Background: The E2s domain of hepatitis E virus (HEV) capsid protein is the major target for antibody response.
Results: Six antigenic sites of the E2s domain were identified by constructing, clustering, and characterizing a tool box containing representative anti-HEV monoclonal antibodies.
Conclusion: The comprehensive functional epitopes of E2s domain were identified.
Significance: This study provided a novel method for the comprehensive characterization of conformational antigenic domains.
Keywords: antigen, hepatitis virus, monoclonal antibody, protein structure, vaccine development, clustering analysis, conformational mAbs, E2s domain, SPSS, tool box
Abstract
The hepatitis E virus (HEV) ORF2 encodes a single structural capsid protein. The E2s domain (amino acids 459–606) of the capsid protein has been identified as the major immune target. All identified neutralizing epitopes are located on this domain; however, a comprehensive characterization of antigenic sites on the domain is lacking due to its high degree of conformation dependence. Here, we used the statistical software SPSS to analyze cELISA (competitive ELISA) data to classify monoclonal antibodies (mAbs), which recognized conformational epitopes on E2s domain. Using this novel analysis method, we identified various conformational mAbs that recognized the E2s domain. These mAbs were distributed into 6 independent groups, suggesting the presence of at least 6 epitopes. Twelve representative mAbs covering the six groups were selected as a tool box to further map functional antigenic sites on the E2s domain. By combining functional and location information of the 12 representative mAbs, this study provided a complete picture of potential neutralizing epitope regions and immune-dominant determinants on E2s domain. One epitope region is located on top of the E2s domain close to the monomer interface; the other is located on the monomer side of the E2s dimer around the groove zone. Besides, two non-neutralizing epitopes were also identified on E2s domain that did not stimulate neutralizing antibodies. Our results help further the understanding of protective mechanisms induced by the HEV vaccine. Furthermore, the tool box with 12 representative mAbs will be useful for studying the HEV infection process.
Introduction
Hepatitis E virus (HEV)4 is a non-enveloped, single-stranded, positive-sense RNA virus (1–3) that is the causative agent of acute hepatitis E (HE) infection, an emerging disease in many developing countries (4–7). The viral genome is ∼7.2 kb in length (1, 2) and contains three open reading frames (ORFs). ORF1 encodes a non-structural protein that is involved in viral replication and protein processing (8). ORF3 overlaps with the other two ORFs and encodes a small protein that participates in viral evasion of the immune system, capsid assembly, and viral release (9–13). ORF2 exclusively encodes a structural protein that is 660 amino acids in length; with the N-terminal 112 residues responsible for the packaging of the viral RNA genome (14–16). The generation of N-terminal truncated virus-like particles (aa 112–608) have identified 3 definitive domains: the S domain (aa 129–319) forms the viral shell; the M domain (aa 320–455) is associated with the S domain and involves the formation of the 2-, 3-, and 5-fold icosahedral symmetries of the HEV capsid; and the P domain (aa 456–606, equivalent to the E2s domain) forms the protrusions that extend outward from the shell (17–21). Based on the high-resolution crystal structure, the E2s domain adopts a twisted anti-parallel β-barrel-fold and maintains a tight dimeric structure (21, 22).
Previous studies demonstrated that the HEV E2s domain forms tight homodimers, which is necessary for host recognition (23, 24). The E2s domain is also the region that contains the immune-dominant epitopes (20, 21, 23). Moreover, the E2s domain was identified as the minimum peptide capable of inducing HEV-neutralizing antibodies (25). Similar to the outer membrane protrusions on other viral surfaces (26–30), the HEV E2s domain harbors the major neutralizing epitopes for protection (31–35). A series of recombinant proteins containing the E2s domain, which included the bacterially expressed truncated proteins pE2 (aa 394–606) (22, 23, 36) and p239 (aa 368–606) (37) and the baculovirus expression system expressed T = 1 virus-like particle (21), protected non-human primates and humans efficiently against HEV infection and liver injury (32, 34). Among the truncated proteins, p239 was successfully used in the only approved HEV vaccine (32).
Thus, studies of the antigenic sites on the E2s domain are necessary to understand the host antibody response to HEV and the molecular mechanisms of HEV infection. Although several epitopes on the E2s domain were identified by various teams using mAbs (25, 33, 37–40), a map of epitopes remains incomplete given the lack of a panel of representative mAbs that represents all potential conformational epitopes. Moreover, a simple but reliable method is required to identify these epitopes. Here, a comprehensive map of epitopes on the E2s domain was generated based on a novel efficient method for clustering the mAb-recognizing conformational epitopes. We used a tool box with representative mAbs selected from a large panel of 96 mAbs targeting the HEV capsid protein. This study provided new data on the function of the HEV E2s domain that will increase our understanding of the mechanisms of HEV vaccine-induced protection.
Experimental Procedures
Cloning, Expression, and Purification of p239 and Mutant Antigens
The gene encoding p239 was described previously (36). Genes for mutant antigens with the surface amino acids replaced by alanine (Ala) were prepared by site-directed mutagenesis using polymerase chain reaction (PCR). Then, the genes were cloned into the pTO-T7 plasmid and transformed into the Escherichia coli ER2566 strain (Invitrogen). The transformant was cultured in LB medium at 37 °C for 4 h and then incubated for an additional 4 h in the presence of 0.2 mm isopropyl thio-β-d-galactoside. The cells were lysed by sonication in the presence of 2% Triton X-100. The sonicate was allowed to stand at 4 °C for 30 min and then centrifuged at 12,000 rpm for 10 min. Then, the precipitant was washed once with 0.2% Triton X-100 and twice with buffer I (200 mm Tris-HCl, pH 8.5, 5 mm EDTA, and 100 mm NaCl). Each wash was followed by centrifugation at 12,000 rpm for 10 min. The pellet was resuspended in 4 m urea buffer (200 mm Tris-HCl, pH 8.5, 5 mm EDTA, 100 mm NaCl, and 4 m urea), allowed to stand for 30 min at room temperature, and centrifuged at 12,000 rpm for 10 min. The supernatant was dialyzed against PBS (pH 7.4) overnight and centrifuged at 12,000 rpm for 10 min. Target proteins (p239 and mutants) were present in the supernatants. The proteins were then purified and characterized according to methods previously described (36, 41).
Antibodies
mAbs were raised against p239 antigens using a standard murine mAb preparation protocol (37).
Indirect ELISA
An indirect enzyme-linked immunosorbent assay (ELISA) was developed to detect the reactivity of HEV antibodies, including mAbs and sera obtained from mice or humans. Briefly, microwell plates were coated at 37 °C for 3 h with 100 μl of each of the purified recombinant antigens at a concentration of 1 μg/ml in carbonate-bicarbonate buffer (pH 9.6). The wells were blocked with 0.5% (w/v) casein in phosphate-buffered saline (PBS) at 37 °C for 2 h, washed, and dried. Antibodies diluted in PBS were added to the plates and incubated at 37 °C for 30 min. After 5 rinses, HRP-conjugated anti-mouse or anti-human IgG Fab antibodies diluted 5000-fold in enzyme dilution buffer were added to detect the bound antibodies. After incubation at 37 °C for 30 min, the plates were washed as described above, and 100 μl of tetramethylbenzidine substrate solution was added to the wells. The reaction was stopped by adding 50 μl of 2 m H2SO4 after incubation at 37 °C for 15 min, and the absorbance was measured at 450 nm with a reference wavelength of 620 nm.
Western Blot
The recombinant p239 proteins with or without boiling were loaded onto SDS-PAGE gel, respectively, and subsequently electroblotted onto nitrocellulose membrane (Whatman). The blot were blocked and reacted with mAbs according to methods previously described (37).
Immune Capture Assay
The plates were coated with 300 μl of mAbs (0.3 μg/ml) diluted in 20 mm phosphate buffer (16.2 mm Na2HPO4, 3.8 mm NaH2PO4, pH 7.4) at 37 °C for 2 h. Then, the plates were washed once with PBST (PBS with 0.05% Tween 20). The plates were subsequently incubated with 350 μl of blocking reagent (PBS containing 2% BSA) at 37 °C for 2 h and washed. A total of 200 μl of HEV suspension was added to the antibody-coated wells and incubated at 37 °C for 2 h. The wells were rinsed five times. Then, 200 μl of TRIzol was added into each well and incubated at 4 °C for 10 min. HEV RNA was extracted, and the viral RNA titers captured by the mAbs were determined with a real-time reverse transcription (RT)-PCR assay.
