Background: Feed-forward loops are utilized in glucocorticoid signaling and can bestow temporal control to gene regulation.
Results: Cooperation with KLF15 enhances low affinity glucocorticoid receptor binding site activity in coherent feed-forward loops controlling amino acid catabolism.
Conclusion: Feed-forward response element composition contributes to temporal diversity of transcriptional regulation by glucocorticoids.
Significance: Cooperative feed-forward regulatory control may underpin glucocorticoid-induced metabolic side effects.
Keywords: DNA-protein interaction, gene regulation, glucocorticoid receptor, systems biology, transcription enhancer, KLF15, binding site affinity, feed-forward loop
Abstract
Combinatorial gene regulation through feed-forward loops (FFLs) can bestow specificity and temporal control to client gene expression; however, characteristics of binding sites that mediate these effects are not established. We previously showed that the glucocorticoid receptor (GR) and KLF15 form coherent FFLs that cooperatively induce targets such as the amino acid-metabolizing enzymes AASS and PRODH and incoherent FFLs exemplified by repression of MT2A by KLF15. Here, we demonstrate that GR and KLF15 physically interact and identify low affinity GR binding sites within glucocorticoid response elements (GREs) for PRODH and AASS that contribute to combinatorial regulation with KLF15. We used deep sequencing and electrophoretic mobility shift assays to derive in vitro GR binding affinities across sequence space. We applied these data to show that AASS GRE activity correlated (r2 = 0.73) with predicted GR binding affinities across a 50-fold affinity range in transfection assays; however, the slope of the linear relationship more than doubled when KLF15 was expressed. Whereas activity of the MT2A GRE was even more strongly (r2 = 0.89) correlated with GR binding site affinity, the slope of the linear relationship was sharply reduced by KLF15, consistent with incoherent FFL logic. Thus, GRE architecture and co-regulator expression together determine the functional parameters that relate GR binding site affinity to hormone-induced transcriptional responses. Utilization of specific affinity response functions and GR binding sites by FFLs may contribute to the diversity of gene expression patterns within GR-regulated transcriptomes.
Introduction
In response to numerous physiologic stimuli, glucocorticoid signaling is activated in vertebrates by the release of corticosteroid hormones, which bind to the glucocorticoid receptor (GR)2 (1). Upon ligand binding, GR translocates to the nucleus and regulates gene expression to control myriad aspects of cell and organism physiology, including glucose production, lipid storage, and amino acid catabolism (2, 3). Key physiologic processes that are modulated by GR signaling require tight control that extends beyond a simple on/off switch enabled by ligand release (4). Reflecting this, GR regulates gene expression both through direct association with glucocorticoid response elements that harbor sequences with varying similarity to canonical high affinity palindromic GR binding sites (5) and through tethering with other transcription factors in a process that does not necessarily require the presence of a canonical, palindromic GR binding site (GBS) (6–8). Regulatory regions that utilize GR binding sites in combination with binding sites for other factors that contribute to the transcriptional response to hormone, often through physical interaction with GR, have been alternately referred to as either glucocorticoid response units or glucocorticoid response elements (GREs) with the latter term appearing more frequently in recent literature (9, 10). Control of GR activity through GREs is exemplified by a well characterized GRE that regulates the expression of PEPCK, which encodes a key enzyme in gluconeogenesis. A combination of physical interactions between GR and co-regulatory transcription factors and GR binding sites allows the glucocorticoid response at PEPCK to integrate multiple non-ligand signals and is implicated in conferring tissue specificity to hormone signaling (11–13). More recent genome-wide studies of GR occupancy within chromatin support a model in which GR binding and activity are frequently controlled through combinatorial interactions with other factors (5, 7, 14). Relatively few studies, however, have dissected the molecular basis for GRE function to the level of mechanistic detail that is available for PEPCK regulation, and the role that specific GR binding sequences play in determining GR occupancy and transcriptional responses in combination with other transcription factors is not fully understood.
We recently characterized novel GRE subtypes that control the transcriptional responses of feed-forward loops (FFLs) comprising GR and the GR-induced transcriptional factor KLF15, which is a major regulator of amino acid catabolism (15, 16). FFLs, which are now recognized as a dominant motif in gene regulation (17–19) and are increasingly implicated in hormone responses (20–22), are formally defined as factor X (in this case GR) regulating the expression of factor Y (in this case KLF15) with both factors exerting simultaneous control over the expression of downstream targets. FFLs can adopt eight possible configurations depending on whether constituent factors enhance or repress their targets within the loop (23); different FFL configurations endow client gene expression with distinct transcriptional dynamics and expression properties (24, 25). We previously showed that GR and KLF15 form two of the most widely used types of FFLs: the type I coherent FFL in which GR and KLF15 both induce the expression of shared target genes and the type I incoherent FFL in which KLF15 represses GR-mediated induction of FFL targets. GREs regulating the expression of genes encoding the amino acid-metabolizing enzymes AASS and PRODH mediated coherent feed-forward regulation by GR and KLF15, and GR occupancy was enhanced by KLF15 expression at these loci. In contrast, a GRE within the MT2A locus, which was strongly induced by GR alone, exhibited reduced activity and GR occupancy when KLF15 was expressed in combination with hormone. The molecular basis for distinct transcriptional outputs resulting from individual GR-KLF15 feed-forward GREs and the role of GR binding sites within these loci, however, were not determined.
In this study, we used site-directed mutagenesis to define response elements within GREs that contribute to coherent (i.e. cooperative) feed-forward regulation of AASS and PRODH by GR and KLF15 in comparison with the MT2A GRE in which KLF15 antagonizes GR action. Immunoprecipitation was used to determine whether GR and KLF15 physically interact. Electrophoretic mobility shift assays using a semidegenerate pool of oligonucleotides and deep sequencing of bound and unbound GR-DNA complexes (Spec-seq) was used to derive a position weight matrix for GR-DNA binding energies (26). Application of this matrix to GR binding regions identified using chromatin immunoprecipitation (ChIP)-seq in conjunction with binding site swaps further defined the role of GR binding site sequence in determining GR-KLF15 GRE transcriptional outputs. We propose a model in which the role of GR binding site affinity in mediating transcriptional responses to glucocorticoid is constrained by GRE architecture, which determines both threshold affinity requirements and response magnitudes.
Experimental Procedures
Cell Culture and Reagents
Beas-2B cells were grown in high glucose DMEM containing l-glutamine and supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin-streptomycin. Wild type and Klf15−/− mouse embryonic fibroblasts (MEFs) were grown in the same basal medium described above and supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% nonessential amino acids in minimum essential medium. All cells were maintained in 5% CO2 at 37 °C. Dexamethasone (dex; D1756) was purchased from Sigma and prepared using sterile 100% ethanol as vehicle. The full-length KLF15 expression plasmid (pcDNA-KLF15) and the KLF15-expressing adenovirus (Ad-KLF15) and Ad-GFP control have been described previously (15, 27) as have the truncated KLF15 expression constructs in the pcDNA vector (28) that were kindly provided by Dr. Claudia Noack. Primary antibodies for co-immunoprecipitation included mouse anti-KLF15 (ab81604) from Abcam; rabbit anti-GR (M-20; sc-1004), rabbit anti-GR (H-300; sc-8992X), and mouse anti-GR (G-5; sc-393232) from Santa Cruz Biotechnology; normal rabbit IgG (12370MI) from Fisher; and mouse anti-GAPDH (G8795) from Sigma. Secondary antibodies were ECL sheep anti-mouse IgG-HRP (95017-332) and ECL donkey anti-rabbit IgG-HRP (95017-330) obtained from VWR. For the MEF ChIP followed by deep sequencing (ChIP-seq) experiment and independent validation via ChIP-qPCR, anti-GR (N-499; a gift from Dr. Keith Yamamoto) and anti-GR (IA-1; a polyclonal rabbit antibody raised against human GR amino acids 84–112 (QPDLSKAVSLSMGLYMGETETKVMGNDLG); a gift from Dr. Miles Pufall), respectively, were used.
Plasmids
The pAASS and pPRODH luciferase reporter constructs have been described (15). The pMT2A reporter used in the present study contains a smaller (482-bp) region of the MT2A locus that exhibited similar activity to the construct described previously (15). All reporters were generated in the pGL3-Promoter vector (Life Technologies). Site-directed mutagenesis of putative GR and KLF binding sites, identified as having ≥95% core binding sequence similarity to the consensus binding site using Matinspector (29) (Genomatix), was performed using the QuikChange II site-directed mutagenesis kit from Agilent Technologies as instructed by the manufacturer. All mutated sequences were verified by sequencing and further analyzed by Matinspector to confirm they were no longer recognized as binding sites for their cognate transcription factor. The sequences of primers used for site-directed mutagenesis, putative GR and KLF binding sites, and mutations introduced into those sites are presented in supplemental Tables S1 and S2. Transfections were performed as described previously (15).
