Abstract
The non-fluorescent pseudomonads, Pseudomonas corrugata (Pcor) and P. mediterranea (Pmed), are closely related species that cause pith necrosis, a disease of tomato that causes severe crop losses. However, they also show strong antagonistic effects against economically important pathogens, demonstrating their potential for utilization as biological control agents. In addition, their metabolic versatility makes them attractive for the production of commercial biomolecules and bioremediation. An extensive comparative genomics study is required to dissect the mechanisms that Pcor and Pmed employ to cause disease, prevent disease caused by other pathogens, and to mine their genomes for genes that encode proteins involved in commercially important chemical pathways. Here, we present the draft genomes of nine Pcor and Pmed strains from different geographical locations. This analysis covered significant genetic heterogeneity and allowed in-depth genomic comparison. All examined strains were able to trigger symptoms in tomato plants but not all induced a hypersensitive-like response in Nicotiana benthamiana. Genome-mining revealed the absence of type III secretion system and known type III effector-encoding genes from all examined Pcor and Pmed strains. The lack of a type III secretion system appears to be unique among the plant pathogenic pseudomonads. Several gene clusters coding for type VI secretion system were detected in all genomes. Genome-mining also revealed the presence of gene clusters for biosynthesis of siderophores, polyketides, non-ribosomal peptides, and hydrogen cyanide. A highly conserved quorum sensing system was detected in all strains, although species specific differences were observed. Our study provides the basis for in-depth investigations regarding the molecular mechanisms underlying virulence strategies in the battle between plants and microbes.
Keywords: comparative genomics, type III secretion system, type VI secretion system, effectors, siderophores, polyketides, non-ribosomal peptides, pith necrosis
Background
Pseudomonas corrugata (Pcor) Robert and Scarlet 1981 emend. Sutra et al. (1997) and P. mediterranea (Pmed) Catara et al. (2002) are two closely related Gammaproteobacteria species belonging to the genus Pseudomonas sensu stricto (De Vos et al., 1985; Kersters et al., 1996). This genus predominantly consists of fluorescent species, but also contains some non-fluorescent species, including, P. stutzeri, P. alcaligenes, P. pseudoalcaligenes, P. mendocina, P. lemoignei, P. ruhlandii, P. frederiksbergensis, P. graminis, P. plecoglossicida, Pcor, and Pmed.
Pcor was the first of the two species discovered as the causal agent of tomato pith necrosis (TPN) (Scarlett et al., 1978; Catara, 2007). It has since been isolated from several other crop plants suffering pith necrosis, including pepper (Lopez et al., 1988), chrysanthemum (Fiori, 1992) and geranium (Magyarosy and Buchanan, 1995). Pcor is ubiquitous and has been isolated from the root environment in different countries (Kovacevich and Ryder, 1991; Ryder and Rovira, 1993; Schisler and Slininger, 1994; Achouak et al., 2000; Walker et al., 2000; Pandey et al., 2001).
Phylogenetic analyses based on 16S rRNA gene sequences placed Pcor within the P. fluorescens branch (Moore et al., 1996; Anzai et al., 2000), whereas, in another study of the combined nucleotide sequences of the rpoD and gyrB genes, the bacterium was included in intrageneric cluster II within the “P. fluorescens complex” (P. syringae and P. putida being the other two) (Yamamoto et al., 2000). A polyphasic approach revealed that two closely related but distinct taxa were present within the Pcor species. The taxon which contained the type strain CFBP2431T maintained the epithet Pcor, while the other strains included were assigned to a novel species, named P. mediterranea (Catara et al., 2002). The two species are phenotypically distinguishable by the ability of Pcor, but not of Pmed, to utilize histamine, 2-ketogluconate and meso-tartrate as a unique carbon source. The two species can also be clearly distinguished by 16S rDNA analysis, by means of DNA-based fingerprinting methods (Catara et al., 2000, 2002) and by multi-locus sequence analysis (MLSA) (Trantas et al., 2015). Multiplex, conventional, or real-time PCR can also be used to screen tomato planting materials for detection of, and discrimination between, the two bacterial species (Catara et al., 2000; Licciardello et al., 2011). Moreover, a phylogenetic analysis of the concatenated sequences of four core “housekeeping” genes (16S rRNA, gyrB, rpoB, and rpoD) of 107 type strains of Pseudomonas species placed Pcor and Pmed in a separate subgroup within the P. fluorescens lineage (Mulet et al., 2010).
Pith necrosis is characterized by the necrosis and hollowing of the parenchymatic tissue of the stem. Usually, the first visible symptom is chlorosis and withering of the youngest leaves, followed by loss of turgor and eventual collapse of the whole plant in later stages of the disease. The disease occurs world-wide in all tomato-growing areas and can cause severe crop losses. Pcor is a demonstrated plant pathogen, but has potential to be used as a biocontrol agent (Catara, 2007). Most strains have been used as biocontrol agents against different plant pathogenic fungi. Many strains, including those responsible for pith necrosis, have demonstrated either in vitro or in vivo antimicrobial activity.
Pcor produces the antimicrobial and phytotoxic cyclic lipopeptides (CLPs) cormycin A, corpeptin A, and corpeptin B (Emanuele et al., 1998; Scaloni et al., 2004). Corpeptins were isolated from the culture filtrates of the Pcor type strain NCPPB2445 (Emanuele et al., 1998). The corpeptin peptide (CP) moiety showed similarity to Pseudomonas peptins such as syringopeptins (SP22s, SP25s), fuscopeptins (FPs), and tolaasins (ToI-A). Since a number of phytopathogenic Pseudomonas spp. produce antimicrobial nonapeptides, Scaloni screened the culture filtrates of a number of Pcor strains and demonstrated that nonapeptide production is strain-dependent (Scaloni et al., 2004). In the same study Cormycin A was also characterized as a novel compound from the culture filtrates of the strain IPVCT 10.3. Cormycin and corpeptins play a pivotal role in Pcor and Pmed virulence. Mutants unable to produce one or both of them are still able to colonize tomato stem parenchymatic tissues but the symptoms associated with pith necrosis are significantly reduced (Licciardello et al., 2012; Strano et al., 2014).
Recent investigations on the interactions of both Pcor and Pmed with plants highlight the pivotal role of quorum sensing in the regulation of virulence traits (Licciardello et al., 2007, 2012, 2009). Both Pcor and Pmed have a quorum sensing system mediated by N-acyl homoserine lactone (AHL-QS) signal molecules. Strains of both species from different geographical locations and plant origins produce the same AHLs at comparable levels (Licciardello et al., 2007, 2012). In plant-associated Pseudomonas the AHL-QS has been shown to have a role in virulence of plant pathogenic species and in the regulation of traits involved in biological control activity (Quiñones et al., 2004, 2005; Venturi, 2006; Wei and Zhang, 2006; Licciardello et al., 2007, 2012; Hosni et al., 2011; Mattiuzzo et al., 2011). Specifically in Pcor and Pmed, AHL-QS and the genetically linked transcriptional regulator RfiA have a role in virulence in tomato (Licciardello et al., 2009, 2012) and in corpeptin production (Strano et al., 2014).