Real-time PCR
HEV RNA was purified from 50 μl of sample and then quantified using a GenMagScript One-Step RT-PCR Kit (Genmagbio, Beijing, China). The 25-μl reaction mixture contained 12.5 μl of 2× One-Step RT-PCR reaction buffer, 0.2 μl of GenMagScript RT Enzyme Mix, 5 μl of viral RNA templates, and 200 nm of primers (forward primer: JVHEVF (5′-GGTGGTTTCTGGGGTGAC-3′) and reverse primer: JVHEVR (5′-AGGGGTTGGTTGGATGAA-3′)) and probes (JVHEVP; 5′-TGATTCTCAGCCCTT CGC-3′). The CFX96TM Real-time System and C1000TM thermal cycler device (Bio-Rad Inc.) were used for all real-time RT-PCR. The reverse transcription reaction was performed at 55 °C for 15 min followed by denaturation at 95 °C for 15 s. The DNA was amplified with 40 PCR cycles of 95 °C (15 s), 53 °C (45 s), and 72 °C (15 s). Real-time RT-PCR data were collected after the reactions were complete, and threshold cycle (Ct) values were calculated by the CFX manager software. For the generation of standard curves, the Ct values were plotted as a function of the input HEV viral RNA copies that were determined by comparison with concentrations of plasmid standards.
Competitive ELISA (cELISA)
The spatial configuration of epitopes recognized by each of the mAbs were investigated using a “cross-blocking ELISA” experiment. Briefly, 96-well microplates were coated with 100 ng/well of p239 antigen and then incubated with a saturating amount of unlabeled mAbs (50 μg/well). Next, HRP-conjugated mAbs were added at selected dilutions (in PBS containing 20% BSA) that resulted in optical density (OD) readings of ∼1.5 in the indirect ELISA. Samples of unlabeled mAbs were added to the p239-coated microplate and incubated for 30 min at 37 °C. Then, 50 μl of the mAb conjugates were added at the desired dilutions and incubated for an additional 30 min at 37 °C. The microplates were rinsed and developed with tetramethylbenzidine substrate as described above. The reaction was stopped by adding 50 μl of 2 m H2SO4, and the OD was measured at 450 nm against 620 nm. The blocking efficiency was assessed quantitatively by comparing OD measurements obtained with mAb conjugates in the presence or absence of competitor antibodies (ODinhibited/ODoriginal, wherein ODoriginal refers to the OD obtained without any competitor antibodies and ODinhibited refers to the OD values obtained in the presence of competing antibodies).
Cluster Analysis of the Cross-blocking Data by SPSS
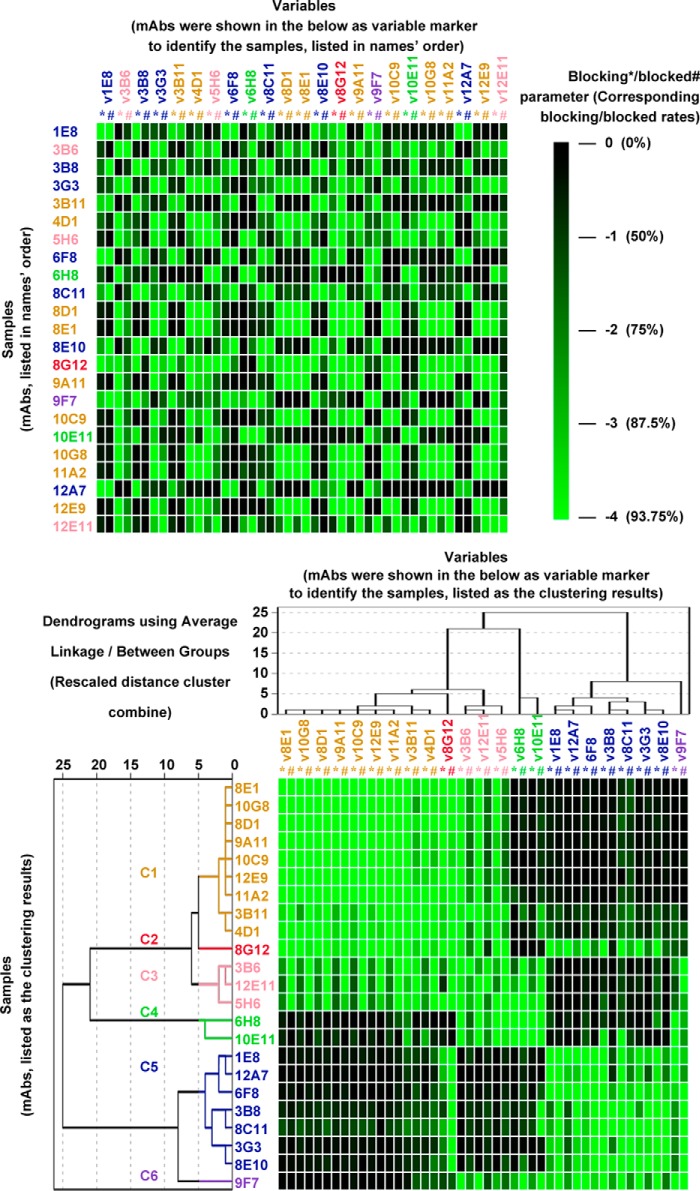
All cELISA data were processed and transformed using the formula log2 (ODinhibited/ODoriginal) before performing clustering analysis with SPSS. Blocking values of one mAb were used to describe the ability of the mAb to block the binding of the other 23 mAbs, including itself. The blocked values represent the extent to which the binding of one mAb could be inhibited by all other mAbs. As shown in Fig. 2, blocking (*) and blocked (#) parameters of each mAb are expressed as continuous values ranging from −4 to 0 (corresponding to R and R′ values ranging from 93.7 to 0%). Values less than −4 were assigned the value of −4. SPSS (Statistical Product and Service Solutions, IBM SPSS statistics 18.0 version) was used for antibody cluster analyses. First, the data to be analyzed were entered using the following parameters. We chose “classify” under the heading of “analysis” followed by “hierarchical cluster analysis.” The antibodies represent the “label cases,” whereas the dates represent the “variables.” We used the “between-group linkage” cluster method and the “cosine” interval method. The cluster results are reported as dendrograms.
FIGURE 2.
SPSS clustering analysis of cELISA data obtained from 23 mAbs directed against the HEV E2s domain. cELISA data from 23 mAbs were processed prior to SPSS analysis. Upper panel, a total of 23 mAbs were used as samples for clustering analysis. Blocking (*) and blocked (#) parameters of each mAb were set as variables. The heat map of cELISA data is shown, with parameters colored continuously from green (−4, corresponding to 93.7% inhibition) to black (0, corresponding to 0% inhibition) according to the scale bar. Samples and variables were arranged in the same order in the heat map. Lower panel, a total of 23 mAbs were classified using SPSS. The dendrogram is shown on the left, with samples and variables re-arranged according to their relative distances. Blocking parameters (*): these values quantitatively indicate the ability of sample mAbs (mAbs shown on the left) to block the binding of variable mAbs in the panel (mAbs shown on the top). Blocked parameters (#): the values quantitatively indicate the extent to which the binding of variable mAbs in the panel (mAbs shown on the top) could be inhibited by sample mAbs (mAbs shown on the left).
P239 Competition Binding Assay
To investigate the competition between selected mAbs and human sera collected after vaccination, a saturating amount of mAbs (50 μg/well) was added to p239-coated plates. Then, we added human serum samples at selected dilutions (in PBS with 20% BSA) that yielded OD readings between 1.0 and 2.0 in the indirect ELISA. Then, 100 μl of HRP-conjugated mouse anti-human antibodies at a 1:5000 dilution (in PBS with 20% BSA) were added and incubated at 37 °C for 2 h. The wells were washed and developed with tetramethylbenzidine as described above. The reactions were stopped by adding 50 μl of 2 m H2SO4. The absorbance was read at 450 nm against a reference wavelength of 620 nm. The blocking rate was calculated as percentage inhibition.
Neutralization Assay
The neutralization activity of selected mAbs against HEV infection was explored using HepG2 cells. HEV was diluted 1:6.5, added to 4-fold serially diluted antibodies in 96-well plates (beginning with a dilution of 1:10), and incubated at 37 °C for 1 h. Antibody concentrations started from 5 μg/μl. After 24 h, the samples were harvested for HEV viral RNA quantification by quantitative polymerase chain reaction. The data were analyzed by non-linear regression (GraphPad Prism, Inc.) to determine the neutralization obtained with a given antibody concentration and the IC50.
