Background: The pathogenic yeast Candida glabrata harbors more than 20 epithelial adhesins (Epas).
Results: Epas are lectins with binding pockets that contain conserved and variable structural features determining ligand binding affinity and specificity.
Conclusion: The functionally diverse Epa family evolved in C. glabrata for efficient host infection.
Significance: Epa-mediated host-ligand binding is a therapeutic target to combat C. glabrata infections.
Keywords: cell adhesion, cell surface, host-pathogen interaction, lectin, structure-function, yeast, Candida glabrata, PA14 domain, epithelial adhesin
Abstract
For host colonization, the human fungal pathogen Candida glabrata is known to utilize a large family of highly related surface-exposed cell wall proteins, the lectin-like epithelial adhesins (Epas). To reveal the structure-function relationships within the entire Epa family, we have performed a large scale functional analysis of the adhesion (A) domains of 17 Epa paralogs in combination with three-dimensional structural studies of selected members with cognate ligands. Our study shows that most EpaA domains exert lectin-like functions and together recognize a wide variety of glycans with terminal galactosides for conferring epithelial cell adhesion. We further identify several conserved and variable structural features within the diverse Epa ligand binding pockets, which affect affinity and specificity. These features rationalize why mere phylogenetic relationships within the Epa family are weak indicators for functional classification and explain how Epa-like adhesins have evolved in C. glabrata and related fungal species.
Introduction
Adhesion of pathogens to host surfaces is a crucial first step for successful tissue invasion and infection. For this purpose, human pathogenic fungi often dispose of large repertoires of cell wall-associated adhesins, as exemplified by Candida albicans and Candida glabrata representing two of the major agents of human fungal infections (1–5). Most known fungal adhesins have a common modular architecture that is thought to contribute to a high variability of the cell surface properties (6). Typically, fungal adhesins are secreted proteins that consist of an N-terminal region for adhesion (A domain)4, a large central segment contained a variable number of highly glycosylated serine- and threonine-rich repeats (B domain), and a C-terminal region carried a glycosylphosphatidylinositol (GPI) anchor required for attachment to the cell wall (7). Therefore, these proteins are also referred to as GPI-anchored cell wall-associated adhesins (8). Detailed analysis of a few selected adhesins from C. albicans and C. glabrata has shown that A domains function by recognizing either host peptide (9, 10) or glycan structures (11, 12) and provided first high resolution insights into the structural features required for ligand binding and discrimination (13, 14). With the exception of these first examples, however, the precise structures and functions of most fungal adhesins are unknown.
The genome of C. glabrata contains an extraordinarily large number of more than 60 sequences that encode typical GPI-anchored cell wall-associated adhesins (5, 15, 16). The largest group comprises the EPA (epithelial adhesin) family that, depending on the strain, contains between 17 (CBS138) and 23 (BG2) members (17, 18). Epa-like adhesins are also present in other species of the Nakaseomyces genus, a group of six yeasts that include C. glabrata and whose genomes have recently been sequenced (19). Remarkably, the two other human pathogenic Nakaseomyces, Candida bracarensis and Candida nivariensis, contain 12 and 9 EPA-like genes, respectively, whereas only a single copy can be found in the nonpathogenic Nakaseomyces delphensis. The other nonpathogenic Nakaseomyces, Nakaseomyces bacillisporus and Candida castellii, contain no Epa-like adhesins that are only distantly related to the Epa family. The best characterized Epa adhesins are C. glabrata Epa1 (CAGL0E06644), Epa6 (CAGL0C00110g), and Epa7 (CAGL0C05643g), which confer adhesion to human epithelial cells (11, 12, 20).
Chip-based glycan array screening in combination with Saccharomyces cerevisiae cells presenting EpaA domains or with proteins purified from Escherichia coli has revealed that Epa1, Epa6, and Epa7 act as lectins with related ligand binding specificities (12, 14). This is in agreement with the idea that Epa adhesins enable C. glabrata to bind to host cells by recognition of mucin-type O-glycans (1, 21). Moreover, Epa1, Epa6, and Epa7 prefer carbohydrate structures with a terminal galactose unit linked via 1–3- or 1–4-glycosidic bonds to glucose, galactose, or their N-acetylated derivatives. However, Epa1 and Epa7 prefer β1–3-and β1–4-linked galactosides, whereas Epa6 is not able to discriminate between α- and β-glycosidic linkages. High resolution crystal structures of the Epa1 A domain (Epa1A) in complex with cognate disaccharide ligands have recently provided the structural basis for ligand binding specificity (14). These structures show that Epa1A is related to the A domain of S. cerevisiae flocculins (Flo adhesins), e.g. Flo5A (gene ID, 856618), and in its core contains a PA14 domain initially characterized in the anthrax-protective antigen (22). For efficient carbohydrate binding, Epa1A utilizes a unique DcisD Ca2+-binding motif that is also found in Flo5A, but not in other C-type lectins (23, 24). In contrast to Flo5A, substrate specificity in Epa1A is not mediated by a flocculin-specific subdomain but by two calcium-binding loops, CBL1 and CBL2, present in the inner part of the ligand binding pocket and by three outer loops. The A domains of all other Epa family members are also PA14 proteins and contain a DcisD motif, suggesting that they act as lectins. However, their ligand-binding patterns and/or precise binding pocket structures are largely unknown.
To determine the diversity of ligand-binding patterns within the Epa family, we here performed a comprehensive functional characterization of the adhesion domains of 17 Epa family members present in the C. glabrata strain CBS138 using large scale glycan array profiling and proteins purified from E. coli. Efficiency of individual A domains to bind to epithelial cells was assessed in vivo by an S. cerevisiae-based expression system, whereas the binding activity of selected A domains was further characterized in vitro by titration with different disaccharide ligands. To obtain new insights into the structural features conferring substrate binding and specificity in the Epa adhesin family, we compared the crystal structures of Epa1A and Epa6A in complex with diverse disaccharide ligands comprised of galactose linked via α- or β-glycosidic bonds to a secondary sugar. Our study shows that the Epa adhesin family recognizes a wide variety of α- and β-linked galactosides as well as nongalactosidic terminal glycans and provides novel insights into the structural motifs that enable ligand discrimination.
Experimental Procedures
Yeast Strains
For isolation of the EPAA domains, the C. glabrata stain CBS138 (American Type Culture Collection ATCC2001) was used. In vivo adhesion assays were performed with the nonadhesive S. cerevisiae strain BY4741 (European Saccharomyces cerevisiae Archive for Functional Analysis) carrying appropriate plasmids (Table 1) and with a triple-auxotrophic derivative of CBS138. Standard methods for yeast culture medium and transformation were used as described previously (25).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype | Source |
|---|---|---|
| pET-28(a)+ | PT7 6×His lacI KanR | Merck, Germany |
| B2445 | YCplac33 | Ref. 54) |
| BHUM1760 | PFLO11-FLO11 (1–30)-SP-3HA-FLO11(214–1360)-TFLO11in YCplac33 | This study |
| BHUM1784 | His6-EPA2(32–262) in pET-28(a)+ | This study |
| BHUM1788 | His6-EPA3(28–266) in pET-28(a)+ | This study |
| BHUM1790 | His6-EPA6(26–271) in pET-28(a)+ | This study |
| BHUM1792 | His6-EPA7(26–271) in pET-28(a)+ | This study |
| BHUM1829 | His6-EPA1(31–271) in pET-28(a)+ | This study |
| BHUM1853 | His6-EPA11(35–290) in pET-28(a)+ | This study |
| BHUM1857 | His6-EPA19(30–269) in pET-28(a)+ | This study |
| BHUM1873 | His6-EPA12(26–292) in pET-28(a)+ | This study |
| BHUM1874 | His6-EPA13(22–261) in pET-28(a)+ | This study |
| BHUM1875 | His6-EPA15(37–277) in pET-28(a)+ | This study |
| BHUM1876 | His6-EPA23(22–280) in pET-28(a)+ | This study |
| BHUM1885 | His6-EPA8(27–317) in pET-28(a)+ | This study |
| BHUM1886 | His6-EPA9(39–305) in pET-28(a)+ | This study |
| BHUM1887 | His6-EPA20(32–290) in pET-28(a)+ | This study |
| BHUM1888 | His6-EPA21(32–280) in pET-28(a)+ | This study |
| BHUM1964 | PPGK1-FLO11(1–25)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1983 | PPGK1-FLO11(1–30)-SP-3HA-EPA1(31–271)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1985 | PPGK1-FLO11(1–30)-SP-3HA-EPA2(32–262)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1987 | PPGK1-FLO11(1–30)-SP-3HA-EPA13(22–261) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1988 | PPGK1-FLO11(1–30)-SP-3HA-EPA23(22–280) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1993 | PPGK1-FLO11(1–30)-SP-3HA-EPA21(32–280)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM1998 | PPGK1-FLO11(1–30)-SP-3HA-EPA7(26–271)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2018 | PPGK1-FLO11(1–30)-SP-3HA-EPA6(26–271)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2020 | PPGK1-FLO11(1–30)-SP-3HA-EPA9(39–305) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2051 | PPGK1-FLO11(1–30)-SP-3HA-EPA8(27–317) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2150 | PPGK1-FLO11(1–30)-SP-3HA-EPA3(28–266)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2152 | PPGK1-FLO11(1–30)-SP-3HA-EPA11(35–290)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2153 | PPGK1-FLO11(1–30)-SP-3HA-EPA12(26–292) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2154 | PPGK1-FLO11(1–30)-SP-3HA-EPA15(37–277) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2155 | PPGK1-FLO11(1–30)-SP-3HA-EPA19(30–269)-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2156 | PPGK1-FLO11(1–30)-SP-3HA-EPA20(32–290) -FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2157 | PPGK1-FLO11(1–30)-SP-3HA-FLO11(214–1360)-TFLO11 in YCplac33 | This study |
| BHUM2495 | His6-EPA10(39–306) in pET-28(a)+ | This study |
| BHUM2496 | His6-EPA22(28–266) in pET-28(a)+ | This study |
Plasmids
All plasmids used in this study are listed in Table 1. Numbering of amino acid residues refers to sequences described UniProt database. To express the EpaA domains in E. coli, the different EPAA domains were amplified with specific primers for each EPAA domain and genomic DNA of C. glabrata strain ATCC2001 as a template and inserted as NdeI/XhoI or NheI/XhoI fragments into the vector pET-28(a)+. To express the different EPAA domains in S. cerevisiae, they were amplified with specific primers for each EPAA domain and genomic DNA of C. glabrata strain ATCC2001 as a template and subsequently inserted as SacII/SacI fragments into plasmid BHUM 2157. To obtain BHUM2157, the FLO11 secretion signal and a 3-fold hemagglutinin tag were amplified using SalI-SS-3HA-EpaXA and 1601-A2-SacI SacII as primers and BHUM1760 as a template. Afterward, the PCR fragment and BHUM1964 were cut using the restriction enzymes SalI and SacI and ligated together to obtain plasmid BHUM2157 carrying the following: (i) the PGK1 promoter; (ii) the FLO11 secretion signal spanning amino acid residues 1–30; (iii) a 3-fold hemagglutinin tag; (iv) the FLO11BC domain encompassing amino acids 214–1360, and (v) the FLO11 terminator.