Whole Cell Extract Preparation and Co-immunoprecipitation
For endogenous co-immunoprecipitation experiments, Beas-2B cells were grown to confluence on three 15-cm dishes. Cells were treated with 100 nm dex in serum-free medium for 4 h to stimulate GR and KLF15 activation (15) after which whole cell extracts were prepared as described previously (30). Protein concentration was quantified using the Protein Assay Dye Reagent Concentrate from Bio-Rad, and samples were aliquoted and stored at −80 °C until further use. Protein extracts (500 μg) were precleared by adding Protein A/G-agarose resin slurry (Thermo Scientific Pierce) and incubating for 30 min at 4 °C on a rotator. The resin was pelleted by brief centrifugation after which the supernatant was transferred to a fresh tube and combined with antibodies against GR or IgG. Protein A/G-agarose resin slurry was then added, and reactions were incubated for 2 h at 4 °C on a rotator. Resulting immune complexes were pelleted and washed. Immunoprecipitated proteins were separated by SDS-PAGE and then transferred to PVDF membranes (Hybond-P, GE Healthcare). Membranes were immunoblotted with antibodies against KLF15, GR, or GAPDH (negative control) as indicated in Fig. 2. Chemiluminescence detection was performed using the ECL Prime Western blotting detection system (GE Healthcare).
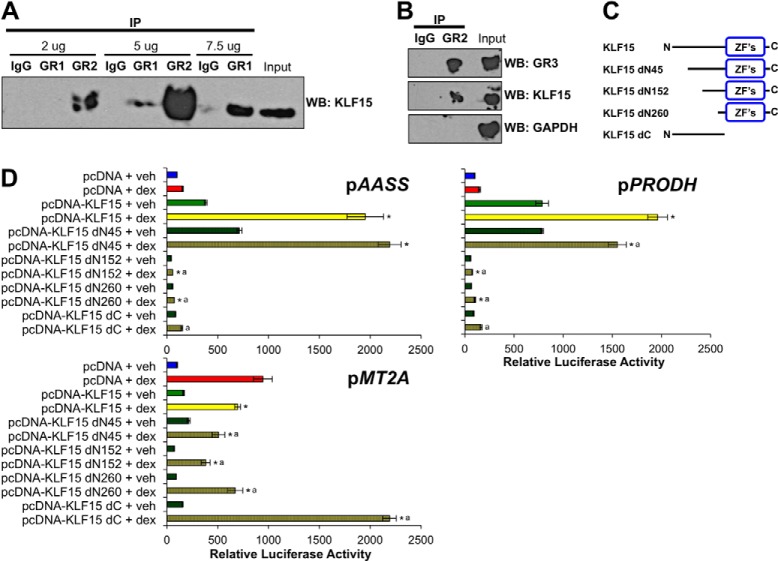
FIGURE 2.
KLF15 physically interacts with GR and utilizes distinct functional domains for cooperation versus repression of GR. A and B, co-immunoprecipitation (IP) of GR and endogenous KLF15 in Beas-2B cells treated with dex and serum-deprived for 4 h. A, whole cell extracts were immunoprecipitated with increasing concentrations of anti-GR (M-20; GR1), a second anti-GR (H-300; GR2), or IgG as a nonspecific control as indicated and then probed with anti-KLF15 by Western blotting (WB). B, whole cell extracts were immunoprecipitated with GR2 antibody or IgG and then probed by Western blotting with anti-KLF15, anti-GR (G-5; GR3) as a positive control, or anti-GAPDH as a negative control as indicated. C, schematic diagram of full-length and mutated KLF15 expression constructs. “ZF's” indicates the three C-terminal Cys2-His2 zinc finger domains. D, relative luciferase activity of pAASS, pPRODH, or pMT2A reporters transfected into Beas-2B cells in combination with full-length or mutated KLF15 expression plasmids or the parent pcDNA vector as indicated. Cells were subsequently treated with dex or vehicle (veh) for 8 h, and luciferase activity was measured. Bars indicate mean reporter activity (+S.D.) relative to activity in the pcDNA + vehicle-treated samples. Each footnoted bar was compared statistically according to the following: *, p ≤ 0.05 versus pAASS pcDNA + dex; a, p ≤ 0.05 versus pAASS pcDNA-KLF15 + dex. D, top right, *, p ≤ 0.05 versus pPRODH pcDNA + dex; a, p ≤ 0.05 versus pPRODH pcDNA-KLF15 + dex. D, bottom, *, p ≤ 0.05 versus pMT2A pcDNA + dex; a, p ≤ 0.05 versus pMT2A pcDNA-KLF15 + dex.
ChIP
MEFs were grown to confluence in 10-cm dishes and treated with 1 μm dex or vehicle in fresh complete medium for 1 h. ChIP was performed as described previously (15) with the following modifications. Cross-linking was performed by adding methanol-free 16% formaldehyde directly to the culture medium to a final concentration of 1% and incubating for 5 min at room temperature on a rocker. Samples were sonicated with a Diagenode Bioruptor at high power in 30-s bursts separated by 30-s incubations in ice water for a total of 23 min. qPCR analysis of DNA obtained by ChIP was performed using Fast SYBR Green qPCR Master Mix (Life Technologies) as described (15). Relative GR occupancy of a putative target region was calculated on a log2 scale and defined as the difference between the CT value for the specific target region relative to the geometric mean of CT values for three negative control regions. Amplification of diluted input DNA established that control and experimental primer efficiencies were well matched (generally <0.5 cycle difference). ChIP-qPCR experiments were performed in biologic quadruplicate and repeated at least once with qualitatively similar results. p values were calculated using t tests as indicated in figures. The primer sequences used are in supplemental Table S3.
ChIP-Sequencing
A minimum of 1 ng of DNA from ChIP samples was used to prepare libraries using the Nugen Ovation Ultralow System V2 1–16 Part Number 0344 according to the manufacturer's instructions. Samples were run on an Illumina HiSeq using 1 × 50 bp end reads. The quality of the ChIP-seq FASTQ sequence files was assessed using the FASTQC software. The genomic locations of these sequences were determined by mapping them to the mouse genome (mm9) using Bowtie2 (31). Peak calling of enriched ChIP regions was performed using MACS2 software by setting the narrow peak range to 300. The resulting peaks were converted to BigWig format for visualization in the UCSC Genome Browser using utility tools implemented by the “HOMER” group. The consistency of replicated sample peaks was qualified using the Irreproducible Discovery Rate algorithm implemented in R (32). Differential binding analysis of transcription factor occupancy with ChIP-Seq was performed using an R Bioconductor package, “DBChIP” (33), and the statistically significant binding sites were annotated using an R Bioconductor package, “ChIPseeker” (34). Data have been deposited in the Gene Expression Omnibus (GSE69947).
Adenoviral Transduction
MEFs were plated in 10-cm dishes so they would be confluent at the time of dex treatment. Cells were transduced with Ad-KLF15 or Ad-GFP as a control at a multiplicity of infection of 50. Approximately 17 h later, cells were treated with 1 μm dex or vehicle in fresh complete medium for 1 h and then assayed by ChIP-qPCR as described above (15).
Spec-seq
Spec-seq was performed as described previously (35) with modifications as detailed below. An initial experiment was performed using the putative consensus sequence AGAACA GGG TGTTCT, randomized library 1 (AGAACN NSN NGTTCT; diversity = 512), randomized library 2 (AGAANN GGG NNNTCT; diversity = 1024), randomized library 3 (AGAACA GGG TGNNNN; diversity = 256), and randomized library 4 (AGAACA GGGC NNNTCT; diversity = 64). The initial total library diversity was ∼1853 with a library composition of 10% positive control sequence + randomized libraries 1–4 + 5% negative control sequence. A fifth library containing 2304 sequences based on DDNACW KKN KGTTCT where D = “not C,” N = “any base,” W = “A or T,” and K = “G or T” was subsequently prepared and analyzed using similar ratios of control sequences. Binding conditions for electrophoretic mobility shift assay (EMSA) were 100 ng of fluorescein amidite-labeled dsDNA + 0/0.5/1/2/4 μm GR DBD protein for each lane and 1× NEB buffer 4. GR DBD was prepared as described previously (36). The EMSA was performed using a 9% 33:1 acrylamide gel and Tris borate-EDTA buffer and run at 200 V for 30 min at 0 °C. The 2 μm protein lane was used for deep sequencing. Sequence data have been deposited in the Gene Expression Omnibus (GSE69386). Bound/unbound fractions resulting from EMSA of these libraries and conditions were used to generate position weight matrices (PWMs) as described. The GR PWM that was generated through this analysis was used to define relative binding energies using the patser program (37). Derived binding affinities are proportional to the exponential function of the calculated energies.