Results
Twenty-three mAbs Recognizing Conformational Epitopes Located on the E2s Domain (aa 459–606) Were Obtained from an Anti-HEV Panel Containing 96 mAbs
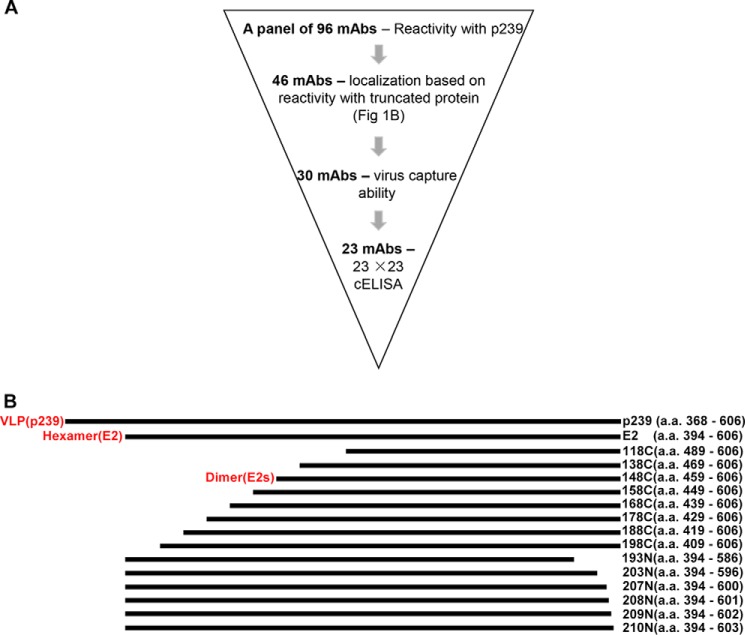
A total of 96 mAbs that recognized the HEV capsid protein were obtained from mice immunized with recombinant truncated ORF2 protein p239. 10-Fold serially diluted mAbs were used to quantitatively determine their reactivities with p239. The results are expressed as EC50 values (ng/ml; a smaller EC50 value is indicative of higher reactivity). A total of 46 mAbs that exhibited significant reactivity with the p239 protein were selected for further analysis. The sorting strategy is summarized in Fig. 1A. The characteristics of the 46 mAbs are summarized in supplemental Table S1.
FIGURE 1.
Generation of a panel of mAbs recognizing the E2s domain of the HEV capsid protein. A, the flow chart describing the sorting procedure for selected mAbs. B, serially truncated versions of pORF2 are represented by bars. The shortest functional dimeric domain was located between residues 459 and 606, which is generally called the E2s domain (equal to 148C). E2, which encodes aa 394–606, primarily exists as a hexamer. p239 self-assembles into virus-like particles (VLP). The serially truncated versions of the E2 protein were constructed by truncating p239 to various lengths either at the N or C terminus. To localize mAb recognition epitopes, the reactivity of mAbs with a set of truncated E2 proteins was determined (supplemental Table S1).
To identify antigenic sites recognized by the 46 mAbs, proteins of various lengths truncated at either the N or C terminus were generated (Fig. 1B). Reactivities between N or C terminally truncated proteins and the 46 mAbs are presented in supplemental Table S1. A total of 16 mAbs recognized linear epitopes; this finding was confirmed using Western blot assays. The remaining 30 mAbs were directed against the E2s domain (aa 459–606), which is essential for HEV-host interactions and disease progression. All 30 mAbs recognized conformation-dependent epitopes. As presented in supplemental Table S1, 23 mAbs were selected based on their capture ability of HEV in vitro, suggesting exposure of the antigenic sites recognized by these mAbs on the surface of the virions.
SPSS Clustering Analyses of cELISA Data Obtained with 23 mAbs Recognizing Conformational Antigenic Sites on the E2s Domain (aa 459–606)
To obtain a comprehensive analysis of conformation-dependent epitopes on the E2s domain, 23 mAbs directed against conformational determinants were selected (Fig. 1). Then pairwise competitive ELISA were performed to improve our understanding of the spatial configuration of the epitopes recognized by these mAbs.
Data obtained from 23 × 23 cELISA included too much information to be analyzed intuitively and accurately using traditional methods (42, 43). Hence, a novel clustering analysis using the statistical software SPSS was established for the classification of mAb epitopes. Distinct from traditional cELISA data analyses, the blocking rates among mAbs were not expressed as percent inhibition but transformed to continuous values ranging from −4 to 0 (corresponding to percent inhibition of 93.7 to 0%, Fig. 2, color scale bar). The antibodies were defined as blocking or blocked parameters to characterize the spatial relationships of one mAb epitope with epitopes recognized by the remaining 22 mAbs. Blocking parameters of one mAb were used to describe the ability of this mAb to block the binding of the 23 mAbs, including itself. Blocked parameters represented the extent to which the binding of one mAb could be blocked by all other mAbs. The 23 mAbs were set as clustered samples, whereas 46 parameters (the blocking and blocked parameters) of each mAb were set as variables for the clustering of these mAbs using SPSS. The results are summarized in Fig. 2.
The heat map in the upper panel of Fig. 2 illustrates the unorganized cELISA data of 23 mAbs without SPSS clustering. Based on this data, it was difficult to categorize cross-reactive mAbs that inhibited one another. In contrast, the heat map in the lower panel shows the same matrix data analyzed by SPSS. The organization of these data allow the sorting of cross-reactive mAbs in a manner that is much more straightforward and accurate, suggesting that this novel method might be efficient for the classification of mAb epitopes. The similarity among mAbs grouped by SPSS clustering analysis was quantitatively evaluated using a dendrogram plot (Fig. 2, lower panel). The distances between the 23 mAbs were calculated and expressed as numbers ranging from 1 to 25. Smaller values indicated closer relationships among the mAb epitopes, and vice versa. Based on the distance values and heat map of 23 mAbs shown in the lower panel of Fig. 2, we noted that mAbs with distance values <5 exhibited similar cELISA heat maps, indicating that the epitopes were recognized by the mAbs with distance values <5. Thus, a rule for classification was created: mAbs with distance values <5 were distributed into the same group. Using this rule, 23 mAbs were sequentially distributed into 6 groups referred to as C1 (9 mAbs, orange), C2 (1 mAb, magenta), C3 (3 mAbs, pink), C4 (2 mAbs, green), C5 (6 mAbs, blue), and C6 (1 mAb, purple). The results suggested that the E2s domain of the HEV capsid protein comprised at least 6 relatively distinct conformation-dependent epitopes.
Selecting a Panel of Representative mAbs That Recognize the HEV E2s Domain
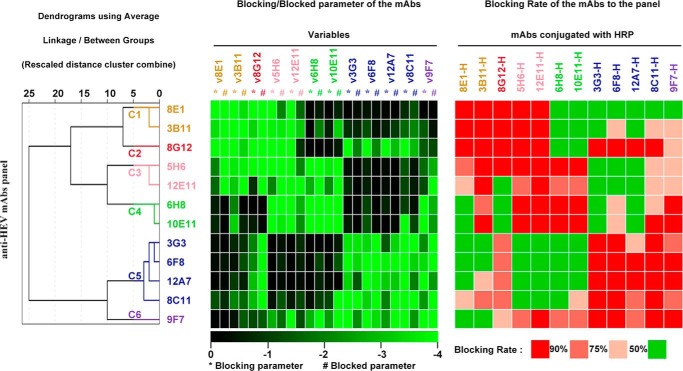
The pattern of cross-inhibition demonstrated that the antibodies that recognized the E2s domain were classified into 6 distinct groups. These groups suggested the presence of 6 independent epitopes on the E2s domain. Further characterization of these mAbs will improve our knowledge of the role that the E2s domain plays in virus-host interactions. However, a panel of 23 mAbs was too complicated for further characterization. We found that the distance values of some of the mAbs in each subgroup were <1 (i.e. 3B11 and 4D1 in group C1) in the clustering tree (Fig. 2, lower panel, the tree on the left). Moreover, mAbs in the same subgroup exhibited highly consistent cELISA profiles, implying that they recognized similar antigenic sites on the E2s domain. A panel of 23 mAbs will provide redundant information because some of them are assigned to the same binding sites. Therefore, representative mAbs with distance values <1 were selected from the subgroups for further study. Finally, 12 representative mAbs were generated as a tool box for further characterization: 8E1 and 3B11 from C1; 8G12 from C2; 5H6 and 12E11 from C3; 6H8 and 10E11 from C4; 3G3, 6F8, 12A7, and 8C11 from C5; and 9F7 from C6 (Fig. 3).
FIGURE 3.