Recombinant Overproduction and Crystallization of EpaA Domains
The wild type EpaA domains were overproduced using a previously developed low temperature protocol (23). The only modification to the protocol was the use of E. coli strain SHuffle T7 express (New England Biolabs GmbH, Frankfurt, Germany) instead of E. coli strain Origami 2. After lysis and clarification of the supernatant, the protein was purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen, Hilden, Germany) and subsequent size exclusion chromatography using HiLoad Superdex 75 PG column (GE Healthcare, Munich, Germany), initially in AM-buffer (20 mm Tris/HCl, pH 8.0, 200 mm NaCl). Some of the EpaA domains interacted strongly with the Superdex 75 PG material under these conditions, resulting in very poor yields. This issue could be solved by adding either 50 mm lactose (AML-buffer) or 10 mm EDTA (AME-buffer) to the AM-buffer.
Initial crystal screening was performed in a 600-nl sitting drop setup using commercially available screens (Qiagen, Hilden, Germany) with a Digitalab Honeybee 963 dispensing system (Genomic Solutions, Huntingdon Cambridgeshire, UK) and yielded several positive conditions at 18 °C. Optimizations of original hits took place in a 2-μl hanging drop setup, which greatly improved crystal size. Drops were composed of 50% protein solution in AML-buffer (15 mg/ml) with 5 mm lactose and 50% reservoir solution for Epa6A. Crystals belonging to space group P212121 were either yielded at 18 °C in conditions containing 80 mm sodium acetate, pH 4.6, 1.6 m ammonium sulfate, 20% glycerol or 10 mm sodium acetate, pH 4.6, 30% PEG 4000, 200 mm ammonium acetate as precipitant.
After size exclusion chromatography of Epa1A with AME-buffer, the protein solution was diluted by a factor of 10 with AM-buffer to reduce EDTA concentration (1 mm). The subsequent crystallization screening was performed as described above, including 5 mm CaCl2 and 5 mm T-Antigen (Dextra Laboratories, Reading, UK) in the protein solution. Crystals belonging to space group P41212 were yielded at 18 °C in conditions containing 100 mm HEPES, pH 7, and 20% PEG 6000. All crystals were flash-frozen in mother liquor supplemented with 20–30% (v/v) glycerol.
Soaking of Pre-grown Epa6A·Lactose Co-crystals
To obtain crystals of Epa6A in complex with either T-antigen, N-acetyld-lactosamine, lacto-N-biose, or α1–3-galactobiose, the respective crystals were grown as described above. The crystals were then soaked in mother liquor supplemented with a small amount of solid carbohydrate (Dextra Laboratories, Reading, UK) for 2–24 h. All crystals were flash-frozen in mother liquor supplemented with 20–30% (v/v) glycerol.
Data Collection, Structure Solution, and Analysis
Datasets for structure solution were recorded either at the BESSY II synchrotron (Berlin, Germany), beamline 14.1, or PETRA III synchrotron (Hamburg), beamline P14 (Table 2). The structures of Epa6A were solved via molecular replacement, using a carefully trimmed model of Epa1A (Protein Data Bank code 4AF9), and for the Epa1A·T-antigen complex, the complete soaking structure was used (Protein Data Bank code 4ASL). Phase solution was performed with PHASER (26); data processing was performed with XDS, XSCALE, PHENIX, and CCP4 (27–29) and refinement with alternating rounds of REFMAC (30) and Coot (31). Secondary structure assignments were performed with STRIDE (32), and manual inspection was done using the Epa1A structure as comparison (Protein Data Bank code 4AF9). Figures of protein structures were generated using the molecular graphics program PyMOL Version 1.4.1 (DeLano Scientific).
TABLE 2.
Data collection statistics for Epa1A and Epa6A complexes
Values for the highest resolution shell are shown in parentheses.
| Epa1A·T-antigen | Epa6A·lactose | Epa6A·T-antigen | Epa6A·N-acetyl-d-lactosamine | Epa6A·lacto-N-biose | Epa6A·α1–3-galactobiose | |
|---|---|---|---|---|---|---|
| PDB code | 4D3W | 4COU | 4COW | 4COY | 4COZ | 4COV |
| X-ray source | 14.1, BESSY | 14.1, BESSY | P14, PETRA III | 14.1, BESSY | P14, PETRA III | 14.1, BESSY |
| Wavelength (Å) | 0.91841 | 0.91841 | 0.826606 | 0.918409 | 0.826606 | 0.918409 |
| Cell dimensions (Å) | a = 55.01, b = 55.01, c = 139.75 | a = 41.3, b = 61.1, c = 106.3 | a = 41.35, b = 61.10, c = 106.61 | a = 41.47, b = 63.33, c = 110.25 | a = 41.28, b = 61.26, c = 106.62 | a = 41.52, b = 63.29, c = 110.30 |
| Resolution (Å) | 24.60-1.50 (1.58-1.50) | 28.77-1.48 (1.56-1.48) | 34.24-2.00 (2.11-2.00) | 41.58-1.80 (1.90-1.80) | 53.12-2.25 (2.37-2.25) | 41.58-1.50 (1.58-1.50) |
| Observed reflections | 384,090 | 174,129 | 109,348 | 146,305 | 120,749 | 257,032 |
| Unique reflections | 35,383 | 45,573 | 18,879 | 27,537 | 13,420 | 47,367 |
| Completeness (%) | 100 (100) | 99.7 (99.9) | 99.8 (99.7) | 99.5 (99.5) | 99.9 (100) | 99.9 (99.9) |
| Multiplicity | 10.9 (10.6) | 3.8 (3.8) | 5.8 (5.7) | 5.3 (5.0) | 9.0 (9.2) | 5.4 (5.2) |
| Mean I/σ(I) | 13.4 (4.2) | 21.1 (2.9) | 11.9 (2.7) | 17.1 (4.4) | 12.1 (3.5) | 20.7 (3.0) |
| Rmerge (%) | 10.1 (50.5) | 4.4 (57.1) | 12.3 (60.4) | 9.2 (40.4) | 17.3 (64.7) | 4.6 (52.0) |
| Rwork/Rfree (%) | 15.8/18.3 | 14.4/17.2 | 17.7/23.0 | 18.7/22.3 | 17.8/22.0 | 15.0/17.6 |
| Total reflections | 33528 | 44337 | 14534 | 26129 | 11930 | 46100 |
| Test set reflections | 1767 | 1173 | 740 | 1378 | 624 | 1223 |
| No. of atoms (total) | 2183 | 2109 | 2056 | 2197 | 2013 | 2219 |
| No. of atoms (water) | 234 | 245 | 198 | 320 | 178 | 338 |
| Mean B value (Å2) | 15.6 | 19.2 | 25.1 | 16.3 | 23.9 | 20.2 |
| Root mean square deviations bond lengths (Å) | 0.012 | 0.008 | 0.010 | 0.010 | 0.008 | 0.007 |
| Root mean square deviations bond angles (°) | 1.611 | 1.295 | 1.319 | 1.384 | 1.209 | 1.134 |
High Throughput Glycan Binding Assays
Recombinant EpaA domains were fluorescently labeled using an AlexaFluor 488 THF kit (Life Technologies, Inc.) and applied to CFG array Version 5.1 chips at protein concentrations of 200 μg/ml. Chip surfaces were repeatedly washed, and the remaining fluorescence was measured and quantified.
Fluorescence Titration Spectroscopy
Fluorescence titrations of EpaA domains against respective carbohydrates were performed as described (23, 24). Binding was followed at an emission wavelength between 340 and 350 nm by excitation of intrinsic tryptophan fluorescence at 280 nm. Fluorescence quench was recorded during titration and fitted using a one-site plus unspecific binding model.
Adhesion of S. cerevisiae and C. glabrata to Human Epithelial Cells
For adhesion tests, the human epithelial cell line Caco-2 (American Type Culture Collection HTB-37) was used together with the S. cerevisiae strain BY4741 carrying plasmids with the appropriate PPGK1-3HA-EPAA-FLO11BC constructs (Table 1). The presence of EpaA domains at the S. cerevisiae cell surface was quantified by immunofluorescence microscopy. For this purpose, cultures of plasmid carrying strains were grown in low fluorescence yeast medium to an optical density at 595 nm of 1, before cells were washed three times in PBS, 1% BSA. Then, cells were incubated with a monoclonal mouse anti-HA antibody (H3663; Sigma, Munich, Germany) at a dilution of 1:1000 in PBS, 1% BSA for 30 min at room temperature. After three wash steps, cells were incubated in darkness with a Cy3-conjugated secondary goat anti-mouse antibody (C2181; Sigma) at a dilution of 1:10,000 in PBS, 1% BSA for 20 min at room temperature. After three further washing steps, a Zeiss Axiovert 200 M microscope was used to visualize S. cerevisiae cells with differential interference contrast and to detect EpaA domains at the cell surface using a rhodamine filter set (AHF Analysentechnik AG, Tübingen, Germany). Cells were photographed with a Hamamatsu Orca ER digital camera (Hamamatsu, Bridgewater, NJ), and pictures were processed and analyzed using the Improvision Volocity software (PerkinElmer Life Sciences, Hamburg, Germany). Fluorescence signals were then quantified using the ImageJ software (33).
