Abstract
Enterococcus faecalis is the most commonly isolated species from endodontic failure root canals; its persistence in treated root canals has been attributed to its ability to resist high pH stress. The goal of this study was to characterize the E. faecalis transcriptome and to identify candidate genes for response and resistance to alkaline stress using Illumina HiSeq 2000 sequencing. We found that E. faecalis could survive and form biofilms in a pH 10 environment and that alkaline stress had a great impact on the transcription of many genes in the E. faecalis genome. The transcriptome sequencing results revealed that 613 genes were differentially expressed (DEGs) for E. faecalis grown in pH 10 medium; 211 genes were found to be differentially up-regulated and 402 genes differentially down-regulated. Many of the down-regulated genes found are involved in cell energy production and metabolism and carbohydrate and amino acid metabolism, and the up-regulated genes are mostly related to nucleotide transport and metabolism. The results presented here reveal that cultivation of E. faecalis in alkaline stress has a profound impact on its transcriptome. The observed regulation of genes and pathways revealed that E. faecalis reduced its carbohydrate and amino acid metabolism and increased nucleotide synthesis to adapt and grow in alkaline stress. A number of the regulated genes may be useful candidates for the development of new therapeutic approaches for the treatment of E. faecalis infections.
Keywords: E. faecalis, alkaline stress, biofilms, transcriptome, genome, sequencing
Introduction
Enterococcus spp. are natural inhabitants of the gastrointestinal tracts of humans and animals (Creti et al., 2004), but they can also be found in water, soil, and vegetables (Burgos et al., 2009). The two most important species, Enterococcus faecium and Enterococcus faecalis, are most frequently implicated in human and animal infections (Aakra et al., 2005). They have been reported to be associated with oral mucosal lesions in immunocompromised patients, periodontitis, and root canal infections (Wahlin and Holm, 1988; Rams et al., 1992; Siqueira and Rôças, 2004).
E. faecalis is the species that is most commonly isolated from the root canals of teeth with failed endodontic treatment (Sundqvist et al., 1998). Moreover, E. faecalis was most often detected as the single species in persistently infected root canals. It has been reported that in teeth with necrotic pulps, E. faecalis has rarely been isolated by culturing (4%), but has frequently been detected by PCR (82%); in fact, it is detected as occurring frequently in teeth with failing endodontic treatment. E. faecalis may occur in low numbers in necrotic pulp, keeping it from being cultured from these samples (Gomes et al., 2006). Moreover, the success rate for the teeth from which E. faecalis was isolated after removal of the earlier root filling was somewhat lower (66%) than the overall success rate of re-treatment (74%) (Sundqvist et al., 1998). Therefore, E. faecalis involved in the etiology of endodontic failure may be attributed to the organism being present in necrotic pulps that survived until chemo-mechanical procedures were performed and intracanal medications were given, which caused ecological changes in the root canals (Love, 2001; Gomes et al., 2003).
Calcium hydroxide is known to be one of the most effective and commonly used endodontic medicaments because of the bactericidal effects derived from its strong alkaline properties; it can provide an extreme alkalinity of approximately pH 12 (Athanassiadis et al., 2007). However, some studies report that calcium hydroxide is not so effective against E. faecalis (Evans et al., 2002; Gomes et al., 2003). The persistence of E. faecalis in treated root canals has been attributed to its ability to resist the high pH of the antimicrobial agents commonly used during treatment (Waltimo et al., 2005). It has even been shown that E. faecalis can directly form biofilms on calcium hydroxide paste in medicated root canals (Distel et al., 2002), and a slight pH increase enhances E. faecalis adhesion to collagen type I (the major organic component of dentine) (Kayaoglu et al., 2005). Moreover, an extreme pH cannot be achieved within dentinal tubules because of the buffering effects of dentin (Haapasalo et al., 2000).
The antimicrobial activity of Ca(OH)2 relates to its release of hydroxide ions (Freeman and Crapo, 1982). The destruction of phospholipids, the structural components of the cellular membrane, can be caused by the induction of lipid peroxidation by hydroxide ions (Halliwell, 1987). Genes are destroyed when hydroxide ions react with bacterial DNA and cause the denaturation of the strands (Imlay and Linn, 1988). It has been reported that the expression of some stress response genes are increased markedly in alkaline media (Appelbe and Sedgley, 2007). An analysis of stress proteins revealed that 37 polypeptides were amplified when subjected to alkaline shock (Flahaut et al., 1997). Although a number of virulence-related traits have been established, no comprehensive genomic or transcriptomic studies have been conducted to investigate how E. faecalis resists alkaline stress. To identify the potential genetic traits or gene regulatory features that allow E. faecalis to survive in alkaline stress, we performed genomic analysis and investigated the growth capacity and transcriptome profiling in pH 10 media, which may ultimately contribute to the development of strategies for the prevention of E. faecalis infections and improve root canal treatment strategies.
Materials and methods
Bacterial strains and growth conditions
E. faecalis (ATCC 33186) was used in all experiments and stored in a 50% (v/v) glycerol solution at −80°C. Bacteria were prepared for all experiments by the inoculation of tryptic soy broth (TSB) containing 1.7% tryptone, 0.3% polypeptone, 0.1% yeast extract (Sangon, Shanghai, China), 0.5% NaCl, 0.25% glucose, and 0.25% dipotassium phosphate, which was incubated aerobically at 37°C for 16–18 h. Under these conditions, the cultures entered stationary phase at an optical density of 1.1 at 600 nm (OD600 nm). The alkaline media (pH 10) was adjusted with maleic acid and K2CO3(Kakinuma and Igarashi, 1990) and sterilized through a 0.22-μm (pore size) filter unit (Millipore, Millex-GP, USA).
Biofilm assay
Confocal microscopy was performed on E. faecalis biofilms grown on glass coverslips. Sterile glass coverslips that were placed on the bottom of six-well culture plates and submerged with 6 mL of alkaline and TSB medium were seeded with a 1:100 dilution of E. faecalis from an overnight culture (OD600 nm = 1.1) and grown for 24 h at 37°C. Just prior to imaging, the biofilms were gently rinsed two times with sterile phosphate-buffered saline, followed by 15 min of staining with 100 μl of the fluorescent stains SYTO9 and PI (Invitrogen, California, USA), which were diluted in PBS in proportions of 1.5:1.5:1000. The slides were visualized by an inverted Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems, Mannheim, Germany). A 488/543-nm double dichroic mirror was used as an excitation beam splitter, and a 405-nm short-pass filter divided green and red fluorescence between the photomultipliers. Identification of viable cells and biofilm quantification was carried out using the Leica confocal software.
