Background: MicroRNAs are noncoding RNAs and have emerged as important regulators in β-cell function.
Results: miR-483 is highly expressed in β-cells but expressed at much lower levels in α-cells, and increased miR-483 induces insulin production in β-cells while repressing glucagon production in α-cells.
Conclusion: miR-483 has opposite effects in α- and β-cells by targeting SOCS3.
Significance: miR-483 is a potential therapeutic target for diabetes.
Keywords: beta cell (B-cell), diabetes, glucagon, insulin, microRNA (miRNA), alpha cells, miR-483
Abstract
Insulin secreted from pancreatic β-cells and glucagon secreted from pancreatic α-cells are the two major hormones working in the pancreas in an opposing manner to regulate and maintain a normal glucose homeostasis. How microRNAs (miRNAs), a population of non-coding RNAs so far demonstrated to be differentially expressed in various types of cells, regulate gene expression in pancreatic β-cells and its closely associated α-cells is not completely clear. In this study, miRNA profiling was performed and compared between pancreatic β-cells and their partner α-cells. One novel miRNA, miR-483, was identified for its highly differential expression in pancreatic β-cells when compared to its expression in α-cells. Overexpression of miR-483 in β-cells increased insulin transcription and secretion by targeting SOCS3, a member of suppressor of cytokine signaling family. In contrast, overexpression of miR-483 decreased glucagon transcription and secretion in α-cells. Moreover, overexpressed miR-483 protected against proinflammatory cytokine-induced apoptosis in β-cells. This correlates with a higher expression level of miR-483 and the expanded β-cell mass observed in the islets of prediabetic db/db mice. Together, our data suggest that miR-483 has opposite effects in α- and β-cells by targeting SOCS3, and the imbalance of miR-483 and its targets may play a crucial role in diabetes pathogenesis.
Introduction
Pancreatic islets consist of two major types of cells, the insulin-producing β-cells (50–70%) and the glucagon-producing α-cells (20–30%), which are physically organized asymmetrically together to maintain a normal glucose homeostasis (1). Insulin and glucagon are two recognized hormones and are secreted in a coordinated manner in pancreatic cells (2). An increase in insulin, triggered by high glucose, suppresses glucagon secretion and a decrease in insulin, in concert with low glucose, stimulates glucagon secretion in healthy individuals (3). In type 2 diabetic patients, β-cell loss and insufficient insulin secretion contribute to hyperglycemia (4, 5). Furthermore, a number of studies have reported that α-cell expansion and elevated glucagon secretion worsen the hyperglycemia in diabetic patients (2, 6–8), suggesting that an imbalanced ratio between β-cell and α-cell mass leads to an imbalanced ratio between insulin and glucagon, which contributes to the development of diabetes. Such imbalance has been well documented in diabetes patients, but the underlying mechanisms that promote this imbalance are not fully understood.
Cell mass remodeling is the result of several processes including proliferation, neogenesis, cell size, and apoptosis (9). In diabetes patients, β-cell loss mainly reflects a combination of impaired β-cell proliferation and increased β-cell apoptosis (10). β-Cell proliferation is stimulated by nutrition and growth factors including glucose, insulin, glucagon-like peptide-1 (GLP-1,) and insulin-like growth factor (IGF-1/2)3 through several intracellular signaling pathways connecting cell surface receptors to proliferation machinery (11, 12). These signaling pathways include the insulin receptor substrate/PI3K/Akt (IRS-PI3K-Akt) pathways, JAK-STAT, MAPK, and calcineurin/nuclear factor of activated T-cells (NFAT) (13, 14). However, high concentrations of glucose, leptin, free fatty acids, reactive oxygen species, or proinflammatory cytokines converge toward a common cell death signaling pathway (JNK/p38 kinase) and cause induction of β-cell apoptosis in the pathogenesis of type 2 diabetes (15).
In contrast, the mechanisms regulating α-cell proliferation and glucagon secretion are poorly understood. It is believed that activated insulin signaling pathway stimulated by high glucose directly or indirectly via β-cell-secreted insulin represses glucagon secretion in α-cells (16, 17). On the other hand, low glucose leads to inactivation of insulin signaling proteins that in turn stimulates glucagon secretion (18, 19). However, activated insulin signaling is considered to induce α-cell proliferation based on the findings from α-cell-specific insulin receptor knock-out (αIRKO) mice and pancreas-specific IRS2 knock-out mice (20, 21). A recent study also demonstrated that insulin stimulates α-cell proliferation through the IR/IRS2/AKT/mTOR (mammalian target of rapamycin) signaling pathway (22). Much remains to be learned about the cross-talk between or among these pathways.
As a key player in gene expression regulation, microRNAs (miRNAs) are endogenous, noncoding RNAs of 21–24 nucleotides that bind to the 3′-UTR of target mRNAs thereby repressing their translation and/or promoting their decay (23, 24). With no exceptions to most eukaryotic cells, recent studies suggested that miRNAs have also emerged as important regulators in β-cell function and proliferation (25, 26). Several miRNAs have been identified to function in controlling and maintaining β-cell proliferation and mass expansion through targeting various cellular signaling pathways. miR-375, the most abundant miRNA detected in pancreatic islets, plays a key role in maintaining normal α-cell and β-cell mass in mice (27). Mice lacking miR-375 have chronic hyperglycemia due to a decreased β-cell mass and increased α-cell numbers (27). Unlike miR-375, miR-7a was reported to inhibit mature β-cell proliferation by targeting the mTOR signaling pathway, and transgenic mice overexpressing miR-7a in β-cells developed diabetes due to impaired insulin secretion and β-cell dedifferentiation (28, 29). Taken together, dysfunctional miRNAs contribute to inappropriate β-cell responses during diabetes pathogenesis.
However, little is known about the functions of specific miRNAs in α-cells and the mechanism for their differential expressions/functions detected in α- and β-cells. In this study, we have performed an miRNA screening to comprehensively assess miRNA expressions in βTC3 in contrast with αTC1-6, which are excellent surrogate systems for equivalent functional studies of primary β- and α-cells (30). We identified a number of miRNAs including miR-483 that were differentially expressed between α- and β-cells. In particular, miR-483 was investigated for its unique higher expression in β-cells than in α-cells. Moreover, the glucose-stimulated miR-483 promoted insulin secretion, insulin transcription, and cell proliferation by targeting the suppressor of cytokine signaling 3 (SOCS3) in β-cells, whereas overexpression of miR-483 inhibits glucagon transcription and secretion in α-cells. Thus, miR-483 has opposite effects in α- and β-cells, and the imbalance of miR-483 expression may contribute to diabetes pathophysiology.
Experimental Procedures
Cell Culture
Pancreatic α-cell line (αTC1-6) and β-cell line (βTC3) were purchased from the American Type Culture Collection (ATCC). Another β-cell line (MIN6) was kindly provided by Dr. Miyazaki (University of Tokyo, Japan) (31). Cells were routinely maintained in DMEM containing 25 mm glucose supplemented with 15% FBS and 1% penicillin-streptomycin (Invitrogen) as described (32).
Mouse Islet Isolation and Flow Cytometric Cell Sorting
Wild type C57BL/6 mice (number 000664), diabetic mice (db/db, BKS.Cg-m+/+leprdb/J, number 000642), and their heterozygous lean controls (db/+) were 8 weeks old and obtained from The Jackson Laboratory (Bar Harbor, ME). All mice were housed in the animal facility of Michigan Technological University on a 12-h light/dark cycle with ad libitum access to water and normal chow. Pancreatic islets were isolated and purified by intra-ductal perfusion of collagenase V (0.5 mg/ml) (Sigma) following the protocol described (33). The purified islets were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin for 24–72 h according to the experiments. All experiments were carried out in accordance with the approval by the Animal Care Committee at the Michigan Technological University.