Results
Characterization of GR and KLF15 Binding Sites within the PRODH, AASS, and MT2A Glucocorticoid Response Elements
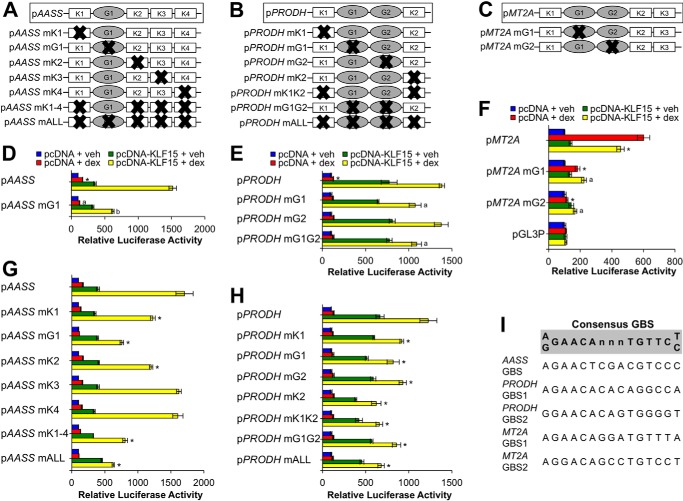
We previously characterized composite GREs that conferred coherent (AASS and PRODH) and incoherent (MT2A) feed-forward transcriptional regulation by GR and KLF15 (15). Sequence analysis using Matinspector revealed that these GREs contain at least one putative binding site for GR and several (two to four) putative sites for KLF15, but the function of these sites is not known. To define the requirement of these putative GR binding sites/response elements (which we will refer to as GBSs) for GR-KLF15 combinatorial regulation of pAASS, pPRODH, and pMT2A, we used site-directed mutagenesis to disrupt the indicated GBS(s) (Fig. 1, A–C and I, and supplemental Table S1) and transfected these mutant reporter constructs into Beas-2B cells in combination with a KLF15 expression plasmid (pcDNA-KLF15) or control vector (pcDNA). Reporter activity was then assayed after 8 h of dex (100 nm) or vehicle treatment (see supplemental Table S1 for sequences of GR binding sites and introduced mutations). Consistent with our prior work, the wild type GR-KLF15 coherent feed-forward GRE constructs pAASS (Fig. 1D) and pPRODH (Fig. 1E) showed a small induction with dex treatment, a stronger (∼5–10-fold) induction in the presence of pcDNA-KLF15, and a greater than additive (∼15-fold) induction with dex + KLF15 co-treatment. Surprisingly, pAASS mG1, which contains a mutation in the only GBS identified within the AASS GRE, exhibited only a modest reduction in the basal response to dex and a significant, yet incomplete abrogation of combinatorial induction by dex + KLF15 (Fig. 1D). A similar pattern of mildly decreased dex responsiveness and reduced induction by dex + KLF15 was observed for the pPRODH mutants harboring either single (mG1 and mG2) or double (mG1G2) GBS mutations (Fig. 1E). These data indicate that the putative GBSs tested contribute to, but are not absolutely required for, combinatorial regulation of AASS or PRODH by GR and KLF15, supporting a model in which GR can cooperate with KLF15 without occupying a canonical GBS match. In contrast, whereas the previously described incoherent GR-KLF15 feed-forward MT2A reporter exhibited strong induction by dex that was diminished in the presence of pcDNA-KLF15, both of the MT2A GBSs were essential for proper co-regulation by GR and KLF15 (Fig. 1F). Specifically, responses to dex and to dex + KLF15 treatment were dramatically reduced in the pMT2A mG1 construct and completely lost in pMT2A mG2. In addition, neither dex nor KLF15 substantially changed the expression of the parent pGL3P construct (Fig. 1F), establishing that the effects of both GR and KLF15 are specific to the cloned response elements. Thus, the GBS requirements differ between the tested coherent and incoherent GREs with mechanisms that do not involve the tested GBSs contributing to coherent feed-forward function of the AASS and PRODH GREs.
FIGURE 1.
Role of GR and KLF15 binding sites in feed-forward glucocorticoid response elements that regulate AASS, PRODH, and MT2A. A–C, schematic diagrams illustrating glucocorticoid response elements of GR-KLF15 feed-forward targets used to generate luciferase reporters. Relative locations of putative GR (G) and KLF15 (K) binding sites are shown; mutated sites are denoted by “X” as appropriate. D–H, luciferase activity of the indicated wild type or mutant reporters transfected into Beas-2B cells together with a KLF15 expression plasmid (pcDNA-KLF15) or empty vector control (pcDNA) prior to treatment with dex or vehicle (veh) for 8 h. Bars indicate mean reporter activation (+S.D.) relative to activity in pcDNA + vehicle-treated samples. The parent pGL3-Promoter (pGL3P) vector is included in F as a control. Statistical comparisons for each panel are as follows. D, *, p ≤ 0.05 versus pAASS pcDNA + veh; a, p ≤ 0.05 versus pAASS pcDNA + dex; b, p ≤ 0.05 versus pAASS pcDNA-KLF15 + dex. E, *, p ≤ 0.05 versus pPRODH pcDNA + veh; a, p ≤ 0.05 versus pPRODH pcDNA-KLF15 + dex. F, *, p ≤ 0.05 versus pMT2A pcDNA + dex; a, p ≤ 0.05 versus pMT2A pcDNA-KLF15 + dex. G, *, p ≤ 0.05 for all compared with pAASS pcDNA-KLF15 + dex. H, *, p ≤ 0.05 for all compared with pPRODH pcDNA-KLF15 + dex. I, 15-mer nucleotide sequences of putative GR binding sites identified within glucocorticoid response elements of AASS, PRODH, and MT2A loci. A consensus GR binding site sequence is shown for comparison (60).
We asked next whether interactions between KLF15 and its cognate binding sites within the AASS and PRODH GREs were required for response element function. We generated a series of pAASS and pPRODH reporters containing mutations of each KLF binding site (KBS) individually or all KBSs together both with and without disruption of the GBSs within each GRE (Fig. 1, A, B, G, and H). We assayed the activity of these mutant reporters following treatment with dex, pcDNA-KLF15, or dex + KLF15 as described above. Individual KBS mutations in pAASS exerted minimal or no effect on the responses to either dex or pcDNA-KLF15. Whereas the pAASS mK3 and mK4 mutants also exhibited induction that mirrored the parent construct with dex + KLF15 co-treatment, the pAASS mK1 and mK2 mutants exhibited a consistent, albeit incomplete abrogation of activity stimulated by dex + KLF15 (Fig. 1G). Surprisingly, even the pAASS constructs harboring mutations in all four KBSs (mK1–4) or mutations in all four putative KLF15 sites and the GBS (mALL) displayed some degree of cooperative induction in the presence of dex + KLF15 although substantially less than that exhibited by the wild type reporter (Fig. 1G). Similarly, the PRODH GRE was resilient to the introduction of mutations in the putative KBSs. As shown in Fig. 1H, mutating KBSs individually, together, or in combination with the GBSs resulted in reduced but not absent co-activation by dex + KLF15. Taken together, the binding site mutation analysis suggests that KLF15 and GR are able to interact with the AASS and PRODH GREs both through typical response elements, but neither GR nor KLF15 appear to require a recognizable binding site to accomplish the majority of the regulation they confer. Our data are also consistent with a potential physical association between GR and KLF15.
KLF15 Physically Interacts with GR and Exhibits Distinct Functional Domain Requirements for Coherent and Incoherent Feed-forward Loops
To examine whether GR associates with KLF15 as suggested by the reporter activity assays, we performed co-immunoprecipitation experiments in Beas-2B cells treated with dex (100 nm) and serum-deprived for 4 h to stimulate GR and endogenous KLF15 activation (15). GR was immunoprecipitated from whole cell extracts with increasing concentrations of GR1 (M-20) or GR2 (H-300) anti-GR antibodies and then probed for KLF15 expression by Western blotting. Fig. 2A illustrates a concentration-dependent increase in KLF15 detection that was observed in samples immunoprecipitated with either of the GR antibodies but not by an IgG control antibody, consistent with a physical interaction between GR and KLF15. Additional experiments in which whole cell extracts were immunoprecipitated with GR2 and then probed with GR3 (G-5) anti-GR or anti-KLF15 antibodies confirmed the presence of both GR and KLF15 in the precipitated complex, whereas the negative control, GAPDH, was not detected (Fig. 2B). Thus, endogenous GR and KLF15 either interact directly or are both present within a multicomponent cellular complex, in either case supporting a model in which molecular association between the two factors contributes to FFL function.
To gain insight into which functional domain(s) of the KLF15 protein may be important for GR-KLF15 combinatorial regulation, we assessed the ability of mutant KLF15 constructs (Fig. 2C), including a series of N-terminal deletions and a truncated C-terminal protein that lacks the zinc finger DNA binding domain (28), to properly co-regulate the wild type pAASS, pPRODH, and pMT2A reporters after co-transfection into Beas-2B cells and treatment with dex (100 nm) for 8 h. As shown in Fig. 2D (top), the KLF15 dN45 construct was able to cooperatively induce pAASS and pPRODH reporter activity to a similar extent as the full-length protein, whereas further deletion of the N-terminal region, which has previously been shown to harbor an activation domain (38), eliminated transcriptional induction of the AASS and PRODH GREs both with and without dex treatment. Indeed, the constructs lacking the activation domain appeared to exhibit gain of function repressive activity in which they reduced the basal and dex-induced activity of the PRODH and AASS constructs. Similarly, all of the N-terminal deletions were capable of combinatorial repression of pMT2A activity, whereas deletion of the C terminus eliminated KLF15 function in both the coherent and incoherent GREs. Taken together, these data indicate that the zinc finger domain is necessary and sufficient for transcriptional repression, whereas KLF15 activation domain function depends on promoter context and is required for cooperation between GR and KLF15.