Selecting a panel of representative mAbs recognizing the HEV E2s domain. A total of 12 mAbs were randomly selected from each group and subgroup where applicable. The 12 mAbs were re-classified using SPSS based on their cELISA profiles as described in the legend to Fig. 2. The dendrogram plot generated from the clustering analysis of the 12 mAbs is presented on the left. The heat map of the cELISA profiles of selected mAbs is presented in the middle (blocking and blocked parameters are colored as described in the legend to Fig. 2). For comparison purposes, the heat map of cELISA data processed by traditional analysis is provided on the right. For the traditional analysis, the percentage inhibition was defined as 5 discontinuous levels (R ≥ 90%, 75% ≤ R <90%, 50% ≤ R <75%, R <50%) with different color legends.
The 12 mAbs were re-clustered with SPSS and a dendrogram was generated (Fig. 3, left panel). Consistently, these mAbs were still classified in the same 6 groups, suggesting that they could be used as representative mAbs for further characterization of mAb epitopes on the E2s domain. The novel analysis of the cELISA profiles (Fig. 3, heat map on the left) was compared with the traditional method (Fig. 3, heat map on the right). In the traditional analysis, the cELISA data were expressed as percent inhibition, which is typically defined as 4 discontinuous levels (level 1, <50%; level 2, 50–75%; level 3, 75–90%; and level 4, >90%). However, the use of this classification system leads to the loss of details during the pretreatment of the data. Additionally, the clustering process of the traditional analysis depends on arbitrary judgments of cross-inhibition profiles between each two mAbs. Hence, the use of traditional analysis to cluster dozens of mAbs based on cELISA data is not easy. For example, mAb 12E11, which exhibited a heat map profile similar to both mAb 5H6 from C3 and mAb 6H8 from C4 (Fig. 3, heat map on the right), was distributed into the same group with both the C3 and C4 mAbs using the traditional analysis method. Thus, mAbs from the C3 and C4 groups were easily included into the same group with the traditional method. However, 12E11 was clearly classified as the same group with 5H6 of group C3 based on the novel analysis method (Fig. 3, left heat map). In this method, continuous values were maintained. The classification was based on a complete analysis of all blocking and blocked parameters of one mAb but not its cross-inhibition with other mAbs of interest. Another example is mAb 9F7. Both novel and traditional methods identified this mAb as a unique group, but it was hard to determine the relationships between this mAb and other groups of mAbs using traditional methods. This situation held for the majority of the mAbs. Therefore, our data supported the finding that the novel classification method with SPSS analysis is an innovative, effective, and reliable method to cluster conformation-dependent mAbs.
Characteristics of the 12 Representative mAbs
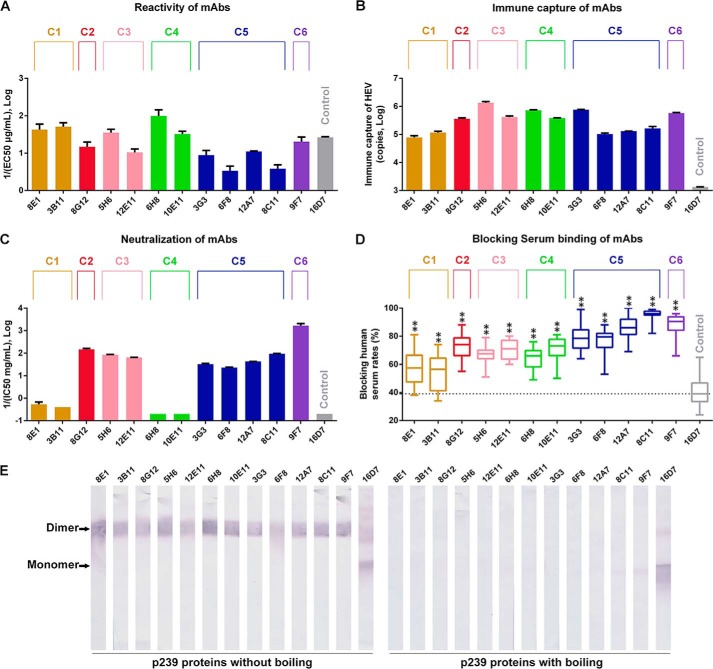
Next, the reactivity of the selected mAbs with the p239 protein was explored by ELISA (Fig. 4A). Despite their different binding affinities, all of the mAbs could bind to the p239 protein. In the Western blot analysis, all selected mAbs could react with the p239 dimer but not with monomer (Fig. 4E). Moreover, all 12 mAbs capture HEV viruses (Fig. 4B), suggesting that these mAbs recognized epitopes that existed on the native HEV capsid protein.
FIGURE 4.
Characterization of 12 typical mAbs. mAb16D7, which recognizes a linear epitope on the HEV capsid protein, was used as the control. A, reactivities of selected mAbs with the p239 protein. Binding affinities were presented as EC50 (μg/ml) values. The reciprocal 1/EC50 values are shown on the y axis, with larger y values representing higher binding affinities. B, HEV viral loads captured by selected mAbs. Viral RNA titers were determined by quantitative PCR. C, neutralization of HEV infection in HepG2 cells by 12 mAbs. Neutralizing efficiencies of 12 mAbs were expressed as IC50 values (mg/ml); the reciprocals (1/IC50) of which are presented on the y axis. Therefore, larger y values represent higher neutralizing activities. D, competition of 12 typical mAbs and 24 human serum samples collected after HEV vaccine inoculation. The blocking efficiencies of 12 mAbs were compared with the blocking efficiency of the control. ** indicates that the increased blocking efficiency of the tested mAb compared with the control is statistically significant. E, analysis of 12 representative mAbs by Western blotting. Samples of p239 with (right) or without boiling (left) were probed with these mAbs. The representative mAbs could react with p239 dimers but not with monomers, suggesting that they were conformation dependent.
The neutralizing capacities of the 12 mAbs were studied in vitro (Fig. 4C) and are reported as IC50 (mg/ml) values. The highest concentration will be designated as the IC50 of an antibody if the first dilution displayed less than or equal to 50% neutralizing activity. The neutralizing abilities of mAbs within the same group were similar to each other. mAbs in C1 (8E1 and 3B11) and C4 (6H8 and 10E11) exhibited minimal inhibition of HEV infection. In contrast, the remaining 8 mAbs in groups C2, C3, C5, and C6 efficiently neutralized HEV infection. mAb 9F7 (C6) was the most efficient neutralizing mAb, with a neutralization efficiency that was ∼10-fold higher than 8G12 (C2), 25-fold higher than the C3 mAbs (5H6 and 12E11), and 50-fold higher than the C5 mAbs (3G3, 6F8, 12A7, and 8C11). These results suggested that antibodies recognizing epitopes similar to the C2, C3, C5, and C6 mAbs efficiently protected humans from HEV.
We also investigated the competition between these mAbs and sera from 24 vaccinated donors to determine whether the epitopes of the 12 mAbs from the 6 groups were similar to those recognized by the vaccinated donors' IgG (Fig. 4D). mAb 16D7, which recognized a linear epitope (aa 428–442 of pORF2) located in the upper portion of the E2s domain (24), was included as a control. All representative mAbs reduced vaccinated sera reactivity by greater than 50%, which was significantly increased compared with the control (p < 0.01). The results further supported the notion that the E2s domain is the major target in the immune response to HEV vaccination or natural infection. Moreover, antibodies recognizing similar epitopes to the 12 representative mAbs existed in human sera after HEV vaccination, suggesting that all 6 of the antigenic sites recognized by the 12 representative mAbs participated in the formation of strong immune dominant epitopes on the E2s domain. The average inhibition activities of mAbs from C2 (72%), C3 (69%), C5 (84%), and C6 (88%) were increased compared with mAbs from C1 (55%) and C4 (67%). When the neutralizing activity of each group was combined, the results suggested that the epitopes recognized by C2, C3, C5, and C6 induced more neutralizing antibodies in sera from patients who received the HEV vaccine. Interestingly, mAb 9F7 from C6 exhibited both the highest neutralization and highest inhibition to sera binding, suggesting that epitopes recognized by C6-like antibodies induce the most efficient protective antibodies in vaccinated donors.