We performed FACS to obtain the purified α- and β-cells from Ins1-mRFP (34) and glucagon-Cre/Rosa26R-YFP (35) mice, respectively. In preparation for sorting, isolated islets were hand-picked and dissociated at 37 °C by adding 0.05% trypsin-EDTA as described previously (36). Digestion was inactivated by the addition of FCS, and dissociated cells were centrifuged and resuspended in PBS containing 10% FBS for sorting. Flow cytometric sorting was performed on a FACSAria (BD Biosciences) using 561 and 488 excitation lines for RFP and YFP, respectively. Sorted α- and β-cells were then collected in lysis buffer for subsequent RNA extraction. On average, the sorted populations were >98% pure with the final yield ranging between 60 and 80%.
MicroRNA Array and Data Analysis
Total RNA was isolated from both βTC3 and αTC1-6 cells using TRIzol (Invitrogen), and the harvested small RNAs were radiolabeled and hybridized to the mouse miRNA array platform developed in our laboratory as described previously (37). The obtained data were clustered using Cluster 3.0 (38) and visualized using Java TreeView (39).
Quantitative Real-time PCR for miRNA and mRNA
Total RNA from islets or cell lines was extracted using the miRNeasy kit (Qiagen) according to the manufacturer's instructions and treated with rDNase I (Sigma). The TaqMan miRNA quantitative real-time PCR detection system (Applied Biosystems) was used for quantification of miR-483, and its expression was normalized to the relative expression of RNU19. For mRNA quantification, cDNAs were generated using the High Capacity cDNA reverse transcription kit (Applied Biosystems), and quantitative real-time PCR was performed using the Power SYBR Green PCR master mix (Applied Biosystems). Real-time PCR was performed on a StepOnePlusTM system (Applied Biosystems) using the following procedure: 10 min at 95 °C, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. All samples were run in duplicate, and the RNA expression was determined using relative comparison method (ΔΔCt), with hypoxanthine guanine phosphoribosyl transferase (Hprt) mRNA as an internal standard. The following are the primers used in the study: pre-insulin, GGGGAGCGTGGCTTCTTCTA (forward) and GGGGACAGAATTCAGTGGCA (reverse); glucagon, AGAAGAAGTCGCCATTGCTG (forward) and CCGCAGAGATGTTGTGAAGA (reverse); Hprt, TCAGTCAACGGGGGACATAAA (forward) and GGGGCTGTACTGCTTAACCAG (reverse).
In Situ Hybridization and Immunohistochemistry Staining
Dissected mouse pancreas were fixed in 4% formaldehyde (pH 7.4) for 24 h at 4 °C and then processed routinely for paraffin embedding. Tissues were cut into 5-μm sections and adhered to glass slides (Superfrost, Fisher Scientific). For in situ hybridization, sections were first deparaffinized and rehydrated and then treated with proteinase K (Roche Applied Science, 40 μg/ml) as described (40). Briefly, a total of 3 pmol of DIG-labeled Locked Nucleic Acid (LNA) probes (Exiqon) were mixed with 200 μl of hybridization buffer and applied onto the slides to hybridize at 37 °C for overnight. Slides were then washed using 2× SSC solution and incubated with alkaline phosphatase-conjugated sheep anti-DIG antibody (Roche Applied Science) at 4 °C overnight. Alkaline phosphatase reaction was carried out with 50 mg/ml NBT/BCIP (4-nitro-blue tetrazolium/5-brom-4-chloro-3′-indolylphosphate) staining solution at room temperature for 1–3 days.
For combined in situ and immunofluorescence staining, fluorescent in situ hybridization was processed as above, except that detection of the DIG-labeled LNA probes was done with peroxidase-conjugated sheep anti-DIG (Roche Applied Science) followed by Tyramide Signal Amplification (TSA)-FITC substrate (PerkinElmer) according to the manufacturer's recommendations. The same slides were blocked in 0.5% BSA for 30 min and incubated with mouse anti-insulin (Sigma) and rabbit anti-glucagon (Sigma) at 4 °C overnight. The next day, the immunodetection was processed with Alexa Fluor 405- or Alexa Fluor 594-conjugated secondary antibodies (Invitrogen) at room temperature for 2 h. Slides were coverslipped using anti-fading mounting medium (Vector Laboratories). The images were captured on an Olympus FluoView FV1000 confocal microscope.
miRNA Transfection and Adenovirus Transduction
MIN6 cells were electroporated with 5 μg of oligonucleotides or 10 μg of plasmids using the Amaxa Nucleofector system (Amaxa Inc.) according to the manufacturer's instructions. 2 days after transfection, cells were treated with low (1 mm) or high (25 mm) glucose without serum for 16 h, and then cell lysates or total RNA was prepared and subjected to analysis by Western blotting or real-time RT-PCR, respectively. For cytokine treatment, cells were incubated with 10 ng/ml cytokine mixture (TNF-α, IL-1β, and IFNγ) in 25 mm glucose medium with 1% fetal bovine serum for the selected time. The following oligonucleotides were used in this study: miR-483 mimic, miRNA mimic negative control (mimic control), anti-miR483, anti-miRNA negative control (anti-control), and siRNA for Socs3. All the oligonucleotides were purchased from Life Technologies.
For overexpression of miR-483 in isolated islets, adenovirus vector containing miR-483 stem-loop precursor sequence (AD-miR483) was constructed using the RAPAd® miRNA adenoviral expression system (Cell Biolabs). Adenovirus containing the GFP was prepared as a control. The isolated islets were partially dispersed, infected with purified adenovirus in RPMI 1640 with 2% FBS for 2 h, and further incubated in the complete RPMI 1640 medium overnight. Infected islets were cultured in the virus-free complete RPMI 1640 medium for an additional 48 h prior to experimentation. Overexpression of miR-483 in islets was confirmed by real-time PCR analysis of miR-483.
Immunoblot Analysis
Cells were lysed in lysis buffer supplemented with protease inhibitors and phosphatase inhibitor cocktails (Sigma). Lysates were resolved on a 10% SDS-PAGE, transferred to Immobilon transfer membrane (Millipore), and incubated with the following antibodies: SOCS3, IRS2 (Cell Signaling), Pdx-1 (EMD Millipore), MafA (Bethyl Laboratories), and β-actin (Sigma). Anti-rabbit and anti-mouse secondary antibodies were purchased from GE Healthcare. Immunoblots were developed using SuperSignal West Pico chemiluminescent substrates (Thermo Scientific) and visualized on a Fuji imager. Protein levels were quantified using the ImageJ software.
Luciferase Assays for miRNA Target Validation
For evaluation of the predicted miR-483 complementary sites at the 3′-UTR of Socs3 gene, the mouse Socs3 3′-UTR containing three miR-483 binding sites (50–450 and 1000–1320 bp) was synthesized and subcloned into the pRLTK vector (Promega). For the luciferase reporter assay, pRLTK reporter constructs (5 μg) were electroporated into MIN6 cells (1 × 106) with miR-483 mimic or control using Amaxa (Lonza). The plasmid PGL-3 containing firefly luciferase (5 μg) was co-transfected together to normalize for transfection efficiency. Luciferase activity was measured with a Dual-Luciferase reporter assay kit (Promega) 2 days after transfection. All the related luciferase assay vectors were provided by Dr. Peter Nelson (University of Kentucky, Lexington, KY) (40).
Insulin and Glucagon Secretion Assay
For quantification of insulin secretion, MIN6 cells were preincubated at 37 °C for 2 h in Krebs-Ringer buffer (KRB) (128.8 mm NaCl, 4.8 mm KCl, 1.2 mm KH2PO4, 1.2 mm MgSO4, 2.5 mm CaCl, 5 mm NaHCO3, 10 mm HEPES, 0.1% BSA) containing 1 mm glucose and then stimulated in the same buffer containing 25 mm glucose for 1 h. The supernatant from each treatment was then collected for insulin assay using a mouse insulin ELISA assay kit (Mercodia). Cells were lysed in acid ethanol solution for total DNA. For glucagon secretion assay, αTC1-6 cells were preincubated in KRB containing 25 mm glucose at 37 °C for 2 h. Subsequently, the cells were stimulated with KRB containing 1 mm glucose for 1 h, and the secreted glucagon in the collected supernatants was measured by a mouse glucagon ELISA assay kit (Wako Chemicals). Both insulin and glucagon values were normalized to total cellular DNA content from the respective lysates.