Binding Site Sequence Does Not Determine GR-KLF15 Cooperation
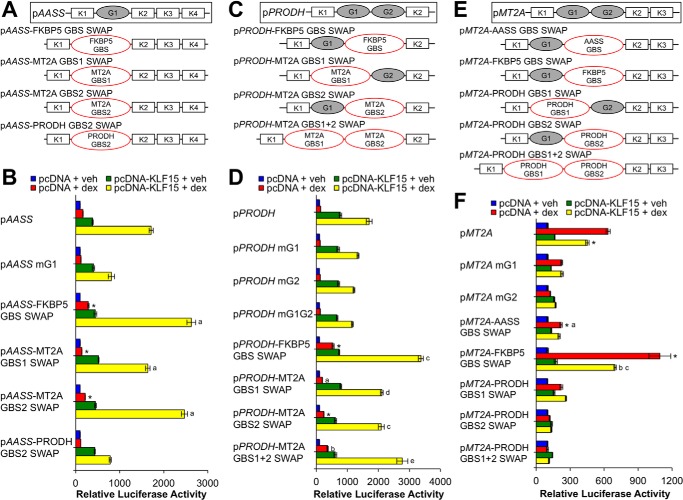
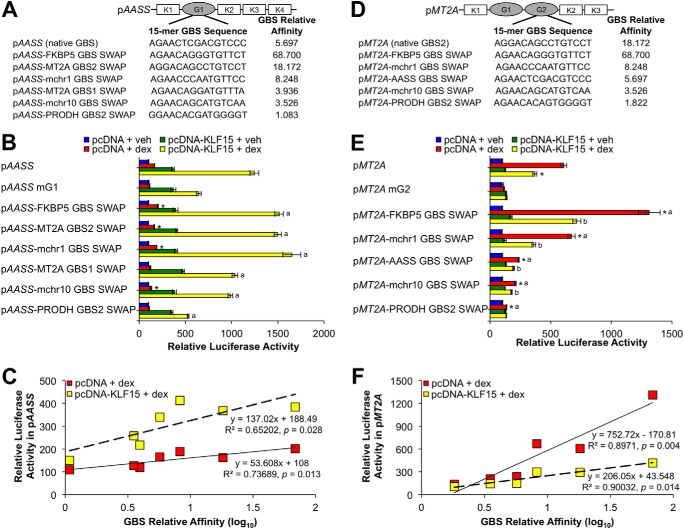
The above data support a model in which architectural features of the PRODH and AASS GREs mediate cooperative interaction between GR and KLF15, presumably through enabling activity of the N-terminal activation domain of KLF15. To determine whether the specific sequences of the GBSs within PRODH and AASS confer activation potential to KLF15, we performed a series of swaps between the GBSs we had defined within the PRODH, AASS, and MT2A feed-forward GREs as well as a GBS within a well characterized intronic GRE for FKBP5 that closely matches the consensus GR binding sequence (39). These additional luciferase reporters are identical to their wild type counterparts except that the native GBS sequence has been replaced as closely as possible with a functional GBS of a distinct co-regulated gene (GBS swaps; see Fig. 3, A, C, and E, and Table S2). We assayed activity of these constructs by transfection and treatment with vehicle or dex (Fig. 3, B, D, and F) and found that replacing the GBSs within the AASS and PRODH GREs with either (or both for PRODH) of the MT2A GBSs or the FKBP5 GBS maintained cooperation between GR and KLF15, indicating that the specific GBS sequences within the AASS and PRODH GREs do not determine GR-KLF15 cooperativity. In contrast, neither the AASS nor PRODH GBSs were repressed by KLF15 in the context of the MT2A GRE, although the level of induction from the AASS GBS was much lower than the wild type site, and the PRODH GBSs had minimal or no detectable activity when used to replace the two MT2A GREs. In contrast, substitution of the MT2A GBS2 with the FKBP5 GBS resulted in hyperactivation of the construct. Inspection of the sequences of the various GBSs characterized in Figs. 1 and 3 indicates that the PRODH and AASS GBSs only weakly match the consensus GR binding sequence (see Fig. 1I), whereas the MT2A GBS2 is a more robust match with the FKBP5 GBS (AGAACA GGG TGTTCT), aligning even more closely with the consensus, suggesting that site affinity characteristics may contribute to the function of the MT2A GRE.
FIGURE 3.
GR binding site sequence is not sufficient to determine cooperative interaction between GR and KLF15. A, C, and E, schematic diagrams of GBS swap reporters in which the wild type GBS sequence was mutated to match the GBS(s) of the gene indicated in the red ellipse(s). Luciferase activity of pAASS (B), pPRODH (D), and pMT2A (F) wild type or GBS swap reporters transiently transfected into Beas-2B cells in combination with pcDNA-KLF15 or pcDNA control before treatment with dex or vehicle (veh) for 8 h is shown. Bars indicate mean reporter activation (+S.D.) relative to activity in pcDNA + vehicle-treated samples. B, *, p ≤ 0.05 versus pAASS mG1 pcDNA + dex; a, p ≤ 0.05 versus pAASS mG1 pcDNA-KLF15 + dex. D, *, p ≤ 0.05 versus pPRODH mG2 pcDNA + dex; a, p ≤ 0.05 versus pPRODH mG1 pcDNA + dex; b, p ≤ 0.05 versus pPRODH mG1G2 pcDNA + dex; c, p ≤ 0.05 versus pPRODH mG2 pcDNA-KLF15 + dex; d, p ≤ 0.05 versus pPRODH mG1 pcDNA-KLF15 + dex; e, p ≤ 0.05 versus pPRODH mG1G2 pcDNA-KLF15 + dex. F, *, p ≤ 0.05 versus pMT2A pcDNA + dex; a, p ≤ 0.05 versus pMT2A mG2 pcDNA + dex; b, p ≤ 0.05 versus pMT2A-FKBP5 GBS swap pcDNA + dex; c, p ≤ 0.05 versus pMT2A pcDNA-KLF15 + dex.
GR ChIP-seq Defines Additional Binding Regions That Mediate Conditional GR Activity
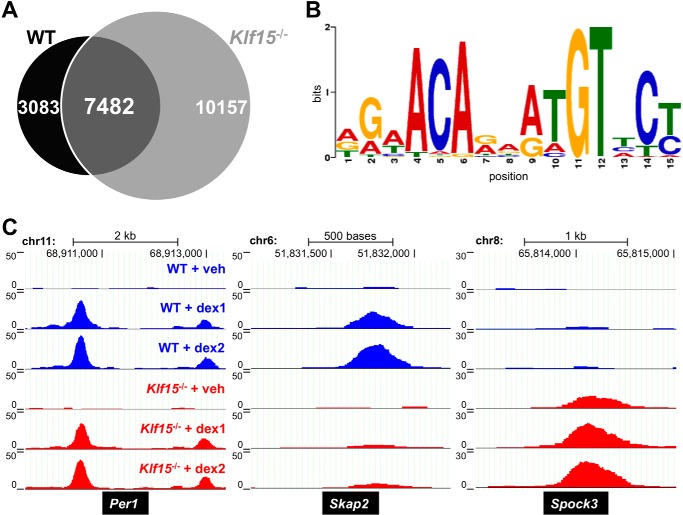
The observation that weak matches for the consensus GR binding site function in cooperation with KLF15 in the AASS and PRODH GREs is congruent with prior observations in which conditional (e.g. cell-specific or requiring a tethering factor) GR binding regions (GBRs) frequently utilize lower affinity or non-canonical GBSs (40). To define the characteristics of GBSs that mediate conditional hormonal responses in greater detail, we used a GR antibody to perform ChIP-seq in MEFs isolated from wild type (WT) and Klf15−/− mice. ChIP samples were prepared in duplicate from WT and Klf15−/− MEFs treated with either vehicle control (EtOH) or 1 μm dex for 1 h. Samples derived from WT MEFs treated with EtOH were combined because of limiting starting material; the remainder of the samples were prepared as independent libraries and subjected to deep sequencing. Approximately 19–27 million reads were obtained per sample, base call accuracy was >99.9% for 97% of base reads, and 97% of sequence reads mapped to the mouse genome. GBRs were identified using MACS2, and r values between samples were computed. Strong concordance (r ∼ 0.9) was observed between biologically equivalent dex-induced samples (e.g. WT dex treatment samples 1 and 2), indicating good technical reproducibility. In contrast, r values between WT and Klf15−/− cells treated with dex were ∼0.8, indicating greater biologic variability in GR binding between the two different MEF lines. Substantial variability was also present in a filtered data set of autosomal binding sites in the upper 80% of read counts that exhibited at least a 2-fold occupancy increase with dex treatment. 10,565 peaks met these criteria in wild type cells, whereas there were 17,639 such sites in Klf15−/− cells with an overlap of 7482 (Fig. 4A). Despite the variability, application of Centrimo (41), a tool within the MEME software suite (42), to the WT, Klf15−/−, and intersection ChIP-seq data sets identified putative GR binding motifs within 49, 42, and 52% of occupied regions, respectively; a de novo match for the consensus GR binding logo (Fig. 4B) was also identified from the WT data set using the MEME-ChIP tool (43).Taken together, these results indicate that the binding patterns defined by ChIP-seq reflect GR occupancy within the tested cell types.
FIGURE 4.
Genome-wide characterization of dex-induced GR occupancy in wild type and Klf15−/− cells. A, Venn diagram depicting binding sites identified by GR ChIP-seq within the upper 80% of read counts that exhibited at least a 2-fold increase in GR occupancy with 1-h dex (1 μm) treatment compared with vehicle (veh). Sites meeting these criteria in wild type MEFs are indicated by the black circle, whereas those in Klf15−/− MEFS are indicated by the light gray circle. Sites shared by both cell types are represented in dark gray. B, GR binding logo generated through analyzing GBRs from WT MEFS with MEME-ChIP. C, examples of GR ChIP-seq peaks with equivalent (left), increased (middle), and decreased (right) GR occupancy in wild type compared with Klf15−/− MEFS as visualized in the UCSC Genome Browser. Chromosome number and approximate chromosomal locations of peaks are indicated at the top of each panel, whereas the corresponding gene symbols are provided in black boxes below each panel. The vertical scale on the left of each panel indicates normalized occupancy as measured by reads per million reads. chr, chromosome.