Here, we present the draft genome sequences of five Pmed and four Pcor strains, including the type strains, in order to investigate mechanisms of pathogenicity and the synthesis of antimicrobial compounds. The selected strains were isolated from different geographical locations in order to cover the genetic heterogeneity of the two species, allowing us to perform in-depth comparative genomic analysis. Our analysis revealed unique features of the pathogenicity of Pcor and Pmed, and significant insights into the antimicrobial activity and production of secondary metabolites in these species.
Materials and methods
Bacterial strains and genomic DNA preparation
We compared the genomes of nine strains: four Pcor strains and five Pmed strains. Table 1 contains the metadata of all strains. Strains were selected based on their geographical origin (Greece, Italy, Spain, and U.K.) and intraspecific diversity based on phenotypic and genotypic characteristics reported in previous studies (Catara et al., 2002; Trantas et al., 2015). The type strains of both species were included. The genome set included seven de novo sequenced strains and two previously sequenced strains: Pcor CFBP5454 (Licciardello et al., 2014b) and Pmed CFBP5447 (Licciardello et al., 2014a). The strains were grown at 28°C either in Nutrient Dextrose agar (NDA) or Lysogeny Broth (LB) agar.
Table 1.
Strains used for the comparative analysis of Pseudomonas corrugata (Pcor) and P. mediterranea (Pmed).
| Strain | Species | Isolation source | Location of isolation | IMG genome ID | References |
|---|---|---|---|---|---|
| TEIC1022 | Pmed | Infected tomato plant | Crete, Greece | 2563366521 | Trantas et al., 2015 |
| TEIC1105 | Pmed | Infected tomato plant | Crete, Greece | 2563366524 | Trantas et al., 2015 |
| CFBP5404 | Pmed | Infected pepper plant | Tenerife, Spain | 2563366522 | Catara et al., 2002 |
| CFBP5444 | Pmed | Infected tomato plant | Sicily, Italy | 2563366526 | Catara et al., 2002 |
| CFBP5447T | Pmed | Infected tomato plant | Sicily, Italy | 2596583589 | Catara et al., 2002 |
| TEIC1148 | Pcor | Infected tomato plant | Crete, Greece | 2563366520 | Trantas et al., 2015 |
| CFBP5403 | Pcor | Infected tomato plant | Tenerife, Spain | 2563366523 | CFBP |
| CFBP5454 | Pcor | Infected tomato plant | Sicily, Italy | 2558309045 | Catara et al., 2002 |
| NCPPB2445T | Pcor | Infected tomato plant | UK | 2563366525 | NCPPB |
TEIC, Technological Educational Institute of Crete Bacterial collection; CFBP, International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, Fera, York, U.K.
DNA was extracted from over-night cultures in LB broth using the DNeasy Blood & Tissue Kit from QIAGEN (UK) according to the manufacturer's instructions.
Phenotypic characterization and In Planta tests
Pcor and Pmed strains were tested in assays that had been previously reported to reveal phenotypic differences between species (Catara et al., 1997, 2002; Sutra et al., 1997; Solaiman et al., 2005). To determine the production of protease, lipase, and gelatinase, strains were seeded on agar plates containing the appropriate substrates as previously described (Lelliott and Stead, 1987; Schaad et al., 1988).
Antimicrobial activity of Pcor and Pmed was assessed on potato dextrose agar (PDA), against two CLP indicator microorganisms (the Gram positive bacterium Bacillus megaterium ITM100 and the yeast Rhodotorula pilimanae ATTC26432) and against two phytopathogenic bacteria (P. syringae pv. tomato PVCT28.3.1 and Xanthomonas campestris pv. campestris PVCT 62.4) (Licciardello et al., 2007, 2012). The antimicrobial activity of culture filtrates of selected strains was assessed by the well-diffusion assay against the CLP indicator strains as described by Licciardello et al. (2012). Culture filtrates were prepared from 4 day-old still bacterial cultures in Improved Minimal Medium (IMM) (Surico et al., 1988). After centrifugation (9000 g, 20 min), the supernatant was passed through a 0.22 μm Millipore filter (Millipore, Billerica, MA, U.S.A.) to obtain cell-free culture filtrates that were 10 × concentrated using the Vacufuge concentrator 5301 (Eppendorf, Milan, Italy). All tests were carried out twice and with three technical replicates each time.
The hypersensitive response (HR) test was performed using a blunt syringe to infiltrate 108 CFU ml−1 of bacteria into leaves of Nicotiana benthamiana. Ten leaf panels were infiltrated per strain. After infiltration, plants were placed at 25°C in a growth chamber and the collapse/necrosis of the mesophyll was assessed daily. Nicotianna benthamiana seeds kindly provided by Prof. N. Panopoulos (University of Crete, Greece).
Artificial inoculations were performed in tomato (commercial hybrid Elpida), and pepper (commercial hybrid Raiko) plants at the stage of three or four true leaves. Inoculations were made with 24–48 h bacterial cultures grown on KB medium. The inoculum was collected with a sterile toothpick and used for inoculation by piercing the stems of the plants with a lateral tilt to a depth of 0.5 cm. The inoculated sites were covered with parafilm and the plants remained at high humidity (90–100%) at ambient temperature (about 23°C) for 48 h. Two days later the plants were transferred to a greenhouse with 60–70% humidity and 15–26°C temperature. Plants were observed for 10–15 days after inoculation (Catara et al., 2002; Trantas et al., 2015).
Library preparation, sequencing, annotation and assembly
Genomic DNA was isolated from the strains TEIC1022, TEIC1105, CFBP5404, CFBP5444, TEIC1148, CFBP5403, and NCPPB2445T using the Gentra Puregene DNA Purification Kit (Qiagen). Illumina DNA library preparation was performed using the NEBNext DNA Library Master Mix kit (New England Biolabs) following the manufacturer's protocol for user supplied barcodes. The quality of the prep prior to sequencing was confirmed on a Bioanalyzer 2100 with a DNA1000 chip (Agilent) and a Qubit Fluorometer using a Quant-iTTM dsDNA BR assay kit (Life Technologies). Genome sequencing was performed on an Illumina MiSeq using V2 chemistry producing 150 base, paired-end reads. Read numbers per strain are reported in Supplementary File 1.
The draft genome sequences of Pcor strain CFBP5454 and Pmed strain CFBP5447T were previously obtained by using Illumina GAIIx technology combining mate-pair and paired-end libraries (CFBP5447) or paired-end libraries (CFBP5454) (Licciardello et al., 2014a,b) and used for comparative analyses. For this study, the 157 and 47 contigs of CFBP5454 and CFBP5447T strains were linked and placed into 84 and 36 scaffolds or super-contigs, respectively. The orientation, order and distance between the contigs were estimated using the insert size between the paired-end and/or mate-pair reads. The analysis was performed using the SSPACE Premium scaffolder version 2.0 (Boetzer et al., 2011). Gaps within scaffolds were automatically closed with GapFiller (Boetzer and Pirovano, 2012).
