Background: Biosynthetic transformation of fatty acid peroxides commonly is catalyzed by cytochromes P450.
Results: A catalase-related hemoprotein in the coral Capnella imbricata converts 8R-hydroperoxy-eicosatetraenoic acid in a P450-type reaction to short-chain aldehydes.
Conclusion: A catalase-related hydroperoxide lyase is identified in Animalia.
Significance: The catalase-related hemoprotein has the catalytic competence of a P450.
Keywords: arachidonic acid (AA) (ARA), catalase, eicosanoid biosynthesis, enzyme mechanism, high performance liquid chromatography (HPLC), nuclear magnetic resonance (NMR), allene oxide synthase, coral, hydroperoxide lyase
Abstract
In corals a catalase-lipoxygenase fusion protein transforms arachidonic acid to the allene oxide 8R,9-epoxy-5,9,11,14-eicosatetraenoic acid from which arise cyclopentenones such as the prostanoid-related clavulones. Recently we cloned two catalase-lipoxygenase fusion protein genes (a and b) from the coral Capnella imbricata, form a being an allene oxide synthase and form b giving uncharacterized polar products (Lõhelaid, H., Teder, T., Tõldsepp, K., Ekins, M., and Samel, N. (2014) PloS ONE 9, e89215). Here, using HPLC-UV, LC-MS, and NMR methods, we identify a novel activity of fusion protein b, establishing its role in cleaving the lipoxygenase product 8R-hydroperoxy-eicosatetraenoic acid into the short-chain aldehydes (5Z)-8-oxo-octenoic acid and (3Z,6Z)-dodecadienal; these primary products readily isomerize in an aqueous medium to the corresponding 6E- and 2E,6Z derivatives. This type of enzymatic cleavage, splitting the carbon chain within the conjugated diene of the hydroperoxide substrate, is known only in plant cytochrome P450 hydroperoxide lyases. In mechanistic studies using 18O-labeled substrate and incubations in H218O, we established synthesis of the C8-oxo acid and C12 aldehyde with the retention of the hydroperoxy oxygens, consistent with synthesis of a short-lived hemiacetal intermediate that breaks down spontaneously into the two aldehydes. Taken together with our initial studies indicating differing gene regulation of the allene oxide synthase and the newly identified catalase-related hydroperoxide lyase and given the role of aldehydes in plant defense, this work uncovers a potential pathway in coral stress signaling and a novel enzymatic activity in the animal kingdom.
Introduction
Lipid mediator biosynthesis commonly involves the dioxygenation of a fatty acid followed by a cytochrome P450-catalyzed rearrangement of the resulting fatty acid peroxide in the next step of the pathway. In higher animals the cyclooxygenases are so coupled with thromboxane synthase (cytochrome P450 CYP5) or prostacyclin synthase (CYP8), thereby producing important mediators of vascular hemostasis (1). In plants, the pair of enzymes is usually a lipoxygenase (LOX),2 forming a fatty acid hydroperoxide, and a member of the P450 CYP74 family with allene oxide synthase (AOS), divinyl ether synthase, or hydroperoxide lyase (HPL) activity (2, 3). These enzymes give rise to products including the plant hormone jasmonic acid via AOS and mediators of plant defense via the HPL (4). In fungi there are many examples of a single participating gene encoding a two-domain protein comprised of an N-terminal heme dioxygenase and a C-terminal cytochrome P450 (5–7). The resulting oxylipin products act as hormone-like signals modulating asexual and sexual spore development and in the production of toxins (7, 8).
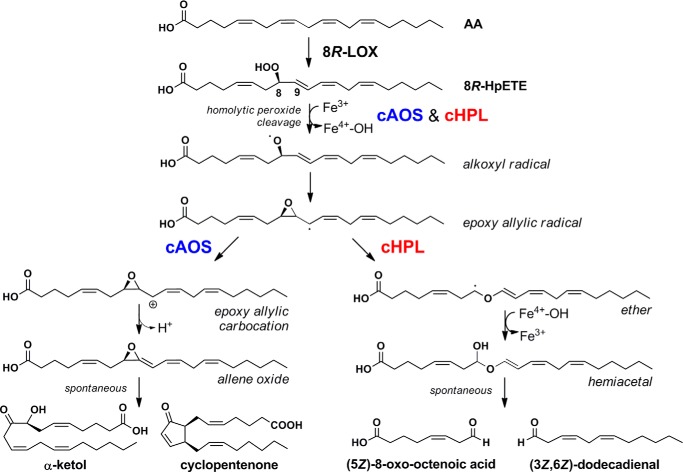
Certain lower animals, best studied in corals, express a different type of fusion protein that consists of an N-terminal catalase-related hemoprotein coupled to a C-terminal LOX domain (9, 10). The prototypical enzyme of this type is the cAOS-LOX fusion protein, which dioxygenates arachidonic acid to an 8R-hydroperoxyeicosatetraenoic acid (8R-HpETE) intermediate followed by its conversion to an allene oxide by the cAOS domain (11). Although the plant AOS and cAOS catalyze similar reactions, the enzymes are structurally unrelated (12–14). The highly labile allene oxide products of both enzyme types readily break down to stable end products, α-ketol and cyclopentenone (6). Moreover, despite the structural relatedness to catalase, cAOS lacks the capability to catalyze the decomposition of hydrogen peroxide to water and molecular oxygen (14, 15).
In our recent study of the soft coral Capnella imbricata we cloned two highly homologous catalase-lipoxygenase fusion proteins (designated a and b) with 88% of amino acid identity (16). Although the catalytically important amino acids of catalase-related and lipoxygenase domains were conserved, the biosynthesis with [1-14C]arachidonic acid led to the formation of different products. Although fusion protein a catalyzed the formation of an α-ketol and cyclopentenone stable end products and thereby can be designated as a cAOS-LOX, incubation with fusion protein b gave rise to unknown polar compounds (16). Herein we present the identification of short-chain compounds formed by the catalase-related hemoprotein b and thereby characterize a fatty acid hydroperoxide lyase pathway in coral arachidonic acid metabolism.
Experimental Procedures
Materials
Arachidonic acid (AA) was purchased from Nu-Chek Prep Inc. (Elysian, MN). 8R-HpETE was synthesized using the Plexaura homomalla 8R-lipoxygenase domain of the cAOS-LOX fusion protein, expressed in Escherichia coli (11). The 18O2-8R-HpETE was prepared from AA by P. homomalla 8R-LOX under an 18O2 (Isotech (Miamisburg, OH)) atmosphere in a degassed buffer (50 mm Tris, pH 8.0). The 18O-labeled water was purchased from the Mound Facility (Miamisburg, OH). Only HPLC grade solvents were used.
Cloning and Expression of Fusion Protein b and the Catalase-related Domain
The ORF of fusion protein b (GenBankTM accession number KF000374) with terminal NheI restriction sites and with an N-terminal His6 tag was PCR-amplified from the fusion protein b construct (16) using forward (5′-ATTCATATGATGGTTTGGAAAAATTTTGGTTACG-3′) and reverse (5′-CAGCTAGCCTAGATTGCAGTTCCG-3′) primers. Subsequently, the ORF of the catalase-related domain with a stop codon and C-terminal His4 tag was PCR-amplified from fusion protein b sequence using forward (5′-ATTCATATGATGGTTTGGAAAAATTTTGGTTACG-3′) and reverse (5′-ATAGCTAGCTTAGTGATGGTGATGGTTCTGTCCTGTTGGAACCAGAG-3′) primers with NdeI and NheI restriction sites, respectively. The amplicons of fusion protein b and the catalase-related domain were cloned into the pET11a expression vector (Stratagene) and sequenced. The His-tagged fusion protein b and catalase-related domain were expressed in E. coli BL21(DE3) cells (Novagen) in a Terrific Broth medium at 20 °C overnight and purified as described previously (17). The protein fractions were dialyzed against an ice-cold 50 mm Tris (pH 8.0) buffer containing 100 mm NaCl by slowly stirring overnight at 4 °C. Purified proteins were stored at −80 °C for further use. Proteins were quantified based on the absorbance at 406 nm (ϵ ∼ 100,000 m−1 cm−1) characteristic for hemoproteins.