For quantification of insulin and glucagon secretion in islets, virus-infected islets were transferred to 1.5-ml Eppendorf tubes and preincubated at 37 °C for 30 min in KRB supplemented with 2.7 mm glucose (for insulin secretion) or 16.7 mm glucose (for glucagon secretion). The islets were then stimulated with KRB containing either 2.7 mm or 16.7 mm glucose for 1 h, and the supernatant from each treatment was collected for insulin and glucagon assay. Islets were also lysed in acid ethanol solution for total DNA isolation, and the total cellular DNA content was used to normalize the insulin and glucagon values.
Cell Apoptosis and Proliferation Assay
Two days after transfection, cells were seeded in 96-well plates (1 × 104 cells/well) and cultured for 16 h with or without cytokines (10 ng/ml) to induce apoptosis. Cytoplasmic histone-associated DNA fragments were quantified by the cell death detection ELISA kit (Roche Applied Science), which detects DNA laddering derived from apoptotic cell death with anti-histone-biotin antibody and anti-DNA-peroxidase antibody. For proliferation assay, the thymidine analog BrdU (10 μm) was added to the culture medium for the last 16 h of incubation prior to fixation, and DNA synthesis was measured using the cell proliferation ELISA kit (Roche Applied Science) according to the protocol.
Statistical Analysis
Data are expressed as means ± S.D. of three independent experiments. Statistical significance was determined by unpaired Student's t test (two-tailed) or one-way analysis of variance with Tukey's post hoc test with differences considered significant at p < 0.05 (marked as *) and p < 0.01 (marked as **).
Results
miR-483 Is Differentially Expressed between Pancreatic α- and β-Cells
To understand what are the major miRNAs that are differentially expressed between pancreatic α- and β-cells and their potential cellular functions, we profiled 553 selected human and mouse miRNAs in αTC1-6 and βTC3 cells, respectively, using our optimized homemade miRNA array platform (37). About 108 miRNAs in total were detectable in the assay analysis. Among these detectable miRNAs, most miRNAs showed approximately equally expression in the two types of pancreatic cells with a -fold change from 0.5 to 1.5 (Fig. 1, A and B). Among the differentially expressed miRNAs, nine, including miR-483, miR-375, miR-99b, and miR-24, were expressed significantly higher in βTC3, whereas 14 miRNAs, including miR-124 and miR-103/107, were expressed much higher in αTC1-6 (Fig. 1B).
FIGURE 1.
miR-483 is highly expressed in pancreatic β-cells but detected at much lower levels in α-cells. A, miRNA array analysis in βTC3 and αTC1-6 cells. Total RNA was isolated, and small RNA was harvested by gel cleaning for radiolabeling. The radiolabeled small RNAs were then hybridized to an array platform containing a collection of mouse/human miRNAs. The identified miR-483 is marked with an oval. B, hierarchical clustering of the identified miRNAs based on their expression profiles using Gene Cluster 3.0 (average linkage and Euclidean distance as similarity measure). The data are the average of three independent experiments ± S.D. The expression of each miRNA was normalized to external and internal controls and median-centered prior to clustering. The expression levels ranged from +0.12 log10 to −0.12 log10. C, validation of miR-483 expression by real-time PCR. miR-483 was highly expressed and glucose-stimulated in βTC3 cells when compared with αTC1-6 cells. The expression of miR-483 was normalized to U6 RNA level, and the data are the average of three independent experiments ± S.D. **, p < 0.01
We carefully examined the short list of candidate miRNAs that potentially have a role in distinguishing the two types of pancreatic cells and found that miR-483 expression was significantly higher in βTC3, but almost undetectable in αTC1-6. Real-time PCR confirmed that the miR-483 level was 100-fold higher in βTC3 than αTC1-6 (Fig. 1C). It is well known that high glucose induces opposite effects in α- and β-cells. We further examined the effect of glucose concentration on miR-483 expression level. High glucose (25 mm) dramatically induced miR-483 level by 2.5-fold in βTC3 when compared with low glucose (1 mm) (Fig. 1C). However, miR-483 level was not significantly induced by high glucose in αTC1-6. Similar to βTC3 cells, miR-483 was also detected in another pancreatic β cell line, MIN6 cells, a currently popular cell line for studying insulin production and signaling, so this cell line was therefore used for further functional studies of miR-483.
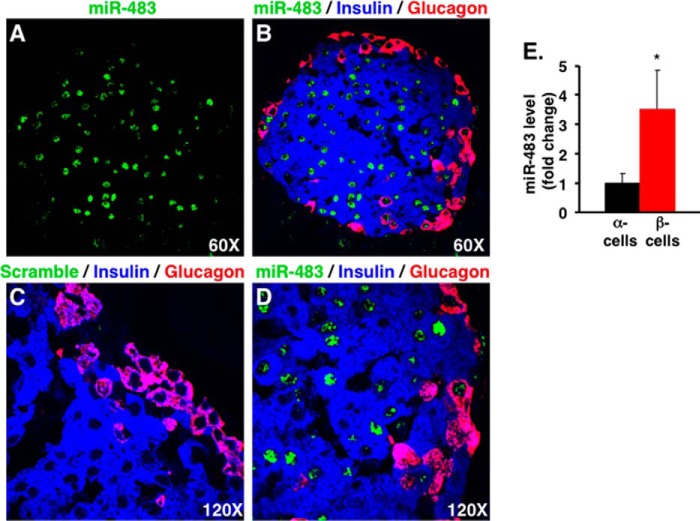
We further examined the expression and localization of miR-483 in primary β- and α-cells in wild type mice. To determine the localization of miR-483 by FISH, mouse pancreas cross sections were prepared for detecting miR-483 using miR-483 LNA probe, followed by immunostaining for co-localization of this miRNA with insulin and glucagon. As shown in Fig. 2, miR-483 was intensely detected in insulin-stained β-cells but detected at much lower levels in glucagon-stained α-cells when compared with negative control scramble (Fig. 2, A–D). Interestingly, miR-483 appeared highly enriched in the nuclei in both β-cells and α-cells, which was consistent with a previous study (41). To validate the expression of miR-483 in purified α- and β-cells, the islets were isolated from Ins1-mRFP and glucagon-Cre/Rosa26R-YFP mice and further sorted out RFP+ β-cells and YFP+ α-cells by high-speed FACS. As expected, real-time PCR confirmed that miR-483 level in purified β-cells was 3.5-fold higher than its expression in purified α-cells (Fig. 2E). Taking together, the data demonstrated that miR-483 was differentially expressed between pancreatic α- and β-cells.
FIGURE 2.