Genome assemblies, gene mining, and alignments
De novo sequence assemblies were performed by CLC Genomics Workbench 7 (http://www.clcbio.com). All genomes were annotated through JGI's auto-annotation pipeline (Markowitz et al., 2014). All BLAST queries (Altschul et al., 1997) were accomplished at JGI (Mavromatis et al., 2009).
Phylogenetic analyses
Whole genome phylogenetic trese were built using Orthologsorter (Farias and Almeida, 2013). All protein-coding genes were first analyzed by OrthoMCL (Li et al., 2003). Among all families found, those with exactly one representative in each genome were taken, except for out-group genomes, which were allowed to have zero or one gene in each family. Next, Muscle (Edgar, 2004) and Gblocks (Castresana, 2000) were used with the default parameters to align each chosen family and remove non-informative columns. Then, all alignments were concatenated, resulting in a complete alignment, which was then processed by RAxML (Stamatakis, 2006) to build the tree, using the PROTCAT model, rapid bootstrapping and a subsequent maximum likelihood (ML) search.
Gene-specific phylogenetic analysis of the following genes was undertaken: fruK (fructose-1-phosphate kinase), gltA (type II citrate synthase), gyrB (b subunit of DNA gyrase), mutL (methyl-directed mismatch repair protein), rpoB (beta subunit of RNA polymerase), rpoD (sigma factor of RNA polymerase). Sequences were extracted from our assemblies of each strain. Sequences for reference strains were retrieved from GenBank. Sequence alignments were carried out using CLUSTALW (Thompson et al., 1994) with the default parameters. The phylogenetic trees were generated in MEGA6 (Tamura et al., 2013). Neighbor-Joining trees were generated according to the p-distance method (Saitou and Nei, 1987). In order to confirm the relationships found with the Neighbor-Joining dendrogram, comparison Maximum Likelihood trees were generated according to the Tamura-Nei model (Tamura and Nei, 1993). The percentage of replicate trees (1500 replicates) in which the associated strains clustered together was estimated (Felsenstein, 1985) and is shown next to the tree nodes.
Results and discussion
Variable phenotypic traits
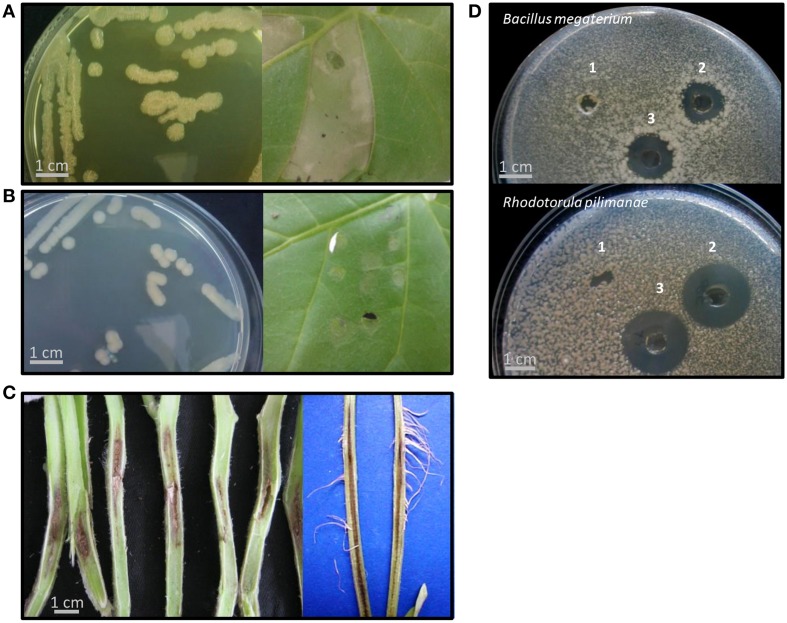
The Pcor species epithet “corrugata” refers to the typical morphology of bacterial colonies on NDA; slightly raised, with a wrinkled-rough surface and undulate margins that produce a yellow diffusible pigment (Scarlett et al., 1978). Pmed strains also show this morphology. Nevertheless, in both species non-pigmented strains with smooth colonies and intermediate phenotypes have been observed (Siverio et al., 1993; Sutra et al., 1997; Catara et al., 2002). The strains assessed in this study (Table 1) were representative of this colony variability. Five of the strains showed the “rough-phenotype” and produced yellow diffusible pigments (R-type). Three strains shared a “smooth phenotype” with whitish-cream, smooth-surfaced colonies that did not produce any pigment (S-type; Figures 1A,B). One strain showed an intermediate phenotype (I-type), growing in smooth, slightly elevated colonies with a low-level of diffusible pigment (Table 2).
Figure 1.
Representative phenotypes of Pseudomonas corrugata (Pcor) and P. mediterranea (Pmed) strains in this study. (A,B) Strains showed either a rough colony morphology with production of a yellow diffusible pigment in the medium (R-phenotype) or smooth colonies that did not produce pigment (S-phenotype). All “R” strains induced a hypersensitive-like response (HR) in Nicotiana benthamiana (Pcor CFBP5454 in A) while the “S” strains Pcor TEIC1148 and Pmed CFBP5444 (the latter in B) did not induce HR. (C) All strains were able to induce tomato pith necrosis. (D) Representative bacterial culture filtrates produced in cyclic lipopeptide (CLP)-inducing conditions were tested for antimicrobial activity against the Gram-positive bacterium Bacillus megaterium and the yeast Rhodotorula pilimanae. Pmed CFBP5444 culture filtrate representative of the “S” phenotype did not show antimicrobial activity. 1, Pmed CFBP5444; 2, Pcor CFBP5454; 3, Pmed CFBP5447. All tests were performed at least three times with identical results.
Table 2.
Features linked with antimicrobial activity of genomes under study.
| Strain | Species | Morphology | Hydrolytic activity | HR | HCN | Antimicrobial activity against | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Casein | Gelatine | Tween 80 | Rp | Bm | Pst | Xcc | |||||
| NCPPB2445T | Pcor | R | W | W | + | + | W | + | + | + | − |
| TEIC1148 | Pcor | S | − | − | − | − | − | + | − | − | − |
| CFBP5403 | Pcor | R | + | + | − | + | + | + | + | + | − |
| CFBP5454 | Pcor | R | + | + | + | + | + | + | + | + | + |
| CFBP5447T | Pmed | R | + | + | W | + | + | + | + | + | − |
| TEIC1022 | Pmed | S | − | − | − | + | − | + | − | − | − |
| TEIC1105 | Pmed | I | W | W | − | + | W | + | + | + | − |
| CFBP5404 | Pmed | R | W | W | − | + | + | + | + | + | − |
| CFBP5444 | Pmed | S | − | − | − | − | − | + | − | − | − |
Species abbreviations are Pcor, Pseudomonas corrugata; Pmed, P. mediterranea; Rp; Rhodotorula pilimanae; Bm, Bacillus megaterium; Pst, P. syringae pv. tomato and Xcc, Xanthomonas campestris pv. campestris. TEIC: Technological Educational Institute of Crete Bacterial collection; CFBP: International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, Fera, York, U.K. Isolate names followed by the letter “T” are type strains; R, “rough-phenotype” producing yellow diffusible pigments (R-type); S, “smooth phenotype” with whitish-cream smooth-surfaced colonies that do not produce any pigment (S-type); I, “intermediate phenotype” producing rather smooth colonies with weak diffusible pigment (I-type); HR, hypersensitive response-like reaction; HCN, production of hydrogen cyanide; +, positive reaction; −, negative reaction or no antimicrobial activity; w, weak-positive reaction.