Chiral analysis of C. imbricata Fusion Protein b-derived 8-HpETE Intermediate
Incubations with 5 nm fusion protein b were performed at room temperature using 100 μm AA in a 50 mm Tris (pH 8.0) buffer containing 100 mm NaCl and 1 mm CaCl2 in the presence of the reducing agent SnCl2 to reduce the LOX-catalyzed 8-HpETEs in situ. Incubations with Gersemia fruticosa 8R-LOX were performed in parallel. Individual LOX products were extracted with EtOAc, and the corresponding methyl esters were prepared using diazomethane. LOX products were isolated on SP-HPLC using a Phenomenex silica 5-μm column (0.46 × 25 cm) with a hexane/isopropyl alcohol solvent system (isocratic: 100:2 by volume) at a flow rate of 1 ml/min at 235 nm. Methyl esters of 8-HETE were analyzed on a Chiralcel OD-H (0.46 × 25 cm) column at a flow rate of 1 ml/min at 235 nm using the same solvent system as described previously. The methylated C. imbricata LOX product was co-chromatographed with the methyl esters of ±8-HETE standard (Cayman Chemical Co.), and the 8R-HETE reference by G. fruticosa 8R-LOX (10) was used to determine the stereoconfiguration of 8-HETE formed by the LOX domain of the C. imbricata fusion protein b.
Incubations with Fusion Protein b and Catalase-related Domain
Incubations with 5 nm fusion protein b were conducted at room temperature using 100 μm AA in a 50 mm Tris (pH 8.0) buffer containing 100 mm NaCl and 1 mm CaCl2 in a quartz cuvette. The change in the absorbance was recorded by repetitive scanning (200–350 nm) using a Lambda-35 UV-visible spectrometer (PerkinElmer Life Sciences). Alternative substrates, eicosapentaenoic acid (20:5ω3), docosahexaenoic acid (22:6ω3), and 5,8,11-eicosatrienoic acid (20:3ω9) (Cayman Chemical Co.), were tested in parallel. Identical incubations with 5 nm catalase-related domain (the presence of CaCl2 in the incubation is not essential) were performed using 100 μm 8R-HpETE as a substrate, and the disappearance of a conjugated diene chromophore (ϵ ∼ 25,000 m−1 cm−1) was recorded at 235 nm. The substrate specificities with 8RS-, 11RS-, and 11R-HpETE were examined in parallel.
To analyze the primary products formed, reactions with 8R-HpETE and the catalase-related domain were stopped with the addition of the reducing agent NaBH4 (0.5 mg of NaBH4 per 1 ml of buffer). The reaction was acidified down to pH 4 and loaded on a 1-cc Oasis HLB cartridge (Waters), eluted with 1 ml of MeOH, and stored at −80 °C for further analysis. Products were taken to dryness and dissolved in a corresponding column solvent before the HPLC analyses.
To analyze the oxygen incorporation from the substrate, incubations with 0.3 mm 18O2-8R-HpETE and 24 μm catalase-related domain were performed in a 1-ml ice-cold incubation buffer either for 30 s or for 2 min on ice. In addition, reactions with 24 μm catalase domain and 2 mm 8R-HpETE in a 0.2-ml ice-cold buffer (50 mm Tris (pH 8.0), 100 mm NaCl) containing 0.1 ml H218O were carried out for 30 s on ice. Control reactions with the unlabeled substrate and water were conducted in parallel. All reactions were stopped by adding NaBH4.
Carbon Monoxide Binding of Ferrous catalase-related Domain
The ferrous state of the catalase-related domain of fusion protein b was generated by the addition of sodium dithionite into a 1-ml quartz cuvette containing 1 μm ferric catalase-related domain in the incubation buffer. A ferrous-CO complex was prepared by bubbling CO gas into the cuvette for 2 min. The change in the chromophore of the Soret band (406 nm; ferric cHPL) to a ferrous-CO complex was observed using UV-visible spectrometer scanning at 200–700 nm.
HPLC Analyses
Non-reduced and NaBH4-reduced products were analyzed by RP-HPLC using a C18 Waters Symmetry 5-μm column (46 × 250 mm) with a CH3CN/H2O/glacial acetic acid solvent system (gradient: 20:80:0.01 to 80:20:0.01 by volume for 30 min; isocratic: 80:20:0.01 by volume for 10 min) at a flow rate of 1 ml/min. UV signals at 205, 220, 235, and 270 nm were recorded using an Agilent 1100 series diode array detector. For the 1H NMR analysis, products formed from 2 mg of substrate were extracted on a 6-cc Agilent Bond Elut C18 column and isolated on RP-HPLC using the same solvent system as described previously. Additional purification was performed on SP-HPLC using a Thomson silica 5-μm column (4.6 × 250 mm) with a hexane/isopropyl alcohol/glacial acetic acid column solvent (100:3:0.01 and 100:10:0.01 by volume for polar and less polar compounds, respectively) at a flow rate of 1 ml/min. Isolated products were dissolved in MeOH and stored at −80 °C until further analyses.
RP-HPLC-MS and NMR Analyses
Products formed from 8R-HpETE by catalase domain were separated on RP-HPLC using a Kinetex C18 2.6-μm column (3 × 100 mm) with CH3CN/H2O/HAc (45:55:0.01, by volume) at 0.4 ml/min, and molecular weights were established from the M-H anions measured by negative ion electrospray LTQ1 iontrap LC-MS instrument.
The products formed from 18O-labeled 8R-HpETE by the catalase-related domain were eluted on RP-HPLC/MS using a Kinetex C18 2.6-μm (3 × 100 mm) column with a MeOH/H2O/HAc solvent system containing 10 mm NH4OAc, 0.1% AcOH, and 100 μm AgBF4 (gradient: 60:40:0.01 to 90:10:0.01 (MeOH/H2O/HAc) by volume with a flow rate of 0.2 ml/min for 5 min; isocratic: 90:10:0.01 by volume with a flow rate of 0.3 ml/min). The molecular weights were established from the formed silver adducts using silver coordination on double bonds and the M-H+ cation measured by positive ion electrospray Quantum3 LC-MS instrument.
1H NMR and 1H,1H COSY NMR spectra were recorded on a Bruker 600-MHz spectrometer at 298 K. The parts/million values are reported relative to residual non-deuterated C6D6 (δ = 7.16) and CDCl3 (δ = 7.24) for non-reduced and NaBH4-reduced products, respectively. All spectra were analyzed on TopSpin 3.0 software (Bruker).
Results
Heterologous Expression of a Novel Catalase-Lipoxygenase Fusion Protein
The C. imbricata catalase-lipoxygenase fusion cDNA including an N-terminal His tag was cloned into the expression vector pET11a, and the 122-kDa protein was expressed in E. coli BL21(DE3) with a 1-mg yield of active protein per liter under the conditions described under “Experimental Procedures.” The catalase-domain (43 kDa) alone was expressed similarly, obtaining the same yield and activity (Fig. 1). The UV-visible spectrum shows the main Soret band at 406 nm, typical of a high spin ferric catalase (Fig. 1). The absorbance shifted to 427 nm in the CO complex of the dithionite-reduced (ferrous) enzyme, a value close to the reported lambda max of the P. homomalla CO complex of ferrous-cAOS (427 nm) (18, 19).