Combined in situ hybridization and immunostaining of mouse pancreas sections. A–D, in situ hybridization was performed on sections with an miR-483 probe (A, B, and D) or a scramble probe (C). The same sections were counterstained with anti-insulin (blue) and anti-glucagon (red) antibodies. The three-color overlay images showed that miR-483 (green) was highly expressed in β-cells (blue) but detected at much lower levels in α-cells (red). Hybridization of scramble probe is negative control (C). Magnification: A and B, 60×; C and D, 120×. E, real-time PCR confirmed the miR-483 expression on sorted α- and β-cells from adult islets purified from glucagon-Cre/Rosa26R-YFP and Ins1-mRFP1 transgenic mice, respectively. The expression of miR-483 was normalized to U6 RNA level, and data are represented as means ± S.D. on n = 3. *, p < 0.05
Overexpression of miR-483 Induces Insulin Secretion and Inhibits Glucagon Secretion through Activation of Insulin Signaling
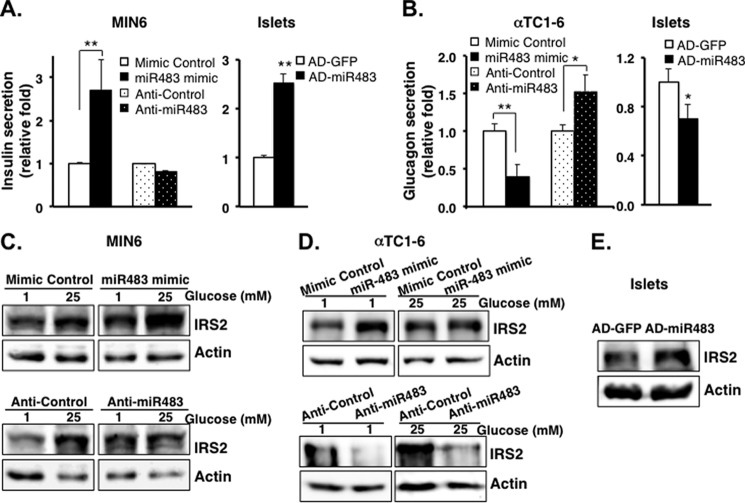
To examine the function of miR-483 in β-cells, we measured insulin secretion in MIN6 cells that were transfected with miR-483 mimic or anti-miR483. Overexpression of miR-483 by miR-483 mimic significantly increased glucose-stimulated insulin secretion by 2.5-fold when compared with the control (Fig. 3A). Inhibition of miR-483 with anti-miR483 oligonucleotides had no significant effect on insulin secretion (Fig. 3A). To validate the effects of miR-483 in isolated primary mouse islets, we generated recombinant adenovirus overexpressing miR-483 (Ad-miR483) and infected freshly isolated primary mouse islets with this adenovirus. After 2 days, we observed a 60-fold increase in miR-483 level when compared with the islets infected with adenoviral GFP control (Ad-GFP). Consistent with the role for miR-483 in MIN6, a significant increase in insulin secretion was obtained in islets infected with Ad-miR483 when compared with the islets infected with Ad-GFP (Fig. 3A).
FIGURE 3.
Overexpression of miR-483 induces insulin secretion but inhibits glucagon secretion via activated IRS2 signaling. A, insulin secretion was measured in MIN6 transfected with miR-483 mimic and control mimic, or anti-miR483 and anti-control at 25 mm glucose, or in isolated islets infected with recombinant adenovirus overexpressed miR-483 (AD-miR483) or control (Ad-GFP) at 16.7 mm glucose. B, glucagon secretion was analyzed in αTC1-6 transfected with miR-483 mimic or anti-miR483 at 1 mm glucose or in isolated islets infected with AD-miR483 at 2.7 mm glucose. Secreted insulin and glucagon levels in the medium were quantified using mouse insulin and glucagon ELISA, respectively, and normalized to total cellular DNA content and then presented as relative -fold. The presented data are the average of three independent experiments ± S.D. *, p < 0.05, **, p < 0.01 versus control. C–E, transfected cells were incubated with 1 or 25 mm glucose in DMEM for 16 h, and the expression of IRS2 and actin was analyzed by Western blot in MIN6 (C) and αTC1-6 (D) and isolated islets (E). All the experiments were repeated 3–6 times.
To examine the functional differences of miR-483 in α-cells, miR-483 mimic was then transfected to αTC1-6 cells to examine the effect of miR-483 on glucagon secretion. When compared with increased insulin secretion in MIN6 cells, overexpressed miR-483 significantly reduced glucagon secretion to almost 2.6-fold in αTC1-6 cells (Fig. 3B). The glucagon secretion was consistently decreased in islets infected with Ad-miR483 when compared with the islets infected with AD-GFP (Fig. 3B). In contrast, inhibition of miR-483 significantly induced glucagon secretion in αTC1-6 cells (Fig. 3B).
Because miR-483 expression is significantly up-regulated by high glucose, we further examined the effect of miR-483 on glucose-stimulated insulin signaling in both cell lines as well as in isolated islets. Overexpression of miR-483 using miR-483 mimic significantly increased the protein level of IRS2 in either cell line, MIN6 (Fig. 3C) or αTC1-6 cells (Fig. 3D), using miR-483 mimic or in infected islets using AD-miR483 (Fig. 3E). The increase of IRS2 was correlated to the increased insulin secretion in β-cells and decreased glucagon secretion in α-cells. In contrast, inhibition of miR-483 dramatically decreased IRS2 expression in both MIN6 and αTC1-6 cells (Fig. 3C). Taken together, the data suggested that miR-483 played a critical role in regulating insulin/glucagon secretion via activating insulin signaling.
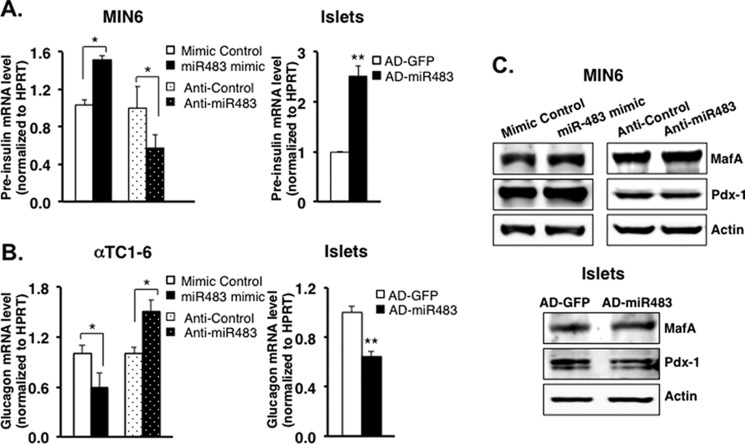
Glucose-stimulated miR-483 Induces Insulin Transcription while Inhibiting Glucagon Transcription
To understand which step of insulin and glucagon expression is critical for the role of miR-483, we further examined the effect of miR-483 on insulin and glucagon transcription. The miR-483 mimic or anti-miR483 was transfected into MIN6 and αTC1-6, followed by real-time PCR for the expression of pre-insulin and glucagon at their transcript levels. As shown in Fig. 4A, high glucose-stimulated pre-insulin mRNA was significantly increased in MIN6 cells transfected with miR-483 mimic when compared with the control. In contrast, transfection with anti-miR483 resulted in an ∼2-fold reduction in pre-insulin mRNA, suggesting that miR-483 promotes insulin gene expression at the transcriptional level under high glucose condition. However, low glucose-induced glucagon mRNA was significantly repressed by overexpression of miR-483 or induced by anti-miR483 in αTC1-6 cells (Fig. 4B). These opposite effects in regulating insulin and glucagon transcription were further confirmed in islets infected with Ad-miR483 (Fig. 4, A and B). We hypothesized that the induction of insulin gene transcription by miR-483 may be involved through the regulation of insulin transcription factors, such as Pdx-1 and MafA. However, expression of both Pdx-1 and MafA has no effect upon miR-483 overexpression or inhibition in MIN6 and islets Fig. 4C).
FIGURE 4.
miR-483 promotes insulin transcription while repressing glucagon transcription. A, MIN6 were transfected with miR-483 mimic or anti-miR483, or isolated islets were infected with AD-miR483. After 48 h, transfected cells were incubated with 25 mm glucose in DMEM (for MIN6) or 16.7 mm glucose (for islets) for 16 h, and total RNA was extracted to analyze the expression of pre-insulin by real-time PCR. B, 48 h after miR-483 mimic transfection in αTC1-6 or AD-miR483 in isolated islets, transfected cells were incubated with 1 mm glucose in DMEM (for αTC1-6) or 2.7 mm glucose (for islets) for 16 h, and total RNA was extracted to analyze the expression of glucagon by real-time PCR. The data were normalized to internal control HPRT, and the results are the average of three independent experiments ± S.D.; **, p < 0.01 versus mimic control. C, the expression of MafA, Pdx-1, and actin was analyzed by Western blot in transfected MIN6 and islets. Results are representative of three independent experiments.