To rigorously define specific GBRs with cell type-conditional occupancy patterns, we applied differential binding analysis, which allows high confidence identification of peaks that are statistically different between multiple ChIP-seq data sets. Using differential binding analysis to compare the dex-treated WT and Klf15−/− ChIP samples, we identified a total of 1088 autosomal peaks with differential binding; 781 of these exhibited at least a 2-fold increase in GR occupancy in one of the two cell types. Examples of peaks with increased, decreased, and equivalent GR occupancy in WT compared with Klf15−/− cells as visualized in the UCSC Genome Browser are shown in Fig. 4C. We focused our subsequent analysis on differential GBRs with increased occupancy in WT versus Klf15−/− MEFs, a pattern that is similar to the regulation of AASS and PRODH by GR and consistent with cooperation between GR and factors that are differentially expressed in WT versus Klf15−/− cells. 324 of 781 differentially occupied, dex-inducible sites exhibited greater than 2-fold higher occupancy in WT versus Klf15−/− cells after dex treatment. ChIP-qPCR analysis validated that increased GR occupancy in WT cells in comparison with the Klf15−/− cells was stable and reproducible at multiple sites identified through ChIP-seq (Fig. 5, A and B). Restoring KLF15 expression via a KLF15-expressing adenovirus (Ad-KLF15), however, did not alter the pattern of GR occupancy at tested sites that exhibited decreased occupancy in Klf15−/− cells, suggesting that differential binding was unlikely to be a direct consequence of Klf15 deficiency.
FIGURE 5.
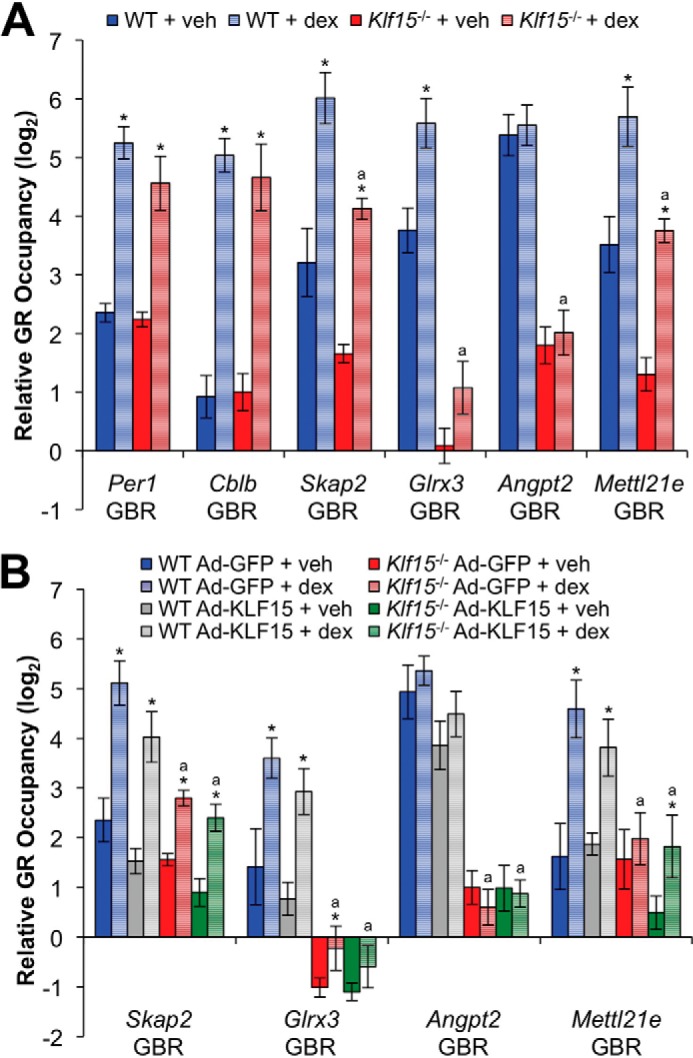
Independent validation of differentially occupied GR binding regions identified by ChIP-seq. A, ChIP-qPCR analysis of GR occupancy within the indicated genes in wild type and Klf15−/− MEF cells treated with dex (1 μm) or vehicle (veh) for 1 h. Relative GR occupancy was calculated as the difference between the CT value for the indicated target and the geometric mean of CT values for three negative control regions not associated with GR occupancy. Bars indicate means (+S.D.) expressed on a log2 scale. *, p ≤ 0.05 versus respective vehicle-treated control; a, p ≤ 0.05 versus WT + dex. B, ChIP-qPCR analysis of GR occupancy within the indicated genes in wild type and Klf15−/− MEFs infected with a KLF15-overexpressing (Ad-KLF15) or control (Ad-GFP) adenovirus for ∼17 h prior to 1-h treatment with dex (1 μm) or vehicle. GR occupancy is expressed as described for A. *, p ≤ 0.05 versus respective vehicle-treated control; a, p ≤ 0.05 versus WT Ad-GFP + dex.
Affinity Characteristics of Differential GR Binding Regions Determined through Spec-seq
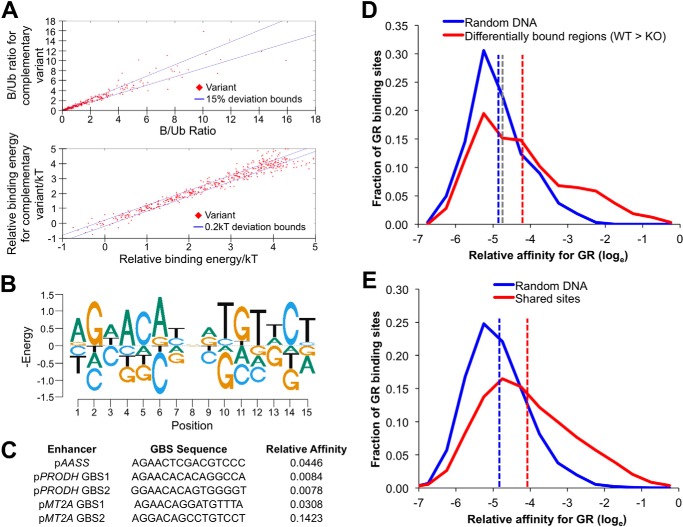
A recent report on cell type-specific estrogen receptor and GR binding sites concluded that GBRs that exhibit “yes/no” cell type specificity generally harbor extremely low affinity GBSs (40). To determine affinity characteristics of the differential GBRs (false discovery rate <0.05) we had identified, we first performed Spec-seq analysis for GR. Spec-seq is a technique that probes a targeted subset of sequence space using EMSAs and deep sequencing to define a PWM for a transcription factor (in this case GR) that includes both positive and negative energies for each residue within the derived binding logo (26, 35). Spec-seq-derived matrices can be used to accurately estimate relative energies of interactions between specific transcription factors and DNA with binding affinity proportional to the inverse of the natural log of the predicted energy. As starting material for this analysis, we used a relatively high affinity GBS found within the FKBP5 locus as a standard and libraries of partially degenerate matches for the FKBP5 GBS, including specific sequences from several GBRs identified in our ChIP-seq experiment. EMSA was performed on these libraries using a purified preparation of the GR DNA binding domain, and ratios of bound to unbound fractions were determined for each sequence within the library through quantitative sequencing. Complementary sequences contained within the libraries (i.e. oligonucleotides that were included in the library in both forward and reverse directions) exhibited highly correlated bound/unbound ratios and derived energies (Fig. 6A) as would be expected for interactions between homodimeric GR and DNA. Regression analysis using all sequences with two or fewer mismatches to the FKBP5 consensus sequence was used to generate an energy PWM (44) that is shown in Table S4; the corresponding logo is depicted in Fig. 6B. Energies calculated using the PWM closely matched actual experimental data for those sequences (r2 = 0.95), indicating that the additivity assumption of the PWM provides a good approximation of the actual binding energies.
FIGURE 6.
Derivation and application of a refined position weight matrix for GR using Spec-seq. A, correlation of bound and unbound fractions (top graph) and derived energies (bottom) for complementary oligonucleotides included within a representative library of binding sites analyzed for GR binding by EMSA and sequencing. B, energy logo derived from an oligonucleotide pool of known and putative GR binding sites and semidegenerate sequences that illustrates the preferred binding sequence identified for GR. The heights of each letter are the energies (in kT elements) for each base adjusted so the sum for each position is 0. The energy scale is negative so the preferred sequence (lowest energy) is on top, and the bases are in order from highest (top) to lowest (bottom) affinity. C, relative binding affinities of native GR binding site sequences in the glucocorticoid response elements mediating pAASS, pPRODH, and pMT2A feed-forward regulation. D, graphs fitted to relative calculated affinities for the peak affinity GR binding site identified within each of the 324 differentially bound regions (red) and shuffled sequences (blue) as described in the text. The red dashed line indicates mean relative affinity of native DNA sites, the blue dashed line indicates mean relative affinity of shuffled sites, and the gray dashed line is the calculated affinity of the PRODH GBS that is required for full activity of the PRODH GRE. E, similar to D above except that the relative calculated affinities for the peak affinity GR binding site within each of the ∼7500 peaks that overlap between WT and Klf15−/− MEFs and shuffled sequences are depicted.