For EpaA-directed adhesion assays to human epithelial cells, confluent monolayers of Caco-2 cells were grown in 75-cm2 tissue culture flasks (Greiner, Frickenhausen, Germany) and split 1:3 every 2nd or 3rd day, depending on the confluence, which did not exceed 80%. Once the cell culture was initiated, a periodic medium change was performed using Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1 mm sodium pyruvate, and 1% gentamicin (Invitrogen, Karlsruhe, Germany). After ∼20 subcultures, cells were seeded into 24-well polystyrene plates (Greiner, Frickenhausen, Germany) and incubated at 37 °C under 5% CO2 for 1–2 days until a confluent monolayer was formed.
Adhesion assays of S. cerevisiae and C. glabrata strains on human epithelial Caco-2 cell lines were performed as described previously (34). Briefly, 24-well polystyrene plates with confluent monolayers of Caco-2 cells were used after removal of the culture medium and addition of 250 μl of fresh pre-warmed DMEM without gentamicin. S. cerevisiae or C. glabrata strains carrying appropriate plasmids were grown in YPD medium to exponential phase at 30 °C and diluted in DMEM, 10% FBS, 1 mm sodium pyruvate to a concentration of ∼6000 cells/ml of medium. 50 μl of these yeast cell suspensions were then added to each well with a confluent layer of Caco-2 cells. Plates were incubated at 37 °C under 5% CO2 for 0, 30, 60, 120, or 180 min, respectively. The complete supernatant containing the nonadherent S. cerevisiae or C. glabrata cells was removed and plated on YPD agar to determine the colony-forming units (CFU). To determine the adherent yeast cells, wells were washed twice with 300 μl of phosphate-buffered saline (PBS) before the epithelial cells together with the attached S. cerevisiae or C. glabrata cells were scratched off the polystyrene surface. The resulting suspension was also plated on YPD plates to determine the CFU of adherent cells. After incubation for 2 days at 30 °C, CFU values for nonadherent and adherent cells were determined using an aCOLyte colony counter (7510 DWS; Synbiosis, Cambridge, UK). The average values for nonadherent and adherent cells were determined based on 10 independent experiments. Outliers were eliminated with the help of the standard deviation, the standard error and a t test. Relative adhesion values (A) were calculated by using Equation 1,
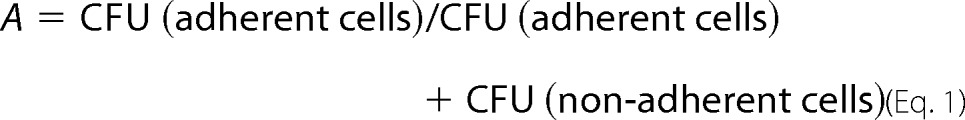 |
Bioinformatic Analysis
Hierarchical clustering analysis of EpaA domains was performed using the Cluster 3.0 software (35–37) and the centroid linkage clustering algorithm. For this purpose, relative glycan binding values (absolute binding values divided by the binding value of the best bound glycan) for each EpaA domain toward the 610 glycans present on the CFG array Version 5.1 were calculated, and clustering results were visualized by using the Java TreeView software (35–37). Pie chart analysis of glycan binding profiles was done as described previously (14).
Figures of protein structures were generated with the molecular graphics software PyMOL Version 1.4.1 (38). To model the structures of further Epa family members, the Modeler 9v7 software was used (39). As templates, the structures of Epa1A (4ASL) and Epa6A (4COU) were used. The respective sequences were collected from the Candida Genome Database (40). To analyze structures for their conserved residues and regions, the Consurf server was used with Epa1A as template (41, 42). The alignment was constructed using T-coffee (43). Molecular evolutionary genetic analysis was performed and visualized using the MEGA6 software (44). Where indicated, 500 bootstrap replications were included.
Data Deposition
The glycan array data from this publication have been deposited at the Consortium for Functional Glycomics and assigned the identifiers cfg_rRequests 2554, 2645, 2737, and 2854. The atomic coordinates and structure factors have been deposited in the Protein Data Bank and assigned the accession codes 4COU, 4COV, 4COW, 4COY, 4COZ, and 4D3W.
Results
Diversity of Epa Family Ligand-binding Patterns
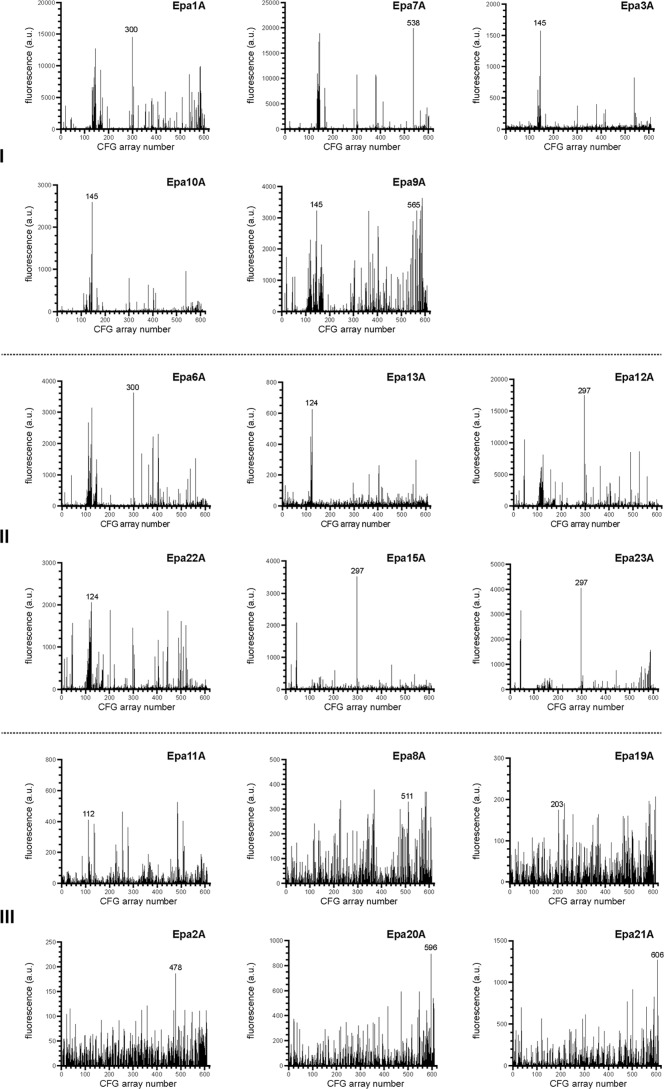
We first determined the carbohydrate specificity profiles of 17 Epa adhesins encoded in the genome of C. glabrata strain ATCC 2001/CBS138 (5, 21). For this purpose, corresponding A domains were purified from E. coli and analyzed for in vitro binding to 610 different carbohydrates using a mammalian glycan array from the CFG. The 17 binding profiles obtained by this approach (Fig. 1) provided 10,370 data points, which were further analyzed by hierarchical clustering using the Cluster 3.0 and TreeView software (35–37). This analysis revealed that each of the EpaA domains possesses its own specific binding profile and that the Epa family can be broadly divided into three separate functional classes (Fig. 2). Class I consists of five EpaA adhesins (Epa1A, Epa7A, Epa3A (CAGL0E06688g), Epa10A (CAGL0A01284g), and Epa9A (CAGL0A01366g)), whose best-bound glycans are β1–3- or β1–4-linked galactosides; class II contains six proteins that best bind to either β1–3- and α1–6-linked (Epa6A, Epa13A (CAGL0L13332g), and Epa22A (CAGL0K00170g)) or sulfated galactosides (Epa12A (CAGL0M00132g), Epa15A (CAGL0J11968g), and Epa23A (CAGL0I00220g)); and class III consists of six members, which best bind to acidic sugars (Epa19A (CAGL0A00110g), Epa2A (CAGL0E06666g), Epa20A (CAGL0E06666g), and Epa21A (CAGL0D06732g)) and sulfated galactosamines (Epa8A (CAGL0C00847g)) or α1–3-linked galactosides (Epa11A (CAGL0L13299g)). Generally, the preferred ligands of class I and class II were bound with apparently medium to high affinity as based on data obtained by the glycan array analysis (Fig. 1), whereas binding values obtained for members of class III indicated mostly weak and hence less specific binding.
FIGURE 1.
Glycan binding profiles. Binding profiles of 17 EpaA domains from C. glabrata strain CBS138 (ATCC 2001) were obtained by using the CFG mammalian glycan array Version 5.1 and fluorescently labeled EpaA proteins purified from E. coli at a concentration of 200 μg/ml. Fluorescence units measured for the different EpaA domains and the 610 different glycans present on the array are shown as black lines. Glycans with unambiguous terminal disaccharide types, which were best bound by the different EpaA domains, are indicated with numbers according to their position on the CFG array. Profiles are depicted according to their classification (roman numbers) obtained by the hierarchical clustering analysis presented in Fig. 2.