FIGURE 1.
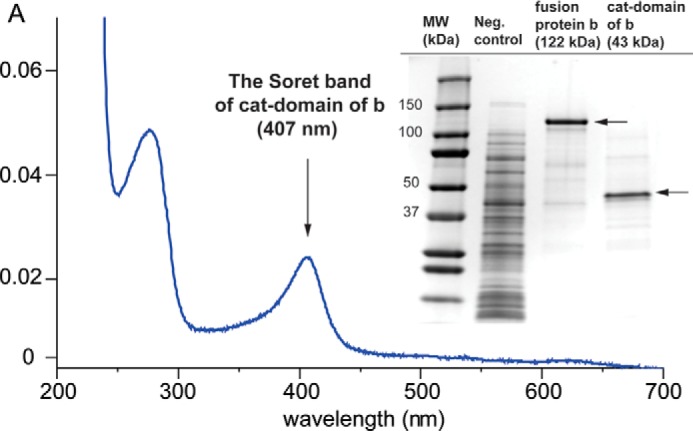
The UV-visible spectrum of catalase-related domain of fusion protein b and the SDS-PAGE gel image of expressed fusion protein b and catalase domain. cat-domain of b, the catalase-related domain of fusion protein b.
Characterization of the Catalytic Specificity of the LOX Domain
The 8-HpETE formed by C. imbricata cHPL-LOX was chromatographed on a Chiralcel OD-H chiral column as the HETE methyl ester derivative as described under “Experimental Procedures.” The retention time of the methyl ester of 8-HETE at 15 min corresponds to the 8R enantiomer formed by G. fruticosa 8R-LOX. The chromatogram of the racemic 8-HETE standard analyzed on chiral SP-HPLC appeared in two peaks, the first peak being 8R and the second peak 8S (10), with retention times at 15 and 18 min, respectively. The methyl ester of 8-HETE co-chromatographed with the racemic 8-HETE standard resulted in an increase of the first peak (data not shown). Therefore, co-elution of the C. imbricata fusion protein b-derived 8-HETE with the references of 8R enantiomer confirms the specificity of the 8R-LOX domain.
HPLC Analyses of Products of the Catalase-related Domain
Incubations of 8R-HpETE with the His-tagged fusion protein b or its catalase-only domain resulted in the disappearance of the conjugated diene chromophore of 8R-HpETE and the formation of end products with weak or negligible UV absorbance (Fig. 2). In addition, C. imbricata fusion protein b converts the substrate analogue 5,8,11-eicosatrienoic acid and naturally occurring eicosapentaenoic acid and docosahexaenoic acid similarly to AA (data not shown). Among arachidonate-derived substrates, the expressed catalase-related domain was specific for 8R-HpETE and did not measurably metabolize 8RS-HpETE (8S being inhibitory) or 11R- and 11RS-HpETE. The kcat and Km of cHPL with 8R-HpETE were determined as 133 ± 5 s−1 and 3.8 ± 0.5 μm, respectively, with a kcat/Km value of 35 μm−1 s−1.
FIGURE 2.
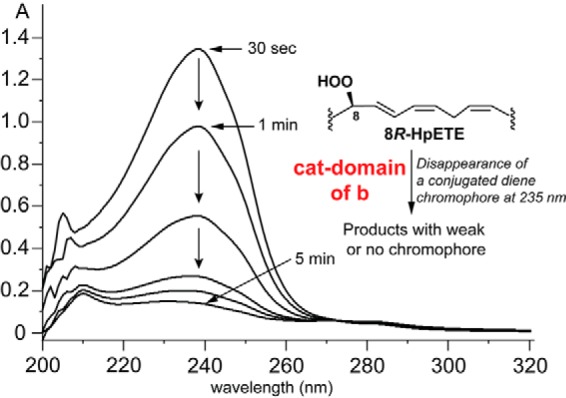
Repetitive UV scans illustrating the transformation of 8R-HpETE by the catalase-related domain of C. imbricata fusion protein b. The transformation is associated with the disappearance of the conjugated diene chromophore of 8R-HpETE and gives a product(s) with weak or no chromophore. cat-domain of b, the catalase-related domain of fusion protein b.
RP-HPLC analysis of the products from 8R-HpETE consistently showed the appearance of two very broad UV-205-nm absorbing areas on the chromatogram followed in both cases by a sharp 220-nm peak (Fig. 3A). It became apparent (as explained below) that the primary enzymatic products were represented by the exceptionally broad peaks labeled 1 and 2 on Fig. 3A and that the sharp peaks labeled 3 and 4 eluting at ∼8 and 37 min were actually formed via non-enzymatic isomerization of 1 and 2, respectively. It was evident at this stage that the catalase domain exhibited a novel activity as no allene oxide-derived α-ketol or cyclopentenone was detected. It was also established that the products were formed by the chain cleavage of 8R-HpETE because after incubation and extraction of an incubation of [1-14C]-AA with fusion protein b, the 14C label was retained only in the early eluting product 3, and product 4 was unlabeled (16).
FIGURE 3.
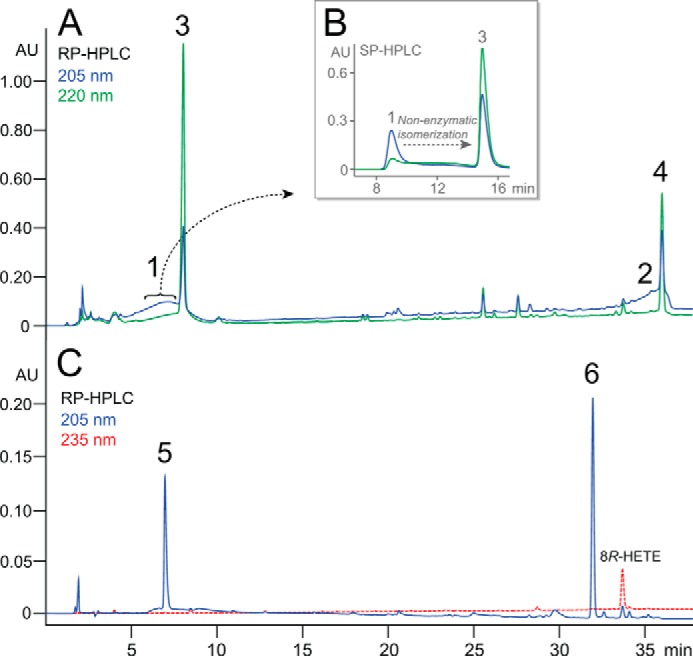
HPLC analyses of products of the catalase domain of b reacting with 8R-HpETE. A, RP-HPLC chromatogram of the product profile monitored at 205 nm (blue line, showing broad peaks 1 and 2) and 220 nm (green line, sharp peaks of products 3 and 4). B, inset, SP-HPLC chromatogram illustrating the non-enzymatic isomerization of isolated product 1 to product 3. C, RP-HPLC chromatogram of the products of in situ NaBH4 reduction of the enzymatic incubation, producing 5 and 6 (blue line) as alcohol derivatives of products 1 and 2, respectively, with 8R-HETE (reduced substrate) eluting at 34 min and detected at 235 nm (red line). The RP-HPLC analyses used a C18 Waters Symmetry 5-μm column (4.6 × 250 mm) at a flow rate of 1 ml/min with a solvent gradient of CH3CN/H2O/glacial acetic acid in the proportions 20:80:0.01 to 80:20:0.01 (by volume) over 30 min then held isocratically for 10 min. UV signals at 205, 220, 235, and 270 nm were recorded using an Agilent 1100 series diode array detector. The SP-HPLC in panel B used a Thomson silica 5-μm column (4.6 × 250 mm) with hexane/isopropyl alcohol/glacial acetic acid solvent (100:10:0.01, by volume) and a flow rate of 1 ml/min. AU, absorbance units.