We applied the new GR PWM to derive affinities of the GBSs within the AASS, PRODH, and MT2A GREs relative to the maximal predicted affinity (Fig. 6C). We found that the AASS and PRODH GBSs have lower affinity than the MT2A GBS2 with calculated affinities of the two PRODH GBSs calculated to be ∼20-fold less than MT2A GBS2 and ∼0.8% of maximal possible affinity, whereas the AASS GBS affinity was about 3-fold less than MT2A GBS2 and ∼5% of maximal affinity. These affinity data support a model in which low to moderate affinity GR binding sites contribute to the function of coherent GR-KLF15 response elements.
To determine GBS requirements for conditional GR binding regions more generally, we applied the new PWM for GR to interrogate 301-bp regions at the center of each of the 324 differentially bound sites with at least 2-fold greater occupancy in WT versus Klf15−/− MEFS after dex treatment. Of these, 181 harbored sites with affinity greater than 1% (i.e. e−4.6) of the maximal possible affinity for the PWM logo. A graph of the distribution of affinities for the entire set relative to maximal predicted affinity is shown in Fig. 6D; the average affinity site is indicated by the vertical red line and was ∼1.5% of maximal affinity. We also calculated the maximal affinity GBS within random 301-base pair sequences created by shuffling the set of 324 DBRs and graphed this affinity distribution. Comparison of these distributions shows that the average maximum affinity GBS within the native DBR sequences is higher than in randomized DNA, with the higher average largely attributable to a greater percentage of sites within the native sequence sites with affinities greater than ∼1.5% of maximal possible affinity. We also compared the calculated maximal affinity GBS found within each of ∼7500 GBRs that are shared between the WT and Klf15−/− MEFs with maximal affinity sites found within shuffled sequences (Fig. 6E). The affinity distributions for these shared sites were strikingly similar to the distributions for sites from binding regions with differential occupancy between WT and Klf15−/− MEFs (Fig. 6, compare E with D). This suggests that binding site affinity is not a primary determinant of differential GR occupancy in WT versus Klf15−/− cells.
Context-dependent Correlation between GBS Affinity and Transcriptional Activity
To determine directly the relationship between GR DNA binding affinity and activity within the GR-KLF15 feed-forward GREs, we performed additional GBS swaps into both pAASS and pMT2A to create an affinity series. For this analysis, we chose two additional sites identified within our ChIP-seq data set with calculated affinities between the strong MT2A GBS and the PRODH GBSs. Relative affinities of these and the GBSs used in the previously created swap constructs are shown in Fig. 7A; the affinity of the pPRODH GBS2, which was the lowest affinity site that we tested in our mutational analysis, was set at 1. We tested each of these constructs in luciferase assays (Fig. 7, B and E) and subsequently graphed the resulting relative luciferase activities as a function of the common log of the relative affinity of the specific GBS present in each swap construct (Fig. 7, C and F). Calculation of the Pearson correlation coefficient and linear fitting showed that binding site affinity was moderately (r2 = 0.73) but significantly (p < 0.05) correlated with AASS GRE transcriptional output with dex treatment alone. The addition of KLF15 increased the slope of the linear relationship but did not substantially alter the strength of correlation. For the MT2A GRE, the correlation between affinity and output was much stronger (r2 = 0.890) with dex treatment alone. As was the case for AASS, the addition of KLF15 modulated the slope but not the strength of correlation, although in this case (consistent with an incoherent FFL), the slope was reduced by KLF15. Taken together, these data indicate that correlations between GR binding site affinity and the transcriptional output of GR-KLF15 FFLs are governed by both response element architecture and KLF15 expression.
FIGURE 7.
GR binding site affinity correlates with transcriptional output in a context-dependent manner. A and D, 15-mer nucleotide sequences and corresponding relative affinities of GR binding sites swapped into the indicated sites of pAASS (A) and pMT2A (D) reporters by site-directed mutagenesis to generate GBS swap constructs. B, luciferase activity of pAASS wild type, mG1, and GBS swap reporters transiently transfected into Beas-2B cells in combination with pcDNA-KLF15 or pcDNA control prior to 8 h of treatment with dex or vehicle (veh). Bars indicate mean reporter activation (+S.D.) relative to activity in pcDNA + vehicle-treated samples. *, p ≤ 0.05 versus pAASS mG1 pcDNA + dex; a, p ≤ 0.05 versus pAASS mG1 pcDNA-KLF15 + dex. C, scatterplot of pAASS wild type and GBS swap reporter activations obtained in B following treatment with pcDNA + dex (relative to pcDNA + vehicle) or pcDNA-KLF15 + dex (expressed relative to pcDNA-KLF15 + vehicle) as a function of the relative affinity (on a log10 scale) of the GBS contained within that construct. E, reporter activity of pMT2A wild type, mG2, and GBS swap constructs in Beas-2B cells transfected and treated as described for B. *, p ≤ 0.05 versus pMT2A pcDNA + dex; a, p ≤ 0.05 versus pMT2A mG2 pcDNA + dex; b, p ≤ 0.05 versus respective activity with pcDNA + dex. F, scatter plot of pMT2A wild type and GBS swap reporter activations obtained in E as a function of the relative affinity (on a log10 scale) of the GBS contained within that construct as described for C.
Discussion
Glucocorticoids can both induce costly metabolic programs and regulate normal circadian physiologic rhythms, but the basis for differential gene expression responses to hormone have not been fully established. Here we show that GR-controlled coherent feed-forward regulation of AASS and PRODH, both of which encode enzymes involved in amino acid metabolism, utilizes low to moderate affinity GR binding sites and a presumptive physical interaction between GR and KLF15 to enable combinatorial regulation. As KLF15 is expressed at very low levels in many cell types and tissues prior to induction by GR, this architecture is congruent with “and” regulatory logic in which control of AASS and PRODH expression by GR through the tested GREs depends on the presence of both ligand-activated GR and time-dependent accumulation of KLF15. These data provide a mechanistic explanation for the distinct timing properties we observed previously for targets of the GR-KLF15 axis where genes that required KLF15 for maximal induction by GR, such as PRODH, exhibited a relative delay in peak expression in comparison with KLF15-repressed targets. Feed-forward response elements that utilize low affinity GR binding sites in combination with requisite cooperation between GR and GR-induced transcription factors may thus enable both graded and time-dependent gene expression responses to hormone.
Although FFLs have only recently been recognized as a regulatory paradigm for GR, relationships among GR binding site affinity, sequence, and transcriptional responses have been intensely studied for years. Proposed associations between these parameters include the notion that binding site affinity correlates directly with transcriptional responses to glucocorticoids (45). Others have proposed that allosteric interactions between GR and individual GBSs (10, 36) or interactions between GR and other DNA-associated factors (40, 46, 47) decouple GR activity at specific GBSs from in vitro binding affinity. Our data support a hybrid paradigm in which both binding site affinity and tethering between GR and other factors are key determinants of the GR response. Specifically, for the MT2A GRE, we observed a striking correlation (r2 = ∼0.897, p < 0.001; see Fig. 7F) between affinity and output over an ∼60-fold range of calculated GR binding affinities and ∼15-fold range in measured activities. Whereas the slope of the linear relationship was altered by the addition of KLF15, the coefficient of correlation was essentially unchanged. Thus, even in the context of a very strong correlation between binding site affinity and the transcriptional response, the activity of additional factors, in this case KLF15, can dramatically impact the transcriptional response to GR activation.
The correlation between transcriptional output and binding site affinity for the AASS GRE albeit still highly significant (r2 = ∼0.737, p < 0.05) was substantially weaker than the correlation observed for MT2A. An additional difference in the relationship between affinity and response between the two GREs was that maximal transcriptional activity of the AASS GRE occurred when a GBS of intermediate affinity was substituted into the locus, whereas for MT2A, transcriptional activity increased throughout the range of tested GBS affinities. Thus response element composition can specify both the coefficient of correlation relating affinity to transcriptional output and the affinity threshold required for peak activity. The mechanisms underlying these properties of the AASS and MT2A GREs and whether these observations can be generalized to other GREs remain to be determined.
Whereas functional roles for both KLF15 and GR binding sites within the AASS and PRODH GREs were clearly established by our extensive mutational analysis, our data do not fully explain how GR and KLF15 associate with these response elements to enable cooperative induction. Specifically, combinatorial disruption of the entire set of putative GR and KLF15 binding sites identified by sequence scanning with Matinspector did not completely abrogate induction of either GRE by dex + KLF15. This raises the possibility that GR and/or KLF15 can associate with additional DNA binding factors that also interact with these regulatory regions. Alternatively, additional binding sites for GR and/or KLF15 may be present within the AASS and PRODH GREs that were not recognized by Matinspector. In that regard, interrogation of these response elements using our new PWM identified several additional putative GBSs with calculated affinities that were similar to the GBSs already analyzed within the PRODH GRE; functional analysis of these sites, which are delineated in supplemental Fig. S1, is underway. Further characterization of KLF15 binding preferences both in vitro and in vivo may similarly facilitate the identification of additional putative KLF binding sites within these loci.
The inability of exogenous KLF15 to restore the GR binding pattern we observed in Klf15−/− MEFs to the WT pattern suggests that multiple mechanisms contribute to establishing GR binding regions within MEFs and presumably other cell types. These potentially include sex-mediated effects on GR binding as well as stable alterations in the expression of compensatory transcription factors and chromatin structure caused by Klf15 deficiency. The importance of cellular context in modulating GR-KLF15 function is further highlighted by the absence of prominent GR binding in WT MEFs within the endogenous AASS and PRODH loci (see the data set submitted to Gene Expression Omnibus (GSE69947)), which we have shown are regulated and occupied by GR and KLF15 in several other cell types (15, 48). Thus, although binding site affinity can contribute to transcriptional regulation by GR, this effect is subordinate to the overall regulatory context, which may confer (or prohibit) GR access to specific regulatory sites irrespective of the underlying binding site sequence.