Locations of the 6 Antigenic Sites and an Overview of the Critical Epitopes for Anti-HEV Antibody Responses
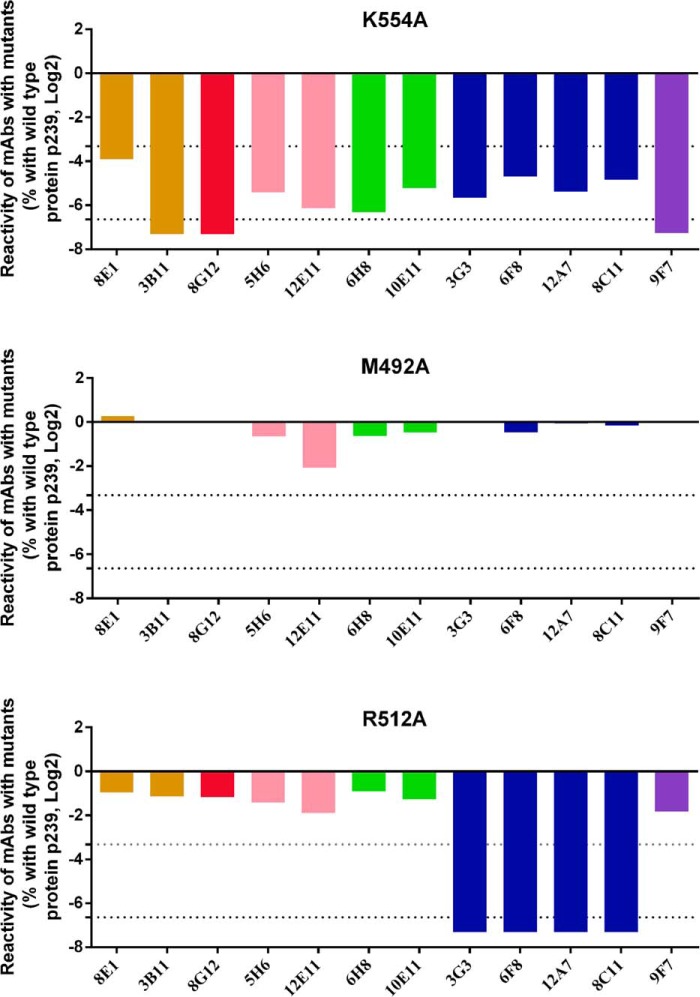
To localize the epitopes recognized by the 12 representative mAbs, the critical binding residues of each mAb were identified using Ala scanning mutagenesis on the surface of the E2s domain. According to the published structure of the E2s domain (Protein Data Bank code 3RKC) (22), 64 non-Ala residues are exposed on the E2s domain surface. Systematically, 64 mutants were generated by replacing the residues of the recombinant p239 protein with Ala. SDS-PAGE was used to verify the dimerization of these mutants. A total of 61 mutants could self-assemble into dimer conformations, whereas three mutants (Y488A, T552A, and L588A) could not form E2s dimers and were excluded from further analysis. Subsequently, the reactions of the 12 representative mAbs with the 61 mutants were investigated using indirect ELISA assays, from which the EC50 of each combination of mAbs and mutants was determined. Native p239 were included as the control. Compared with the control wells, the mutations that resulted in a >10-fold reduction (mean effect) or 100-fold reduction (mean significant effect) in reactivity with the mAbs were selected and defined as critical binding residues for each mAb. Among the 61 mutations, 6 mutations (D481A, D522A, K554A, G589A, P592A, and T563A) that reduced the binding of mAbs from at least 4 groups by >10-fold, such as the K554A mutant presented in the upper panel of Fig. 5, were identified as being involved in large structural rearrangements of the E2s domain and were excluded from further analysis. A total of 27 of the 61 mutations (i.e. M492A shown in Fig. 5, middle panel) failed to abrogate the binding of mAbs from any group. The remaining 28 residues (i.e. R512A shown in Fig. 5, lower panel), which abrogated the binding of mAbs from less than or equal to 2 groups, were identified as the specific residues for mAb binding. The specific residues for each mAb were selected as the key binding residues for localization of the epitopes recognized by the corresponding mAb.
FIGURE 5.
Reactivity of 12 mAbs with three representative alanine scanning mutants: R512A (upper panel), M492A (middle panel), and K554A (lower panel). The reactivity of the mAbs with proteins (including mutants and native p239) were determined by indirect ELISA and are expressed as the EC50 of each combination of mAbs and proteins. The influence that one mutation exerted on the reactivity of the selected mAbs was evaluated by the influence-index defined by the formula: log2 (EC50, p239/EC50, mutant). Compared with the native p239 protein, 6 mutations (i.e. K554A) reduced the reactivities of mAbs from more than 4 groups. A total of 27 mutations (i.e. M492A) did not abrogate the binding of mAbs from each group. A total of 28 mutations (i.e. R512A) abrogated the binding of mAbs from less than or equal to 2 groups.
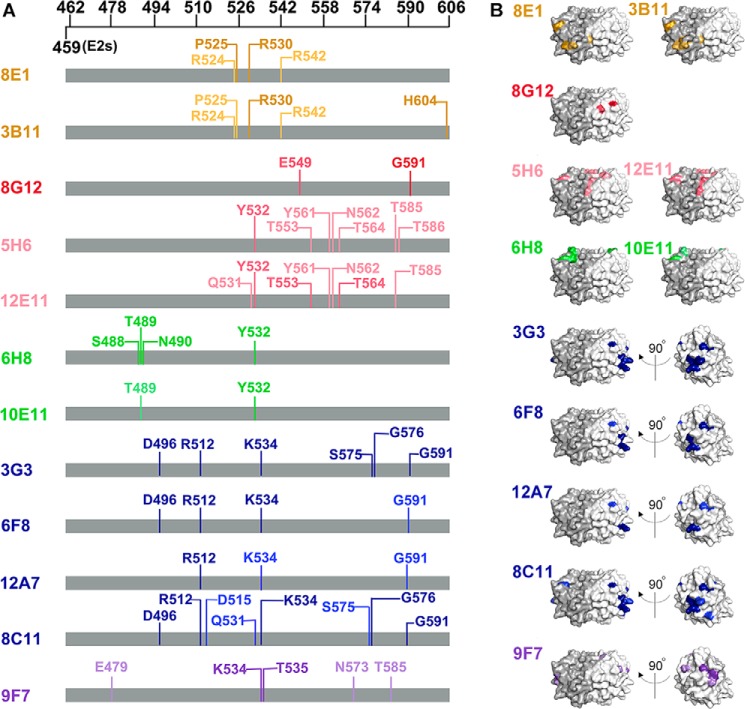
Next, key binding residues of each mAb were marked on the sequence of the E2s domain (Fig. 6A). The key binding residues that resulted in a >10-fold reduction in mAb binding are indicated with a light color, and those that resulted in a >100-fold reduction in mAb binding are denoted with a dark color. As expected, key binding residues of mAbs from the same group were similar to each other. In contrast, mAbs from different groups recognized epitopes comprising different residues. The binding sites of each mAb were composed of discontinuous residues spanning large regions on the E2s domain, confirming that the epitopes recognized by the 12 mAbs were conformation dependent. However, whether these discontinuous residues were close enough to generate an epitope on the E2s tertiary structure remains unclear. To visualize the positions of the essential binding residues of each mAb, corresponding substituted residues were indicated on the structure of the E2s domain using the same color code for each mAb (Fig. 6B). Indeed, we found that the identified residues for each mAb were sufficiently close to form an epitope on the surface of the E2s domain. Moreover, observations of the key binding residues on the E2s domain structure were consistent with those on the sequence of p239; epitopes recognized by mAbs from the same group were similar, whereas those recognized by mAbs from different groups were distinct or in one case partially overlapped.
FIGURE 6.
Identification of key epitope residues of 12 representative mAbs directed against the E2s domain by alanine scanning mutagenesis. Activities of selected mAbs with mutants were determined by indirect ELISA. Mutations that could reduce the reactivity of one mAb by at least 10-fold compared with the native p239 protein were defined as key binding residues for this antibody. A, key binding amino acids of each mAb were mapped onto the E2s sequence using light and dark colors depending on the levels of activity reduction (light, 10 to 100-fold reduction; dark, >100-fold reduction). B, key binding sites of each mAb were mapped onto the structure of the E2s domain. Epitopes of mAbs from different groups are indicated in different colors: C1 (3B11 and 8E1, orange), C2 (8G12, red), C3 (5H6 and 12E11, pink), C4 (6H8 and 10E11, green), C5 (3G3, 6F8, 12A7, and 8C11, blue), and C6 (9F7, purple). Similarly, mutations that reduced the reactivity 10- to 100-fold are depicted in light colors, whereas mutations that reduced the reactivity >100-fold are indicated by dark colors.