SOCS3 Is One of the Targets of miR-483 and Plays Opposite Functions in α- and β-Cells
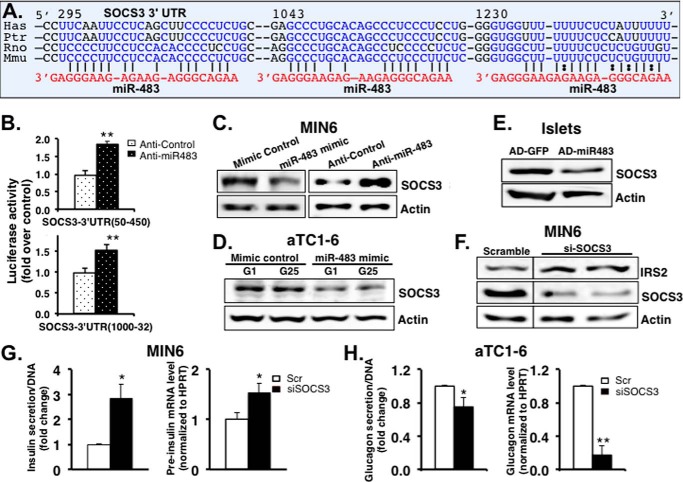
To understand which target genes are negatively regulated by miR-483 in activating insulin production and/or inhibiting glucagon production, we predicted candidate target genes using miRNA target prediction programs. Among many candidates, SOCS3 was shown to have two additional miR-483 complementary sites on its 3′-UTR at the 1043- and 1230-bp regions (Fig. 5A), in addition to the one reported at the 295-bp region of the 3′-UTR in a study with the liver Hepa1-6 cells (42). These two complementary sites are highly conserved among its homologs in various mammalian species including humans and mice, suggesting a critical role of miR-483 in regulating SOCS3, which may function in pancreatic cells (Fig. 5A).
FIGURE 5.
SOCS3 is one of the targets of miR-483 in pancreatic β-cells. A, bioinformatic prediction of the interaction between miR-483 and the 3′-UTRs of SOCS3 of various species. Has, Ptr, Rno, and Mmu represent human, chimpanzee, rat, and mouse, respectively. The predicted three binding sites are indicated in blue. B, luciferase assay confirmed that miR-483 inhibited the luciferase reporter activity, in which the SOCS3 3′-UTR fragments (50–450 and 1000–1320 bp) were fused with a Renilla luciferase reporter gene in pRLTK, respectively. The reporter constructs were co-transfected into MIN6 cells with either anti-miR483 or anti-control. The pGL3 firefly luciferase plasmid was co-transfected for detection of transfection efficiency. 48 h after transfection, luciferase activity was assayed using the Dual-Luciferase reporter assay kit. Data represent means ± S.E. of three independent experiments. **, p < 0.01. C–E, overexpression of miR-483 down-regulates SOCS3 protein expression in MIN6 (C), αTC1-6 (D), and isolated islets (E) validated by Western blots. Data are representative of three independent experiments. F, silencing of SOCS3 activated the expression of IRS2. 48 h after transfection with siRNA against SOCS3 (#1 and #2) or Scramble (Scr) in MIN6 cells, cell lysates were prepared for Western blots to detect the expression of IRS2, SOCS3, and actin. Results are representative of three independent experiments. G, silencing of SOCS3 increased insulin secretion and insulin transcription. H, silencing of SOCS3 reduced glucagon secretion and transcription. Secreted insulin and glucagon levels in the medium were quantified using mouse insulin or glucagon ELISA and normalized to total cellular DNA contents. Total RNA was extracted to analyze the expression of insulin or glucagon by real-time PCR. The data were normalized to internal control HPRT. The presented data are the average of three independent experiments ± S.D. *, p < 0.05, **, p < 0.01.
To examine a potential physical interaction between miR-483 and the three predicted binding sites at the 3′-UTR of SOCS3 transcript, we synthesized two segments of mouse SOCS3 3′-UTR (SOCS3-3′-UTRI between 50 and 450 bp and SOCS3-3′-UTRII between 1000 and 1332 bp, respectively) and subcloned them into the pRLTK vector (Promega). The reporter constructs were co-electroporated into MIN6 cells with anti-miR483 inhibitor or control using Amaxa (Lonza). When compared with the control, introduction of anti-miR483 increased reporter activities of both SOCS3-3′-UTRI and SOCS3-3′-UTRII by 60%, suggesting that inhibition of miR-483 abolished the repression of miR-483 on the luciferase reporter activities (Fig. 5B).
To further determine whether SOCS3 is a potential target of miR-483, we examined the effect of miR-483 on SOCS3 protein level in MIN6, αTC1-6, and isolated islets. Overexpression of miR-483 using miR-483 mimic decreased SOCS3 protein level in both MIN6 and αTC1-6 when compared with the control (Fig. 5, C and D). AD-miR483-infected islets also showed a reduction in SOCS3 expression when compared with the AD-GFP control (Fig. 5E). In contrast, inhibition of miR-483 using anti-miR483 significantly increased SOCS3 protein level (Fig. 5C). Taken together, these data demonstrated that SOCS3 is a direct target of miR-483 in both α-cells and β-cells.
In the insulin-sensitive peripheral tissues, including white adipose tissue, liver, and muscle, SOCS3 inhibits insulin action through binding IRS2 and causing IRS2 ubiquitination and degradation (43–46). To validate the function of SOCS3 in MIN6 and αTC1-6 cells, we silenced SOCS3 by transfecting the cells with SOCS3 siRNA (Applied Biosystems). As shown in Fig. 5F, significant reduction of SOCS3 expression was observed in cells expressing either SOCS3 siRNA-1 or siRNA-2. As expected, silencing of SOCS3 significantly increased IRS2 expression by almost 3-fold in MIN6 (Fig. 5F). Moreover, SOCS3 silencing led to an increase in both insulin secretion and insulin transcription (Fig. 5G), which is consistent with a previous study in which SOCS3 negatively regulated insulin secretion and insulin transcription in β-cell lines and SOCS3 transgenic mice (47–49). In contrast, silencing of SOCS3 significantly inhibited both glucagon secretion and transcription in αTC1-6 cells (Fig. 5H).
miR-483 Protects against Cytokine-mediated Apoptosis in β-Cells
Studies have shown that SOCS3 is induced by inflammatory cytokines and can inhibit excessive cell growth and induce apoptosis as part of maintaining cell stability (50–52). We hypothesized that miR-483 may be involved in protecting β-cell growth by targeting SOCS3. A cytokine mixture with 10 ng/ml each of TNFα, IL-1β, and IFNγ was incubated with MIN6 cells to induce apoptosis. Although overexpression of miR-483 had no significant effect on apoptosis, silencing of miR-483 by anti-miR483 increased cytokine-induced apoptosis in MIN6 cells (Fig. 6A). Consistently, the cell proliferation rate was significantly decreased by anti-miR483 when cells were exposed to cytokine (Fig. 6B), suggesting that miR-483 is necessary to inhibit target protein SOCS3 to protect β-cells against cytokine-induced apoptosis. However, such profound protection was not observed in αTC1-6 cells. Overexpression or inhibition of miR-483 had no significant effect on apoptosis (Fig. 6C) and cell proliferation under both normal growth conditions and cytokine treatment in αTC1-6 cells (Fig. 6D). Taken together, pancreatic β- and α-cells have distinct characteristics in the expressions of miR-483 and its target gene SOSC3 as well as their responses to cytokines in terms of cell apoptosis and proliferation.
FIGURE 6.
Silencing of miR-483 increases cytokine-induced apoptosis and represses cell proliferation in MIN6 cells. A and B, miR-483 mimic or anti-miR483 and their controls were transfected into MIN6. After 48 h, cells were incubated with or without cytokine (10 ng/ml) for an additional 16 h. The ratio of apoptotic cells was determined in cells treated with cytokine versus cells without cytokine treatment. Cell proliferation was measured by BrdU incorporation after incubation with or without cytokine (10 ng/ml) for 16 h. C and D, miR-483 has no significant effect on cytokine-induced apoptosis and cell proliferation in αTC1-6 cells. αTC1-6 cells were transfected with miR-483 mimic or anti-miR483 and their controls. 48 h after transfection, cell apoptosis and cell proliferation were measured in transfected cells after incubation with or without cytokine (10 ng/ml) for 16 h. The results are the average of three independent experiments ± S.D. *, p < 0.05 versus control.