Analysis of site affinities within ChIP-seq data further supports a model in which both site affinity and regulatory context determine the GR response. Indeed, based on the PWM for GR that we derived from Spec-seq, ∼40% of randomly shuffled 301-bp sequences contain a GBS with an affinity greater than ∼0.8% of the maximal possible site affinity, similar to the affinity of the GBSs within the PRODH locus. Thus, sites that are capable of mediating responses to GR occur frequently within random DNA, indicating that their activity is likely tempered/controlled by additional factors. Interestingly, these data contrast with prior studies where the average calculated peak GR binding affinity within native 100-bp regions that were occupied by GR specifically in A549 cells appeared to be <0.1% of maximal affinity (40). Variations between algorithms that interrogate DNA using highly specified motifs versus identifying sequence matches using the Spec-seq-defined PWM for GR, which reduces mismatch penalties through assigning biochemically derived energy differences for all four bases at each position within the logo, likely accounts for these differences. Although the Spec-seq logo was generated based on in vitro EMSA analysis performed with a purified preparation of the GR DNA binding domain, the calculated affinity range for biologically active GBSs using this logo was ∼125, which appears more plausible than much larger operational affinity ranges obtained by others (40); the close match of affinity to output that we observed also suggests that the calculated relative binding affinities reflect biologic properties of the sites.
The affinity distributions derived from random DNA sequences in comparison with GR-occupied regions suggests a model in which there is both positive and negative selection for GR binding site sequences and their function. Inspection of the affinity distributions in Fig. 6, C and D, shows that a subset of GBRs contain GBSs that rarely occur in random DNA, indicating as reported by others a strong positive selection for a subset of GR binding sites within the GR cistrome (40, 49). In contrast, the middle portion of the GBR peak affinity distribution has significant overlap with sites found in shuffled DNA, although the frequency of sites with affinities between ∼1 and 10% of maximal affinity (i.e. e−4.5–e−2.2) is modestly higher within GBRs than random DNA, suggesting a component of positive selection for such sites. As sequences with calculated affinities within this range can mediate responses to GR, evolutionary pressure likely extends to DNA in the neighborhood of randomly occurring low-moderate affinity GR binding sequences, potentially resulting in the selection of nearby sequences that enable the activity of cis-acting activators or repressors of GR.
Although in our affinity studies we selected for GR dimer-bound sequences, there has recently been compelling evidence to suggest that GR associates with chromatin and regulates transcription as a monomer. Two recent studies used “ChIP-exo” in which exonuclease digestion is used to generate footprints from immunoprecipitated chromatin to identify regions potentially occupied by monomeric GR within genomic DNA (50, 51) exemplified in the study from Starick et al. (51) by the discovery of a combinatorial site for monomeric GR and TEA Domain (TEAD) transcription factors. Interestingly, the calculated affinity of the GR dimer for the native 15-base sequence, GGAATG GAA TGTTCT, which encompasses several putative GR/TEAD “combisites,” was ∼3.5% of the maximal possible affinity for the GR dimer. This is well within the range of affinities that were functional in our transfection-based assays of GR activity. But is GR working as a monomer or dimer at these sites? Although the presence of a potential transcriptionally active binding site for the dimer at the location of these footprint-protected GR “half-sites” does not preclude monomeric occupancy by GR, a statistical mechanical model in which the monomer and dimer states are in dynamic equilibrium could potentially reconcile these seemingly contradictory data. In this model, one half of the dimer has a greater overall dwell time with a half-site than the other, resulting in a preferential protection of that half-site from exonuclease digestion. Thus, dimeric GR may not appear as the dominant species at some genomic regions in the time regime of ChIP-exo. This model is bolstered by studies showing that the dynamics of genomic association influence the ChIP-exo footprint (52). An intriguing corollary to this model is that a combination of strong and weak half-sites, together comprising a low-moderate affinity site for a GR dimer, may allow greater positional freedom for dimeric GR to associate with other transcription factors. Alternatively, GR may occupy selected individual binding regions as either a monomer or as a dimer in a process that could vary between chromosomes within the same cell and/or depend on cellular context. In either case, it will be interesting to determine whether genomic regions in which GR occupancy had been previously attributed to GR tethering interactions with other factors or monomeric binding (53) harbor sequences with calculated affinity for GR that is greater than or equal to the affinity of PRODH GBS1.
Gene regulation through GR-driven FFLs utilizes sequence information from both GR binding sites and binding sites for the second component of the FFL, thus expanding the available information content that directs transcriptional response to hormone. In that regard, the utilization of submaximal affinity binding sites within the coherent GR-KLF15 FFLs we analyzed is strikingly reminiscent of elegant work by Granner and co-workers (9, 54, 55) in which low affinity GR binding sites and interactions between GR and lineage-restricted transcription factors were implicated in restraining regulation of gluconeogenesis by GR to the liver. Thus, within the context of a GRE, relatively weak matches for the consensus palindromic GR logo appear to participate in both specifying the operational cell type and establishing the response dynamics of metabolic gene induction by GR. Although GREs and gene expression kinetics are subject to many additional layers of regulation beyond that enabled by the GBSs (56–59), our data support a model in which sites with submaximal binding affinity for GR are used both to optimize the utilization of potentially costly metabolic pathways and to endow clients of GR-driven FFLs with preferred expression dynamics. A molecular prediction based on this model is that GREs with time-dependent increases in occupancy following exposure to ligand contain GBSs of lower average affinity than GREs that achieve rapid peak GR binding; “slow occupancy” sites are also predicted to contain active sites for other factors that are themselves regulated by GR. Future studies that combine genome-wide investigation of the temporal properties of GR occupancy with detailed interrogation of specific time-dependent GREs can be used to test these hypotheses.
Author Contributions
S. K. S. designed, performed, and interpreted experiments and co-wrote the manuscript. Z. Z. designed, performed, and interpreted experiments. V. K. performed and interpreted experiments and reviewed the manuscript. L. Z. performed protein purification for Spec-seq and reviewed the manuscript. M. A. P. reviewed the manuscript and assisted in data interpretation. M. K. J. reviewed the manuscript. T. L. P interpreted ChIP-seq data. G. D. S. designed and interpreted experiments and reviewed the manuscript. A. N. G. conceived the project, designed and interpreted experiments, and co-wrote the manuscript.
Supplementary Material
Acknowledgments
ChIP-sequencing was performed at the University of Colorado Cancer Center, which is supported by National Institutes of Health Grant P30-CA046934 from the NCI. We thank David Granas for technical assistance. We thank Keith Yamamoto for the generous gift of the GR antibody and for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL109557 (to A. N. G.) and HG000249 (to G. D. S). This work was also supported by institutional funds from National Jewish Health. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and Tables S1–S4.
- GR
- glucocorticoid receptor
- GBS
- GR binding site
- dex
- dexamethasone
- seq
- sequencing
- Spec
- specificity
- GRE
- glucocorticoid response element
- FFL
- feed-forward loop
- DBR
- differential binding region
- PWM
- position weight matrix
- PEPCK
- phosphoenolpyruvate carboxykinase
- MEF
- mouse embryonic fibroblast
- qPCR
- quantitative PCR
- KLF
- Krüppel-like factor
- KBS
- KLF binding site
- GBR
- GR binding region
- ChIP-seq
- ChIP followed by deep sequencing
- AASS
- aminoadipic semialdehyde synthase
- PRODH
- proline dehydrogenase.