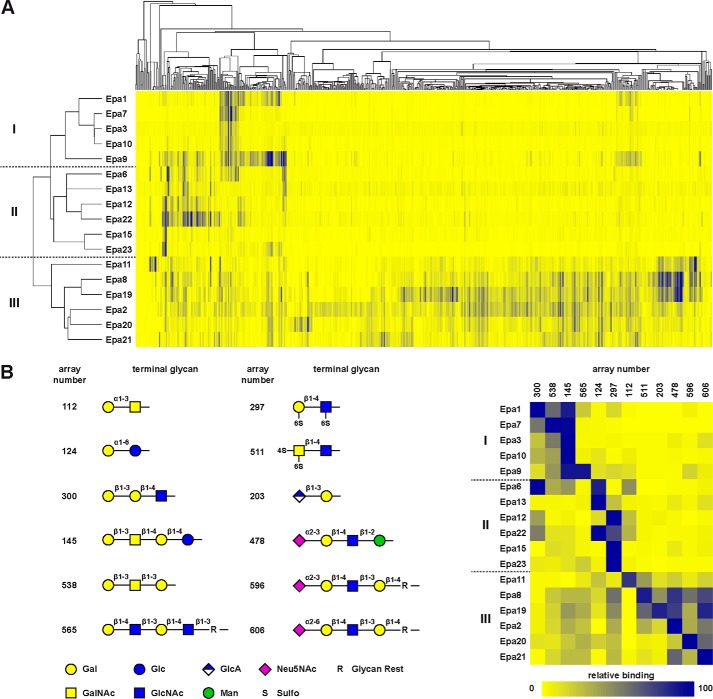
FIGURE 2.
Glycan profile-based relationship of the Epa family. A, hierarchical clustering analysis. The 17 Epa paralogs from C. glabrata strain CBS138 were clustered according to the relative in vitro binding strength of their A domains purified from E. coli toward a total of 610 different glycans present on the CFG array Version 5.1 using Cluster 3.0 and Java TreeView software. Relationships among the 17 Epa adhesins as given by their binding profiles are shown by the tree on the left; glycans are clustered as shown on the top according to the similarity of their recognition by different Epa paralogs. Relative binding strength is shown in color with values ranging from 0 (yellow) to 100 (dark blue). Classification is indicated by roman numbers and dotted lines. B, comparison of relative binding strength of best bound glycan ligands. The left part shows the terminal residues of all glycan structures with unambiguous terminal disaccharide types best bound by the individual EpaA domains. Numbers label the different polyglycan types according to their position on the CFG array. The right side shows the relative binding strength of these glycans to all EpaA domains. CFG array numbers are indicated. Relative binding is shown in color as in A.
To further assess and compare the different ligand recognition patterns, we also classified the 17 EpaA adhesins with respect to the best-bound terminal sugars in combination with the linkage type to the penultimate carbohydrate moiety. This analysis also leads to a tripartite classification of the Epa family that is comparable with the one obtained by unbiased hierarchical clustering (Fig. 3). It also shows the following within class I: (i) Epa7A and Epa3A are highly specific for terminal galactose-linked via a β1–3 glycosidic bond to the penultimate sugar moiety (Galβ1–3); (ii) Epa1A best binds to Galβ1–3 and Galβ1–4; (iii) Epa10A recognizes Galβ1–3 and to some degree also Galα1–4; and (iv) Epa9A is able to bind Galβ1–3 and Galβ1–4 as well as Galα1–3 and Galα1–4. In class II, the preferred ligands contain terminal α-linked galactose (Epa6A, Epa13A, Epa12A, and Epa22A) or terminal (6S)-galactose units connected by a β-type linkage (Epa12A, Epa22A, Epa15A, and Epa23A). Finally, EpaA proteins of class III prefer nongalactosides as terminal residues (Epa8A, Epa19A, Epa2A, Epa20A, and Epa21A) with the exception of Epa11A that binds to both nongalactosides and Galα1–3 with comparable strength.
FIGURE 3.
Ligand binding profiles of EpaA domains. Pie charts show the glycan types, for which the CFG array binding signals obtained by the different EpaA domains exceeded either 50 or 20%, respectively, of the signal of the best binder and for which terminal disaccharide types are unambiguous. The different glycan types are color-coded according the terminal (primary) carbohydrate moiety and the linkage type to the following (second) carbohydrate unit as indicated on the right side. The areas shown correspond to the logarithm of the fluorescence signal over noise for the best binder as exemplified by the two circles at the lower right. Functional classes are indicated by roman numbers.
In summary, our comprehensive glycan profiling shows that the 17 Epa members encoded in the genome of strain CBS138 (i) all appear to be well folded and functional in vitro, (ii) all possess individual ligand-binding patterns, (iii) together cover a wide variety of glycosidic ligands containing terminal α- and β-linked galactosides as well as nongalactosidic sugars, and (iv) display highly variable ligand binding affinities.
Epithelial Cell Adhesion by EpaA Domains
We next directly compared in vivo functionality of the different EpaA domains by measuring their adhesion activity to human colorectal and epithelial Caco-2 cells. For this purpose, all 17 EPAA domains analyzed in vitro were individually expressed in nonadhesive S. cerevisiae cells using a flocculin gene-based expression system as described previously (14). Successful expression and presentation of the EpaA domains on the cell surface were first monitored by fluorescence microscopy. Here, we found significant amounts of A domains in the case of 15 Epa paralogs (Fig. 4A). Corresponding S. cerevisiae strains were further assayed for in vivo epithelial cell adhesion along with appropriate control strains and a C. glabrata strain (Fig. 4B). As expected, strains that lack an EpaA domain did not adhere to Caco-2 cells. The 15 A domain presenting strains exhibited variable epithelial cell adhesion and roughly divide the Epa adhesins into three groups. The first group consisting of Epa1A, Epa7A, and Epa6A conferred very efficient cell adhesion, which was between 13- and 17-fold higher than the activity measured in the absence of an A domain, but did not fully reach the adhesion strength observed for C. glabrata (Fig. 4B). Epa9A, Epa12A, Epa15A, Epa23A, and Epa8A compose a second group and mediate adhesion with values that are between 3- and 6-fold higher than the control measurement. All other EpaA domains constitute a third class and confer adhesion that is less than 3-fold better than the control. In summary, we find that of the 15 Epa adhesins tested about 20% (3/15) mediate efficient and 35% (5/15) confer mediocre epithelial cell adhesion, whereas the other members of the Epa family (7/15) mediate only very weak binding to Caco-2 cells.
FIGURE 4.
In vivo binding of EpaA domains. A, expression of different EpaA domains using a heterologous S. cerevisiae expression system. S. cerevisiae strain BY4741 carrying either of the plasmids B2445 (control), BHUM2157 (no A), BHUM1983 (Epa1A), BHUM1985 (Epa2A), BHUM2150 (Epa3A), BHUM2018 (Epa6A), BHUM1988 (Epa7A), BHUM2051 (Epa8A), BHUM2020 (Epa9A), BHUM2152 (Epa11A), BHUM2153 (Epa12A), BHUM1987 (Epa13A), BHUM2154 (Epa15A), BHUM2155 (Epa19A), BHUM2156 (Epa20A), BHUM1993 (Epa21A) or BHUM1988 (Epa23A) was grown to logarithmic phase before EpaA domains were visualized at the cell surface by immunofluorescence microscopy using anti-HA primary and Cy3-conjugated secondary antibodies. Specific signals were quantified, and the values obtained are shown in percentage relative to the strain expressing the carrier domain alone (no A) set to 100. Scale bar corresponds to 10 μm. B, epithelial cell adhesion conferred by 15 EpaA domains. Adhesion was determined using S. cerevisiae strains shown in A or a C. glabrata strain (triple-auxotrophic derivative of CBS138) after 2 h of incubation with a monolayer of Caco-2 cells. Adhesion of S. cerevisiae strains expressing no adhesin (control) or only the carrier domain (no A) is shown in black. For S. cerevisiae strains, EpaA-specific adhesion is indicated by the white part of bars that results from subtracting the value obtained by the strain expressing the carrier domain alone (no A) from the total adhesion. Error bars indicate standard deviation obtained by 10 independent experiments. The functional classification is shown at the bottom. DIC, differential interference contrast.
Quantification of Ligand Binding by Epa1A, Epa6A, and Epa7A
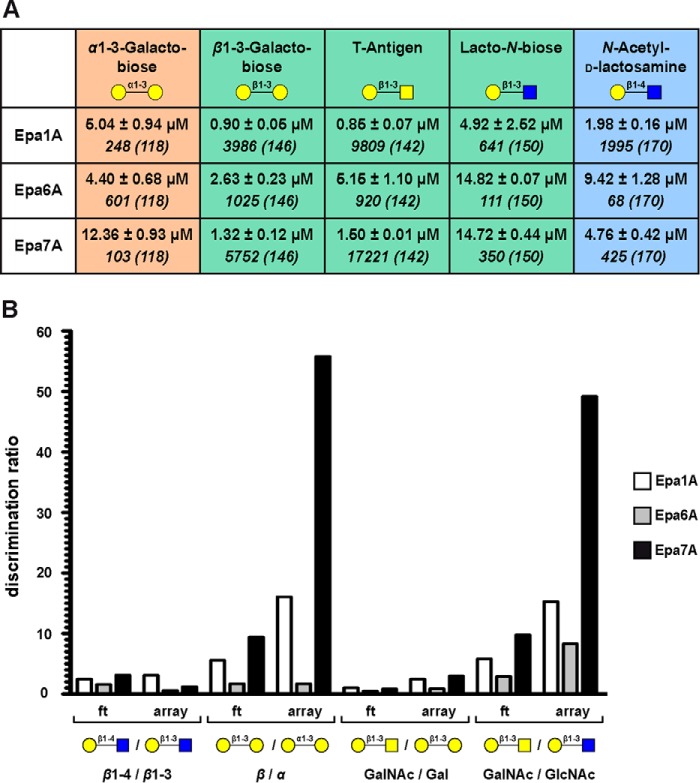
We next wanted to quantify the ligand binding specificities for a number of selected EpaA domains in more detail. For this purpose, we focused on Epa1A, Epa6A, and Epa7A, because (i) these adhesins are closely related at the protein sequence level (12, 14), and (ii) we here found these adhesins to confer the most efficient epithelial cell binding (Fig. 4B). We performed fluorescence titration analysis and determined the in vitro binding constants (KD values) for these proteins with five different disaccharides, α1–3-galactobiose (Galα1–3Gal), β1–3-galactobiose (Galβ1–3Gal), T-antigen (Galβ1–3GalNAc), lacto-N-biose (Galβ1–3GlcNAc), and N-acetyl-d-lactosamine (Galβ1–4GlcNAc). Comparison of the different KD values not only reveals the differences in ligand binding affinities but also provides the discrimination ratios with respect to β1–4-linked versus β1–3- and β- versus α-linked disaccharides and with regard to the nature of the sugar moiety attached to galactose, namely Gal, GalNAc, or GlcNAc. We found that Epa1A and Epa7A only minimally discriminate between β1–4- and β1–3-linkages as exemplified by the comparison of Galβ1–4GlcNAc and Galβ1–3GlcNAc, although the affinity of Epa1A for these ligands is generally 2.4–3.0-fold higher (Fig. 5A). However, Epa1A and Epa7A strongly discriminate between β- and α-glycosidic linkages as illustrated by the Galβ1–3Gal/Galα1–3Gal ratios of 5.6 and 9.4, respectively. This efficient β/α discrimination is further emphasized by the glycan array values obtained for these disaccharides with even higher ratios of 16 (Epa1A) and 56 (Epa7A). Moreover, both proteins exhibit similarly high affinities toward Galβ1–3Gal with KD values of about 1 μm, but Epa7A binds the Galα1–3Gal isomer 2.5-fold less efficient than Epa1A. With respect to the nature of the second hexose moiety, we further find that Epa1A and Epa7A clearly prefer Gal and GalNAc over GlcNAc. In contrast to Epa1A and Epa7A, Epa6A is almost unspecific with regard to discriminating between different types of glycosidic linkages and significantly less specific with respect to discriminating between Gal, GalNAc, and GlcNAc at the secondary position of disaccharides (Fig. 5B). Remarkably, however, the overall affinity of Epa6A to the diverse ligands is comparable with Epa1A and Epa7A.