Increased Expression of miR-483 in the Islets of Diabetic Mice
To examine the change of miR-483 expression during the development of diabetes, we harvested the pancreases from 10-week-old db/db or control lean mice (db/+) and prepared them for in situ hybridization. When compared with the control mice, miR-483 expression increased in the islets of prediabetic mice in correspondence with the increase of the β-cell mass (53) (Fig. 7A). Real-time PCR further confirmed that this increase of miR-483 was almost 2-fold in isolated islets from prediabetic mice when compared with control mice (Fig. 7B).
FIGURE 7.
Increased expression of miR-483 in the islets of diabetic mice. A, in situ hybridization of pancreatic sections of 10-week-old heterozygous control mice (db/+, left panel) and diabetic mice (db/db, right panel) using DIG-labeled probes for miR-483. B, real-time PCR confirmed increased miR-483 and repressed SOCS3 expression in freshly isolated islets of db/db mice at the age of 10 weeks when compared with control mice. The expression of miR-483 was normalized to U6 RNA level, and the data are shown as means ± S.D. of three independent experiments. **, p < 0.01 versus control.
The isolated islets were further analyzed for the expression of SOCS3 by real-time PCR. When compared with control lean mice, SOCS3 mRNA level declined 8-fold in prediabetic islets, correlating with the increase of miR-483 in prediabetic islets (Fig. 7C). Thus, our in vitro data for functions of miR-483 were also supported by the in vivo changes of miR-483 and SOCS3 in prediabetic mice.
Discussion
In this study, we first observed that the expression of miR-483 was much higher in pancreatic β-cells than in α-cells. Glucose-stimulated miR-483 stimulated IRS2-mediated insulin signaling, leading to increased insulin secretion in β-cells. Moreover, miR-483 promoted insulin transcription by directly targeting SOCS3. In contrast, overexpression of miR-483 repressed low glucose-stimulated glucagon secretion and transcription in α-cells. In addition, miR-483 serves as a β-cell survival factor to suppress cytokine-induced apoptosis in β-cells while serving no significant effect in α-cells. Increased miR-483 is shown in the islets of prediabetic mice, suggesting that miR-483 may facilitate compensatory β-cell mass expansion during the early stage of diabetes.
miRNAs typically recognize their target mRNAs through complementary sites of their seed region to the binding sites located on the 3′-UTR of the target mRNA. However, we revealed that the 3′-UTR of SOCS3 contains three atypical miR-483 recognition sites that are not perfectly base-paring with the seed region of miR-483 (Fig. 5A). We demonstrated that these atypical miR-483 recognition sites were valid and sufficient to mediate the suppression of SOCS3 by miR-483. These atypical recognitions between miRNAs and their targets are also supported by previous studies with numerous biochemical and structural findings on miRNA-target interactions (54, 55).
miR-483 has been reported to be expressed in mouse liver and adipocyte tissues and regulates fat metabolism by targeting SOCS3 (42). SOCS3 belongs to the SOCS family of inhibitory proteins, which can interfere with insulin signaling in muscle, liver, and adipose tissue by inhibition of tyrosine phosphorylation of IRS2 (56). In pancreatic β-cells, SOCS3 is induced by various cytokines and complexes with insulin receptor, leading to reduced insulin receptor autophosphorylation and impaired signaling through the IRS2/PI3K pathway (52). In addition, in the transgenic mice with β-cell-specific overexpression of SOCS3, SOCS3 inhibits pre-insulin transcription and β-cell proliferation through the JAK/STAT signaling pathway (48, 49). Although SOCS3 is an established regulator of the JAK-STAT pathway, its own regulation in β-cells is poorly understood. In our study, miR-483 protects β-cell function by inhibiting cytokine-induced SOCS3, suggesting that miR-483 may play a positive acute phase response to inflammatory cytokine and, in turn, influences SOCS3-JAK/STAT-insulin network.
An miRNA profiling was also recently processed in sorted human α- and β-cells (30). Consistent with our finding, results form their study confirmed that human β-cells contain higher miR-483 than α-cells (30). A pivotal observation in our work is that miR-483 is stimulated by high glucose and exerts an opposite effect on insulin and glucagon release. First, glucose-stimulated miR-483 induces insulin production and release from β-cells, which in turn suppresses glucagon release from α-cells. Insulin has been considered as a negative regulator of glucagon release (3, 16). Second, miR-483-activated IRS2 expression and insulin signaling inhibit glucagon secretion. In addition, our study showed that highly expressed miR-483 protects β-cell growth against cytokine-induced apoptosis, suggesting that these cells, although derived from common precursor cells, evolved into closely related types of cells with distinct functions. Taken together, the differentially expressed miR-483 plays a crucial role in the maintenance of α- and β-cell mass and function.
Over one-third of mammalian miRNAs are located within the introns of protein-coding genes, referred to as intronic miRNAs (57). miR-483 is an intronic miRNA and is located within the intron of the Igf2 gene (42). IGF-2 was detected at a relatively high level in islets, and reduced IGF-2 expression was observed in the impaired islets of the adult Goto-Kakizaki rat model of type 2 diabetes (58). The transgenic mice with β-cell-specific overexpression of IGF-2 showed an increase in insulin mRNA level and β-cell mass (59). Moreover, the persistent presence of circulating IGF-II largely prevented β-cell apoptosis in the early postnatal period (60, 61). It is possible that miR-483 is not only transcriptionally linked to IGF-2 expression, but also coordinately regulated under different physiological conditions. Interestingly, miR-483 was found to be highly enriched in the nucleus of both α-cells and β-cells. In support, a number of miRNAs have been identified to directly bind to the gene promoter, potentially inducing overexpression or down-regulation of target genes (62, 63). More recently, overexpressed miR-483 in the pediatric kidney cancer cells localized in the nuclei and enhanced the transcription of IGF-2 transcripts by binding to its 5′-UTR (41). However, the interaction between nuclear miR-483 and IGF-2 in α- and β-cells still needs to be further examined under physiological conditions. Furthermore, insulin gene is clustered closely together with IGF-2 gene, suggesting that miR-483 and IGF-2, together with insulin, might be controlled by a common regulatory mechanism. Therefore, identification of the coordinated interaction between miR-483 and IGF-2 will be important for understanding the differential expression of miR-483 between α- and β-cells and establishing a functional relationship between them.
Two studies have recently reported miRNA profiling in the sorted human α- and β-cells or α- and β-cell lines by the miRNA PCR array method (30, 64). Among the identified miRNAs in these studies, most of them, including miR-483, miR-375, and miR-99b, expressed at a relatively higher level in β-cells, as confirmed in our array. Other miRNAs, such as miR-124 and miR-103/107, expressed relatively higher in α-cells in our array. miR-124 has been shown to regulate pancreas development and insulin exocytosis and induce neural differentiation and proliferation (65, 66). miR-103/107 has been demonstrated to induce insulin resistance and glucose intolerance in both liver and adipocytes (67). Therefore, the data suggest that pro- or anti-proliferative miRNAs co-exist in both α-cells and β-cells and function interactively in maintaining the identity and functions of α- and β-cells. It will be of interest to dissect the functional interactions of these miRNAs and their homeostatic roles in the development of diabetes.
In summary, we characterized the distinct expression and functions of miR-483 between α- and β-cells. miR-483 can be stimulated by glucose and insulin, and activated miR-483 is required for glucose-stimulated IRS2 signaling and protects β-cells against apoptosis. The balance of miR-483 and its targets would not only protect β-cell function, but also help to preserve β-cell mass. Future dissection of the interaction of miR-483 with the SOCS3-mediated functional pathway may extend our understanding on the potential therapeutic role of miR-483.
Author Contributions
X. T. performed the experiments shown in Fig. 1, coordinated the study, and wrote the paper. R. M. designed, performed, and analyzed the experiments shown in Figs. 3–7. Y. M. designed and performed the experiments shown in Figs. 2 and 7. S. Z. constructed vectors and performed the luciferase reporter assay shown in Fig. 5. Y. Z., and C. X. performed the fluorescence-activated cell sorting analysis shown in Fig. 2. G. G. provided Ins1-mRFP mice. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Peter Nelson for providing luciferase reporter plasmids, Dr. Guiliang Tang for critical reading and editing of this manuscript, and the undergraduate students Austin Burke and Pedro Rodrigues for technical assistance.