The appearance of pleiotropic mutants after sub-culturing or re-isolation from soil occurred and was observed as a conversion from rough to smooth colony morphology. This phenomenon was also associated with either loss of exoenzyme activity (i.e., inability to hydrolyse casein, gelatine, lecithin, and Tween 80), or modification of antagonistic activity and hypersensitive response (HR) induction (Siverio et al., 1993; Gustine et al., 1995; Barnett et al., 1999). In particular, we observed that the three S-type strains lacked caseinase and gelatinase activity, whereas the R- and I-type strains maintained the ability to hydrolyze casein and gelatin. On the other hand, only three R- strains were able to hydrolyze Tween 80 (lipase test; Table 2). It is noteworthy that at least one of the strains in this study (CFBP5444) was isolated from plants as a natural S phenotype.
An important trait of Pcor is that it elicits a plant defense response in non-host plants; namely, the production of the phytoalexin medicarpin in white clover callus and an HR-like response in tobacco leaves (Gustine et al., 1990, 1995). Tobacco HR-like response is not induced by all the strains of the two species (Sutra et al., 1997; Catara et al., 2002). We tested the ability of suspensions of Pcor and Pmed to induce an HR-like response in N. benthamiana leaves. Within 24 h of infiltration, seven out of the nine strains caused the infiltrated leaf panels to collapse, and after a further 24 h these leaf panels turned necrotic. Leaf panels inoculated with the S-type strains Pcor TEIC1148 and Pmed CFBP5444 did not show any symptoms (Figures 1A,B). However, no genes encoding components of the T3SS was found in any of the Pcor and Pmed genomes (see results for P. corrugata and P. mediterranea lack of T3SS). In a study carried out on Pcor CFBP5454 (which gives an HR-like response) it was observed that the bacterial population titre in infiltrated N. benthamiana leaf panels decayed rapidly after inoculation. On the other hand when leaf panels were infiltrated with non-producing CLP mutants (which does not give an HR-like response) the population titre was maintained (Strano et al., 2014). This suggested a role for CLPs in eliciting plant defense responses (Strano et al., 2014).
Artificial inoculations
Tomato and pepper seedlings were Pcor-inoculated at the developmental stage of three-to-four true leaves and within 15 days showed the typical symptoms of pith necrosis disease (Figure 1C). Similar symptoms were observed on plants infected with Pmed strains. In all infected plants, the pith necrosis extended 1.5–3 cm on both sides of the initial infection point. Secondary acquired aerial roots appeared on some artificially inoculated tomato plants. Collectively, our results indicated no significant difference of pathogenic severity between Pcor and Pmed strains on tomato plants.
Genome-wide sequence data
The genomic sequences of nine Pcor and Pmed strains (Table 1) were obtained using Illumina MiSeq or GAIIx technology. This produced between 1,209,534 and 11,800,202 paired-end reads (Table 3 and Supplementary File 1). De novo genome assembly for the nine strains was performed independently for each strain using the CLC Genomics Workbench (CLCbio, Aarhus, Denmark). The assemblies contain 84–442 contigs with an N75 ranging from 16,551 to 120,020. The total number of nucleotides assembled in contigs ranged from 6,084,011 to 6,266,776 for the Pcor strains and from 6,231,327 to 6,461,985 for Pmed strains.
Table 3.
Short table of genomic statistics for Pseudomonas corrugata and P. mediterranea strains under study.
| P. corrugata | P. mediterranea | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CFBP5403 | CFBP5454 | NCPPB2445 | TEIC1148 | CFBP5404 | CFBP5444 | CFBP5447 | TEIC1022 | TEIC1105 | |
| Total number of reads | 11,800,202 | 8,420,692 | 1,519,930 | 8,608,470 | 1,209,534 | 1,394,308 | 24,635,686 | 1,276,402 | 23,866,648 |
| Average read length | 35 | 70.6 | 145 | 35 | 145 | 145 | 51 | 145 | 44 |
| Total bases | 413,007,070 | 594,525,036 | 220,389,850 | 301,296,450 | 175,382,430 | 202,174,660 | 1,256,357,465 | 185,078,290 | 1,040,233,200 |
| DNA, total number of bases in contigs | 6,166,582 | 6,195,615 | 6,084,011 | 6,266,776 | 6,281,542 | 6,298,862 | 6,270,666 | 6,231,327 | 6,299,426 |
| Ambiguous bases | 39,282 | 2299 | 3749 | 31,312 | 3143 | 3499 | 18 | 11,988 | 1,651 |
| Percentage of ambiguous bases | 0.64 | 0.04 | 0.06 | 0.50 | 0.05 | 0.06 | 0.00 | 0.19 | 0.03 |
| DNA coding number of bases | 5,471,555 | 5,491,990 | 5,391,923 | 5,567,198 | 5,564,235 | 5,572,667 | 5,573,461 | 5,507,149 | 5,573,467 |
| DNA G+C number of bases | 3,706,298 | 3,747,457 | 3,686,460 | 3,763,425 | 3,846,554 | 3,853,935 | 3,862,577 | 3,813,571 | 385,5401 |
| GC content (%) | 60.1 | 60.5 | 60.6 | 60.1 | 61.2 | 61.2 | 60.7 | 61.2 | 61.2 |
| Total number of contigs | 442 | 84 | 104 | 432 | 150 | 89 | 47 | 101 | 91 |
| N75 | 17,365 | 48,599 | 85,837 | 16,551 | 11,1354 | 118,717 | 447,675 | 83,242 | 120,020 |
| N50 | 29,882 | 79,475 | 202,143 | 32,179 | 188,131 | 229,164 | 662,701 | 142,363 | 230,696 |
| Genes total number | 5912 | 5570 | 5463 | 5845 | 5652 | 5638 | 5533 | 5594 | 5643 |
| Coding density (%) | 87.9 | 88.5 | 88.4 | 88.4 | 88.4 | 88.3 | 88.4 | 88.1 | 88.3 |
| Completeness (%) | 97.02 | 99.69 | 99.68 | 99.14 | 99.69 | 99.68 | 99.69 | 99.45 | 99.69 |
| Contamination (%) | 0.80 | 0.42 | 0.45 | 0.47 | 0.48 | 0.79 | 0.48 | 0.43 | 0.79 |
| Protein coding genes | 5809 | 5443 | 5313 | 5737 | 5515 | 5484 | 5786 | 5450 | 5481 |
| RNA genes | 103 | 127 | 150 | 108 | 137 | 154 | 135 | 144 | 162 |
