani P. L., Baccarani M., Dalla Favera R., Pileri S. A. (2007) Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J. Clin. Invest. 117, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang W. K., Xia D. (2013) Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. J. Biol. Chem. 288, 36624–36635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pye V. E., Beuron F., Keetch C. A., McKeown C., Robinson C. V., Meyer H. H., Zhang X., Freemont P. S. (2007) Structural insights into the p97-Ufd1-Npl4 complex. Proc. Natl. Acad. Sci. U.S.A. 104, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maruyama T., Fukushima H., Nakao K., Shin M., Yasuda H., Weih F., Doi T., Aoki K., Alles N., Ohya K., Hosokawa R., Jimi E. (2010) Processing of the NF-κB2 precursor p100 to p52 is critical for RANKL-induced osteoclast differentiation. J. Bone Miner. Res. 25, 1058–1067 [DOI] [PubMed] [Google Scholar]
- 44. Solan N. J., Miyoshi H., Carmona E. M., Bren G. D., Paya C. V. (2002) RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 277, 1405–1418 [DOI] [PubMed] [Google Scholar]
- 45. Xiao G., Fong A., Sun S. C. (2004) Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J. Biol. Chem. 279, 30099–30105 [DOI] [PubMed] [Google Scholar]
- 46. Qu Z., Qing G., Rabson A., Xiao G. (2004) Tax deregulation of NF-κB2 p100 processing involves both β-TrCP-dependent and -independent mechanisms. J. Biol. Chem. 279, 44563–44572 [DOI] [PubMed] [Google Scholar]
- 47. Meyer H. J., Rape M. (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang J., Chang C. C., Lombardi L., Dalla-Favera R. (1994) Rearranged NFKB2 gene in the HUT78 T-lymphoma cell line codes for a constitutively nuclear factor lacking transcriptional repressor functions. Oncogene 9, 1931–1937 [PubMed] [Google Scholar]
- 49. Yilmaz Z. B., Kofahl B., Beaudette P., Baum K., Ipenberg I., Weih F., Wolf J., Dittmar G., Scheidereit C. (2014) Quantitative dissection and modeling of the NF-κB p100-p105 module reveals interdependent precursor proteolysis. Cell Rep 9, 1756–1769 [DOI] [PubMed] [Google Scholar]