FIGURE 5.
In vitro binding of selected EpaA domains. A, dissociation constants (KD values) for binding of Epa1A, Epa6A, and Epa7A to five different galactosides (α1–3-galactobiose = Galα1–3Gal; β1–3-galactobiose = Galβ1–3Gal; T-antigen = Galβ1–3GalNAc; lacto-N-biose = Galβ1–3GlcNAc; and N-acetyl-d-lactosamine = Galβ1–4GlcNAc) were determined by fluorescence titration analysis and are shown in regular letters, including values for standard deviations. Binding values obtained for Epa1A, Epa6A, and Epa7A and the five galactosides on glycan arrays (corresponding array numbers are indicated in parentheses) are shown below KD values in italic letters. B, discrimination ratios with respect to different linkage types (β1–4/β1–3 and β/α) and with regard to the nature of the sugar moiety attached to galactose (Gal/GalNAc and GalNAc/GlcNAc) were calculated from the corresponding association constants (1/KD) obtained by fluorescence titration (ft) analysis or from the glycan array fluorescence signals (array) shown in A.
Structural Basis for Ligand Discrimination by Epa1A and Epa6A
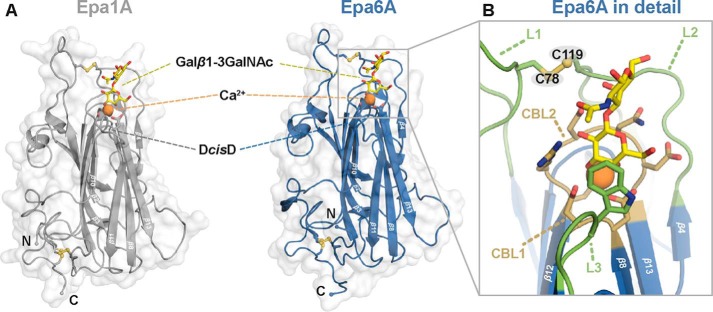
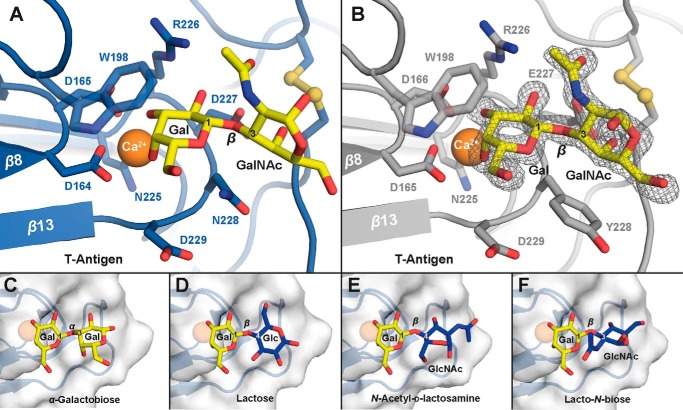
To better understand the different binding patterns obtained with members of the Epa1 subfamily, we aimed at obtaining crystal structures of Epa1A and Epa6A in complex with different galactosides assayed for in vitro binding. For Epa1A, we have previously solved the structure in complex with different disaccharides (Galβ1–3GalNAc, PDB code 4ASL; and Galβ1–3Glc, PDB code 4AF9) by soaking the crystals obtained before by co-crystallization with lactose (14). Here, we obtained a structure of Epa1A in complex with the T-antigen (Galβ1–3GalNAc, PDB code 4D3W, Table 2) by co-crystallization in a new space group at a resolution of 1.5 Å, which does not significantly differ from the previous structure (root mean square deviation of 0.31 Å for 172 Cα positions). This finding not only underscores the fact that EpaA domains are structurally quite rigid, it also validates the previously used soaking protocol. Therefore, we used the same crystallization approach to determine the structures of Epa6A in complex with lactose and Galβ1–3GalNAc, which were obtained at a resolution of 1.56 and 2.15 Å (Table 2; PDB codes 4COU and 4COW), respectively. The overall structural fold of Epa6A is highly comparable with Epa1A with a root mean square deviation of 0.37 Å for 177 Cα positions (Fig. 6). The structure is composed of a β-sandwich that is derived from the PA14 domain of the B. anthracis-protective antigen (22). It harbors two consecutive aspartate residues connected by a cis-peptide bond, as has been found in Epa1A and the A domains of the S. cerevisiae flocculins Flo5A and Lg-Flo1A (14, 23, 45). Ligand binding in Epa6A is achieved by coordination via a Ca2+ ion, which is fixed by the DcisD motif and an asparagine residue in CBL2 (Fig. 6B). In addition, the binding pocket harbors residues of the calcium-binding loop CBL2 and a tryptophan residue situated in a flexible loop above the pocket, which both mediate crucial interactions and are responsible for specificity and affinity. The binding pocket is also shielded from the surrounding solvent by the flexible loops L1 and L2, which are connected by a highly conserved disulfide bridge formed by Cys-78 and Cys-119. Finally, the overall orientation of Galβ1–3GalNAc in the Epa6A binding pocket is comparable with the orientation of the same ligand in Epa1A (Fig. 7, A, B and D). However, the precise ligand interaction patterns of the two proteins clearly differ with respect to two residues in CBL2. In the case of Epa6A, ligand binding involves Asp-227 and Asn-228, whereas the corresponding positions in Epa1A are represented by Glu-227 and Tyr-228 (Fig. 7, A and B).
FIGURE 6.
Overall structural features of Epa6A and Epa1A. A, overall fold of Epa6A (blue) shows a β-sandwich comparable with that of Epa1A (gray). The complex structures of Epa6A and Epa1A show a bound T-antigen ligand (Galβ1–3GalNAc, yellow) complexed by a calcium ion (orange) via a DcisD motif that is highly conserved in fungal adhesins. B, selectivity and affinity in ligand binding is achieved by two calcium binding loops (CBL1 and CBL2; brown) in combination with three flexible loops, L1 to L3 (green). L1 and L2 are connected by a disulfide bridge via Cys-78 and Cys-119, which shields the binding pocket from surrounding solvent. L3 contains a tryptophan residue (green) that is highly conserved in Epa adhesins and is essential for ligand binding.
FIGURE 7.
Ligand discrimination by Epa6A and Epa1A binding pockets. A, binding pocket of Epa6A (blue) in complex with the T-antigen (yellow) and residues conferring binding affinity and specificity. A calcium ion (orange) is complexed via a DcisD motif and Asn-225. Interaction between Epa6A and the galactose moiety is accomplished by residues in CBL2 and Trp-198 of loop L3. B, binding mode of T-antigen to the pocket of Epa1A (gray) is similar to that of T-antigen to Epa6A, but the interaction pattern is different. The SigmaA-weighted 2Fobs − Fcalc electron density highlights the orientation of the carbohydrate (contoured at 2.0 σ). C–F, structures of the Epa6A binding pocket in complex with four different galactosides. Although the orientation of the terminal galactose moiety is fixed in all four complexes, the binding pose of the second hexose is more variable and depends on its type and the linkage to galactose.