| Protein coding genes with function prediction | 4812 | 4621 | 4516 | 4753 | 4608 | 4598 | 4804 | 4611 | 4606 |
| Protein coding genes without function prediction | 997 | 822 | 797 | 984 | 907 | 886 | 982 | 839 | 875 |
| Protein coding genes for enzymes | 1307 | 1355 | 1341 | 1331 | 1339 | 1339 | 1312 | 1337 | 1339 |
| w/o enzymes but with candidate KO based enzymes | 147 | 19 | 26 | 68 | 17 | 11 | 130 | 24 | 13 |
| Protein coding genes connected to transporter classification | 832 | 792 | 777 | 810 | 790 | 791 | 809 | 785 | 791 |
| Protein coding genes connected to KEGG pathways | 1497 | 1564 | 1546 | 1536 | 1565 | 1570 | 1523 | 1538 | 1570 |
| Protein coding genes connected to KEGG Orthology (KO) | 2733 | 2850 | 2807 | 2808 | 2833 | 2828 | 2760 | 2804 | 2828 |
| Protein coding genes connected to MetaCyc pathways | 1272 | 1325 | 1311 | 1300 | 1313 | 1313 | 1287 | 1312 | 1313 |
| Protein coding genes with COGs | 3839 | 4012 | 3927 | 3923 | 3965 | 3979 | 3927 | 3949 | 3978 |
| Chromosomal cassettes | 717 | 532 | 508 | 697 | 556 | 562 | 613 | 560 | 559 |
| Biosynthetic clusters | 39 | 46 | 38 | 45 | 49 | 50 | 44 | 46 | 43 |
| Genes in biosynthetic clusters | 289 | 386 | 332 | 331 | 397 | 410 | 389 | 405 | 369 |
| COG clusters | 2033 | 2051 | 2046 | 2053 | 2037 | 2039 | 2026 | 2026 | 2039 |
| KOG clusters | 841 | 841 | 852 | 852 | 837 | 839 | 840 | 845 | 839 |
| Pfam clusters | 2601 | 2566 | 2551 | 2597 | 2,547 | 2,551 | 2,577 | 2,547 | 2,550 |
| TIGRfam clusters | 1345 | 1362 | 1355 | 1369 | 1369 | 1372 | 1365 | 1362 | 1372 |
For the full table refer to the supporting material (Supplementary File 1). TEIC, Technological Educational Institute of Crete Bacterial collection; CFBP, International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, Fera, York, U.K. Isolate names followed by the letter “T” are type strains.
We used CheckM to assess the completeness and the overall quality of the assemblies (Parks et al., 2015). CheckM was utilized to measure percentage of ambiguous bases, coding density, completeness and contamination (Table 3). We determined that all assemblies were of good quality by assessing them against the thresholds described in Parks et al. (2015). The coding densities of the genomes are within the normal range of 85–90% in bacterial genomes (Ussery et al., 2009).
General genome features
The basic subsystems of the sequenced genomes were predicted with the RAST server (Overbeek et al., 2014) and the IMG automated annotation pipeline (Markowitz et al., 2014). In particular, we identified differences in proteins predicted to participate in virulence, disease and defense; the number of genes from these families ranged from 106 in strain Pmed TEIC1022–127 in strain Pmed CFBP5447T (Table 4). However, it is unclear whether these differences represented true differences between strains or if they were due to low quality draft genomes and errors in their assembly.
Table 4.
Organismal subsystem prediction overviews estimated by the RAST server.
| Protein family\Strains (No of total proteins) | NCPPB2445T (3957) | CFBP5404 (3961) | CFBP5403 (3920) | CFBP5454 (3993) | TEIC1148 (3947) | CFBP5447T (4146) | CFBP5444 (3985) | TEIC1022 (3938) | TEIC1105 (3977) |
|---|---|---|---|---|---|---|---|---|---|
| Cofactors, Vitamins, Prosthetic Groups, and Pigments | 347 | 349 | 359 | 351 | 352 | 371 | 348 | 355 | 348 |
| Cell, Wall, and Capsule | 194 | 184 | 206 | 203 | 197 | 197 | 184 | 184 | 184 |
| Virulence, Disease, and Defense | 117 | 107 | 123 | 120 | 123 | 127 | 107 | 106 | 107 |
| Potassium metabolism | 31 | 32 | 31 | 31 | 31 | 32 | 32 | 32 | 32 |
| Photosynthesis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Miscellaneous | 51 | 55 | 51 | 51 | 51 | 55 | 55 | 54 | 55 |
| Phages, Prophages, and Transposable elements | 14 | 27 | 23 | 27 | 12 | 30 | 35 | 28 | 30 |
| Membrane Transport | 212 | 198 | 209 | 216 | 210 | 193 | 200 | 185 | 200 |
| Iron acquisition and metabolism | 28 | 30 | 31 | 28 | 28 | 32 | 30 | 30 | 30 |
| RNA Metabolism | 188 | 188 | 183 | 185 | 184 | 193 | 190 | 191 | 190 |
| Nucleosides and Nucleotides | 127 | 127 | 124 | 128 | 128 | 129 | 127 | 127 | 127 |
| Protein Metabolism | 272 | 252 | 246 | 248 | 245 | 270 | 250 | 255 | 250 |
| Cell division and Cell cycle | 34 | 35 | 35 | 34 | 34 | 37 | 35 | 35 | 35 |
| Motility and Chemotaxis | 130 | 156 | 135 | 130 | 131 | 130 | 157 | 129 | 157 |
| Regulation and Cell signaling | 117 | 116 | 120 | 116 | 121 | 120 | 117 | 119 | 116 |
| Secondary Metabolism | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| DNA Metabolism | 122 | 126 | 117 | 145 | 131 | 163 | 125 | 125 | 125 |
| Regulons | 8 | 9 | 9 | 9 | 8 | 10 | 8 | 9 | 8 |
| Fatty Acids, Lipids, and Isoprenoids | 216 | 218 | 211 | 217 | 212 | 228 | 218 | 230 | 218 |
| Nitrogen Metabolism | 95 | 85 | 101 | 96 | 95 | 87 | 86 | 87 | 86 |
| Dormancy and Sporulation | 4 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 |
| Respiration | 138 | 134 | 142 | 144 | 141 | 143 | 135 | 134 | 135 |
| Stress Response | 193 | 185 | 188 | 193 | 203 | 196 | 188 | 190 | 187 |
| Metabolism of Aromatic Compounds | 130 | 130 | 136 | 122 | 126 | 138 | 131 | 134 | 131 |
| Amino Acids and Derivatives | 614 | 619 | 597 | 612 | 610 | 631 | 620 | 623 | 619 |
| Sulfur Metabolism | 54 | 75 | 53 | 49 | 49 | 73 | 75 | 75 | 75 |
| Phosphorus Metabolism | 42 | 33 | 44 | 43 | 44 | 35 | 35 | 34 | 35 |
| Carbohydrates | 473 | 480 | 436 | 484 | 470 | 515 | 486 | 486 | 486 |
TEIC, Technological Educational Institute of Crete Bacterial collection; CFBP, International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, Fera, York, U.K. Isolate names followed by the letter “T” are type strains.