This work was supported by National Institutes of Health Grant DK084166 (to X. T.). The authors declare that they have no conflicts of interest with the contents of this article.
- IGF-2
- insulin-like growth factor 2
- IRS2
- insulin receptor substrate 2
- miRNA
- microRNA
- miR
- microRNA
- SOCS3
- the suppressor of cytokine signaling 3
- DIG
- digoxigenin
- KRB
- Krebs-Ringer buffer
- Ad
- adenovirus
- HPRT
- hypoxanthine guanine phosphoribosyl transferase.
REFERENCES
- 1. Cabrera O., Berman D. M., Kenyon N. S., Ricordi C., Berggren P. O., Caicedo A. (2006) The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. U.S.A. 103, 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breckenridge S. M., Cooperberg B. A., Arbelaez A. M., Patterson B. W., Cryer P. E. (2007) Glucagon, in concert with insulin, supports the postabsorptive plasma glucose concentration in humans. Diabetes 56, 2442–2448 [DOI] [PubMed] [Google Scholar]
- 3. Cooperberg B. A., Cryer P. E. (2010) Insulin reciprocally regulates glucagon secretion in humans. Diabetes 59, 2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Porte D., Jr., Kahn S. E. (2001) β-Cell dysfunction and failure in type 2 diabetes: potential mechanisms. Diabetes 50, Suppl. 1, S160–S163, 10.2337/diabetes.50.2007.S160 [DOI] [PubMed] [Google Scholar]
- 5. Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. (2003) β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes 52, 102–110 [DOI] [PubMed] [Google Scholar]
- 6. D'Alessio D. (2011) The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obes. Metab. 13, Suppl. 1, 126–132, 10.1111/j.1463-1326.2011.01449.x [DOI] [PubMed] [Google Scholar]
- 7. Yoon K. H., Ko S. H., Cho J. H., Lee J. M., Ahn Y. B., Song K. H., Yoo S. J., Kang M. I., Cha B. Y., Lee K. W., Son H. Y., Kang S. K., Kim H. S., Lee I. K., Bonner-Weir S. (2003) Selective β-cell loss and α-cell expansion in patients with type 2 diabetes mellitus in Korea. J. Clin. Endocrinol. Metab. 88, 2300–2308 [DOI] [PubMed] [Google Scholar]
- 8. Deng S., Vatamaniuk M., Huang X., Doliba N., Lian M. M., Frank A., Velidedeoglu E., Desai N. M., Koeberlein B., Wolf B., Barker C. F., Naji A., Matschinsky F. M., Markmann J. F. (2004) Structural and functional abnormalities in the islets isolated from type 2 diabetic subjects. Diabetes 53, 624–632 [DOI] [PubMed] [Google Scholar]
- 9. Montanya E., Téllez N. (2009) Pancreatic remodeling: β-cell apoptosis, proliferation and neogenesis, and the measurement of β-cell mass and of individual β-cell size. Methods Mol. Biol. 560, 137–158 [DOI] [PubMed] [Google Scholar]
- 10. Kassem S. A., Ariel I., Thornton P. S., Scheimberg I., Glaser B. (2000) β-Cell proliferation and apoptosis in the developing normal human pancreas and in hyperinsulinism of infancy. Diabetes 49, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 11. Heit J. J., Karnik S. K., Kim S. K. (2006) Intrinsic regulators of pancreatic β-cell proliferation. Annu. Rev. Cell Dev. Biol. 22, 311–338 [DOI] [PubMed] [Google Scholar]
- 12. Kulkarni R. N. (2005) New insights into the roles of insulin/IGF-I in the development and maintenance of β-cell mass. Rev. Endocr. Metab. Disord. 6, 199–210 [DOI] [PubMed] [Google Scholar]
- 13. Kulkarni R. N., Mizrachi E. B., Ocana A. G., Stewart A. F. (2012) Human β-cell proliferation and intracellular signaling: driving in the dark without a road map. Diabetes 61, 2205–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernal-Mizrachi E., Kulkarni R. N., Scott D. K., Mauvais-Jarvis F., Stewart A. F., Garcia-Ocaña A. (2014) Human β-cell proliferation and intracellular signaling part 2: still driving in the dark without a road map. Diabetes 63, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandrup-Poulsen T. (2001) β-Cell apoptosis: stimuli and signaling. Diabetes 50, Suppl. 1, S58–S63, 10.2337/diabetes.50.2007.S58 [DOI] [PubMed] [Google Scholar]
- 16. Ravier M. A., Rutter G. A. (2005) Glucose or insulin, but not zinc ions, inhibit glucagon secretion from mouse pancreatic α-cells. Diabetes 54, 1789–1797 [DOI] [PubMed] [Google Scholar]
- 17. Diao J., Asghar Z., Chan C. B., Wheeler M. B. (2005) Glucose-regulated glucagon secretion requires insulin receptor expression in pancreatic α-cells. J. Biol. Chem. 280, 33487–33496 [DOI] [PubMed] [Google Scholar]
- 18. Zhou H., Tran P. O., Yang S., Zhang T., LeRoy E., Oseid E., Robertson R. P. (2004) Regulation of α-cell function by the β-cell during hypoglycemia in Wistar rats: the “switch-off” hypothesis. Diabetes 53, 1482–1487 [DOI] [PubMed] [Google Scholar]
- 19. Hope K. M., Tran P. O., Zhou H., Oseid E., Leroy E., Robertson R. P. (2004) Regulation of α-cell function by the β-cell in isolated human and rat islets deprived of glucose: the “switch-off” hypothesis. Diabetes 53, 1488–1495 [DOI] [PubMed] [Google Scholar]
- 20. Cantley J., Choudhury A. I., Asare-Anane H., Selman C., Lingard S., Heffron H., Herrera P., Persaud S. J., Withers D. J. (2007) Pancreatic deletion of insulin receptor substrate 2 reduces β and α cell mass and impairs glucose homeostasis in mice. Diabetologia 50, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 21. Kawamori D., Kurpad A. J., Hu J., Liew C. W., Shih J. L., Ford E. L., Herrera P. L., Polonsky K. S., McGuinness O. P., Kulkarni R. N. (2009) Insulin signaling in α cells modulates glucagon secretion in vivo. Cell Metab. 9, 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z., Kim W., Chen Z., Shin Y. K., Carlson O. D., Fiori J. L., Xin L., Napora J. K., Short R., Odetunde J. O., Lao Q., Egan J. M. (2011) Insulin and glucagon regulate pancreatic α-cell proliferation. PLoS One 6, e16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broderick J. A., Zamore P. D. (2011) MicroRNA therapeutics. Gene. Ther. 18, 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guay C., Roggli E., Nesca V., Jacovetti C., Regazzi R. (2011) Diabetes mellitus, a microRNA-related disease? Transl. Res. 157, 253–264 [DOI] [PubMed] [Google Scholar]
- 26. Mao Y., Mohan R., Zhang S., Tang X. (2013) MicroRNAs as pharmacological targets in diabetes. Pharmacol. Res. 75, 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poy M. N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. (2009) miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y., Liu J., Liu C., Naji A., Stoffers D. A. (2013) MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes 62, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Latreille M., Hausser J., Stützer I., Zhang Q., Hastoy B., Gargani S., Kerr-Conte J., Pattou F., Zavolan M., Esguerra J. L., Eliasson L., Rülicke T., Rorsman P., Stoffel M. (2014) MicroRNA-7a regulates pancreatic β cell function. J. Clin. Invest. 124, 2722–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klein D., Misawa R., Bravo-Egana V., Vargas N., Rosero S., Piroso J., Ichii H., Umland O., Zhijie J., Tsinoremas N., Ricordi C., Inverardi L., Domínguez-Bendala J., Pastori R. L. (2013) MicroRNA expression in α and β cells of human pancreatic islets. PLoS One 8, e55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., Oka Y., Yamamura K. (1990) Establishment of a pancreatic β cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127, 126–132 [DOI] [PubMed] [Google Scholar]
- 32. Tang X., Muniappan L., Tang G., Ozcan S. (2009) Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. RNA 15, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brissova M., Fowler M., Wiebe P., Shostak A., Shiota M., Radhika A., Lin P. C., Gannon M., Powers A. C. (2004) Intraislet endothelial cells contribute to revascularization of transplanted pancreatic islets. Diabetes 53, 1318–1325 [DOI] [PubMed] [Google Scholar]