Therefore, we conclude that the 6 groups of mAbs corresponded to 6 distinct antigenic sites (Fig. 7A). C1 epitopes recognized by mAbs from the C1 group are depicted in orange and are located at the bottom of the E2s dimer. C2 epitopes recognized by the C2 group mAb were located around the E2s dimerization interface (red). C3 epitopes recognized by the C3 group mAbs were localized on the top of the E2s domain and are located at the interface of the two monomers (pink). C4 epitopes recognized by the C4 group mAbs interacted with residues on the top of the E2s dimer (green). C5 epitopes recognized by the C5 group mAbs were located in the groove zone (the zone has been described by Tang et al. (22)) on the E2s dimer (blue), which was consistent with published crystallographic data. C6 epitopes recognized by the C6 group mAb were located around the groove zone and partially overlapped with antigenic sites recognized by C5 and C3 mAbs (purple).
FIGURE 7.
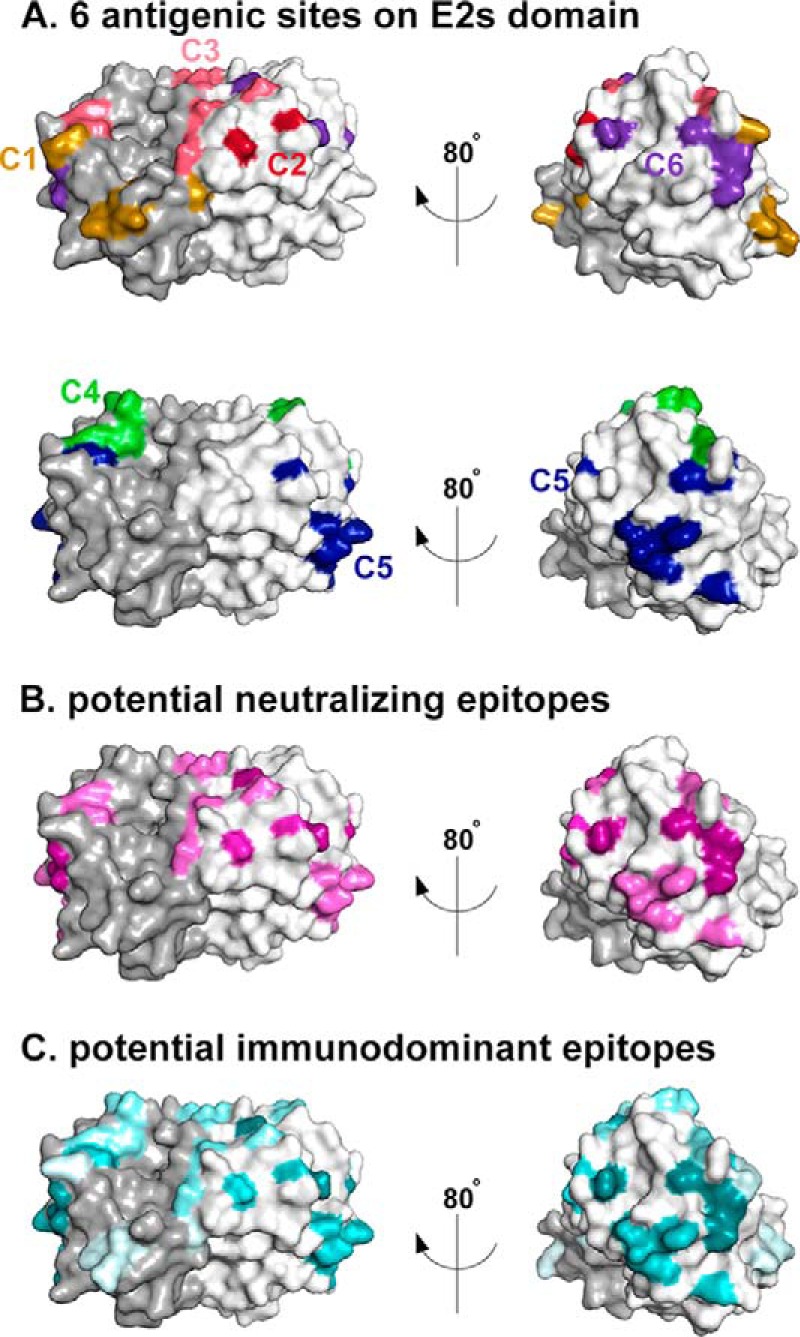
Six potential conformational and functional antigenic sites on the E2s domain identified using representative mAbs. A, in the upper panel, the antigenic sites recognized by mAbs in the C1, C2, C3, and C6 groups are marked on the structure of the E2s domain with the same color codes used in Fig. 6: C1 (orange), C2 (red), C3 (pink), and C6 (purple). In the lower panel, the antigenic sites recognized by mAbs from the C4 and C5 groups are highlighted on the structure of the E2s domain with the same color codes used in Fig. 6: C4 (green) and C5 (blue). B, antigenic sites recognized by neutralizing mAbs in the C2, C3, C5, and C6 groups are marked in magenta based on the neutralizing activity of the corresponding antibodies. The higher the neutralizing activity is, the darker the color. C, antigenic sites recognized by selected mAbs were highlighted in cyan based on the relative abundance of the corresponding antibodies in vaccinated human sera. The darker the color, the more abundant the antibody.
Combining the potential locations of the antigenic sites with the characterization of mAbs from the 6 groups, structure-based functional studies were performed to identify functional antigenic sites on the E2s domain. The key residues of neutralizing epitopes recognized by mAbs from the C6, C2, C3, and C5 groups were colored dark magenta to light magenta according to the average neutralization activity of their corresponding representative mAbs (from high to low) (Fig. 7B). The relatively comprehensive neutralizing antigenic sites were presented on the E2s domain. As presented in Fig. 7B, the majority of the colored residues were located in two regions: the top of the E2s domain located at the interface of the two monomers, including the residues constructing C3 and a portion of the C4 antigenic sites, and on the monomer sides of the E2s domain around the “groove zone,” including the residues constructing the C5 and C6 antigenic sites and a portion of the C2 antigenic site. Additionally, the critical residues of each antigenic site recognized by mAbs from the 6 groups were colored from dark cyan to light cyan based on the blocking ratios of their corresponding mAbs to the vaccinated donors' sera (from high to low) to visualize the major surfaces that participated in stimulating host antibody responses (Fig. 7C). Most of the neutralizing antigenic sites were also denoted with a dark color in Fig. 5C, including the C2, C5, and C6 groups. In particular, the region of antigenic site C6, which was located at the top of the groove zone and in the junction of the other 3 neutralizing antigenic sites, was strongly colored in both Fig. 7, B and C. This result indicated that the residues in this region play an important role in the stimulation of host neutralizing antibody responses during HEV vaccination.
Discussion
The HEV capsid protein encoded by ORF2 is responsible for the immunogenicity of HEV. Therefore, recombinant capsid proteins have been widely used to study HEV immunology given the lack of a reliable cell culture system for HEV. Previous studies have demonstrated that the E2s domain of the capsid protein contains immune-dominant epitopes. Further refinement of the epitope sites in this domain is of great significance because the identification of viral B cell epitopes is important for the development of diagnostic tools and vaccines and increasing our understanding of viral infection and immune responses at the molecular level. Although several crystallographic studies were performed to explore epitope residues, a comprehensive scanning of conformational epitope sites on E2s is still lacking. This study provides new data on the antigenic structure of the HEV E2s domain and suggests the presence of 6 conformation-dependent epitopes.
In general, B cell epitopes are identified and localized using monoclonal antibodies with pepscan analysis, serially truncated recombinant proteins, and competitive ELISA. The first two methods were reported to efficiently determine the locations of mAbs recognizing linear epitopes; however, these methods are not suitable for the localization of conformational epitopes. Competitive ELISA has been used to identify the spatial relationship between conformational mAbs. In this study, we primarily used the first two methods to identify the location of epitopes recognized by 46 mAbs with high reactivity (described in supplemental Table S1) and found that the linear epitopes recognized by 16 mAbs could be accurately divided into four independent peptides located in the range of aa 403–457. The remainder of the conformational mAbs were directed toward unknown epitopes within the E2s domain (aa 459 to 606).