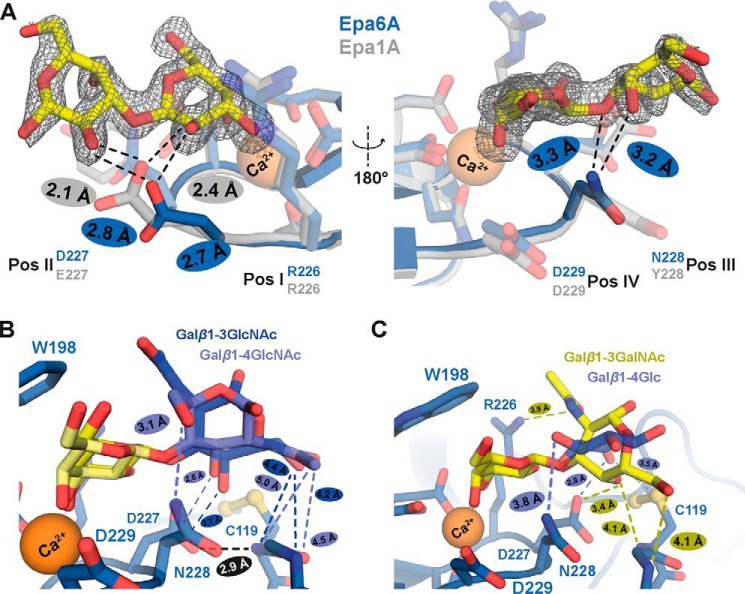
To better understand how individual EpaA domains discriminate between different ligands, we solved the structures of Epa6A in complex with three further glycans, Galα1–3Gal (PDB code 4COV), Galβ1–4GlcNAc (PDB code 4COY), and Galβ1–3GlcNAc (PDB code 4COZ) (Fig. 7, C, E, and F; Table 2). This set of high resolution structures allows delineation of the structural features, which contribute to the differences of specific ligand binding. First, a comparison of the Epa6A·Galα1–3Gal complex with the corresponding model of an Epa1A·Galα1–3Gal complex reveals that the CBL2 region seems to determine the efficiency by which these two proteins are able to discriminate between α- and β-linked carbohydrates (Fig. 8A). Although a shorter aspartate residue (Asp-227) can be found at position II of its CBL2 region, Epa1A harbors a glutamate (Glu-227) at this position. This suggests that the shorter residue in Epa6A enables more efficient binding of α-linked glycosides than Epa1A due to reduced steric hindrance (Fig. 8A). In addition, a hydrogen bond is observed between Galα1–3Gal and the CBL2 position III (Asn-228) of Epa6A. This ligand interaction is absent in the Epa1A·Galα1–3Gal complex, suggesting that the efficient binding of α-linked galactosides is further fostered by interaction with CBL2 position III. This conclusion is also supported by our previous finding that a change of the CBL2 region of Epa1A to its Epa6A counterpart is sufficient to increase the binding to α-linked galactosides (14).
FIGURE 8.
Role of CBL2 in conferring ligand discrimination by Epa1A and Epa6A. A, structural basis for α-/β-discrimination by Epa1A and Epa6A. Shown is a superposition of the structure of Epa1A (gray) with the structure of the Epa6A·α-galactobiose complex (blue). Higher selectivity of Epa6A toward α-linked galactosides is achieved by the sterically preferred Asp residue at CBL2 position II (Asp-227) in comparison with the sterically more demanding Glu residue at the corresponding position of Epa1A (Glu-227). In addition, a hydrogen bond is observed between α-galactobiose and the CBL2 position III (Asn-228) of Epa6A, which is absent in the Epa1A that carries a Tyr at this position (Tyr-228). B, ligand discrimination by Epa6A. Comparison of Galβ1–3GlcNAc (dark blue) and Galβ1–4GlcNAc (light blue) bound to Epa6A reveals divergent ligand-binding patterns. Whereas the backbone of residue Cys-119 of Epa6A interacts with the 6-OH group of the secondary sugar in the case of Galβ1–3GlcNAc, the same residue of Epa6A contacts the N-acetyl group of the GlcNAc moiety of Galβ1–4GlcNAc. C, ligand discrimination by Epa6A. Comparison of Galβ1–3GalNAc (yellow) with Galβ1–4Glc (light blue) bound to Epa6A reveals highly deviating binding patterns. The secondary carbohydrate is rotated by 90° and different amino acid residues of Epa6A are used for interaction.
Our structural analysis further reveals that the binding poses of lactose, N-acetyl-d-lactosamine, and lacto-N-biose bound to the Epa6A domain (Fig. 7, D–F) show different orientations of the second hexoses in the outer binding site despite a common recognition of the galactose moiety within the inner subsite. This is facilitated by alternative H-bonding interactions between Asp-227 and the 4-OH group of GlcNAc in Galβ1–3GlcNAc, the 3-OH of GlcNAc in Galβ1–4GlcNAc, and the 6-OH of the Glc moiety in lactose, respectively (Fig. 8, B and C). Like the corresponding position II residue in Epa1A, the side chain conformation of Asp-227 in Epa6A is stabilized by an H-bond to the peptide group of Cys-119 in loop L2 (Fig. 8B). Interestingly, the bulky N-acetyl moiety is found to be well ordered independent of its contacts either with the backbone of Cys-119 in the Epa6A·Galβ1–4GlcNAc structure or with the indole group of Trp-198 in the Epa6A·Galβ1–3GlcNAc structure. A comparison of the binding mode of the β1,4- and β1,3-linked Gal-GlcNAc disaccharides reveals that the orientation of the GlcNAc moiety is rotated by nearly 180° (Fig. 8B). This leads to an exchange of the interaction pattern regarding the Cys-119 backbone. Although the 6-OH group of the β1,3-linked glycan is interacting with that residue, the β1,4-linked glycan interacts via its N-acetyl moiety. The latter is also forming an additional salt bridge with Asn-228 by its 6-hydroxyl group. Thus, the detailed binding patterns of β1,4- and β1,3-linked Gal-GlcNAc disaccharides clearly differ, even though the involved amino acid residues are nearly conserved. Finally, comparison of Epa6A·Galβ1–3GalNAc with Epa6A·Galβ1–4Glc reveals highly deviating binding patterns. Here, the secondary carbohydrate is rotated by 90°, and different amino acid residues are employed for interaction (Fig. 8C). This strongly suggests that the energetic contribution of the second hexose to the overall binding of the Gal-based ligands appears to be independent of the mode of its accommodation in the outer subsite.
Discussion
We here have performed a comprehensive in vitro and in vivo functional analysis of the large family of epithelial adhesins of the human pathogenic yeast C. glabrata. Our study reveals that most Epa paralogs possess individually tailored ligand binding properties. Moreover, our data permit us to directly compare the structural and functional relationships of this large family of medically relevant fungal virulence factors. As shown in Fig. 9, such a comparison clearly reveals that the phylogenetic relationship, as based on primary sequence and overall three-dimensional structural features of the A domains, does not markedly correlate with a functional classification that is based on the ligand-binding patterns. Members of the functional class II, for instance, are widely scattered on the phylogenetic tree and can be found in several distantly related branches (Fig. 9B). Thus, the A domains of functionally closely related members, such as Epa6 and Epa13 or Epa1 and Epa3, are structurally quite diverse. Vice versa, phylogenetically closely related adhesins, such as Epa6 and Epa7 or Epa3 and Epa22, possess markedly distinct ligand binding specificities. Our study therefore suggests that functionally related Epa variants might have repeatedly developed independently.
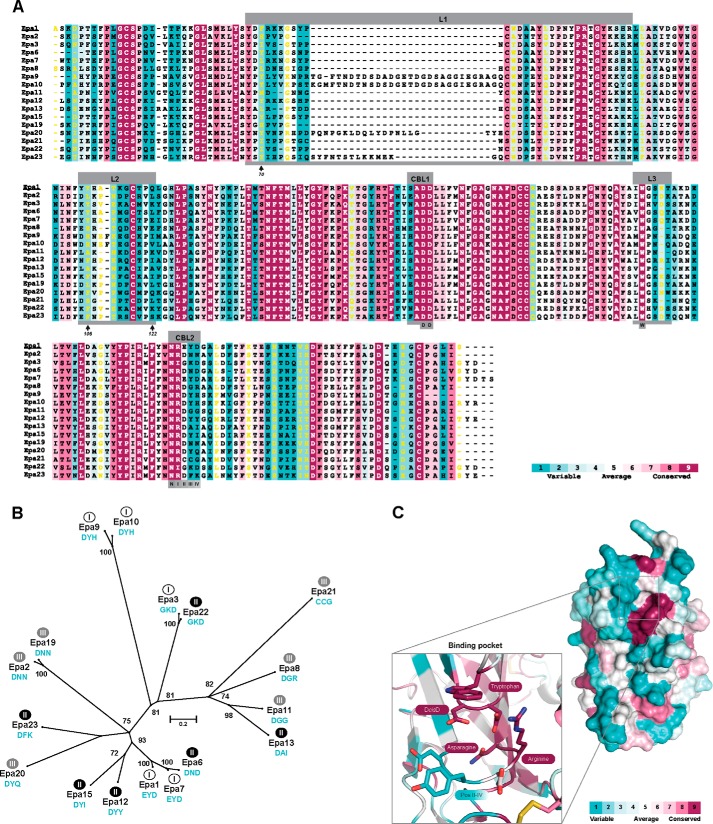
FIGURE 9.
Phylogeny and structural conservation of C. glabrata EpaA domains. A, conservation and variability of individual residues of Epa family A domains. Shown is a structure-based sequence alignment of the 17 Epa paralogs from C. glabrata CBS138 as analyzed in this study. The alignment was generated using a local copy of the T-Coffee software implemented with 3DCoffee (43, 52) in combination with the three-dimensional structure of Epa1A (14) and was further processed using the ConSurf server (41, 53). The degree of conservation/variability of individual residues is color-coded according to the ConSurf-server, and yellow letters show minor reliability. Positions of the loops L1, L2, and L3, which form the outer binding pocket, and the calcium-binding loops CBL1 and CBL2, which constitute the inner binding pocket, are indicated above the sequences. Indicated below the sequences are positions I–IV of CBL2, the positions of the DcisD (DD) motif, and the Asn residue of CBL2 that form the DD-N structural motif, as well as positions of the Trp residue of loop L3 and the Arg residue of CBL2 (position I), which constitute the W-R corner of the binding pocket. Arrows indicate three further residues (70, 106, 122) discussed in the text. B, phylogenetic tree of EpaA domains. The tree was created with the MEGA6 software using 500 bootstrap replications (44) and is based on the structure-guided multiple sequence alignment described in A. Bootstrapping values above 70% are indicated. Functional classification obtained by glycan array analysis is indicated by white (class I), black (class II), or gray (class III) circles. Sequence motifs of CBL2 positions II–IV are shown in turquoise. A bar refers to phylogenetic distances. C, conservation of surface properties of EpaA domains. A structural model of Epa1A is shown on the right, which depicts conserved and variable surface residues. The degree of conservation/variability is color-coded and was obtained by using the ConSurf server (41, 53) and the multiple sequence alignment shown in A. The ligand binding pocket is presented on the left and shows highly conserved residues, including the DcisD motif of CBL1 and an asparagine of CBL2, which both confer coordination of the Ca2+ ion, as well as a tryptophan residue of loop L3 and an arginine residue at position I of CBL2, which form a specific corner of the inner binding pocket. In contrast, residues at positions II–IV of CBL2 are highly variable.