Genome sequencing
Bacteria were incubated aerobically in TSB at 37°C for 24 h. Cells (30 ml) were collected by centrifugation (4000 rpm, 15 min) at 4°C, and the pellets were immediately frozen in liquid nitrogen and kept at −80°C prior to DNA extraction. Genomic DNA extraction was carried out with a bacterial genomic DNA purification kit (EdgeBio, Maryland, USA) according to the manufacturer's instructions. The genome library was constructed by DNA fragmentation with Covaris; both ends of the DNA fragments were fixed with complementary primers to form a “bridge” for subsequent PCR amplification and the generation of single stranded DNA. Sequencing was carried out on an Illumina HiSeq 2000 platform (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China).
RNA extraction and sequencing
Biofilm cells were grown for 24-h in TSB OR in pH 10 medium and were harvested by centrifugation (4000 rpm, 15 min) at 4°C. Cells were then resuspended with TRIzol (Invitrogen, Carlsbad, USA) and disrupted with a Mini-Bead beater (Biospec, California, USA). Total RNA was isolated according to the manufacturer's instructions for TRIzol-chloroform extraction (Invitrogen, California, USA). Genomic DNA was removed with Turbo DNase. Ribosomal RNA (rRNA) was removed with a Ribo-Zero Magnetic kit (G+/G-Bacteria) (Epicentre, Wisconsin, USA). The cDNA library was constructed with extracted mRNA according to a TruseqTM RNA sample prep kit (Illumina, California, USA). The sample was purified and ligated to the genomic adapters provided by Illumina. The purified samples were amplified by 15-cycle PCR. After ligation, the sample was loaded on a 2% agarose E-gel (Invitrogen). The targeted fragments were excised from the gel and purified using Certified Low Range Ultra Agarose (Bio-Rad). The sample was quantified using TBS380 Picogreen (Invitrogen). Sequencing was carried out on the Illumina HiSeq 2000 platform (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China).
Data analysis
The raw image data were converted to sequence data through Base Calling. Then, the raw sequence data were filtered with SeqPrep and Sickle software to obtain clean data. The clean reads from each sample were mapped to the reference genome that was obtained from preceding genome sequencing by Bowtie software (Langmead et al., 2009). The predicted protein sequence from the open reading frames (ORFs) were annotated with NR, string, and a gene database using blastp. Based on their homology to protein families, proteins predicted for E. faecalis were assigned parental (i.e., level 2) Gene Ontology (GO) terms (http://www.geneontology.org/). Deduced proteins with homologs in other organisms were used to determine the Clusters of Orthologous Groups of proteins (COG) item; the COG functional categories were assigned with the STRING database (String v9.0). Based on the KEGG (Kyoto Encyclopedia of Genes and Genomes database), all of the genes were mapped to the KEGG gene database (GENES) by using the BLAST algorithm (blastx/blastp2.2.24+) to find the potential biological pathway of the genes (Camacho et al., 2009).
Differentially expressed genes (DEGs) were obtained based on the RPKM/FPKM [Reads/(Fragments) Per Kilobase of exon model per Million] followed by a multiple hypothesis testing, False Discovery Rate (FDR) control (Trapnell et al., 2010) to correct for the p-value. Genes with an FDR value ≤ 0.05 and |logFC| ≥ 1 were assigned as differentially expressed. Differences in gene expression were also analyzed by the R package, EdgeR (Robinson et al., 2010).
Results
Biofilm formation of E. faecalis by CLSM
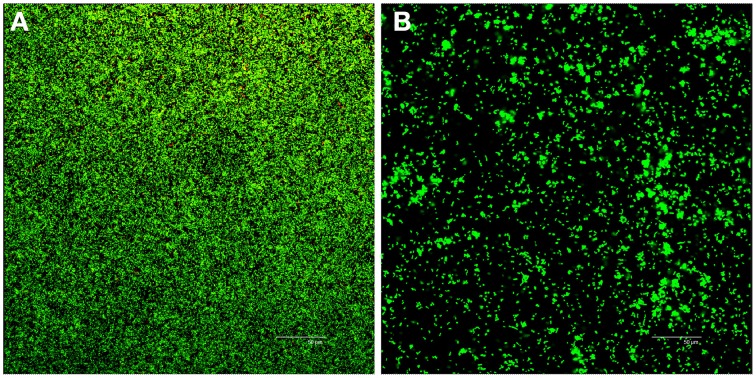
Confocal laser scanning microscopy (CLSM) analysis of 24-h-old biofilms in the control group (pH 7.4) showed a dense and compact biofilm with a complex architecture (Figure 1A); in alkaline media (pH 10), CLSM showed poor biofilms that were mainly composed of isolated microcolonies with sparse cells on the glass surface (Figure 1B). The viable bacteria and biofilm biomass were significantly reduced in pH 10 media compared with the control group (Independent-samples T-test, P < 0.05).
Figure 1.
Confocal analysis of 24-h biofilms of E. faecalis in TSB and pH 10 media. Live bacteria are green, and dead cells are visualized in red. Panels (A,B) are representative biofilm projections of E. faecalis in control group (TSB) and pH 10 media, respectively.
Genomic analysis
Because there is no sequence data publicly available for E. faecalis ATCC33186, genome sequencing was performed before transcriptional analysis to obtain a detailed account of the genetic expression and to validate the performance of our RNA-Seq. The genome sequence was determined by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shang-hai, China), using Illumina HiSeq2000 paired-end sequencing technology (300-bp library). A total of 433,968,680 high-quality base pairs (4,339,345 high-quality reads, with an average read length of 91 bp), giving 148.7-fold coverage of the genome, were assembled into 73 contigs and 63 scaffolds using SOAPdenovo software (http://soap.genomics.org.cn/). The genome of E. faecalis ATCC33186 consists of 63 scaffolds of 2,918,140 bp and had an average G+C content of 37.36%. There are a total of 2824 putative open reading frames (with an average size of 896 bp) predicted by Glimmer 3.0. The detailed sequence information of the genome is listed in Table 1.
Table 1.
General information of the genome from E. faecalis ATCC33186.
| Coding DNA Sequence (CDS) | 2824 |
| Genome length | 2918140 bp |
| GC content in gene region (%) | 38 |
| Gene average length | 896 bp |
| GC content in non-coding sequence (%) | 33 |
| N rate* | 0% |
N rate is the percentage of unknown bases. In the process of sequence assembly, the unknown bases between contigs were filled with N.
Using the published DNA sequence of V583(NC_004668.1), OG1RF(NC_017316.1), and ATCC 29212(NZ_CP008816.1) as reference, E. faecalis ATCC33186 shares 2362, 2284, and 2402 ORFs, respectively, as well as the 3 rRNA genes and 41 tRNA genes (Table 2). Their genome size and GC content were similar (ranging from 2.7 to 3.2 Mb and from 37.4 to 37.8%, respectively).