To identify conformation-dependent epitopes recognized by these mAbs, pairwise competitive ELISA were performed because scanning with synthetic peptides or serially truncated recombinant proteins cannot identify discontinuous epitopes. For the first time, we report the development of a novel method using SPSS cluster analysis to analyze cELISA data and identify distinct or overlapping epitopes. Instead of the discontinuous levels of inhibition (<50%, 50 to 75%, 75 to 90%, and >90%) that are typically used in the traditional analysis of cELISA data, percentage inhibitions were translated into continuous parameters ranging from −4 to 0 (corresponding to 93% to 0% inhibition) in our novel analysis. Moreover, all blocking and blocked parameters of one mAb were set as the variable parameters to support the classification of this mAb. This method is more reliable than traditional analyses, wherein conclusions are mainly based on personal judgments of cross-inhibition profiles. Our method is especially advantageous for the analysis of cELISA data obtained from a large number of antibodies. Hence, the novel analysis method established in this study can be widely applied to studies interested in mapping conformational epitopes and studying their spatial configurations.
A total of 23 conformational mAbs targeting the E2s domain were divided into 6 independent groups based on SPSS analysis, suggesting that at least 6 distinct epitopes were present on the E2s domain. A total of 12 of these mAbs were selected from the 6 groups to serve as representative mAbs assigned to the E2s domain. Consistently, these mAbs were also classified into 6 groups by SPSS. Further characterization of these mAbs revealed that mAbs in the same group were present at similar levels in vaccinated human sera, exhibited comparable neutralization activities, and recognized almost the same epitope residues. In contrast, mAbs from different groups were distinct in those aspects, although some of them recognized shared epitope residues. These results clearly support the notion that SPSS cluster analysis of cELISA data is reliable for the classification of conformation-dependent mAbs.
It is noteworthy that mAb 9F7 from C6 was the most efficient neutralizing antibody in our panel. The C6 epitope was located around the groove zone and overlapped the C3 and C5 epitopes. These results revealed the identification of a novel immune-dominant neutralizing epitope that could induce abundant antibodies with high neutralizing activity. This finding will provide insights into the development of antibody-based drugs for the treatment of HEV. Undoubtedly, the antigenic sites identified by solving the crystal structure of Ag-Ab complexes are the most definitive. However, crystallography is not always the best solution for the mapping of conformational epitopes due to its time-consuming nature and sophisticated procedures, especially when exploring multiple epitopes. In this study, we accomplished the mapping of antigenic sites by using 6 groups of mAbs with alanine scanning of key epitope residues. Potential sites that were capable of inducing protective immune responses against HEV were successfully identified in this study. Therefore, representative mAbs directed to these antigenic sites are good candidates for further characterization by crystallography.
The HEV E2s domain (aa 459–606) is a crucial target for research on HEV protective immunology and is well known as the minimum peptide capable of inducing HEV-neutralizing antibodies (25). To date, several neutralizing antibodies were reported to be reactive with the E2s domain (21, 22, 37, 39, 44), but the identification of neutralizing antibodies failed to keep pace with the determination of their recognized epitope. Indeed, only four of these neutralizing antibodies were reported with their potential epitopes (21, 22, 37, 44). Among them, only one epitope, recognized by mAb 8C11, was clearly determined using an Ab-Ag complex crystal structure (22). Additionally, it should be noted that all these studies were based on the use of 1 to 2 mAbs. Thus, a comprehensive mapping of the antigenic sites on the E2s domain is still of great importance. In the current study, the tool box used for further characterization was generated from a comprehensive and large panel of diversified mAbs recognizing at least 3 independent linear epitopes (supplemental Table S1) located at aa 403–457 and 6 conformational antigenic sites on the E2s domain (Fig. 7). As presented in Fig. 7, the identified residues targeted by the 12 representative mAbs covered most of the E2s domain surface that is exposed on virions. Therefore, the selected mAbs in the tool box recognized almost all of the potential antigenic sites on the E2s domain. Notably, the reported neutralizing epitopes recognized by MAB272 (21), MAB1323 (21), and Fab224 (44) overlapped with the antigenic sites C2, C3, and C5/C6 determined in this study, respectively; the reported neutralizing mAb 8C11 was involved in this study and was selected in the tool box. This finding further supports the notion that this study demonstrated a relatively comprehensive picture of anti-HEV neutralizing targets based on the selected tool box.
All of the antigenic sites previously identified on the E2s domain were recognized by neutralizing antibodies. However, this study identified two non-neutralizing antigenic sites (C1 and C4). The presence of these two non-neutralizing antigenic sites on the E2s domain proved that not all of the antibodies generated against the E2s domain can efficiently inhibit viral infection. This finding also improved our understanding of the distribution of the major neutralizing epitopes on the E2s domain. Indeed, the critical residues involved in inducing neutralizing antibody responses against the E2s domain were clearly defined. We can easily imagine that the stability of the conformation of these regions is quite important for anti-HEV vaccination strategies, and thus the mAbs in the tool box recognizing these regions will be useful for quality analysis of the HEV vaccine. The E2s domain not only plays a critical role in anti-HEV vaccination but also mediates virus-host recognition and interactions. Therefore, it is worth mentioning that the tool box can also support research into the HEV infection process, such as the identification of the HEV RBS (receptor-binding sites).
In summary, this study has provided a tool box with 12 representative mAbs recognizing 6 conformational antigenic sites on the E2s domain using the SPSS clustering function for the analysis of cELISA data for multiple mAbs. Based on this tool box, we have accomplished a comprehensive screening of conformation-dependent epitopes on the E2s domain and demonstrated a relatively complete picture of the distribution of the neutralizing epitopes. This study provides insights that increase the understanding of the protective mechanisms induced by the HEV vaccine. Importantly, this work has established a general method for the comprehensive characterization of conformation-dependent antigenic domains.
Author Contributions
M. Z. and Z. Z. Z. designed the research. M. Z. wrote the paper. M. Z., Z. Z. Z., and N. S. X. analysis all the data. M. Z. screened mAbs and performed the cELISA experiment. X. J. L. performed Ala mutagenesis experiment. Z. M. T. and W. C. performed virus capture experiment. Z. M. T., F. Y., and S. L. W. performed neutralizing activity experiment. K. Z., S. L. W., and F. Y. performed reactivity experiment and blocking sera binding experiment. All authors read and approved the final manuscript.
Supplementary Material
This work was supported in part by Chinese National High-tech R&D Program (863 program) Grant 2011AA02A101, Xiamen Science and Technology Platform Project Grant 3502Z20131001, and Xiamen Science and Technology Plan Project Grant 3502Z201410045. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- HEV
- hepatitis E virus
- aa
- amino acid(s)
- cELISA
- competitive ELISA
- OD
- optical density.