In addition, we scanned the Pcor and Pmed genomes for the presence of genes necessary for the biosynthesis of ethylene and auxin. We queried the genomes with the gene coding for ethylene-forming enzyme (efe) to assess their capability for ethylene biosynthesis. Similarly, we scanned for the presence of the auxin biosynthesis genes coding for indole-3-acetamide synthase (iaaM), indole-3-acetamide hydrolase (iaaH) and indoleacetate-lysine ligase (iaaL). No significant similarities were found.
Two non-fluorescent phytopathogenic pseudomonads in the P. fluorescens clade
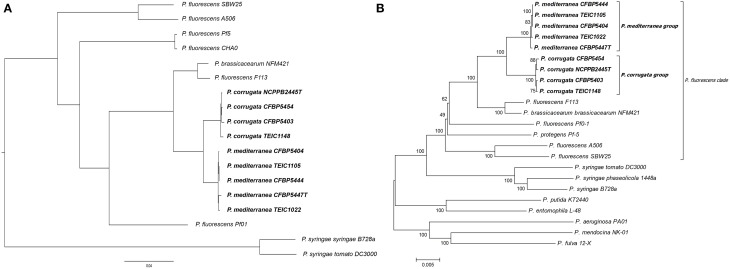
Based on previous 16S rDNA sequence analysis, both Pcor and Pmed have been included in the P. fluorescens group, while the closest species, in terms of genetic distance, has been reported to be Pseudomonas brassicacearum (Anzai et al., 2000; Catara et al., 2002). Here, we generated a whole genome phylogenetic tree from all nine Pcor and Pmed strains, in addition to the closely related Pseudomonas species P. brassicacearum subsp. brassicacearum NFM421, P. fluorescens A506, P. fluorescens CHA0, P. fluorescens F113, P. fluorescens Pf0-1, P. fluorescens Pf-5, P. fluorescens SBW25, and finally, as out-groups, P. syringae pv. syringae B728a and P. syringae pv. tomato DC3000 (Figure 2A). We identified 3826 ortholog families producing, after concatenation, a whole alignment with 1,218,944 columns. All nodes of the phylogenetic tree had 100% bootstrap support, except the node separating strains Pmed CFBP5447 and TEIC1022, which had 98% bootstrap support.
Figure 2.
(A) Phylogenetic tree of Pseudomonas species. Prefixes Pcor, Pmed, Pbra, and Pflu correspond to species Pseudomonas corrugata, P. mediterranea, P. brassicacearum, and P. fluorescens, respectively. All nodes had 100% bootstrap support, except the one separating Pmed CFBP5447 and Pmed TEIC1022, with 98% bootstrap support. The tree was generated using whole genome alignment of orthologous proteins following the approach described in Material and Methods, and was drawn using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). (B) Phylogenetic tree of the examined Pseudomonas strains based on the concatenated partial sequences of fruK, gltA, gyrB, mutL, rpoB, rpoD. Isolate names followed by the letter “T” are type strains. TEIC strains came from the Technological Educational Institute of Crete Collection. CFBP strains came from the French Collection of Plant-associated Bacteria, part of the International Center for Microbial Resources. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The bootstrap consensus tree was inferred from 1500 replicates (Felsenstein, 1985) and is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. The analysis involved 23 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 12,292 positions in the final data set. Evolutionary analyses were conducted in MEGA 6 (Tamura et al., 2013).
Because of the lack of whole genomic sequences for all Pseudomonas species, as well as the poor assembly quality of some analyzed genomes, we also used MLSA for six independent loci, to determine the phylogenetic relationships of the nine strains for which genomes were assembled. The selected loci derived from sequences spanning the coding regions of fruK, gltA, gyrB, mutL, rpoB, and rpoD. Beyond the Pcor and Pmed strains sequenced in this study, several other strains belonging to different phylogenetic clades were selected. The phylogenetic analysis showed that the Pcor and Pmed strains cluster in distinct monophyletic clades within the larger P. fluorescens clade, clearly differentiated from P. brassicacearum subsp. brassicacearum NFM421T and P. fluorescens F113 (Figure 2B). P. brassicacearum has been described as a major root-associated bacterium of Arabidopsis thaliana and Brassica napus, which may co-exist with Pcor and Pmed in agricultural niches (Ortet et al., 2011). P. fluorescens F113 is a plant growth-promoting rhizobacterium that has biocontrol activity against fungal plant pathogens; it was recently transferred to the P. brassicacearum group (Redondo-Nieto et al., 2013). Our phylogenetic results are congruent with MLSA data from Mulet et al. (2010), which grouped Pcor in the P. fluorescens lineage along with Pmed, P. kilonensis, P. thivervalensis, and P. brassicacearum (called the P. corrugata subgroup). This is also in accordance with earlier MLSA data by Trantas et al. (2015), which described the genetic relatedness of Pcor and Pmed to P. brassicacearum subsp. brassicacearum and P. fluorescens. Furthermore, in a whole genome based phylogeny, the P. fluorescens strain F113 was grouped with P. brassicacearum strain Q8r1-96 and the P. fluorescens strains Wood1R and Q2-87 (Redondo-Nieto et al., 2013). All cited species and strains in this P. corrugata subgroup, whose taxonomic position need to be clarified, were isolated from the rhizosphere or from agricultural soils and mostly studied as bio-control strains of soil-borne plant diseases. Notably, Pcor and Pmed are the only non-fluorescent and the only plant pathogenic species placed in this group, to date.