Table 2.
Gene features of ATCC33186, compared to V583, OG1RF, and ATCC 29212.
| ATCC33186 | V583 | OG1RF | ATCC 29212 | |
|---|---|---|---|---|
| Size (bp) | 2,918,140 | 3,218,031 | 2,739,625 | 2,939,973 |
| GC content (%) | 37.4 | 37.5 | 37.8 | 37.5 |
| Gene | 2868 | 3257 | 2640 | 3053 |
| rRNA | 3* | 12 | 12 | 12 |
| tRNA | 41* | 68 | 58 | 61 |
| ORFs | 2824 | 3113 | 2559 | 2908 |
| Compared to V583 | Compared to OGIRF | Compared to ATCC 29212 | ||
| ORFs common to both strains | 2362 | 2284 | 2402 | |
| Unique ORFs | 462 | 540 | 422 | |
| Similar to known proteins | 412 | 456 | 358 | |
| Conserved hypotheticals | 46 | 79 | 60 | |
| No database match | 4 | 5 | 4 | |
The big difference in number of rRNA and tRNA compared with the reported strains partly attribute to the absence of completely assembled chromosome of strain ATCC33186 in the present study.
Gene ontology assignments
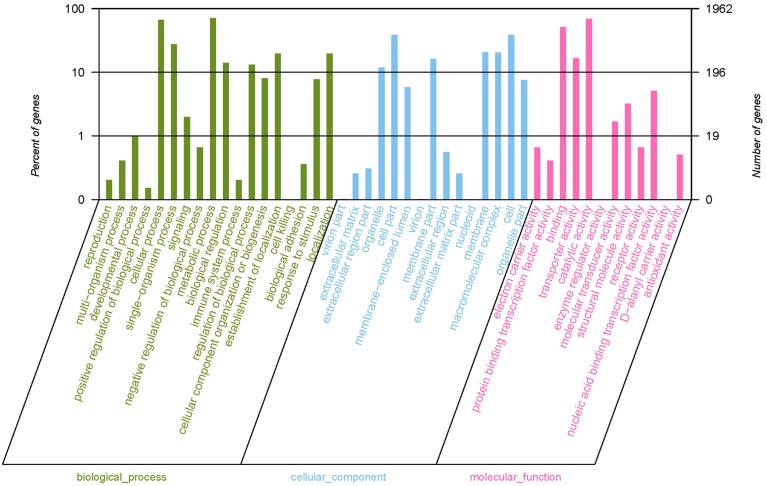
Gene ontology (GO) provided a structured and controlled vocabulary for describing gene products in three categories: molecular functions, cellular components and biological processes, which are helpful to understand the distribution of gene functions of the species at the macro level. We added GO terms using Blast2GO (Conesa et al., 2005), which is based on the automated annotation of all of the unique sequences in the GenBank nr protein database from NCBI. Among the 2824 assembled genes, 1962 were successfully annotated by GO assignments, belonging to one or more sub-categories of GO terms. Among the annotated unigenes, 1813 sequences were assigned to a molecular function category, including catalytic activity (1357 genes), binding (1003 genes), transporter activity (324 genes), nucleic acid binding transcription factor activity (100) and others (Figure 2). Additionally, 980 unigenes are divided into a cellular component category, including cells (759 unigenes), cell parts (759 unigenes), macromolecular complexes (399 unigenes), membrane part (319 unigenes), membranes (407 unigenes), organelles (232 unigenes), and others (Figure 2). In addition, 1722 unigenes are involved in the biological process category, including cellular processes (1297 unigenes), metabolic processes (1399 unigenes), single-organism processes (541 unigenes), establishment of localizations (387 unigenes), localizations (387 unigenes), and others (Figure 2). Because some sequences were assigned to more than one GO term, the total number of GO terms obtained in our dataset was bigger than the total number of the unique sequences.
Figure 2.
Gene ontology (GO) terms for the transcriptome sequences of E. faecalis. Most of the annotated sequences can be divided into three major categories, including biological process (A), cellular component (B), and molecular function (C). The vertical scale on the left indicated the the percentage of each sub-categories of GO terms to the total 1962 genes which were successfully annotated by GO assignments. The vertical scale on the right side indicated the number of each sub-categories of GO terms. Because some genes were assigned to more than one GO term, the sum of the percentage of all sub-categories of GO terms was more than one and the total number of GO terms was bigger than the total number of the unique sequences.
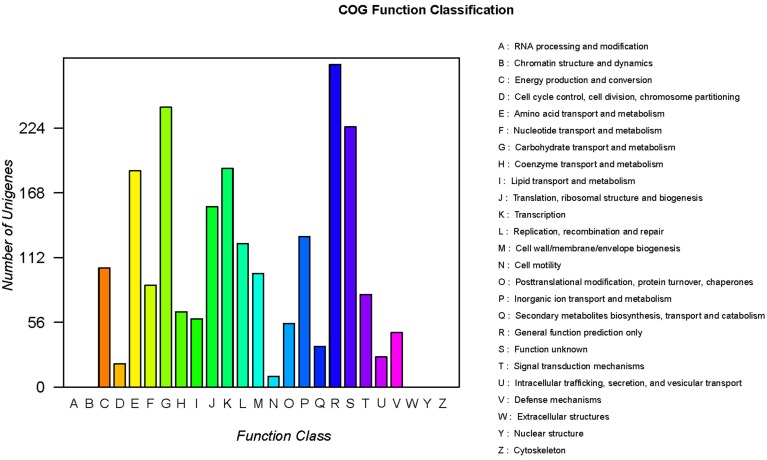
COG annotation
COG classification of the unigenes is important for functional annotation and evolutionary studies (Tatusov et al., 2001). Assignments of COG were used to predict and classify the possible functions of the unique sequences. Based on the sequence homology, a total of 2264 DNA-seqs were finally mapped on 20 different COG categories (Figure 3). With the exception of the R or S categories of COG, which are “function unknown” (225 unigenes) or “general function prediction only” (279 unigenes), others were related to the normal physiological metabolism of cells, such as regulation, transport and cell processing. The largest COG group was “carbohydrate transport and metabolism” (242 unigenes), followed by “amino acid transport and metabolism” (187 unigenes), “transcription” (189 unigenes), “translation, ribosomal structure and biogenesis” (156 unigenes), and others (Figure 3). In summary, these terms account for a large fraction of the overall assignments in the E. faecalis transcriptome dataset.
Figure 3.
Clusters of the orthologous group (COG) classifications of the E. faecalis sequences.