REFERENCES
- 1. Sapolsky R. M., Romero L. M., Munck A. U. (2000) How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21, 55–89 [DOI] [PubMed] [Google Scholar]
- 2. Quax R. A., Manenschijn L., Koper J. W., Hazes J. M., Lamberts S. W., van Rossum E. F., Feelders R. A. (2013) Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 9, 670–686 [DOI] [PubMed] [Google Scholar]
- 3. Kadmiel M., Cidlowski J. A. (2013) Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yosef N., Regev A. (2011) Impulse control: temporal dynamics in gene transcription. Cell 144, 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reddy T. E., Pauli F., Sprouse R. O., Neff N. F., Newberry K. M., Garabedian M. J., Myers R. M. (2009) Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chinenov Y., Gupte R., Rogatsky I. (2013) Nuclear receptors in inflammation control: repression by GR and beyond. Mol. Cell. Endocrinol. 380, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Langlais D., Couture C., Balsalobre A., Drouin J. (2012) The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol. Cell 47, 38–49 [DOI] [PubMed] [Google Scholar]
- 8. Waterman W. R., Xu L. L., Tetradis S., Motyckova G., Tsukada J., Saito K., Webb A. C., Robinson D. R., Auron P. E. (2006) Glucocorticoid inhibits the human pro-interleukin lβ gene (ILIB) by decreasing DNA binding of transactivators to the signal-responsive enhancer. Mol. Immunol. 43, 773–782 [DOI] [PubMed] [Google Scholar]
- 9. Imai E., Stromstedt P. E., Quinn P. G., Carlstedt-Duke J., Gustafsson J. A., Granner D. K. (1990) Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol. Cell. Biol. 10, 4712–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson L. C., Kuchenbecker K. M., Schiller B. J., Gross J. D., Pufall M. A., Yamamoto K. R. (2013) The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat. Struct. Mol. Biol. 20, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J. C., Strömstedt P. E., Sugiyama T., Granner D. K. (1999) The phosphoenolpyruvate carboxykinase gene glucocorticoid response unit: identification of the functional domains of accessory factors HNF3β (hepatic nuclear factor-3β) and HNF4 and the necessity of proper alignment of their cognate binding sites. Mol. Endocrinol. 13, 604–618 [DOI] [PubMed] [Google Scholar]
- 12. Scott D. K., Strömstedt P. E., Wang J. C., Granner D. K. (1998) Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol. Endocrinol. 12, 482–491 [DOI] [PubMed] [Google Scholar]
- 13. Klemm D. J., Roesler W. J., Liu J. S., Park E. A., Hanson R. W. (1990) In vitro analysis of promoter elements regulating transcription of the phosphoenolpyruvate carboxykinase (GTP) gene. Mol. Cell. Biol. 10, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uhlenhaut N. H., Barish G. D., Yu R. T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hubner N., Evans R. M. (2013) Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sasse S. K., Mailloux C. M., Barczak A. J., Wang Q., Altonsy M. O., Jain M. K., Haldar S. M., Gerber A. N. (2013) The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol. Cell. Biol. 33, 2104–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gray S., Wang B., Orihuela Y., Hong E. G., Fisch S., Haldar S., Cline G. W., Kim J. K., Peroni O. D., Kahn B. B., Jain M. K. (2007) Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 5, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eichenberger P., Fujita M., Jensen S. T., Conlon E. M., Rudner D. Z., Wang S. T., Ferguson C., Haga K., Sato T., Liu J. S., Losick R. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerstein M. B., Kundaje A., Hariharan M., Landt S. G., Yan K. K., Cheng C., Mu X. J., Khurana E., Rozowsky J., Alexander R., Min R., Alves P., Abyzov A., Addleman N., Bhardwaj N., Boyle A. P., Cayting P., Charos A., Chen D. Z., Cheng Y., Clarke D., Eastman C., Euskirchen G., Frietze S., Fu Y., Gertz J., Grubert F., Harmanci A., Jain P., Kasowski M., Lacroute P., Leng J., Lian J., Monahan H., O'Geen H., Ouyang Z., Partridge E. C., Patacsil D., Pauli F., Raha D., Ramirez L., Reddy T. E., Reed B., Shi M., Slifer T., Wang J., Wu L., Yang X., Yip K. Y., Zilberman-Schapira G., Batzoglou S., Sidow A., Farnham P. J., Myers R. M., Weissman S. M., Snyder M. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Penn B. H., Bergstrom D. A., Dilworth F. J., Bengal E., Tapscott S. J. (2004) A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 18, 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasse S. K., Gerber A. N. (2015) Feed-forward transcriptional programming by nuclear receptors: regulatory principles and therapeutic implications. Pharmacol. Ther. 145, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang B., Hsu H. K., Hsu P. Y., Bonneville R., Chen S. S., Huang T. H., Jin V. X. (2012) Hierarchical modularity in ERα transcriptional network is associated with distinct functions and implicates clinical outcomes. Sci. Rep. 2, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chinenov Y., Coppo M., Gupte R., Sacta M. A., Rogatsky I. (2014) Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics 15, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangan S., Alon U. (2003) Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U.S.A. 100, 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan S., Bren A., Dekel E., Alon U. (2008) The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol. Syst. Biol. 4, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alon U. (2007) Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 [DOI] [PubMed] [Google Scholar]
- 26. Stormo G. D., Zuo Z., Chang Y. K. (2015) Spec-seq: determining protein-DNA-binding specificity by sequencing. Brief. Funct. Genomics 14, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haldar S. M., Lu Y., Jeyaraj D., Kawanami D., Cui Y., Eapen S. J., Hao C., Li Y., Doughman Y. Q., Watanabe M., Shimizu K., Kuivaniemi H., Sadoshima J., Margulies K. B., Cappola T. P., Jain M. K. (2010) Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Sci. Transl. Med. 2, 26ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noack C., Zafiriou M. P., Schaeffer H. J., Renger A., Pavlova E., Dietz R., Zimmermann W. H., Bergmann M. W., Zelarayán L. C. (2012) Krueppel-like factor 15 regulates Wnt/β-catenin transcription and controls cardiac progenitor cell fate in the postnatal heart. EMBO Mol. Med. 4, 992–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Werner T. (2000) Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol. Biol. 132, 337–349 [DOI] [PubMed] [Google Scholar]
- 30. Smith C. L., Wolford R. G., O'Neill T. B., Hager G. L. (2000) Characterization of transiently and constitutively expressed progesterone receptors: evidence for two functional states. Mol. Endocrinol. 14, 956–971 [DOI] [PubMed] [Google Scholar]
- 31. Langmead B., Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Q., Brown J. B., Huang H., Bickel P. J. (2011) Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 [Google Scholar]
- 33. Liang K., Keles S. (2012) Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics 28, 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu G., Wang L. G., He Q. Y. (2015) ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- 35. Zuo Z., Stormo G. D. (2014) High-resolution specificity from DNA sequencing highlights alternative modes of Lac repressor binding. Genetics 198, 1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hertz G. Z., Stormo G. D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 [DOI] [PubMed] [Google Scholar]
- 38. Prosdocimo D. A., Anand P., Liao X., Zhu H., Shelkay S., Artero-Calderon P., Zhang L., Kirsh J., Moore D., Rosca M. G., Vazquez E., Kerner J., Akat K. M., Williams Z., Zhao J., Fujioka H., Tuschl T., Bai X., Schulze P. C., Hoppel C. L., Jain M. K., Haldar S. M. (2014) Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J. Biol. Chem. 289, 5914–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hubler T. R., Scammell J. G. (2004) Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gertz J., Savic D., Varley K. E., Partridge E. C., Safi A., Jain P., Cooper G. M., Reddy T. E., Crawford G. E., Myers R. M. (2013) Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 52, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bailey T. L., Machanick P. (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Machanick P., Bailey T. L. (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stormo G. D. (2013) Modeling the specificity of protein-DNA interactions. Quant. Biol. 1, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bain D. L., Yang Q., Connaghan K. D., Robblee J. P., Miura M. T., Degala G. D., Lambert J. R., Maluf N. K. (2012) Glucocorticoid receptor-DNA interactions: binding energetics are the primary determinant of sequence-specific transcriptional activity. J. Mol. Biol. 422, 18–32 [DOI] [PubMed] [Google Scholar]
- 46. Motlagh H., Anderson J., Li J., Hilser V. (2015) Disordered allostery: lessons from glucocorticoid receptor. Biophys. Rev. 7, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearce D., Matsui W., Miner J. N., Yamamoto K. R. (1998) Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J. Biol. Chem. 273, 30081–30085 [DOI] [PubMed] [Google Scholar]
- 48. Haldar S. M., Jeyaraj D., Anand P., Zhu H., Lu Y., Prosdocimo D. A., Eapen B., Kawanami D., Okutsu M., Brotto L., Fujioka H., Kerner J., Rosca M. G., McGuinness O. P., Snow R. J., Russell A. P., Gerber A. N., Bai X., Yan Z., Nosek T. M., Brotto M., Hoppel C. L., Jain M. K. (2012) Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc. Natl. Acad. Sci. U.S.A. 109, 6739–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. So A. Y., Cooper S. B., Feldman B. J., Manuchehri M., Yamamoto K. R. (2008) Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc. Natl. Acad. Sci. U.S.A. 105, 5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lim H. W., Uhlenhaut N. H., Rauch A., Weiner J., Hübner S., Hübner N., Won K. J., Lazar M. A., Tuckermann J., Steger D. J. (2015) Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 25, 836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Starick S. R., Ibn-Salem J., Jurk M., Hernandez C., Love M. I., Chung H. R., Vingron M., Thomas-Chollier M., Meijsing S. H. (2015) ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 25, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sung M. H., Guertin M. J., Baek S., Hager G. L. (2014) DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell 56, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schiller B. J., Chodankar R., Watson L. C., Stallcup M. R., Yamamoto K. R. (2014) Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 15, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stafford J. M., Wilkinson J. C., Beechem J. M., Granner D. K. (2001) Accessory factors facilitate the binding of glucocorticoid receptor to the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 276, 39885–39891 [DOI] [PubMed] [Google Scholar]
- 55. Mitchell J., Noisin E., Hall R., O'Brien R., Imai E., Granner D. (1994) Integration of multiple signals through a complex hormone response unit in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 8, 585–594 [DOI] [PubMed] [Google Scholar]
- 56. Miranda T. B., Morris S. A., Hager G. L. (2013) Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol. Cell. Endocrinol. 380, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. John S., Sabo P. J., Thurman R. E., Sung M. H., Biddie S. C., Johnson T. A., Hager G. L., Stamatoyannopoulos J. A. (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chodankar R., Wu D. Y., Schiller B. J., Yamamoto K. R., Stallcup M. R. (2014) Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc. Natl. Acad. Sci. U.S.A. 111, 4007–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen S., Sarlis N. J., Simons S. S., Jr. (2000) Evidence for a common step in three different processes for modulating the kinetic properties of glucocorticoid receptor-induced gene transcription. J. Biol. Chem. 275, 30106–30117 [DOI] [PubMed] [Google Scholar]
- 60. La Baer J., Yamamoto K. R. (1994) Analysis of the DNA-binding affinity, sequence specificity and context dependence of the glucocorticoid receptor zinc finger region. J. Mol. Biol. 239, 664–688 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.