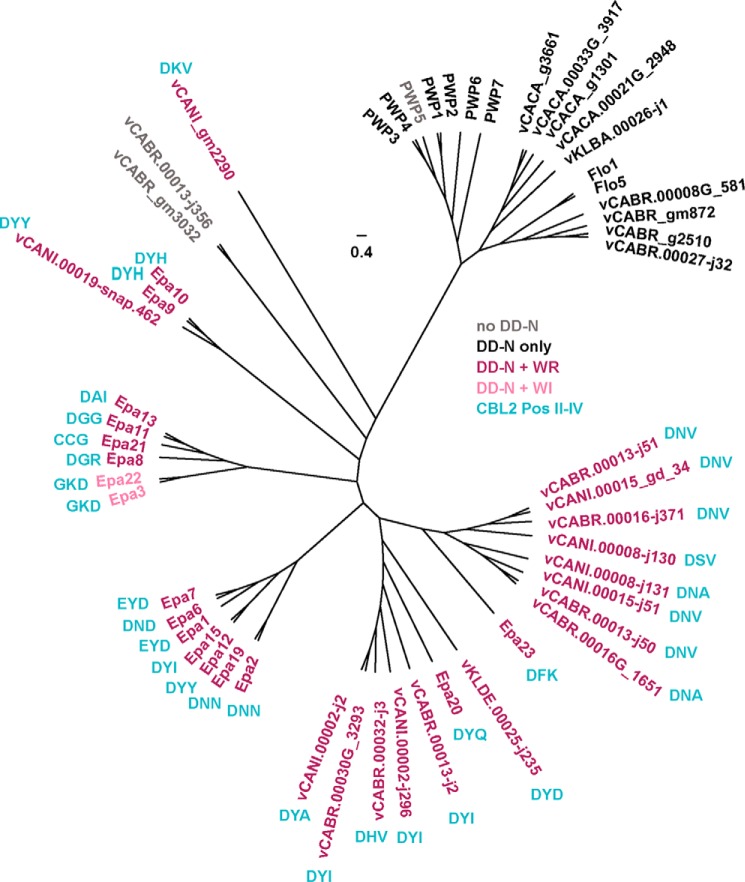
How could the Epa family have evolved? A scenario becomes evident when including a comparison of the variability/conservation of amino acid residues located on the protein surfaces of the A domains of different Epa members or within their ligand binding pockets (Fig. 9, A and C). This analysis reveals that although all EpaA domains have conserved PA14/Flo5-like cores, their surface composition is highly variable. Important exceptions, however, are found in the ligand binding pockets and include the highly conserved DcisD motif of CBL1 and an asparagine of CBL2, both of which confer coordination of the Ca2+ ion, and we would like to refer to these as the “DD-N” signature (Fig. 9, A and C). In addition, highly conserved surface features include a tryptophan from loop L3 and an arginine at position I of CBL2 (14), which form a corner of the inner binding pocket, and we refer to them as the “W-R” corner signature. Together, the DD-N and W-R signatures constitute an invariable core of the binding pocket and are essential for the efficient binding of the terminal hexose moiety in most C. glabrata EpaA domains (Fig. 9C). It is interesting to note that the DD-N calcium-binding signature can be found in over 85% of the estimated 200 Epa-like domains that are currently present in the known fungal genome sequences.5 In contrast, the W-R signature is highly restricted to the Epa-like adhesins of the “glabrata group” of Nakaseomyces species, including C. glabrata, C. bracarensis, C. nivariensis, and N. delphensis (19), and can be found in only very few Epa-like orthologs from other fungi. Therefore, the W-R signature is a hallmark of the glabrata group and might have evolved in a close ancestor of this group of Nakaseomyces.
Apart from the conserved core module, the binding pockets of the different C. glabrata EpaA domains also contain three highly variable residues that are located within the CBL2 region (Fig. 9C). We have previously defined these residues as CBL2 positions II–IV and suggested that they significantly contribute to the ligand binding specificity of different Epa members (14). Our current functional analysis, which reveals distinct binding patterns for the 17 Epa paralogs from C. glabrata strain CBS138, supports this hypothesis, because the CBL2 regions of nine members carry unique sequence motifs at positions II–IV (Fig. 9, A and B). Moreover, four pairs of Epa adhesins exist with two members each that carry identical CBL2 sequence motifs (Epa1 and Epa7; Epa2 and Epa19; Epa3 and Epa22; and Epa9 and Epa10). In the case of three of these pairs, both members belong to the same functional class. Thus, our study further emphasizes a strong correlation between CBL2 sequences and functional specificity in the case of 15 Epa family members. Moreover, 12 of the 16 Epa-like orthologs found in C. bracarensis, C. nivariensis, and N. delphensis, which contain a DD-W-NR core structure, carry CBL2 motifs not present in C. glabrata (Fig. 10), suggesting that their binding specificities differ from C. glabrata Epa adhesins. Thus, specific adaptation of the CBL2 motifs of Epa adhesins might have evolved after the DD-N and W-R core motifs and account for the differences in host specificities observed for the different members of the glabrata group of Nakaseomyces (19, 46). It is important to point out, however, that residues outside the CBL2 region must also contribute to ligand binding specificity, given the fact that we found four cases in which two different Epa proteins have identical CBL2 motifs but distinct ligand-binding patterns. The identity of such residues remains to be determined, but they might well reside within loops L1, L2 or L3, which also form part of the binding pocket (14) or that can interact with residues of the CBL2 region (see below).
FIGURE 10.
Phylogenetic analysis of Epa family members and related PA14 domain-containing adhesins. A structure-guided alignment of the Epa family and Epa-related orthologs from C. glabrata, C. bracarensis, C. nivariensis, N. delphensis, N. bacillisporus, and C. castellii was carried out together with other PA14 domain-harboring adhesins (Pwp proteins from C. glabrata and Flo adhesins from S. cerevisiae) using appropriate sequences (19, 40) and the three-dimensional structure of Epa1A (14) as described for Fig. 9A. Molecular evolutionary genetic analysis was then performed and visualized using the MEGA software (44). PA14-like A domains in the tree are colored with respect to the following structural motifs: no DD-N (gray), only DD-N (black), DD-N and W-R (purple), or DD-N and W-I (pink). For A domains containing W-R or W-I motifs, the residues at CBL2 positions II–IV are shown in turquoise. Details on sequence motifs are described in Fig. 9A and in the text.
How exactly do structural features of the binding pockets of different Epa adhesins determine specific ligand binding? Our study contributes to answering this challenging question by providing a set of crystal structures from Epa1 and Epa6, two structurally closely related but functionally distinct Epa variants. Here, we have found compelling evidence that the residues at positions II and III of the CBL2 region are directly determining the efficiency with which Epa1 and Epa6 are able to discriminate between α- and β-linked glycosides. However, our study also suggests that residues outside of the CBL2 region must account for ligand discrimination efficiency, as exemplified by Epa1 and Epa7. Although both proteins carry identical CBL2 motifs, Epa7 significantly better discriminates between α- and β-linked 1–3-galactobiose as well as between Galβ1–3GalNAc and Galβ1–3GlcNAc, respectively. Close to their binding site, these two Epa variants only differ by single residues in loops L1 and L2 (Epa1A, Phe-70 and Gln-1; Epa7A, Leu-70 and Glu-122). These residues could indirectly affect the conformation of CBL2 and the fine structure of the binding pocket. However, our previous studies on the flocculin Flo5A have demonstrated that it is often difficult to rationalize such long range effects on the specificity profile, because changes in loop dynamics and hydration can elude structural analysis (23). Nevertheless, our study indicates that residues at specific positions within the L1 and L2 loops are crucial for determining the precise ligand binding specificity, e.g. by conferring fine-tuning of the binding pocket in the case of EpaA domains with identical CBL2 motifs.
A similar discrepancy is found for the domain pair Epa3A and Epa22A, which differ by only 20 of overall 230 amino acids. Here, the Epa3A domain can be clearly assigned to group I by its binding specificity to Galβ1–3-linked glycans, whereas the Epa22A domain exerts a broad specificity profile that is characteristic for group II EpaA domains (Fig. 3). Interestingly, the CBL1 and CBL2 regions of Epa3 and Epa22 are identical, including the replacement of the otherwise conserved W-R core motif by W-I (Figs. 9A and 10). The only significant difference close to the carbohydrate-binding site is found for residue 106 in loop L2, which is a tyrosine in Epa3 but a phenylalanine in Epa22. L2 is folded around this residue that makes additional contacts with CBL2. Furthermore, all group I EpaA domains harbor a tyrosine or at least histidine (Epa10) in this position, whereas group II and III members have replacements by Phe, Leu, Ala, or Ser (Fig. 9A). Accordingly, one may infer that indirect effects on the conformational space occupied by CBL2 as well as L1–L3 loops affect the specificity profiles of EpaA domains. For comparison, in the flocculin Flo5 from S. cerevisiae such long range effects of single site mutations were likewise found to broaden the specificity from primary mannose to glucose ligands as well (23).
A further open question concerns the precise function of the EpaA domains of class III. Our glycan profiling suggests that these adhesins are able to bind diverse nongalactosides with low affinity and therefore might contribute to host cell adhesion by acting as low specificity lectins. However, it might well be that these Epa adhesins are able to bind with high affinity and specificity to yet unknown glycan structures that are not present on the arrays used in this study. This hypothesis is supported by the fact that all class III members contain the DD-N and W-R core motifs as well as individual CLB2 motifs that are not found in other Epa family members or related adhesins of the glabrata group of Nakaseomyces (Fig. 10). Clearly, more complex glycan arrays will be required to address this issue in the future (47).