KEGG pathway mapping
The DNAseqs were mapped to the KEGG database to define their metabolic pathways. Pathway-based annotation and analysis helps to further understand the biological function of the genes. According to the KEGG results, 1532 unigenes were classified into 144 KEGG metabolic pathways, representing compound biosynthesis, degradation, utilization and metabolism (Table S1). Many metabolic-related pathways, including fructose and mannose metabolism, pyrimidine metabolism, and fatty acid metabolism, and many biosynthesis pathways, such as the biosynthesis of secondary metabolites, aminoacyl-tRNA biosynthesis, peptidoglycan biosynthesis, fatty acid biosynthesis and so on, were predicted in the KEGG database (Table S1). As in the above GO, one annotated unigene was assigned to one or more of the KEGG pathways.
KEGG pathway analysis and COG analysis are helpful for predicting potential genes and their functions at the whole transcriptome level. The predicted metabolic pathways, together with COG analysis, are useful for further investigations of gene functions in future studies.
Transcriptome sequences assembly and analysis
Whole genome mRNA sequencing is an attractive method for monitoring global changes in gene expression while overcoming many of the pitfalls of traditional DNA microarrays (Marioni et al., 2008). To identify the genes important for E. faecalis-tolerating alkaline stress, we searched for genes that were differentially expressed in pH 10 media. Total RNA was isolated from E. faecalis biofilm cells grown for 24-h in TSB or pH 10 media (see the Materials and Methods Section). For the first set of samples, the mRNA was selectively enriched through a single step of rRNA depletion. These samples were fragmented and used to obtain cDNA libraries that were sequenced on an Illumina HiSeq 2000 platform. The detailed sequence information of the transcriptome is listed in Table S2. A total of 1,415,985,842 and 1,810,228,366 bases from 14,350,108 and 18,494,702 Illumina clean reads with a mean read length of 101 bp were obtained from bacterial samples cultured with TSB or pH 10 media, respectively. Of these, 13,910,120 (96.93%) and 18,130,016 (98.03%) clean reads from the control and pH 10 groups were mapped to the reference genome, respectively. Approximately 6,223,986 (43.37%) and 5,288,486 (28.59%) clean reads mapped to CDS.
Differentially expressed genes
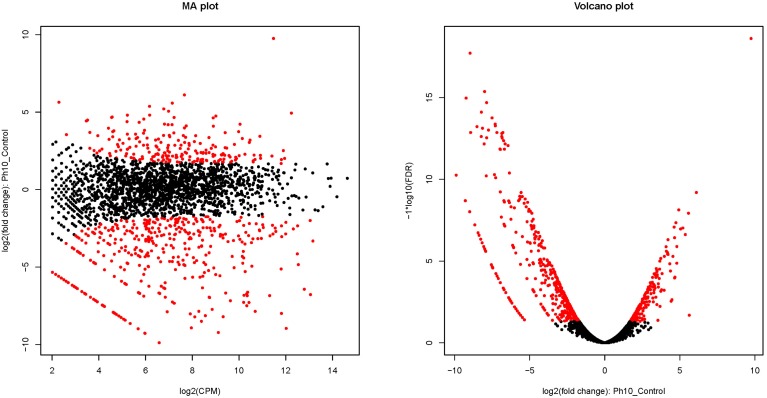
Primary sequence analysis and annotations of all of the unigenes provided us with a great deal of useful information to understand the transcriptome and to further define the DEGs that are induced by alkaline stress. The transcriptome sequencing results revealed 613 genes that were differentially expressed (DEGs) for E. faecalis grown in pH 10 medium (Figure 4). Among these DEGs, 211 genes were found to be differentially up-regulated genes (DUGs), and 402 genes were identified as differentially down-regulated genes (DDGs). The data, including the gene ID, gene length, FRKM in the control and pH 10 groups, log2FC1 (pH 10/Control), FDR, up- or down-regulation (pH 10/control) and annotation of all of the DEGs are provided in Table S3.
Figure 4.
Differentially expressed genes of E. faecalis cultured in pH 10 media. Red dots indicate differentially expressed genes. Black-colored dots were not considered as significantly differentially expressed. In the figure of the MA plot, the X-axis shows the average count of reads per million reads based on a log2 scale, and the Y-axis shows the fold-change values between the control and alkaline group based on a log2 scale. In the figure of a volcano plot, the X-axis shows the fold-change values between the control and alkaline groups based on a log2 scale, and the Y-axis shows the FDR value of differentially expressed genes based on a -log10 scale.
Up-regulated genes (DUGs)
GOG analysis showed the DEGs in 20 predicted pathways. The number of up-regulated genes in each functional category is shown in Table 3. Among the 211 up-regulated genes, the genes encoding amino acid transport and metabolism represented the highest percentage (34 genes; around 16% of all up-regulated genes), while genes related to nucleotide transport and metabolism around 11% (24 genes) of the genes induced by the alkaline stress. Moreover, genes encoding proteins involved in inorganic ion transport and metabolism (16 genes), cell wall/membrane/envelope biogenesis (9 genes), coenzyme transport and metabolism (8 genes), carbohydrate transport and metabolism (7 genes), energy production and conversion (6 genes), and cell cycle control, cell division, chromosome partitioning (2 genes) were up-regulated. In addition, genes classified in the category of genes of unknown function represent a considerable part of the induced genes (26 genes). The majority of up-regulated genes (15 genes) related to amino acid transport and metabolism are genes encoding amino acid or peptide ABC transporter (Table S4). Table S4 gives the full list of genes found to be induced (up-regulated), with fpkm value. Genes predicted to encode hypothetical proteins are excluded from this list.
Table 3.
Number of genes that were differentially expressed in alkaline stress.
| COG category | Genes | DUGs | DDGs |
|---|---|---|---|
| Energy production and conversion | 104 | 6 | 24 |
| Cell cycle control, cell division, chromosome partitioning | 20 | 2 | 0 |
| Amino acid transport and metabolism | 187 | 34 | 11 |
| Nucleotide transport and metabolism | 87 | 24 | 1 |
| Carbohydrate transport and metabolism | 243 | 7 | 41 |
| Coenzyme transport and metabolism | 64 | 8 | 7 |
| Lipid transport and metabolism | 58 | 1 | 4 |
| Translation, ribosomal structure and biogenesis | 156 | 8 | 8 |
| Transcription | 193 | 8 | 15 |
| Replication, recombination and repair | 125 | 5 | 10 |
| Cell wall/membrane/envelope biogenesis | 96 | 9 | 6 |
| Cell motility | 9 | 2 | 0 |
| Posttranslational modification, protein turnover, chaperones | 56 | 2 | 10 |
| Inorganic ion transport and metabolism | 127 | 16 | 12 |
| Secondary metabolites biosynthesis, transport and catabolism | 35 | 1 | 5 |
| General function prediction only | 292 | 25 | 25 |
| Function unknown | 343 | 26 | 62 |
| Signal transduction mechanisms | 70 | 2 | 6 |
| Intracellular trafficking, secretion, and vesicular transport | 27 | 3 | 1 |
| Defense mechanisms | 47 | 8 | 1 |
Down-regulated genes (DDGs)
The numbers of down-regulated genes in each functional category are shown in Table 3. Among the down-regulated genes, those with unknown function are the dominant category (62 genes; 15%), followed by genes involved in carbohydrate transport and metabolism (41 genes; 10%), genes involved in energy production and conversion (24 genes), general function prediction only (25 genes), and genes involved in transcription (15 genes), amino acid transport and metabolism (11 genes), posttranslational modification (10 genes), and translation, ribosomal structure and biogenesis (8 genes). Table S5 gives the full list of genes found to be decreased (down-regulated), with fpkm value. Genes predicted to encode hypothetical proteins are excluded from this list.