Once it became apparent that the broad peaks 1 and 2 were a consistent feature of the product profile, product 1 was collected from the initial RP-HPLC run and re-chromatographed on SP-HPLC, (Fig. 3B). This clearly showed that product 1 was converting to 3 and, indeed, the elevated baseline between the peaks reflected this transformation in progress (Fig. 3B). Initial characterization of the stable products 3 and 4 showed that they each displayed a conjugated enone chromophore (lambda max 225 nm in acetonitrile). The facile isomerization of 1 and 2 would be readily explained by a double bond isomerization to form the conjugated enone systems of 3 and 4. To prevent this from occurring, an incubation of 8R-HpETE with the catalase domain was terminated by the addition of NaBH4 in situ, and the resulting products were analyzed by RP-HPLC (Fig. 3C); this revealed two new products, designated 5 and 6, with stable chromatographic characteristics and displaying only 205-nm UV absorbance.
LC-MS and NMR Analyses
Negative ion electrospray LC-MS analysis of product 3 gave an M-1 ion at m/z 155, corresponding to the predicted mass of an oxo-octenoic acid, whereas product 4 was undetectable. Product 3 was identified by NMR (Table 1). The doublet at 9.27 ppm (H8) represents the C-8 aldehydic proton, which is coupled to H7 at 5.81 ppm with a J5,6 coupling of 15.6 Hz to H6 (5.89 ppm) defining the 6,7-trans double bond and the conjugated enone system (Fig. 4A). These data and the remaining signals established the structure of 3 as (6E)-8-oxo-octenoic acid. Due to volatility and solubility issues, the purification of product 4 for NMR analysis was unsuccessful, and its structure was deduced as the corresponding conjugated C12 aldehyde, 1-oxo-2E,6Z-dodecene, from its UV spectral characteristics and by silver-coordinated LC-MS and NMR analyses of the corresponding C12 NaBH4-reduced fragment.
TABLE 1.
1H NMR chemical shifts (δ) and coupling constants (Hz) of C8-oxo acid (product 3)
NMR analysis was conducted using C6D6 solvent.
| Hydrogens | Chemical shift (δ) | Coupling constant (Hz) | Multiplicity | Number of protons |
|---|---|---|---|---|
| H8 | 9.27 | J8,7 = 7.6 | d | 1 |
| H7 | 5.81 | J7,6 = 15.6 (trans) | dd | 1 |
| J7,8 = 7.6 | ||||
| H6 | 5.89 | J6,5 = 6.5 | dt | 1 |
| J6,7 = 15.6 (trans) | ||||
| H5 | 1.52 | J5,4 = 14.9 | dd | 2 |
| J5,6 = 7.2 | ||||
| H4 | 0.88 | J4,3 = 7.8 | m | 2 |
| J4,5 = 15.4 | ||||
| H3 | 1.18 | J3,2 = 15.5 | m | 2 |
| J3,4 = 7.7 | ||||
| H2 | 1.85 | J3,4 = 14.6 | t | 2 |
FIGURE 4.
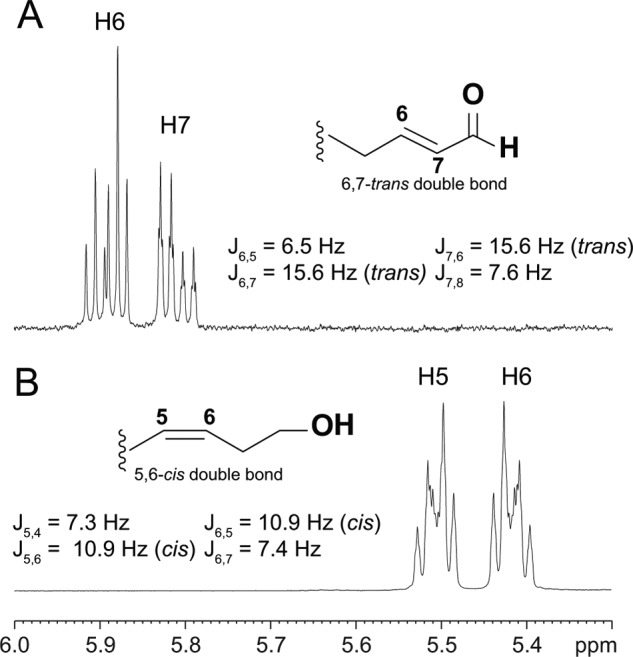
1H NMR one-dimensional spectra of double bond region of (6E)-8-oxo-octenoic acid (A) and (5Z)-8-hydroxy acid (B). Coupling constants between H6 and H7 (A) and H5 and H6 (B) describe the stereo configuration of the corresponding double bond. The (6E)-8-oxo-octenoic acid (A) represents isomerized product from primary (6E)-8-oxo-octenoic acid, which is analyzed as C8-hydroxy acid derivative (B). C8-oxo acid (A) and C8-hydroxy acid (B) were analyzed in C6D6 and CDCl3 solvents, respectively.
Identification of Products 5 and 6
The NaBH4-reduced products 5 and 6 (Fig. 3C) were identified by NMR (Table 2). In the spectrum of product 5 (Table 2, left), the triplet at 3.64 ppm had an area of two, representing the geminal C-8 hydrogens, and indicating a primary alcohol. The same applies to the hydroxyl of 6 (H-1 at 3.64 ppm; Table 2, right). Most significantly, in each case the primary alcohols were coupled to a CH2 moiety, which in turn is coupled to a cis double bond (10.9 Hz; cis); Fig. 4B. Together with the rest of the NMR data, the structures of 5 and 6 were thus established as (5Z)-8-hydroxy-octenoic acid and (3Z,6Z)-dodecadien-1-ol, respectively. It follows that the primary products of the catalase domain detected as the broad peaks at 205 nm, products 1 and 2, were the corresponding aldehydes, (5Z)-8-oxo-octenoic acid and (3Z,6Z)-1-oxo-dodecadienal (Scheme 1). The results establish that the C. imbricata catalase domain of fusion protein b displays a novel hydroperoxide lyase activity and that it can be designated as a cHPL, catalase-related hydroperoxide lyase.
TABLE 2.
1H NMR chemical shifts (δ) and coupling constants (Hz) of C12-hydroxy acid (left) and C12-alcohol (right)
NMR analysis was conducted using a CDCl3 solvent (left and right).
| C8-hydroxy acid (product 5) |
C12 alcohol (product 6) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hydrogens | Chemical shift (δ) | Coupling constant (Hz) | Multiplicity | Number of protons | Hydrogens | Chemical shift (δ) | Coupling constant (Hz) | Multiplicity | Number of protons |
| H8 | 3.64 | J8,7 = 12.9 | t | 2 | H12 | 0.87 | J12,11 = 6.7 | t | 3 |
| H7 | 2.31 | J7,6 = 6.8 | q | 2 | H9 | 1.35 | J9,8 = 7.1 | m | 2 |
| J7,8 = 13.2 | J9,10 = 10.6 | ||||||||
| H6 | 5.42 | J6,5 = 10.9 (cis) | m | 1 | H8 | 2.03 | J8,7 = 7.3 | q | 2 |
| J6,7 = 7.4 | J8,9 = 7.2 | ||||||||
| H5 | 5.51 | J5,4 = 7.3 | m | 1 | H7 | 5.38 | J7,6 = 10.9 (cis) | m | 1 |
| J5,6 = 10.9 (cis) | J7,8 = 7.3 | ||||||||
| H4 | 2.13 | J4,3 = 14.6 | q | 2 | H6 | 5.31 | J6,7 = 10.6 (cis) | dt | 1 |
| J4,5 = 7.3 | J6,5 = 7.1 | ||||||||
| H3 | 1.71 | J3,2 = 7.3 | m | 2 | H5 | 2.81 | J5,6 = 7.3 | t | 2 |
| J3,4 = 14.7 | J5,4 = 7.3 | ||||||||
| H2 | 2.35 | J3,2 = 14.6 | t | 2 | H4 | 5.52 | J4,5 = 7.4 | dt | 1 |
| J4,3 = 10.7 (cis) | |||||||||
| H3 | 5.38 | J3,4 = 10.7 (cis) | m | 1 | |||||
| J3,2 = 7.3 | |||||||||
| H2 | 2.43 | J2,1 = 13.4 | q | 2 | |||||
| J2,3 = 6.8 | |||||||||
| H1 | 3.64 | J1,2 = 6.5 | t | 2 | |||||
SCHEME 1.