- 34. Piccand J., Meunier A., Merle C., Jia Z., Barnier J. V., Gradwohl G. (2014) Pak3 promotes cell cycle exit and differentiation of β-cells in the embryonic pancreas and is necessary to maintain glucose homeostasis in adult mice. Diabetes 63, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herrera P. L. (2000) Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127, 2317–2322 [DOI] [PubMed] [Google Scholar]
- 36. Xu C. R., Li L. C., Donahue G., Ying L., Zhang Y. W., Gadue P., Zaret K. S. (2014) Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J. 33, 2157–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang X., Gal J., Zhuang X., Wang W., Zhu H., Tang G. (2007) A simple array platform for microRNA analysis and its application in mouse tissues. RNA 13, 1803–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 39. Saldanha A. J. (2004) Java Treeview: extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 40. Wang W. X., Rajeev B. W., Stromberg A. J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P. T. (2008) The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 28, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu M., Roth A., Yu M., Morris R., Bersani F., Rivera M. N., Lu J., Shioda T., Vasudevan S., Ramaswamy S., Maheswaran S., Diederichs S., Haber D. A. (2013) The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 27, 2543–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma N., Wang X., Qiao Y., Li F., Hui Y., Zou C., Jin J., Lv G., Peng Y., Wang L., Huang H., Zhou L., Zheng X., Gao X. (2011) Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483–5p as an IGF2 partner. Mol. Cell Endocrinol. 333, 96–101 [DOI] [PubMed] [Google Scholar]
- 43. Rui L., Yuan M., Frantz D., Shoelson S., White M. F. (2002) SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 277, 42394–42398 [DOI] [PubMed] [Google Scholar]
- 44. Shi H., Tzameli I., Bjørbaek C., Flier J. S. (2004) Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J. Biol. Chem. 279, 34733–34740 [DOI] [PubMed] [Google Scholar]
- 45. Ueki K., Kondo T., Kahn C. R. (2004) Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol. 24, 5434–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ueki K., Kondo T., Tseng Y. H., Kahn C. R. (2004) Central role of suppressors of cytokine signaling proteins in hepatic steatosis, insulin resistance, and the metabolic syndrome in the mouse. Proc. Natl. Acad. Sci. U.S.A. 101, 10422–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rønn S. G., Hansen J. A., Lindberg K., Karlsen A. E., Billestrup N. (2002) The effect of suppressor of cytokine signaling 3 on GH signaling in β-cells. Mol. Endocrinol. 16, 2124–2134 [DOI] [PubMed] [Google Scholar]
- 48. Laubner K., Kieffer T. J., Lam N. T., Niu X., Jakob F., Seufert J. (2005) Inhibition of preproinsulin gene expression by leptin induction of suppressor of cytokine signaling 3 in pancreatic β-cells. Diabetes 54, 3410–3417 [DOI] [PubMed] [Google Scholar]
- 49. Lindberg K., Rønn S. G., Tornehave D., Richter H., Hansen J. A., Rømer J., Jackerott M., Billestrup N. (2005) Regulation of pancreatic β-cell mass and proliferation by SOCS-3. J. Mol. Endocrinol. 35, 231–243 [DOI] [PubMed] [Google Scholar]
- 50. Emanuelli B., Peraldi P., Filloux C., Chavey C., Freidinger K., Hilton D. J., Hotamisligil G. S., Van Obberghen E. (2001) SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-α in the adipose tissue of obese mice. J. Biol. Chem. 276, 47944–47949 [DOI] [PubMed] [Google Scholar]
- 51. Karlsen A. E., Rønn S. G., Lindberg K., Johannesen J., Galsgaard E. D., Pociot F., Nielsen J. H., Mandrup-Poulsen T., Nerup J., Billestrup N. (2001) Suppressor of cytokine signaling 3 (SOCS-3) protects β-cells against interleukin-1β- and interferon-γ-mediated toxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 12191–12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emanuelli B., Glondu M., Filloux C., Peraldi P., Van Obberghen E. (2004) The potential role of SOCS-3 in the interleukin-1β-induced desensitization of insulin signaling in pancreatic β-cells. Diabetes 53, Suppl. 3, S97–S103, 10.2337/diabetes.53.suppl_3.S97 [DOI] [PubMed] [Google Scholar]
- 53. Wang Q., Brubaker P. L. (2002) Glucagon-like peptide-1 treatment delays the onset of diabetes in 8 week-old db/db mice. Diabetologia 45, 1263–1273 [DOI] [PubMed] [Google Scholar]
- 54. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shin C., Nam J. W., Farh K. K., Chiang H. R., Shkumatava A., Bartel D. P. (2010) Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jorgensen S. B., O'Neill H. M., Sylow L., Honeyman J., Hewitt K. A., Palanivel R., Fullerton M. D., Öberg L., Balendran A., Galic S., van der Poel C., Trounce I. A., Lynch G. S., Schertzer J. D., Steinberg G. R. (2013) Deletion of skeletal muscle SOCS3 prevents insulin resistance in obesity. Diabetes 62, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Monteys A. M., Spengler R. M., Wan J., Tecedor L., Lennox K. A., Xing Y., Davidson B. L. (2010) Structure and activity of putative intronic miRNA promoters. RNA 16, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cornu M., Yang J. Y., Jaccard E., Poussin C., Widmann C., Thorens B. (2009) Glucagon-like peptide-1 protects β-cells against apoptosis by increasing the activity of an IGF-2/IGF-1 receptor autocrine loop. Diabetes 58, 1816–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Devedjian J. C., George M., Casellas A., Pujol A., Visa J., Pelegrín M., Gros L., Bosch F. (2000) Transgenic mice overexpressing insulin-like growth factor-II in β cells develop type 2 diabetes. J. Clin. Invest. 105, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petrik J., Arany E., McDonald T. J., Hill D. J. (1998) Apoptosis in the pancreatic islet cells of the neonatal rat is associated with a reduced expression of insulin-like growth factor II that may act as a survival factor. Endocrinology 139, 2994–3004 [DOI] [PubMed] [Google Scholar]
- 61. Hill D. J., Strutt B., Arany E., Zaina S., Coukell S., Graham C. F. (2000) Increased and persistent circulating insulin-like growth factor II in neonatal transgenic mice suppresses developmental apoptosis in the pancreatic islets. Endocrinology 141, 1151–1157 [DOI] [PubMed] [Google Scholar]
- 62. Place R. F., Li L. C., Pookot D., Noonan E. J., Dahiya R. (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 105, 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim D. H., Saetrom P., Snøve O., Jr., Rossi J. J. (2008) MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 105, 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barbagallo D., Piro S., Condorelli A. G., Mascali L. G., Urbano F., Parrinello N., Monello A., Statello L., Ragusa M., Rabuazzo A. M., Di Pietro C., Purrello F., Purrello M. (2013) miR-296–3p, miR-298–5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics 14, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baroukh N., Ravier M. A., Loder M. K., Hill E. V., Bounacer A., Scharfmann R., Rutter G. A., Van Obberghen E. (2007) MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J. Biol. Chem. 282, 19575–19588 [DOI] [PubMed] [Google Scholar]
- 66. Lovis P., Gattesco S., Regazzi R. (2008) Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 389, 305–312 [DOI] [PubMed] [Google Scholar]
- 67. Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M. H., Stoffel M. (2011) MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 [DOI] [PubMed] [Google Scholar]