Stress response of E. faecalis toward exposure to alkaline stress
Exposure to alkaline repressed universal stress protection genes, including orf02602, orf00603 orf02216, orf02700. The gene (orf01945) encoding Gsp62 and a cold shock protein cspA (orf00260) also showed a significantly decreased expression in alkaline stress. The stress- and starvation inducible gls24 operon (orf02321-6) was significantly down-regulated. And gls24-like gene (orf02786) within the pathogenicity island (PAI) were down-regulated significantly and might possibly contribute to the decrease of E. faecalis during growth in pH 10. An organic hydroperoxide resistance gene, ohr (Gsp65; orf01411), the NADH peroxidase (orf00385) and the superoxide dismutase gene sodA (orf01421) were also down-regulated in alkaline stress. An ohr mutant has previously been shown to be less resistant to the oxidative stress generated by 20 mM Tertiary- Butylhydroperoxide, suggesting that Ohr may be implicated in oxidative stress resistance in E. faecalis (Rincé et al., 2001).
Discussion
E. faecalis are most frequently implicated in human and animal infections (Aakra et al., 2005). It is most commonly isolated from the root canals of teeth with failed endodontic treatment (Sundqvist et al., 1998), which attribute to its resistance to alkaline stress (Bystrom et al., 1985; Evans et al., 2002). Our study also confirmed that E. faecalis could survive and form biofilms at pH 10 in vitro. Although certain stress response of E. faecalis in alkaline media have been studied (Flahaut et al., 1997; Appelbe and Sedgley, 2007), the global gene regulation and interplay during adaptation to alkaline environment remain largely unaddressed. With the present study we provide novel information about the gene regulation relevant to such an adaptation process. The transcriptome sequencing results revealed that 613 genes were differentially expressed (DEGs) for E. faecalis grown in pH 10 medium; 211 genes were found to be differentially up-regulated and 402 genes differentially down-regulated.
A comparison of the ATCC33186 genome with the genome features of the published DNA sequence of V583(NC_004668.1), OG1RF(NC_017316.1) and ATCC 29212(NZ_CP008816.1) showed only minor variations: OG1RF consists of 2,739,625 bp with an average G+C content of 37.8%, ATCC 29212 consists of 2,939,973 bp with an average G+C content of 37.5% and V583 consists of 3,218,031 bp with an average G+C content of 37.5%. And ATCC33186 shared 2362, 2284 and 2402 ORFs with V583, OG1RF, and ATCC 29212, respectively, but only shared 3 rRNA genes and 41 tRNA genes. This difference may attribute to only the draft genome of strain ATCC33186 presented in the study, the missing rRNA and tRNA may be much less if the circular chromosomes are completely assembled.
The majority of up-regulated genes (15 genes) related to amino acid transport and metabolism are genes encoding amino acid or peptide ABC transporter. It indicated that the supply and transport of amino acids may be increased in pH 10 media. It has also reported that genes encoding putative amino acid ABC transporter were highly up-regulated in E. faecalis exposure to human urine (Vebø et al., 2010). Previous studies on the biosynthetic capacities and nutritional requirements of E. faecalis have shown that all strains require histidine, isoleucine, methionine, and tryptophan for growth and that arginine, glutamate, glycine, leucine, or valine was essential for the growth of some strains (Murray et al., 1993; Bourgogne et al., 2008). Moreover, E. faecalis is capable of utilizing certain amino acids as energy and carbon sources (Deibel, 1964). Because our data show that the transcription of all genes encoding ABC-transporters were up-regulated, it is possible that E. faecalis meets its survival demand by uptaking these essential peptides and amino acids when growing in alkaline stress.
Fourteen genes encoding proteins involved in nucleotide transport and metabolism displayed increased expression under alkaline conditions. The genes for pyrCDEF (orf00246, orf00247, orf00248, and orf00252) participated in pyrimidine metabolism, and all of these genes were significantly up-regulated in alkaline stress. The carAB (orf00250 and orf00251) operons and proABC (orf02358, orf023587, orf00819), which are involved in arginine and proline metabolism, also showed significant up-regulation under alkaline stress. Koo et al. (2011) also reported that the increase of the carA expression level had a positive correlation with thymidine production. The repression of the carAB operon leads to Lactobacillus plantarum FB331 growth inhibition in the presence of arginine (Nicoloff et al., 2004). The significant up-regulation of these genes may promote pyrimidine biosynthetic in the alkaline stress. The capacity for de novo pyrimidine biosynthesis is required for the virulence of some bacteria (Ghim et al., 1999).
The ade gene (orf00396) encoding adenine deaminase and add gene (orf01977) encoding adenosine deaminase were markedly up-regulated in alkaline stress. Adenine deaminase (ADE) catalyzes the conversion of adenine to hypoxanthine and ammonia (Kamat et al., 2011). The transcription of guaD (orf00812) encoding guanine deaminase, guaC (orf00814) encoding guanosine monophosphate (GMP) reductase and guaB (orf02514) encoding inosine 5′-monophosphate dehydrogenase (IMPDH) were markedly enhanced in alkaline stress. IMPDH catalyzes the pivotal step in guanine nucleotide biosynthesis (Mandapati et al., 2014). GMP reductase catalyzes the reductive deamination of GMP to inosine monophosphate (IMP). GMP first binds to the enzyme, followed by NADPH binding, and NADP(+) dissociates first, followed by IMP release. GMP reductase also plays an important role in the conversion of the nucleoside and nucleotide derivatives of guanine to adenine nucleotides. In addition, as a member of the purine salvage pathway, it plays a critical role in reutilization of free intracellular bases (Li et al., 2006; Martinelli et al., 2011). Additionally, the gmk gene (orf01089) encoding guanylate kinase was also up-regulated. Guanylate kinase is involved in the reversible transfer of phosphate groups from ATP to GMP. GMP and ATP serve as the most effective phosphate acceptors and donor, respectively (Gupta et al., 2014). This means nucleotide metabolism pathway is enhanced in alkaline stress.