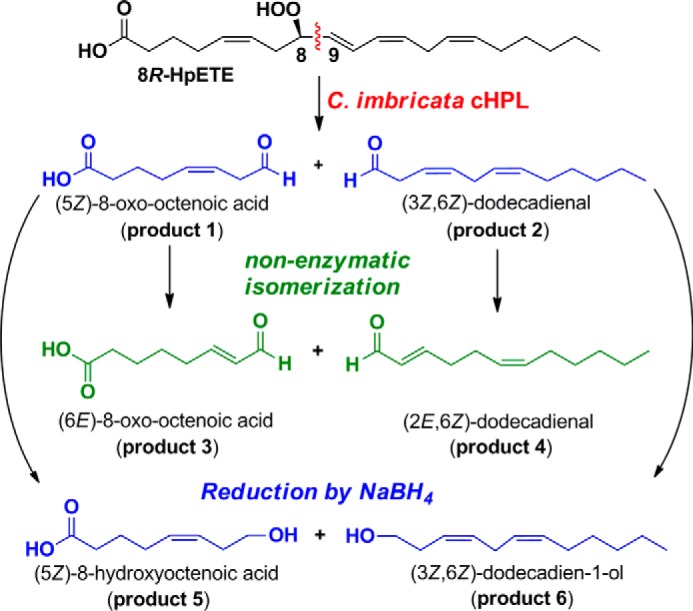
The representation of product formation by cHPL. Primary products (product 1 and 2; blue) were identified as alcohol derivatives (product 5 and 6; blue). The secondary products (isomerized; product 3 and 4) are presented as green.
Isotopic Analyses of Products Formed by cHPL
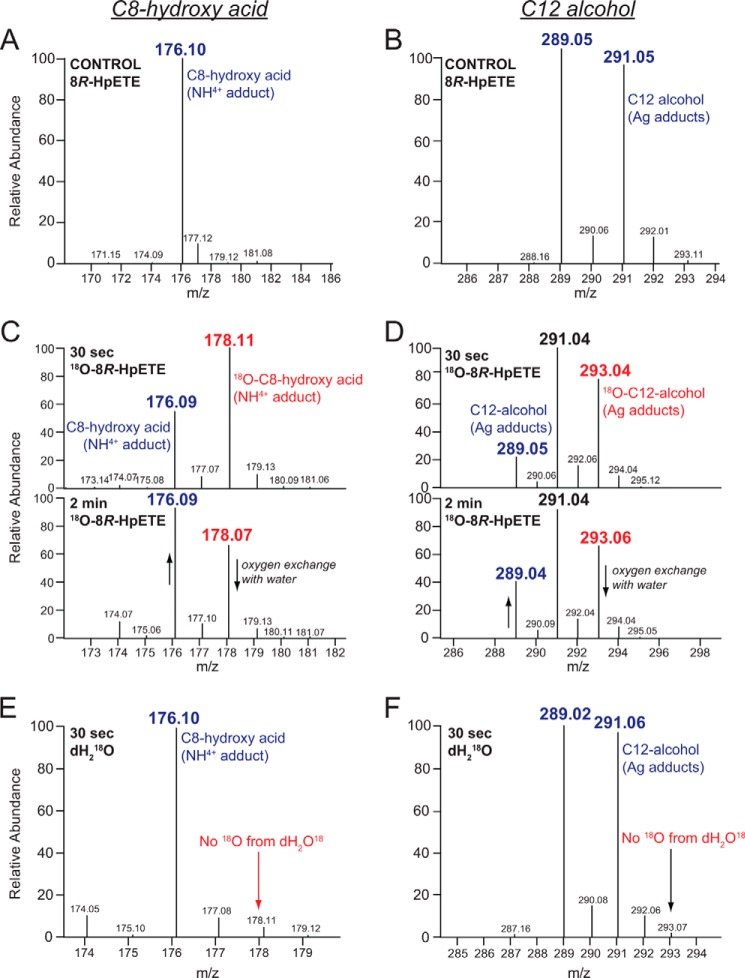
Incubations with 18O2-labeled 8R-HpETE were conducted to define the oxygen incorporation in products and describe the reaction mechanism of cHPL. As aldehydes readily exchange oxygen with water, 30-s incubations with 18O2-labeled 8R-HpETE and cHPL were conducted on ice, and the primary aldehydes were analyzed after in situ reduction with NaBH4 to the corresponding alcohols. The incorporation of 18O in the C8-hydroxy acid and C12-alcohol was determined by LC-MS analyses using a silver coordination methodology that produces ionization of any alkene-containing molecule (20). The two isotopes of silver (107 and 109) were almost equally represented, giving doublets of ions for the silver adducts of (5Z)-8-hydroxy acid and (3Z,6Z)-dodecadien-1-ol at 265/267 and 289/291, respectively, and 267/269 and 291/293 for the corresponding 18O-labeled ions. Additional ions were observed for the ammonium adducts ([M+18]+) with the carboxyl of the C8-hydroxy acid with predicted m/z for the unlabeled and 18O-labeled species of 176 and 178, respectively.
LC-MS analyses revealed that incubations with 18O-labeled 8R-HpETE resulted in the formation of 18O-incorporated C8-hydroxy acid (having m/z of 267/269, respective to silver adducts, and m/z of 178, respective to the NH4+ adduct) (Fig. 4C) and C12-alcohol (m/z of 291/293 respective to silver adducts) (Fig. 5D). The C8-hydroxy acid and C12-alcohol formed in a H218O-containing buffer (Fig. 5, E and F, respectively) did not contain any oxygen-18 isotope and were identical to the control incubations (Fig. 5, A and B, respectively). The altered ratios of 16O and 18O in C8-hydroxy acid and C12-alcohol after longer incubation (2 min instead of 30 s) illustrate the time dependence in oxygen exchange between the aldehydes and water (Figs. 5C and 4D). These results establish that the hydroperoxy oxygens of 8R-HpETE are retained in the two aldehydic fragments, a mechanism consistent with the conversion of 8R-HpETE to a hemiacetal derivative that spontaneously breaks down to the two-chain cleavage products (21).
FIGURE 5.
LC-MS analysis of 18O incorporation in the NaBH4-reduced products of C. imbricata cHPL. The left panels illustrate the NH4+ adduct ions of the C8-hydroxy acid (panels A, C, and E) and the right panels show the doublet of Ag+ adduct ions of the C12 alcohol (panels B, D, and F). A and B, the ion profiles from 30-s incubation of cHPL with unlabeled 8R-HpETE. C and D, after incubation of cHPL with [18O2]8R-HpETE for 30 s or 2 min, illustrating 18O incorporation and the partial loss of 18O in the 2 min incubation. E and F, the ion profiles from the incubation of cHPL with unlabeled 8R-HpETE in the presence of 18O-labeled water. For clarity, only NH4+ adducts of the C8-hydroxy acid are shown. The 16O- and 18O-containing products are presented as blue and red, respectively.