REFERENCES
- 1. Reyes G. R., Purdy M. A., Kim J. P., Luk K. C., Young L. M., Fry K. E., Bradley D. W. (1990) Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science 247, 1335–1339 [DOI] [PubMed] [Google Scholar]
- 2. Tam A. W., Smith M. M., Guerra M. E., Huang C. C., Bradley D. W., Fry K. E., Reyes G. R. (1991) Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reyes G. R., Yarbough P. O., Tam A. W., Purdy M. A., Huang C. C., Kim J. S., Bradley D. W., Fry K. E. (1991) Hepatitis E virus (HEV): the novel agent responsible for enterically transmitted non-A, non-B hepatitis. Gastroenterol. Jpn. 26, 142–147 [DOI] [PubMed] [Google Scholar]
- 4. Reyes G. R. (1993) Hepatitis E virus (HEV): molecular biology and emerging epidemiology. Prog. Liver Dis. 11, 203–213 [PubMed] [Google Scholar]
- 5. Teshale E. H., Hu D. J., Holmberg S. D. (2010) The two faces of hepatitis E virus. Clin. Infect. Dis. 51, 328–334 [DOI] [PubMed] [Google Scholar]
- 6. RH P., SU E. (2005) Prevention. in Viral Hepatitis (Thomas H. C., Lemon S., Zuckerman A. J., eds) 3rd Ed., pp. 635–645, Blackwell Publishing, Malden, MA [Google Scholar]
- 7. World Health Organization South-East Asia Region (2012) Viral Hepatitis in the WHO South-East Asia Region. Know it. Confront it. Hepatitis affects everyone, everywhere. World Health Organization, New Delhi, India: http://apps.searo.who.int/PDS_DOCS/B4752.pdf [Google Scholar]
- 8. Koonin E. V., Gorbalenya A. E., Purdy M. A., Rozanov M. N., Reyes G. R., Bradley D. W. (1992) Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U.S.A. 89, 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korkaya H., Jameel S., Gupta D., Tyagi S., Kumar R., Zafrullah M., Mazumdar M., Lal S. K., Xiaofang L., Sehgal D., Das S. R., Sahal D. (2001) The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276, 42389–42400 [DOI] [PubMed] [Google Scholar]
- 10. Nagashima S., Takahashi M., Jirintai S., Tanaka T., Nishizawa T., Yasuda J., Okamoto H. (2011) Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J. Gen. Virol. 92, 2838–2848 [DOI] [PubMed] [Google Scholar]
- 11. Yamada K., Takahashi M., Hoshino Y., Takahashi H., Ichiyama K., Nagashima S., Tanaka T., Okamoto H. (2009) ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90, 1880–1891 [DOI] [PubMed] [Google Scholar]
- 12. Emerson S. U., Nguyen H. T., Torian U., Burke D., Engle R., Purcell R. H. (2010) Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J. Virol. 84, 9059–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi M., Yamada K., Hoshino Y., Takahashi H., Ichiyama K., Tanaka T., Okamoto H. (2008) Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 153, 1703–1713 [DOI] [PubMed] [Google Scholar]
- 14. Li T. C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M. A., Uchida T., Takeda N., Miyamura T. (1997) Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71, 7207–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mushahwar I. K., Dawson G. J., Reyes G. R. (1996) Hepatitis E virus: molecular biology and diagnosis. Eur. J. Gastroenterol. Hepatol. 8, 312–318 [PubMed] [Google Scholar]
- 16. Reyes G. R., Huang C. C., Tam A. W., Purdy M. A. (1993) Molecular organization and replication of hepatitis E virus (HEV). Arch. Virol. Suppl. 7, 15–25 [DOI] [PubMed] [Google Scholar]
- 17. Xing L., Kato K., Li T., Takeda N., Miyamura T., Hammar L., Cheng R. H. (1999) Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 265, 35–45 [DOI] [PubMed] [Google Scholar]
- 18. Li T. C., Takeda N., Miyamura T., Matsuura Y., Wang J. C., Engvall H., Hammar L., Xing L., Cheng R. H. (2005) Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 79, 12999–13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xing L., Li T. C., Mayazaki N., Simon M. N., Wall J. S., Moore M., Wang C. Y., Takeda N., Wakita T., Miyamura T., Cheng R. H. (2010) Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 285, 33175–33183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guu T. S., Liu Z., Ye Q., Mata D. A., Li K., Yin C., Zhang J., Tao Y. J. (2009) Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U.S.A. 106, 12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamashita T., Mori Y., Miyazaki N., Cheng R. H., Yoshimura M., Unno H., Shima R., Moriishi K., Tsukihara T., Li T. C., Takeda N., Miyamura T., Matsuura Y. (2009) Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 106, 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang X., Yang C., Gu Y., Song C., Zhang X., Wang Y., Zhang J., Hew C. L., Li S., Xia N., Sivaraman J. (2011) Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc. Natl. Acad. Sci. U.S.A. 108, 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S., Tang X., Seetharaman J., Yang C., Gu Y., Zhang J., Du H., Shih J. W., Hew C. L., Sivaraman J., Xia N. (2009) Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 5, e1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He S., Miao J., Zheng Z., Wu T., Xie M., Tang M., Zhang J., Ng M. H., Xia N. (2008) Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 89, 245–249 [DOI] [PubMed] [Google Scholar]
- 25. Meng J., Dai X., Chang J. C., Lopareva E., Pillot J., Fields H. A., Khudyakov Y. E. (2001) Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288, 203–211 [DOI] [PubMed] [Google Scholar]
- 26. Prasad B. V., Hardy M. E., Dokland T., Bella J., Rossmann M. G., Estes M. K. (1999) X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290 [DOI] [PubMed] [Google Scholar]
- 27. Dong J., Dong L., Méndez E., Tao Y. (2011) Crystal structure of the human astrovirus capsid spike. Proc. Natl. Acad. Sci. U.S.A. 108, 12681–12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pokidysheva E., Zhang Y., Battisti A. J., Bator-Kelly C. M., Chipman P. R., Xiao C., Gregorio G. G., Hendrickson W. A., Kuhn R. J., Rossmann M. G. (2006) Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124, 485–493 [DOI] [PubMed] [Google Scholar]
- 29. Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 [DOI] [PubMed] [Google Scholar]
- 31. Im S. W., Zhang J. Z., Zhuang H., Che X. Y., Zhu W. F., Xu G. M., Li K., Xia N. S., Ng M. H. (2001) A bacterially expressed peptide prevents experimental infection of primates by the hepatitis E virus. Vaccine 19, 3726–3732 [DOI] [PubMed] [Google Scholar]
- 32. Zhu F. C., Zhang J., Zhang X. F., Zhou C., Wang Z. Z., Huang S. J., Wang H., Yang C. L., Jiang H. M., Cai J. P., Wang Y. J., Ai X., Hu Y. M., Tang Q., Yao X., Yan Q., Xian Y. L., Wu T., Li Y. M., Miao J., Ng M. H., Shih J. W., Xia N. S. (2010) Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 376, 895–902 [DOI] [PubMed] [Google Scholar]
- 33. Shrestha M. P., Scott R. M., Joshi D. M., Mammen M. P., Jr., Thapa G. B., Thapa N., Myint K. S., Fourneau M., Kuschner R. A., Shrestha S. K., David M. P., Seriwatana J., Vaughn D. W., Safary A., Endy T. P., Innis B. L. (2007) Safety and efficacy of a recombinant hepatitis E vaccine. N. Engl. J. Med. 356, 895–903 [DOI] [PubMed] [Google Scholar]
- 34. Bryan J. P., Tsarev S. A., Iqbal M., Ticehurst J., Emerson S., Ahmed A., Duncan J., Rafiqui A. R., Malik I. A., Purcell R. H. (1994) Epidemic hepatitis E in Pakistan: patterns of serologic response and evidence that antibody to hepatitis E virus protects against disease. J. Infect. Dis. 170, 517–521 [DOI] [PubMed] [Google Scholar]
- 35. Purdy M. A., McCaustland K. A., Krawczynski K., Tam A., Beach M. J., Tassopoulos N. C., Reyes G. R., Bradley D. W. (1992) Expression of a hepatitis E virus (HEV)-trpE fusion protein containing epitopes recognized by antibodies in sera from human cases and experimentally infected primates. Arch. Virol. 123, 335–349 [DOI] [PubMed] [Google Scholar]
- 36. Li S. W., Zhang J., He Z. Q., Gu Y., Liu R. S., Lin J., Chen Y. X., Ng M. H., Xia N. S. (2005) Mutational analysis of essential interactions involved in the assembly of hepatitis E virus capsid. J. Biol. Chem. 280, 3400–3406 [DOI] [PubMed] [Google Scholar]
- 37. Zhang J., Gu Y., Ge S. X., Li S. W., He Z. Q., Huang G. Y., Zhuang H., Ng M. H., Xia N. S. (2005) Analysis of hepatitis E virus neutralization sites using monoclonal antibodies directed against a virus capsid protein. Vaccine 23, 2881–2892 [DOI] [PubMed] [Google Scholar]
- 38. Schofield D. J., Glamann J., Emerson S. U., Purcell R. H. (2000) Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 74, 5548–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He J., Kuschner R. A., Dewar V., Voet P., Asher L. V., Vaughn D. W. (2007) Characterization of monoclonal antibodies to hepatitis E virus (HEV) capsid protein and identification of binding activity. J. Biomed. Sci. 14, 555–563 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J., Li S. W., Wu T., Zhao Q., Ng M. H., Xia N. S. (2012) Hepatitis E virus: neutralizing sites, diagnosis, and protective immunity. Rev. Med. Virol. 22, 339–349 [DOI] [PubMed] [Google Scholar]
- 41. Wei M., Zhang X., Yu H., Tang Z. M., Wang K., Li Z., Zheng Z., Li S., Zhang J., Xia N., Zhao Q. (2014) Bacteria expressed hepatitis E virus capsid proteins maintain virion-like epitopes. Vaccine 32, 2859–2865 [DOI] [PubMed] [Google Scholar]
- 42. Schofield D. J., Purcell R. H., Nguyen H. T., Emerson S. U. (2003) Monoclonal antibodies that neutralize HEV recognize an antigenic site at the carboxyterminus of an ORF2 protein vaccine. Vaccine 22, 257–267 [DOI] [PubMed] [Google Scholar]
- 43. Luo T. R., Minamoto N., Hishida M., Yamamoto K., Fujise T., Hiraga S., Ito N., Sugiyama M., Kinjo T. (1998) Antigenic and functional analyses of glycoprotein of rabies virus using monoclonal antibodies. Microbiol. Immunol. 42, 187–193 [DOI] [PubMed] [Google Scholar]
- 44. Xing D., Yeh C. I., Shapley R. M. (2009) Spatial spread of the local field potential and its laminar variation in visual cortex. J. Neurosci. 29, 11540–11549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.