The majority proteins of down-regulated genes involved in energy production and conversion encoded are related to ATP synthase. The transcript level of the atpE gene (orf00031) and atpABCDFG (orf01626-33), encoding F0F1 ATP synthase subunits, were significantly down-regulated in pH 10 media. ATP hydrolysis relies on FoF1-ATPase, which in E. faecalis, is composed of the Fo membrane proton channel and the F1 cytoplasmic catalytic site (Kakinuma, 1998; Aslangul et al., 2006). The AtpE protein is a V/A-type H+-transporting ATPase subunit E, and the H+-ATPase functions to pump protons out of the cell to form an electrochemical proton gradient (Heefner, 1982). Additionally, the nhaC gene (orf01548), encoding a Na+/H+ antiporter was significantly down-regulated in pH 10 media. According to Kakinuma and Igarashi (1990), the proton motive force is dramatically decreased and the activity of the H+-ATPase is very low at pHs above 8, and at pH 10, the proton potential proceeds to zero. Under these conditions, the Na+/H+ antiporter, which relies on H+−ATPase, does not operate. Therefore, the transcription of the genes decreased, causing an impairment of cytoplasmic alkalization activity, which can create conditions for cells survival in alkaline stress.
Most DEGs encoding proteins involved in carbohydrate transport and metabolism displayed decreased expression under alkaline conditions. 15 genes celABC (orf01842, orf02475, orf02476, orf02813), manXYZ (orf01871-4, orf01078-81) and scrA (orf00808), bglF (orf01289) involved in encoding the IIABCD component of the phosphotransferase system (PTS) were significantly down-regulated under alkaline stress. These include the predicted lactose/galactose and gluconate PTS systems, the latter is part of a predicted metabolic pathway that facilitates gluconate uptake and catabolism via the mannonate route (Deutscher et al., 2006). This is consistent with a recent investigation that showed that genes encoding PTS were down-regulated in other stress conditions (Vebø et al., 2010). PTS is a distinct method used by bacteria for sugar uptake where the source of energy is phosphoenolpyruvate (PEP). It is involved in transporting many sugars into bacteria, including glucose, mannose, fructose, and cellobiose. PTS sugars can differ between bacterial groups, with each group evolving to mirror the most suitable carbon sources available in the environment. This implies that substrates besides glucose might play a role for growth of E. faecalis in alkaline stress. Moreover, gnd (orf01075) encoding 6-phosphogluconate dehydrogenase-like protein, uxuA (orf01082) encoding mannonate dehydratase, bglA (orf01290-1, orf01293) encoding 6-phospho-beta-glucosidase, gpmB (orf01292) encoding the phosphoglycerate mutase family protein and fruK (orf01488) encoding 1-phosphofructokinase were down-regulated in alkaline stress. This suggested that the glycolytic pathway of E. faecalis becomes slack during growth in alkalis; the cells initiate use of less preferred carbon and energy sources, especially fructose and mannose. The operons orf00241, orf01641, orf01875, and orf02474, encoding glycosyl hydrolase family proteins, were all down-regulated, implying that glycan degradation may also be decreased.
E. faecalis is equipped with two pathways for glycerol dissimilation. Glycerol can either be phosphorylated first by glycerol kinase and then oxidized by glycerol-3-phosphate oxidase (the glpK pathway) or first be oxidized by glycerol dehydrogenase and then phosphorylated by dihydroxyacetone kinase (the dhaK pathway) (Bizzini et al., 2010). In our study, the GlpF operon (orf00557) encoding glycerol uptake facilitator protein, glxK (orf01600) encoding glycerate kinase and dhaKL (orf02382-3) encoding dihydroxyacetone kinase were significantly down-regulated, suggesting that glycerol metabolism was reduced. The enzymes of glycerol metabolism are crucial for the pathogenicity of M. pneumoniae but also for other essential, yet to be identified, functions in the M. pneumoniae cell (Hames et al., 2009). The down-regulation of Glp and glxK may influence other functions or the pathogenicity of E. faecalis.
However, the metE gene (orf01358) encoding methionine synthase was significantly down-regulated under alkaline stress. It has been demonstrated that the stabilized MetE enzymes alone were responsible for the accelerated growth and higher survival observed in E. coli cells at elevated temperatures (Mordukhova and Pan, 2013). This down-regulation may reduce the synthesis of cysteine and methionine, so cells grow slower. Orf01979-encoding lipase/acylhydrolase and orf02404-encoding peptidase T, which cleaves the N-terminal amino acid of tripeptides, were also down-regulated in alkaline stress. The hydrolytic enzymes lipase and peptidase were found to be involved in the life cycle of Enterococcus species (Fisher and Phillips, 2009). This may also explains the slower biofilm growth in alkaline stress.
In our study, E. faecalis down-regulated stress protection genes in response to alkaline stress. Inactivation of gls24 and glsB (orf02322 and orf02323, respectively) has been reported to have a pleiotrophic effect on cell morphology and stress tolerance in E. faecalis (Giard et al., 2000). In our previous study, we also have observed cells under alkaline stress developed irregular shapes with pits on the cell wall surface that resemble separation anomalies (Ran et al., 2013). However, E. faecalis induces gls24 gene and some other stress protection genes in response to other different stress conditions (Giard et al., 1997; Vebø et al., 2009, 2010). Proteomic analyses with systematic exposure to various stresses have previously identified six genes encoding general stress response proteins (GSPs) which were up-regulated in E. faecalis by a wide variety of environmental stimuli (Rince et al., 2000). The reasons for the contrary results may be that the time of E. faecalis exposure to the stress conditions does not exceed 60 min in these studies, but in our study E. faecalis grown in alkaline stress for 24 h before analysis, the survival cells had adapted to the alkaline environment, so most of the differently regulated stress response genes were down-regulated which indicated the sensitivity to alkaline stress might decrease and its stress tolerance in alkaline stress might increase.
Conclusion
In this study, de novo transcriptome sequencing of E. faecalis using the Illumina 2000 was performed for the first time. A total of 18,494,702 high-quality transcriptome reads were obtained, giving rise to an average of 101 bp per read. Compared to the control group, many genes involved in carbohydrate transport and metabolism and energy production and conversion were significantly down-regulated, but genes involved nucleotide and amino acid transport and metabolism were significantly up-regulated, which might contribute to adaptation and survival of E. faecalis in alkaline stress. Most interesting is mostly differently expressed stress response genes were down-regulated in alkaline stress. This study provides new insights into the adaptive process of E. faecalis to growth and persistence in alkaline stress, and represent a first step in the process to identify proteins that might be important to the survival of E. faecalis in high pH environments. It is hoped that future studies can incorporate the rapid advances underway in metabolomics and proteomics. Extrapolation of the present data to the clinical situation can only be speculative. However, the present data may contribute toward better understanding the survival strategies of E. faecalis following prolonged exposure to pH levels similar to those encountered in root canals receiving intracanal Ca(OH)2 treatment.