Discussion
Currently, although individual lipoxygenases have been described in corals (22–25), only 8R-LOX seems to be associated with a catalase-related/LOX fusion protein (9, 10, 16). The structural analysis of the P. homomalla cAOS domain of the cAOS-LOX fusion protein revealed that conservation of the catalase core structure (14) and the most significant catalase-related residues are also present in cHPL, thus expanding the diversity of heme-containing catalase-related enzymes in the fatty acid hydroperoxide metabolism. Illustrating the independence of both domains, the separately expressed P. homomalla AOS-LOX domains each retained activity (11), and a similar functional independence of the two domains of the cHPL-LOX was shown here. The covalent linkage of cHPL with 8R-LOX probably helps to target the fusion protein into membranes, assuring the close proximity of the component domains to the newly released arachidonic acid substrate, as demonstrated with cAOS-LOX (26).
We determined the catalytic efficiency (kcat/Km) of the C. imbricata cHPL as 35 μm−1 s−1, which is similar to the P. homomalla cAOS, 31 μm−1 s−1 (11). The catalytic efficiency of the plant cytochrome P450 HPL (CYP74) can be highly variable, depending on the inclusion of detergents. For example, for a Medicago truncatula CYP74 HPL with its optimal substrate 13S-hydroperoxy-C18:3ω3, the kcat/Km varied from 3 to 151 μm−1 s−1 in the presence and absence of detergent, respectively (27). Although the enzymatic efficiency of HPLs are comparable, the turnover numbers reported for the CYP74 HPLs are an order of magnitude higher than for the C. imbricata cHPL (28–31).
Reaction Mechanism of cHPL
Our evidence indicates that the coral cHPL catalyzes the C8-oxo acid and C12-aldehyde formation from 8R-HpETE in a route precedented for cytochrome P450 HPL enzymes. It is generally agreed that plant P450 CYP74 family enzymes homolytically cleave the fatty acid hydroperoxide moiety to form an epoxyallylic carbon radical intermediate from which a number of biosynthetic outcomes are known (3). These include allene oxide synthase and divinyl ether synthase transformations (2, 3, 32). In the third possibility, the HPL route, the rearrangement leads to a covalently complete yet unstable final product, a full-length fatty acid hemiacetal, which through spontaneous decomposition gives the two short-chain aldehydic products (21, 33). The equivalent pathway for the coral cHPL is illustrated in Scheme 2. The ferric cHPL enzyme catalyzes a homolytic cleavage of the 8R-hydroperoxy group with oxidation of the heme to Compound II (Fe4+-OH). The fatty acid alkoxyl radical cyclizes to an epoxy allylic radical. At this point the steps promoted by cAOS or cHPL diverge. cAOS stabilizes the carbocation formation and catalyzes the hydrogen abstraction, which results in the formation of allene oxide (Scheme 2, left side). cHPL stabilizes the epoxy allylic radical and initiates the cleavage of C8-C9-bond in the epoxy group, which results in an allylic ether radical formation. The oxygen rebound from Compound II gives rise to a hemiacetal intermediate. Our isotopic analysis and results by Grechkin and co-workers (21, 33) clearly demonstrate that water is not involved in this process and that both of the original hydroperoxide oxygens are retained in the products. Notably, the chain cleavage in the P450 and cHPL type of fatty acid hydroperoxide lyase occurs within the original pentadiene of the fatty acid hydroperoxide, between C8 and C9 in the case of coral cHPL.
SCHEME 2.
The cAOS (left) and cHPL (right) pathways in soft coral C. imbricata.
Technical Challenges in the HPLC Analysis of 3Z-Aldehydes
In our study we observed exceptionally adverse chromatographic behavior of the two primary cHPL products, each containing an aldehydic moiety with a 3Z double bond. On RP-HPLC each primary product ran as an abnormal broad band rather than as a distinct peak (Fig. 3), and poor performance was observed also on SP-HPLC (Fig. 3B). Although we could attribute this anomalous performance partly to isomerization to the conjugated enal (the corresponding 2E-aldehydic isomer), our impression is that this is not the only issue. The broad bands on RP-HPLC do not show the characteristic enal chromophore of the 2E-aldehydes, suggesting that some adverse interaction of the 3Z-aldehydes occurs with the HPLC solvent and stationary phase. One of us has prior experience with the HPLC of an authentic sample of a plant CYP74B lyase product (3Z-nonenal) (31), and the same phenomenon was evident, suggesting it is general to the 3Z-aldehydes. As volatile molecules, 3Z-aldehydes are commonly detected among short-chain fragments of fatty acids, and there is very substantial literature on their analysis. However, the chromatographic analyses are typically restricted to gas chromatography in which the chromatographic behavior is quite normal. To the best of our knowledge the adverse HPLC characteristics of the 3Z-aldehydes have not been generally recognized or reported.
A Contrasting Route to Fatty Acid Chain Cleavage
The coral cHPL is unique in the animal kingdom. The mechanism and products of transformation are distinct from unrelated enzymatic and non-enzymatic lyase reactions that produce chain cleavage between the hydroperoxide-bearing carbon and the CH2 moiety in the α position outside the original pentadiene. To give an example of the latter involving the identical fatty acid hydroperoxide substrate, 8R-HpETE, whereas cHPL gives rise to C8 and C12 aldehydes, there is an uncharacterized lyase activity in starfish oocytes that cleaves the carbon chain between C7 and C8, producing a C7-aldehyde acid and C13 aldehyde (34). Non-enzymatic transformation by free heme favors a similar pathway (e.g. Refs. 35 and 36). This alternative type of lyase reaction is known to be initiated by LOX enzymes and is facilitated under anaerobic conditions (37, 38). Examples include the reactions of porcine leukocyte 12-LOX (39) and rabbit leukocytes (40) and the lipoxygenase-lyase pathway in the moss Physcomitrella patens (41).
Biological Significance of HPL Metabolism
The AA-derived C8-oxo acid and C12-aldehyde with their isomerized derivatives are potential biologically active signal mediators in corals. In plants, the HPL-derived green leaf volatiles are shown as mediators in abiotic and biotic stress (42). The (3Z)-hexenal and 12-oxo-cis-dodecenoic acid formed by 13-HPL have the capability to isomerize non-enzymatically (43) or by the isomerases (44, 45) to biologically active forms, 2E-hexenal and 12-oxo-trans-dodecenoic acid (also known as traumatin), respectively. Traumatin plays an important role in plant wound healing (46), and 2(E)-hexenal induces the expression of HPL, AOS, LOX (47, 48), and other stress genes in plants (49). Moreover, the electrophilic α,β-unsaturated carbonyl group of aldehydes might interact with nucleophiles in biological systems (50, 51), and in plants, antimicrobial properties of oxylipins including 2(E)-hexenal have been presented by Prost et al. (52).
As mentioned in the Introduction, our previous studies detected what we termed the “unknown polar compounds” in C. imbricata that were identified here as aldehydic products of the cHPL-LOX. These products and the gene expression of the cHPL-LOX were measureable in coral samples under baseline conditions and after wounding (16) and thermal-induced stress (a change in sea water temperature from 23 °C to 28 °C or 31 °C) (53). In contrast to the increased gene expression of the cAOS-LOX fusion protein induced both by wounding and thermal stress, the gene expression of the cHPL-LOX was stable during these test conditions. There are few possible interpretations of this apparent lack of enhancement of the cHPL-LOX activity; (i) in contrast to its plant counterpart, the cHPL may not be involved in a response to mechanical injury, and (ii) given that plant oxylipin synthesis is activated within seconds or a few minutes of wounding (54, 55), a short-lived increase in the cHPL-LOX products may occur immediately after wounding, which has not been analyzed (16, 53). Activation of the plant HPL pathway is associated with changes in the expression of other defense genes (47, 49, 56), and equivalent experiments to explore effects of the coral lyase products are required to elucidate the role of cHPL-LOX pathway.