The core and the pan-genome of P. corrugata and P. mediterranea species
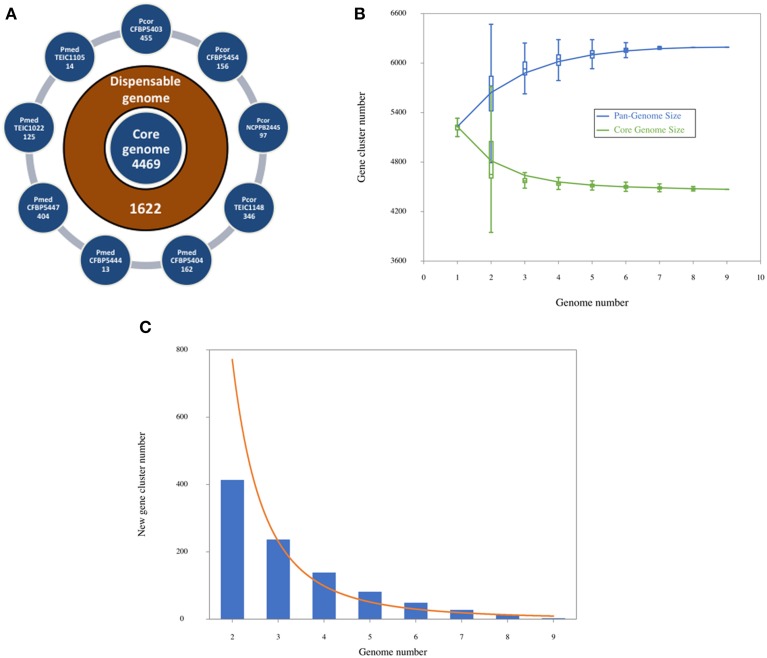
We determined the number of shared and strain specific genes based on the clusters found by OrthoMCL (Li et al., 2003), taking protein-coding genes from all nine Pcor and Pmed as input (Figure 3A). Each external circle shows the number of specific (singleton) genes of each genome. The core genome size is 4469 genes (the number of families with at least one gene in each genome) and the dispensable genome size (sometimes called accessory genome) is 1622 genes. Core-genome and pan-genome rarefaction curves are shown in Figure 3B. The core genome size of the nine genomes presented here is larger than the core genome of the P. fluorescens clade earlier reported (Loper et al., 2012), exemplifying the heterogeneity of the clade. On the other hand, the new gene curve (Figure 3C) reached a plateau relatively early, which suggests that there is respective similarity between the nine sequenced strains.
Figure 3.
(A) Numbers of shared and specific genes between Pseudomonas corrugata and P. mediterranea based on clusters. External circles show singletons of each genome. The intermediate circle shows the number of dispensable genes, and the inner one shows the core genome size, given by the number of clusters with at least one gene of each genome. (B) Core-genome and pan-genome sizes according to the number of genomes considered in the dataset. For each k in X axis, all possible combinations of k genomes (among 9 of this study) are taken and, for each one of these combinations, pan and core numbers are plotted. (C) Number of new genes observed upon adding a new genome at each step in the dataset.
Proteome comparison
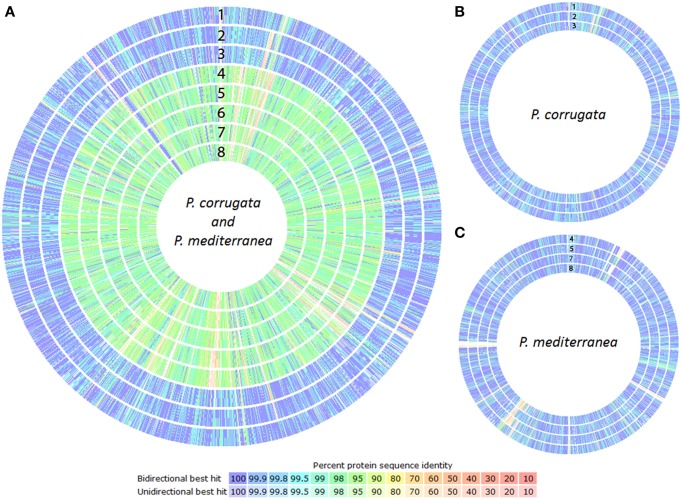
According to the phylogenetic trees constructed either with an MLSA or a whole-genome approach, the Pcor and Pmed species were identified to be closely related, and both had similar evolutionary distance from strains of P. fluorescens and P. brassicacearum. The proteomes of the nine sequenced strains were compared against the proteome of P. brassicacearum subsp. brassicacearum NFM421 (Supplementary File 1), using the RAST proteome comparison pipeline. Proteome similarities within both the Pcor and Pmed sequenced strains (Figure 4A), as well as intraspecific similarities between Pcor (Figure 4B) and Pmed (Figure 4C) strains are present.
Figure 4.
Proteome comparison of the Pseudomonas corrugata (Pcor) and P. mediterranea (Pmed) strains based on RAST genome annotation. Each proteome is presented as a closed circle with similarity for each protein against the reference genome designated by colored lines. (A) Pcor and Pmed interspecific proteome comparison against Pcor NCPPB2445 type strain, (B) Pcor intraspecific proteome comparison against Pcor NCPPB2445 type strain, (C) Pmed intraspecific proteome comparison against Pmed CFBP5447 type strain. 1, CFBP5403; 2, CFBP5454; 3, TEIC1148; 4, CFBP5404; 5, CFBP5444; 6, CFBP5447T; 7, TEIC1022, and 8, TEIC1105. The white gaps and the low similarity areas in intraspecies proteome comparisons correspond to phage genome integrations.
Protein secretion and translocation systems
Protein and DNA translocation into neighboring cells and secretion to the extracellular environment is achieved by several secretion/translocation systems in Gram negative bacteria. These secretion systems have been classified into six types, from the type I secretion system through to the type VI secretion system (T1SS-T6SS) (Tampakaki et al., 2010).
We screened the genomes of Pcor and Pmed type strains for the evidence of these secretion/translocation systems, using the gene clusters of P. fluorescens F113 as sequence queries. Our data revealed that both Pcor and Pmed genomes encode a wide variety of secretion systems, including six T1SSs, two T2SSs, one T4SS (detected only partially in Pcor strains), five T5aSSs, three T5bSS, one T5dSS (which is detected only in Pmed strains) and three T6SSs (Table 5). Surprisingly, no T3SS-encoding locus was detected in any of the examined strains.
Table 5.
The full repertoire of protein secretion/translocation systems of Pseudomonas corrugata (Pcor) and P. mediterranea (Pmed) type strains.