Nucleotide sequence accession number
The draft genome sequence of E. faecalis ATCC33186 and the RNA-seq sequencing data have been deposited in NCBI database under accession number SRP056562 (DNA sequence) and SRP056563 (RNA sequence).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This investigation was supported by grants from the National Natural Science Funds of China (No. 81271133) and the Science and Technology Commission of Shanghai Municipality (No. 11495811000). We acknowledge Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. for their kindly help in sequencing and bioinformatics analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00795
KEGG summary of E. faecalis sequences (XLS).
The detail sequence information of the transcriptome.
The data of all of the gene transcription information (XLS).
The list of genes found to be induced (up-regulated) in alkaline stress.
The list of genes found to be decreased (down-regulated) in alkaline stress.
References
- Aakra A., Vebø H., Snipen L., Hirt H., Aastveit A., Kapur V., et al. (2005). Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 49, 2246–2259. 10.1128/AAC.49.6.2246-2259.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbe O. K., Sedgley C. M. (2007). Effects of prolonged exposure to alkaline pH on Enterococcus faecalis survival and specific gene transcripts. Oral Microbiol. Immunol. 22, 169–174. 10.1111/j.1399-302X.2007.00340.x [DOI] [PubMed] [Google Scholar]
- Aslangul E., Massias L., Meulemans A., Chau F., Andremont A., Courvalin P., et al. (2006). Acquired gentamicin resistance by permeability impairment in Enterococcus faecalis. Antimicrob. Agents Chemother. 50, 3615–3621. 10.1128/AAC.00390-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiadis B., Abbott P. V., Walsh L. J. (2007). The use of calcium hydroxide, antibiotics and biocides as antimicrobial medicaments in endodontics. Aust. Dent. J. 52, S64–S82. 10.1111/j.1834-7819.2007.tb00527.x [DOI] [PubMed] [Google Scholar]
- Bizzini A., Zhao C., Budin-Verneuil A., Sauvageot N., Giard J. C., Auffray Y., et al. (2010). Glycerol is metabolized in a complex and strain-dependent manner in Enterococcus faecalis. J. Bacteriol. 192, 779–785. 10.1128/JB.00959-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A., Garsin D. A., Qin X., Singh K. V., Sillanpaa J., Yerrapragada S., et al. (2008). Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. 10.1186/gb-2008-9-7-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos M. J., López R. L., Abriouel H., Omar N. B., Galvez A. (2009). Multilocus sequence typing of Enterococcus faecalis from vegetable foods reveals two new sequence types. Foodborne Pathog. Dis. 6, 321–327. 10.1089/fpd.2008.0169 [DOI] [PubMed] [Google Scholar]
- Bystrom A., Claesson R., Sundqvist G. (1985). The antibacterial effect of camphorated paramonochlorophenol, camphorated phenol and calcium hydroxide in the treatment of infected root canals. Endod. Dent. Traumatol. 1, 170–175. 10.1111/j.1600-9657.1985.tb00652.x [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Creti R., Imperi M., Bertuccini L., Fabretti F., Orefici G., Di Rosa R., et al. (2004). Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 53, 13–20. 10.1099/jmm.0.05353-0 [DOI] [PubMed] [Google Scholar]
- Deibel R. H. (1964). Utilization of arginine as an energy source for the growth of Streptococcus faecalis. J. Bacteriol. 87, 988–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J., Francke C., Postma P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031. 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel J. W., Hatton J. F., Gillespie M. J. (2002). Biofilm formation in medicated root canals. J. Endod. 28, 689–693. 10.1097/00004770-200210000-00003 [DOI] [PubMed] [Google Scholar]
- Evans M., Davies J. K., Sundqvist G., Figdor D. (2002). Mechanisms involved in the resistance of Enterococcus faecalis to calcium hydroxide. Int. Endod. J. 35, 221–228. 10.1046/j.1365-2591.2002.00504.x [DOI] [PubMed] [Google Scholar]
- Fisher K., Phillips C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757. 10.1099/mic.0.026385-0 [DOI] [PubMed] [Google Scholar]
- Flahaut S., Hartke A., Giard J. C., Auffray Y. (1997). Alkaline stress response in Enterococcus faecalis: adaptation, cross-protection, and changes in protein synthesis. Appl. Environ. Microbiol. 63, 812–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. A., Crapo J. D. (1982). Biology of disease: free radicals and tissue injury. Lab. Invest. 47, 412–426. [PubMed] [Google Scholar]
- Ghim S. Y., Kim C. C., Bonner E. R., D'Elia J. N., Grabner G. K., Switzer R. L. (1999). The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 181, 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giard J. C., Hartke A., Flahaut S., Boutibonnes P., Auffray Y. (1997). Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res. Microbiol. 148, 27–35. 10.1016/S0923-2508(97)81897-9 [DOI] [PubMed] [Google Scholar]
- Giard J. C., Rince A., Capiaux H., Auffray Y., Hartke A. (2000). Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182, 4512–4520. 10.1128/JB.182.16.4512-4520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes B. P., Pinheiro E. T., Sousa E. L., Jacinto R. C., Zaia A. A., Ferraz C. C., et al. (2006). Enterococcus faecalis in dental root canals detected by culture and by polymerase chain reaction analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 102, 247–253. 10.1016/j.tripleo.2005.11.031 [DOI] [PubMed] [Google Scholar]
- Gomes B. P., Souza S. F., Ferraz C. C., Teixeira F. B., Zaia A. A., Valdrighi L., et al. (2003). Effectiveness of 2% chlorhexidine gel and calcium hydroxide against Enterococcus faecalis in bovine root dentine in vitro. Int. Endod. J. 36, 267–275. 10.1046/j.1365-2591.2003.00634.x [DOI] [PubMed] [Google Scholar]
- Gupta S., Yadav S., Singh N., Verma A., Siddiqi I., Saxena J. K. (2014). Purification and characterization of guanylate kinase, a nucleoside monophosphate kinase of Brugia malayi. Parasitology 141, 1341–1352. 10.1017/S0031182014000675 [DOI] [PubMed] [Google Scholar]
- Haapasalo H. K., Sirén E. K., Waltimo T. M., Ørstavik D., Haapasalo M. P. (2000). Inactivation of local root canal medicaments by dentine: an in vitro study. Int. Endod. J. 33, 126–131. 10.1046/j.1365-2591.2000.00291.x [DOI] [PubMed] [Google Scholar]
- Halliwell B. (1987). Oxidants and human disease: some new concepts. FASEB J. 1, 358–364. [PubMed] [Google Scholar]