Author Contributions
T. T., H. L., A. R. B., and N. S. designed the study and wrote the paper. H. L. and T. T. cloned the constructs, and T. T. performed the experiments. MS experiments and analysis were conducted by T. T. and W. M. C. NMR analysis was conducted by T. T. and W. E. B. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-074888 (to A. R. B.). This work was also supported by the Institutional Research Funding IUT19–9 of the Estonian Ministry of Education and Research and the Estonian Science Foundation Grant 9410 (both to N. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- LOX
- lipoxygenase
- AA
- arachidonic acid
- AOS
- allene oxide synthase
- cAOS
- catalase-related AOS
- HPL
- hydroperoxide lyase
- cHPL
- catalase-related HPL
- 8R-H(p)ETE
- 8R-hydro(pero)xyeicosatetraenoic acid
- RP-HPLC
- reversed phase HPLC
- SP-HPLC
- straight phase HPLC.
REFERENCES
- 1. Tanabe T., Ullrich V. (1995) Prostacyclin and thromboxane synthases. J. Lipid Mediat. Cell Signal. 12, 243–255 [DOI] [PubMed] [Google Scholar]
- 2. Grechkin A. N. (2002) Hydroperoxide lyase and divinyl ether synthase. Prostaglandins Other Lipid Mediat. 68, 457–470 [DOI] [PubMed] [Google Scholar]
- 3. Brash A. R. (2009) Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry 70, 1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feussner I., Wasternack C. (2002) The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297 [DOI] [PubMed] [Google Scholar]
- 5. Garscha U., Jernerén F., Chung D., Keller N. P., Hamberg M., Oliw E. H. (2007) Identification of dioxygenases required for Aspergillus development: studies of products, stereochemistry, and the reaction mechanism. J. Biol. Chem. 282, 34707–34718 [DOI] [PubMed] [Google Scholar]
- 6. Hoffmann I., Jernerén F., Oliw E. H. (2013) Expression of fusion proteins of Aspergillus terreus reveals a novel allene oxide synthase. J. Biol. Chem. 288, 11459–11469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brodhun F., Feussner I. (2011) Oxylipins in fungi. FEBS J. 278, 1047–1063 [DOI] [PubMed] [Google Scholar]
- 8. Tsitsigiannis D. I., Keller N. P. (2007) Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15, 109–118 [DOI] [PubMed] [Google Scholar]
- 9. Koljak R., Boutaud O., Shieh B. H., Samel N., Brash A. R. (1997) Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science 277, 1994–1996 [DOI] [PubMed] [Google Scholar]
- 10. Lõhelaid H., Järving R., Valmsen K., Varvas K., Kreen M., Järving I., Samel N. (2008) Identification of a functional allene oxide synthase-lipoxygenase fusion protein in the soft coral Gersemia fruticosa suggests the generality of this pathway in octocorals. Biochim. Biophys. Acta 1780, 315–321 [DOI] [PubMed] [Google Scholar]
- 11. Boutaud O., Brash A. R. (1999) Purification and catalytic activities of the two domains of the allene oxide synthase-lipoxygenase fusion protein of the coral Plexaura homomalla. J. Biol. Chem. 274, 33764–33770 [DOI] [PubMed] [Google Scholar]
- 12. Lee D. S., Nioche P., Hamberg M., Raman C. S. (2008) Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455, 363–368 [DOI] [PubMed] [Google Scholar]
- 13. Li L., Chang Z., Pan Z., Fu Z. Q., Wang X. (2008) Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 105, 13883–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oldham M. L., Brash A. R., Newcomer M. E. (2005) The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc. Natl. Acad. Sci. U.S.A. 102, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tosha T., Uchida T., Brash A. R., Kitagawa T. (2006) On the relationship of coral allene oxide synthase to catalase: a single active site mutation that induces catalase activity in coral allene oxide synthase. J. Biol. Chem. 281, 12610–12617 [DOI] [PubMed] [Google Scholar]
- 16. Lõhelaid H., Teder T., Tõldsepp K., Ekins M., Samel N. (2014) Up-regulated expression of AOS-LOXa and increased eicosanoid synthesis in response to coral wounding. PloS ONE 9, e89215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brash A. R., Niraula N. P., Boeglin W. E., Mashhadi Z. (2014) An ancient relative of cyclooxygenase in cyanobacteria is a linoleate 10S-dioxygenase that works in tandem with a catalase-related protein with specific 10S-hydroperoxide lyase activity. J. Biol. Chem. 289, 13101–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abraham B. D., Sono M., Boutaud O., Shriner A., Dawson J. H., Brash A. R., Gaffney B. J. (2001) Characterization of the coral allene oxide synthase active site with UV-visible absorption, magnetic circular dichroism, and electron paramagnetic resonance spectroscopy: evidence for tyrosinate ligation to the ferric enzyme heme iron. Biochemistry 40, 2251–2259 [DOI] [PubMed] [Google Scholar]
- 19. Bandara D. M., Sono M., Bruce G. S., Brash A. R., Dawson J. H. (2011) Coordination modes of tyrosinate-ligated catalase-type heme enzymes: magnetic circular dichroism studies of Plexaura homomalla allene oxide synthase, Mycobacterium avium ssp. paratuberculosis protein-2744c, and bovine liver catalase in their ferric and ferrous states. J. Inorg. Biochem. 105, 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Havrilla M. C., Hachey D. L. P., N. A. (2000) Coordination (Ag+) ion spray-mass spectrometry of peroxidation products of cholesterol linoleate and cholesterol arachidonate: high-performance liquid chromatography-mass spectrometry analysis of peroxide products from polyunsaturated lipid autoxidation. J. Am. Chem. Soc. 122, 8042–8055 [Google Scholar]
- 21. Grechkin A. N., Hamberg M. (2004) The “heterolytic hydroperoxide lyase” is an isomerase producing a short-lived fatty acid hemiacetal. Biochim. Biophys. Acta 1636, 47–58 [DOI] [PubMed] [Google Scholar]
- 22. Eek P., Järving R., Järving I., Gilbert N. C., Newcomer M. E., Samel N. (2012) Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J. Biol. Chem. 287, 22377–22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mortimer M., Järving R., Brash A. R., Samel N., Järving I. (2006) Identification and characterization of an arachidonate 11R-lipoxygenase. Arch. Biochem. Biophys. 445, 147–155 [DOI] [PubMed] [Google Scholar]
- 24. Järving R., Lõokene A., Kurg R., Siimon L., Järving I., Samel N. (2012) Activation of 11R-lipoxygenase is fully Ca2+-dependent and controlled by the phospholipid composition of the target membrane. Biochemistry 51, 3310–3320 [DOI] [PubMed] [Google Scholar]
- 25. Brash A. R., Boeglin W. E., Chang M. S., Shieh B. H. (1996) Purification and molecular cloning of an 8R-lipoxygenase from the coral Plexaura homomalla reveal the related primary structures of R- and S-lipoxygenases. J. Biol. Chem. 271, 20949–20957 [DOI] [PubMed] [Google Scholar]