In summary, our study indicates that C. glabrata has developed an extensive array of functionally diverse lectin-like adhesins that might be crucial as a whole for efficient host invasion and dissemination. Importantly, our study permits us to assess the potential host-ligand binding capacity of C. glabrata without knowing the precise EPA gene expression patterns before or during infection. These patterns and underlying regulatory mechanisms are likely to be highly complex and appear to involve both global mechanisms through silencing of sub-telomeric regions as well as gene-specific mechanisms (48, 49). As such, our study might contribute to the development of novel antimycotics (50), for example the design of anti-adhesive multivalent carbohydrates (51), to effectively combat emerging fungal pathogens of the C. glabrata clade (19, 46).
Author Contributions
R. D., M. K., M. M. R., P. K., H. S., S. R., L. O. E., and H. U. M. conceived and designed the experiments. R. D., M. K., M. M. R., P. K., and H. S., performed the experiments. R. D., M. K., M. M. R., P. K., H. S., S. R., L. O. E., and H. U. M. analyzed the data. R. D., M. K., L. O. E., and H. U. M. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Diana Kruhl, Cornelia Hillenbrand, Lisa Ludewig, Lisanne Heim, and Marcel Dörflinger for technical support; David F. Smith and Jamie Heimburg-Molinaro of the Consortium for Functional Glycomics for performing in vitro screening of glycan specificity; and the staff of Beamline 14.1 at the BESSY II synchrotron (Berlin, Germany) and Beamline P14 at the PETRA III synchrotron (Hamburg, Germany).
This work was supported by Deutsche Forschungsgemeinschaft Grants ES 152/7, MO 825/3, SFB 987, and GRK 1216, the International Max-Planck Research School for Environmental, Cellular and Molecular Microbiology, and the Marburg Center for Synthetic Microbiology. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4COU, 4COV, 4COW, 4COY, 4COZ, and 4D3W) have been deposited in the Protein Data Bank (http://wwpdb.org/).
R. Diderrich, M. Kock, M. Maestre-Reyna, P. Keller, H. Steuber, S. Rupp, L.-O. Essen, and H.-U. Mösch, unpublished results.
- A domain
- adhesion domain
- Epa
- epithelial adhesin
- CBL
- calcium binding loop
- Galα
- galactose linked via a α-glycosidic bond
- Galβ
- galactose linked via a β-glycosidic bond
- Galα1–3Gal
- α1–3-galactobiose
- Galβ1–3Gal
- β1–3-galactobiose
- Galβ1–3GalNAc
- T-antigen
- Galβ1–3GlcNAc
- lacto-N-biose
- Galβ1–4GlcNAc
- N-acetyl-d-lactosamine
- CFG
- Consortium for Functional Glycomics.
REFERENCES
- 1. Castano I., De Las Penas A., Cormack B. P. (2006) in Molecular Principles of Fungal Pathogenesis (Heitman J., Filler S. G., Edwards J. E., Jr., Mitchell A. P., eds) pp. 163–175, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2. Perlroth J., Choi B., Spellberg B. (2007) Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45, 321–346 [DOI] [PubMed] [Google Scholar]
- 3. Pfaller M. A., Diekema D. J. (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borg-von Zepelin M., Kunz L., Rüchel R., Reichard U., Weig M., Gross U. (2007) Epidemiology and antifungal susceptibilities of Candida spp. to six antifungal agents: results from a surveillance study on fungaemia in Germany from July 2004 to August 2005. J. Antimicrob. Chemother. 60, 424–428 [DOI] [PubMed] [Google Scholar]
- 5. de Groot P. W., Bader O., de Boer A. D., Weig M., Chauhan N. (2013) Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot. Cell 12, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verstrepen K. J., Reynolds T. B., Fink G. R. (2004) Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2, 533–540 [DOI] [PubMed] [Google Scholar]
- 7. Frieman M. B., McCaffery J. M., Cormack B. P. (2002) Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6-glucan cross-linked cell wall protein. Mol. Microbiol. 46, 479–492 [DOI] [PubMed] [Google Scholar]
- 8. Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N. (2007) A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71, 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr., Filler S. G. (2007) Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y., Filler S. G. (2011) Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10, 168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cormack B. P., Ghori N., Falkow S. (1999) An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285, 578–582 [DOI] [PubMed] [Google Scholar]
- 12. Zupancic M. L., Frieman M., Smith D., Alvarez R. A., Cummings R. D., Cormack B. P. (2008) Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 68, 547–559 [DOI] [PubMed] [Google Scholar]
- 13. Salgado P. S., Yan R., Taylor J. D., Burchell L., Jones R., Hoyer L. L., Matthews S. J., Simpson P. J., Cota E. (2011) Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 108, 15775–15779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maestre-Reyna M., Diderrich R., Veelders M. S., Eulenburg G., Kalugin V., Brückner S., Keller P., Rupp S., Mösch H.-U., Essen L.-O. (2012) Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. U.S.A. 109, 16864–16869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., Munro C. A., Rheinbay E., Grabherr M., Forche A., Reedy J. L., Agrafioti I., Arnaud M. B., Bates S., Brown A. J., Brunke S., et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai C., Mavrianos J., Chauhan N. (2011) Candida glabrata Pwp7p and Aed1p are required for adherence to human endothelial cells. FEMS Yeast Res. 11, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaur R., Domergue R., Zupancic M. L., Cormack B. P. (2005) A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8, 378–384 [DOI] [PubMed] [Google Scholar]
- 18. de Groot P. W., Kraneveld E. A., Yin Q. Y., Dekker H. L., Gross U., Crielaard W., de Koster C. G., Bader O., Klis F. M., Weig M. (2008) The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7, 1951–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gabaldón T., Martin T., Marcet-Houben M., Durrens P., Bolotin-Fukuhara M., Lespinet O., Arnaise S., Boisnard S., Aguileta G., Atanasova R., Bouchier C., Couloux A., Creno S., Almeida Cruz J., Devillers H., et al. (2013) Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics 14, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Domergue R., Castaño I., De Las Peñas A., Zupancic M., Lockatell V., Hebel J. R., Johnson D., Cormack B. P. (2005) Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308, 866–870 [DOI] [PubMed] [Google Scholar]
- 21. de Groot P. W., Klis F. M. (2008) The conserved PA14 domain of cell wall-associated fungal adhesins governs their glycan-binding specificity. Mol. Microbiol. 68, 535–537 [DOI] [PubMed] [Google Scholar]
- 22. Rigden D. J., Mello L. V., Galperin M. Y. (2004) The PA14 domain, a conserved all-β domain in bacterial toxins, enzymes, adhesins, and signaling molecules. Trends Biochem. Sci. 29, 335–339 [DOI] [PubMed] [Google Scholar]
- 23. Veelders M., Brückner S., Ott D., Unverzagt C., Mösch H.-U., Essen L.-O. (2010) Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. U.S.A. 107, 22511–22516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ielasi F. S., Decanniere K., Willaert R. G. (2012) The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: structural and functional study of the carbohydrate-binding domain. Acta Crystallogr. D Biol. Crystallogr. 68, 210–217 [DOI] [PubMed] [Google Scholar]
- 25. Guthrie C., Fink G. R. (eds) (1991) Guide to Yeast Genetics and Molecular Biology, Vol. 194, pp. 1–863, Academic Press, San Diego: [PubMed] [Google Scholar]
- 26. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Heinig M., Frishman D. (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32, W500–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dieterich C., Schandar M., Noll M., Johannes F. J., Brunner H., Graeve T., Rupp S. (2002) In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology 148, 497–506 [DOI] [PubMed] [Google Scholar]
- 35. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 37. Saldanha A. J. (2004) Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 38. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.4.1, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 39. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chap. 5, Unit 5.6, 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inglis D. O., Arnaud M. B., Binkley J., Shah P., Skrzypek M. S., Wymore F., Binkley G., Miyasato S. R., Simison M., Sherlock G. (2012) The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 40, D667–D674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N. (2013) ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53, 199–206 [Google Scholar]
- 43. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 44. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sim L., Groes M., Olesen K., Henriksen A. (2013) Structural and biochemical characterization of the N-terminal domain of flocculin Lg-Flo1p from Saccharomyces pastorianus reveals a unique specificity for phosphorylated mannose. FEBS J. 280, 1073–1083 [DOI] [PubMed] [Google Scholar]
- 46. Bolotin-Fukuhara M., Fairhead C. (2014) Candida glabrata: a deadly companion? Yeast 31, 279–288 [DOI] [PubMed] [Google Scholar]
- 47. Geissner A., Anish C., Seeberger P. H. (2014) Glycan arrays as tools for infectious disease research. Curr. Opin. Chem. Biol. 18, 38–45 [DOI] [PubMed] [Google Scholar]
- 48. Gallegos-García V., Pan S. J., Juárez-Cepeda J., Ramírez-Zavaleta C. Y., Martin-del-Campo M. B., Martínez-Jiménez V., Castaño I., Cormack B., De Las Peñas A. (2012) A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata. Genetics 190, 1285–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Halliwell S. C., Smith M. C., Muston P., Holland S. L., Avery S. V. (2012) Heterogeneous expression of the virulence-related adhesin Epa1 between individual cells and strains of the pathogen Candida glabrata. Eukaryot. Cell 11, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brown G. D., Denning D. W., Levitz S. M. (2012) Tackling human fungal infections. Science 336, 647. [DOI] [PubMed] [Google Scholar]
- 51. Wittmann V., Pieters R. J. (2013) Bridging lectin binding sites by multivalent carbohydrates. Chem. Soc. Rev. 42, 4492–4503 [DOI] [PubMed] [Google Scholar]
- 52. O'Sullivan O., Suhre K., Abergel C., Higgins D. G., Notredame C. (2004) 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 340, 385–395 [DOI] [PubMed] [Google Scholar]
- 53. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
- 54. Gietz R. D., Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]