- Hames C., Halbedel S., Hoppert M., Frey J., Stülke J. (2009). Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191, 747–753. 10.1128/JB.01103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heefner D. L. (1982). Transport of H+, K+, Na+ and Ca++ in Streptococcus. Mol. Cell. Biochem. 44, 81–106. 10.1007/BF00226893 [DOI] [PubMed] [Google Scholar]
- Imlay J. A., Linn S. (1988). DNA damage and oxygen radical toxicity. Science 240, 1302–1309. 10.1126/science.3287616 [DOI] [PubMed] [Google Scholar]
- Kakinuma Y. (1998). Inorganic cation transport and energy transduction in Enterococcus hirae and other streptococci. Microbiol. Mol. Biol. Rev. 62, 1021–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma Y., Igarashi K. (1990). Amplification of the Na+-ATPase of Streptococcus faecalis at alkaline pH. FEBS Lett. 261, 135–138. 10.1016/0014-5793(90)80654-2 [DOI] [PubMed] [Google Scholar]
- Kamat S. S., Holmes-Hampton G. P., Bagaria A., Kumaran D., Tichy S. E., Gheyi T., et al. (2011). The catalase activity of diiron adenine deaminase. Protein Sci. 20, 2080–2094. 10.1002/pro.748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayaoglu G., Erten H., Ørstavik D. (2005). Growth at high pH increases Enterococcus faecalis adhesion to collagen. Int. Endod. J. 38, 389–396. 10.1111/j.1365-2591.2005.00957.x [DOI] [PubMed] [Google Scholar]
- Koo B. S., Hyun H. H., Kim S. Y., Kim C. H., Lee H. C. (2011). Enhancement of thymidine production in E. coli by eliminating repressors regulating the carbamoyl phosphate synthetase operon. Biotechnol. Lett. 33, 71–78. 10.1007/s10529-010-0413-7 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wei Z., Zheng M., Gu X., Deng Y., Qiu R., et al. (2006). Crystal structure of human guanosine monophosphate reductase 2 (GMPR2) in complex with GMP. J. Mol. Biol. 355, 980–988. 10.1016/j.jmb.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Love R. M. (2001). Enterococcus faecalis–a mechanism for its role in endodontic failure. Int. Endod. J. 34, 399–405. 10.1046/j.1365-2591.2001.00437.x [DOI] [PubMed] [Google Scholar]
- Mandapati K., Gorla S. K., House A. L., Mckenney E. S., Zhang M., Rao S. N., et al. (2014). Repurposing cryptosporidium inosine 5′-monophosphate dehydrogenase inhibitors as potential antibacterial agents. ACS Med. Chem. Lett. 5, 846–850. 10.1021/ml500203p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J. C., Mason C. E., Mane S. M., Stephens M., Gilad Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517. 10.1101/gr.079558.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli L. K., Ducati R. G., Rosado L. A., Breda A., Selbach B. P., Santos D. S., et al. (2011). Recombinant Escherichia coli GMP reductase: kinetic, catalytic and chemical mechanisms, and thermodynamics of enzyme-ligand binary complex formation. Mol. Biosyst. 7, 1289–1305. 10.1039/c0mb00245c [DOI] [PubMed] [Google Scholar]
- Mordukhova E. A., Pan J. G. (2013). Evolved cobalamin-independent methionine synthase (MetE) improves the acetate and thermal tolerance of Escherichia coli. Appl. Environ. Microbiol. 79, 7905–7915. 10.1128/AEM.01952-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Ross R. P., Heath J. D., Dunny G. M., Weinstock G. M. (1993). Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175, 5216–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff H., Arsène-Ploetze F., Malandain C., Kleerebezem M., Bringel F. (2004). Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186, 6059–6069. 10.1128/JB.186.18.6059-6069.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rams T. E., Feik D., Young V., Hammond B. F., Slots J. (1992). Enterococci in human periodontitis. Oral Microbiol. Immunol. 7, 249–252. 10.1111/j.1399-302X.1992.tb00034.x [DOI] [PubMed] [Google Scholar]
- Ran S., He Z., Liang J. (2013). Survival of Enterococcus faecalis during alkaline stress: changes in morphology, ultrastructure, physiochemical properties of the cell wall and specific gene transcripts. Arch. Oral Biol. 58, 1667–1676. 10.1016/j.archoralbio.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Rince A., Flahaut S., Auffray Y. (2000). Identification of general stress genes in Enterococcus faecalis. Int. J. Food Microbiol. 55, 87–91. 10.1016/S0168-1605(00)00180-X [DOI] [PubMed] [Google Scholar]
- Rincé A., Giard J. C., Pichereau V., Flahaut S., Auffray Y. (2001). Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183, 1482–1488. 10.1128/JB.183.4.1482-1488.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Mccarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira J. F., Jr., Rôças I. N. (2004). Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 97, 85–94. 10.1016/S1079-2104(03)00353-6 [DOI] [PubMed] [Google Scholar]
- Sundqvist G., Figdor D., Persson S., Sjögren U. (1998). Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 85, 86–93. 10.1016/S1079-2104(98)90404-8 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Natale D. A., Garkavtsev I. V., Tatusova T. A., Shankavaram U. T., Rao B. S., et al. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vebø H. C., Snipen L., Nes I. F., Brede D. A. (2009). The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS ONE 4:e7660. 10.1371/journal.pone.0007660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vebø H. C., Solheim M., Snipen L., Nes I. F., Brede D. A. (2010). Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS ONE 5:e12489. 10.1371/journal.pone.0012489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlin Y. B., Holm A. K. (1988). Changes in the oral microflora in patients with acute leukemia and related disorders during the period of induction therapy. Oral Surg. Oral Med. Oral Pathol. 65, 411–417. 10.1016/0030-4220(88)90354-4 [DOI] [PubMed] [Google Scholar]
- Waltimo T., Trope M., Haapasalo M., Ørstavik D. (2005). Clinical efficacy of treatment procedures in endodontic infection control and one year follow-up of periapical healing. J. Endod. 31, 863–866. 10.1097/01.don.0000164856.27920.85 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KEGG summary of E. faecalis sequences (XLS).
The detail sequence information of the transcriptome.
The data of all of the gene transcription information (XLS).
The list of genes found to be induced (up-regulated) in alkaline stress.
The list of genes found to be decreased (down-regulated) in alkaline stress.