- 26. Gilbert N. C., Niebuhr M., Tsuruta H., Bordelon T., Ridderbusch O., Dassey A., Brash A. R., Bartlett S. G., Newcomer M. E. (2008) A covalent linker allows for membrane targeting of an oxylipin biosynthetic complex. Biochemistry 47, 10665–10676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hughes R. K., Yousafzai F. K., Ashton R., Chechetkin I. R., Fairhurst S. A., Hamberg M., Casey R. (2008) Evidence for communality in the primary determinants of CYP74 catalysis and of structural similarities between CYP74 and classical mammalian P450 enzymes. Proteins 72, 1199–1211 [DOI] [PubMed] [Google Scholar]
- 28. Noordermeer M. A., Van Dijken A. J., Smeekens S. C., Veldink G. A., Vliegenthart J. F. (2000) Characterization of three cloned and expressed 13-hydroperoxide lyase isoenzymes from alfalfa with unusual N-terminal sequences and different enzyme kinetics. Eur. J. Biochem. 267, 2473–2482 [DOI] [PubMed] [Google Scholar]
- 29. Matsui K., Ujita C., Fujimoto S., Wilkinson J., Hiatt B., Knauf V., Kajiwara T., Feussner I. (2000) Fatty acid 9- and 13-hydroperoxide lyases from cucumber. FEBS Lett. 481, 183–188 [DOI] [PubMed] [Google Scholar]
- 30. Matsui K., Miyahara C., Wilkinson J., Hiatt B., Knauf V., Kajiwara T. (2000) Fatty acid hydroperoxide lyase in tomato fruits: cloning and properties of a recombinant enzyme expressed in Escherichia coli. Biosci. Biotechnol. Biochem. 64, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 31. Tijet N., Schneider C., Muller B. L., Brash A. R. (2001) Biogenesis of volatile aldehydes from fatty acid hydroperoxides: molecular cloning of a hydroperoxide lyase (CYP74C) with specificity for both the 9- and 13-hydroperoxides of linoleic and linolenic acids. Arch. Biochem. Biophys. 386, 281–289 [DOI] [PubMed] [Google Scholar]
- 32. Hamberg M. (1989) Fatty acid allene oxides. J. Am. Oil Chem. Soc. 66, 1445–1449 [Google Scholar]
- 33. Grechkin A. N., Brühlmann F., Mukhtarova L. S., Gogolev Y. V., Hamberg M. (2006) Hydroperoxide lyases (CYP74C and CYP74B) catalyze the homolytic isomerization of fatty acid hydroperoxides into hemiacetals. Biochim. Biophys. Acta 1761, 1419–1428 [DOI] [PubMed] [Google Scholar]
- 34. Brash A. R., Hughes M. A., Hawkins D. J., Boeglin W. E., Song W. C., Meijer L. (1991) Allene oxide and aldehyde biosynthesis in starfish oocytes. J. Biol. Chem. 266, 22926–22931 [PubMed] [Google Scholar]
- 35. Gardner H. W. (1989) Oxygen radical chemistry of polyunsaturated fatty acids. Free Radic. Biol. Med. 7, 65–86 [DOI] [PubMed] [Google Scholar]
- 36. Labeque R., Marnett L. J. (1988) Reaction of hematin with allylic fatty acid hydroperoxides: identification of products and implications for pathways of hydroperoxide-dependent epoxidation of 7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene. Biochemistry 27, 7060–7070 [DOI] [PubMed] [Google Scholar]
- 37. Garssen G. J., Vliegenthart J. F., Boldingh J. (1971) An anaerobic reaction between lipoxygenase, linoleic acid, and its hydroperoxides. Biochem. J. 122, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garssen G. J., Vliegenthart J. F., Boldingh J. (1972) The origin and structures of dimeric fatty acids from the anaerobic reaction between soya-bean lipoxygenase, linoleic acid, and its hydroperoxide. Biochem. J. 130, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Glasgow W. C., Harris T. M., Brash A. R. (1986) A short-chain aldehyde is a major lipoxygenase product in arachidonic acid-stimulated porcine leukocytes. J. Biol. Chem. 261, 200–204 [PubMed] [Google Scholar]
- 40. Lam B. K., Linh Y. L., Ho H. Y., Wong P. Y. (1987) Hydroperoxide lyase in rabbit leukocytes: conversion of 15-hydroperoxyeicosatetraenoic acid to 15-keto-pentadeca-5,8,11,13-tetraenoic acid. Biochem. Biophys. Res. Commun. 149, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 41. Wichard T., Göbel C., Feussner I., Pohnert G. (2004) Unprecedented lipoxygenase/hydroperoxide lyase pathways in the moss Physcomitrella patens. Angew. Chem. Int. Ed. Engl. 44, 158–161 [DOI] [PubMed] [Google Scholar]
- 42. Matsui K. (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 9, 274–280 [DOI] [PubMed] [Google Scholar]
- 43. Noordermeer M. A., Van Der Goot W., Van Kooij A. J., Veldsink J. W., Veldink G. A., Vliegenthart J. F. (2002) Development of a biocatalytic process for the production of C6-aldehydes from vegetable oils by soybean lipoxygenase and recombinant hydroperoxide lyase. J. Agric. Food Chem. 50, 4270–4274 [DOI] [PubMed] [Google Scholar]
- 44. Noordermeer M. A., Veldink G. A., Vliegenthart J. F. (1999) Alfalfa contains substantial 9-hydroperoxide lyase activity and a 3Z:2E-enal isomerase. FEBS Lett. 443, 201–204 [DOI] [PubMed] [Google Scholar]
- 45. Takamura H., Gardner H. W. (1996) Oxygenation of (3Z)-alkenal to (2E)-4-hydroxy-2-alkenal in soybean seed (Glycine max L.). Biochim. Biophys. Acta 1303, 83–91 [DOI] [PubMed] [Google Scholar]
- 46. Zimmerman D. C., Coudron C. A. (1979) Identification of traumatin, a wound hormone, as 12-oxo-trans-10-dodecenoic acid. Plant Physiol. 63, 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bate N. J., Rothstein S. J. (1998) C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 16, 561–569 [DOI] [PubMed] [Google Scholar]
- 48. Gomi K., Yamasaki Y., Yamamoto H., Akimitsu K. (2003) Characterization of a hydroperoxide lyase gene and effect of C6-volatiles on expression of genes of the oxylipin metabolism in citrus. J. Plant Physiol. 160, 1219–1231 [DOI] [PubMed] [Google Scholar]
- 49. Kishimoto K., Matsui K., Ozawa R., Takabayashi J. (2005) Volatile C6-aldehydes and Allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 46, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 50. Farmer E. E., Davoine C. (2007) Reactive electrophile species. Curr. Opin. Plant Biol. 10, 380–386 [DOI] [PubMed] [Google Scholar]
- 51. Mueller M. J., Berger S. (2009) Reactive electrophilic oxylipins: pattern recognition and signalling. Phytochemistry 70, 1511–1521 [DOI] [PubMed] [Google Scholar]
- 52. Prost I., Dhondt S., Rothe G., Vicente J., Rodriguez M. J., Kift N., Carbonne F., Griffiths G., Esquerré-Tugayé M. T., Rosahl S., Castresana C., Hamberg M., Fournier J. (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139, 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lõhelaid H., Teder T., Samel N. (2015) Lipoxygenase-allene oxide synthase pathway in octocoral thermal stress response. Coral Reefs 34, 143–154 [Google Scholar]
- 54. D'Auria J. C., Pichersky E., Schaub A., Hansel A., Gershenzon J. (2007) Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 49, 194–207 [DOI] [PubMed] [Google Scholar]
- 55. Glauser G., Dubugnon L., Mousavi S. A., Rudaz S., Wolfender J. L., Farmer E. E. (2009) Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J. Biol. Chem. 284, 34506–34513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bate N. J., Sivasankar S., Moxon C., Riley J. M., Thompson J. E., Rothstein S. J. (1998) Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol. 117, 1393–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]