| Secretion system | P. corrugata NCPPB2445T | P. mediterranea CFBP5447T | P. fluorescens F113 | |
|---|---|---|---|---|
| T1SS | PcorDraft_00553-00551 | PmedDraft_01363-01365 | PSF113_0209-0211 | |
| PcorDraft_00252 | PmedDraft_02514 | PSF113_0530 | ||
| PcorDraft_00971-00969 | PmedDraft_00943-00941 | PSF113_1508-1510 | ||
| PcorDraft_05030-05032 | PmedDraft_04444-04442 | PSF113_2734-2736 | ||
| PcorDraft_00188-00190 | PmedDraft_04408-04410 | PSF113_2945-2947 | ||
| PcorDraft_04243-04245 | PmedDraft_05697-05699 | PSF113_3005-3007 | ||
| PcorDraft_03958-03956 | PmedDraft_00478-00480 | PSF113_3303-3305 | ||
| T2SS | Xcp | PcorDraft_04315-04305 | PmedDraft_01591-01607 | PSF113_0437-0447 |
| Putative substrates | PcorDraft_04316 PcorDraft_03149 PcorDraft_03269 PcorDraft_02349 |
PmedDraft_01594 PmedDraft_05894 PmedDraft_04897 PmedDraft_05316 PmedDraft_00632 |
PSF113_0435, PSF113_0610, PSF113_2337, PSF113_3279 |
|
| Hxc | PcorDraft_00325-00347 | PmedDraft_04580-04583 | PSF113_3690-3700 | |
| T3SS | SPI-I | not detected in Pcor strains | not detected in Pmed strains | PSF113_1778-1801 |
| Hrp1 | not detected in Pcor strains | not detected in Pmed strains | PSF113_5585-5610 | |
| T4SS | GI-like | not detected in strain CFBP5447 (partially only in TEIC1148 and CFBP5403) | PmedDraft_00600-00584 | PSF113_3314-3334 |
| T5aSS | PcorDraft_02455 | PmedDraft_03164 | PSF113_2779 | |
| PcorDraft_02456 | PmedDraft_03163 | PSF113_2780 | ||
| PcorDraft_04540 | PmedDraft_05328 | PSF113_4399 | ||
| PcorDraft_04609 | PmedDraft_01659 | PSF113_5339 | ||
| PcorDraft_00187 | PmedDraft_00248 | PSF113_5848 | ||
| T5bSS | PcorDraft_03404-03405 | PmedDraft_00300-00301 | PSF113_0792-0793 | |
| PcorDraft_02193-02192 | PmedDraft_00959-00960 | PSF113_1489-1490 | ||
| T5dSS | not detected in Pcor strains | PmedDraft_00929 | PSF113_1517 | |
| T6SS | I | PcorDraft_01669-01644 | PmedDraft_03358-03331 | PSF113_5785-5808 |
| II | PcorDraft_04761-04784 | PmedDraft_05238-05225 | PSF113_2407-2422 | |
| III | PcorDraft_05211-05196 | not detected in strains CFBP5447 and TEIC1022 | PSF113_5815-5833 | |
| Putative substrates | PcorDraft_05064 (hcp) | PmedDraft_03683 (hcp) | ||
| PcorDraft_00287 (vgrG) | PmedDraft_02478 (vgrG) | |||
| PcorDraft_01038 (vgrG) | PmedDraft_03454 (vgrG) | |||
| PcorDraft_00092 (vgrG) | PmedDraft_04237 (vgrG) | |||
| PcorDraft_04873 (vgrG) | PmedDraft_03810 (vgrG) | |||
| PcorDraft_03798 (vgrG) | PmedDraft_04962 (vgrG) | |||
TEIC, Technological Educational Institute of Crete Bacterial collection; CFBP, International Center for Microbial Resources, French Collection for Plant-associated Bacteria, INRA, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, Fera, York, U.K. Isolate names followed by the letter “T” are type strains. Xcp, extracellular protein; Hpx, homolog of Xcp; SPI-I and SPI-II, T3SS homologs.
Type II secretion system
The type II secretion system (T2SS) is used by Gram negative bacteria to secrete proteins into the extracellular environment, and it serves as a major virulence mechanism in several bacterial species. It is well-conserved and is composed of a set of 11 to 12 proteins (Whitchurch et al., 2004). Pseudomonas T2SSs could be divided in two main clusters, named Xcp (extracellular protein) and Hpx (homolog to Xcp) (Redondo-Nieto et al., 2013). Similar to the closely related species P. fluorescens F113, two T2SS-encoded loci, one related to Xcp cluster T2SSs and the other related to Hxc cluster T2SSs, are present in the genome of both Pcor and Pmed species (Table 5). The genetic organization of the Pcor and Pmed Hxc cluster is highly similar to the Hxc clusters of P. fluorescens F113 and P. aeruginosa PAO1 (Ball et al., 2002; Redondo-Nieto et al., 2013). Furthermore, at least five predicted Pcor and Pmed proteins could potentially be Xcp substrates (Table 5), based on reported similarities with known Xcp effectors.
P. corrugata and P. mediterranea lack a T3SS
The type III secretion system (T3SS) is a bacterial molecular nano-syringe related to the bacterial flagellum and is required for the translocation of virulence proteins. It is composed of approximately 25 proteins (Tampakaki et al., 2010; Skandalis et al., 2012), and has evolved into seven different families: Ysc, Hrp1, Hrp2, SPI-1, SPI-2, Rhizobiaceae and Chlamydia (Pallen et al., 2005; Troisfontaines and Cornelis, 2005).
An in-depth investigation of the proteomes of other Pseudomonas species revealed 37, 32, 47, and 57 T3SS-associated protein families in P. brassicacearum, P. fluorescens, P. syringae pv. syringae, and in P. syringae pv. tomato, respectively.
The predicted coding sequences from Pcor and Pmed genomes were translated and screened with several Hrp/Hrc homologs from P. syringae, P. brassicacearum, Xanthomonas, and Erwinia T3SSs, including a second rhizobial type T3SS cluster, which has been identified in several P. syringae strains (Gazi et al., 2012). From the 4,469 gene families of Pcor and Pmed species, only five that belong to the bacterial flagellum machinery revealed similarity to T3SS.
We used the amino acid sequences of all type III effector proteins (T3Es) obtained from the PPI Home web page (http://www.pseudomonas-syringae.org/) to screen for T3Es in the predicted proteomes of Pcor and Pmed. We identified only one T3E-like protein, similar to HopPmaJ (HopJ), conserved in all examined strains. BLASTP revealed that this protein is conserved in all pseudomonads in P. fluorescens group including many non-pathogenic strains. However, as far as we are aware there is not confirmation for the T3SS-dependent secretion of this particular protein.
The absence of a Hrp gene cluster from the genomes of Pcor and Pmed strains is a unique feature of these pathogens, since the majority of the known plant, insect and animal pathogenic pseudomonads rely on the presence of a functional T3SS to manipulate immunity and to colonize their hosts (Tampakaki et al., 2004). However, P. aeruginosa isolates that lack a T3SS have become established in hosts by relying on the initial infection and suppression of host immunity by T3SS-positive isolates (Czechowska et al., 2014). The absence of genes encoding a T3SS in Pcor and Pmed strains, in combination with the induction of an HR-like response in Nicotiana plants, potentially indicates the presence of additional virulence mechanisms, which probably include the secretion of apoplastic effectors. These theories remain to be investigated and our work provides the basis for future studies on the characterization of Pcor and Pmed virulence strategies.
Similar virulence strategies have been described for some Gram positive plant pathogens (Eichenlaub and Gartemann, 2011). For example, Clavibacter michiganensis relies on small secreted proteins (e.g., Chp7: CMS_2989 and Pat1: pCM2_0054) to confer virulence. We used the amino-acid sequences of these proteins as baits for screening the Pcor and Pmed predicted proteomes. No similar protein-coding genes were found in either species.
Type VI secretion system
The T6SS was initially identified as a protein secretion apparatus involved in virulence of Vibrio cholerae on Dictyostelium (Mougous et al., 2006) and P. aeruginosa on mouse models (Pukatzki et al., 2006). Furthermore, gene clusters coding for the type VI secretion system (T6SS) are present in the majority of plant pathogenic and symbiotic Gram negative bacterial genomes (Sarris et al., 2012), which indicates its importance in the life cycles of these species. Although the exact role of the T6SS in plant colonization is not clear, it is noteworthy that there are multiple T6SS clusters in single strains.
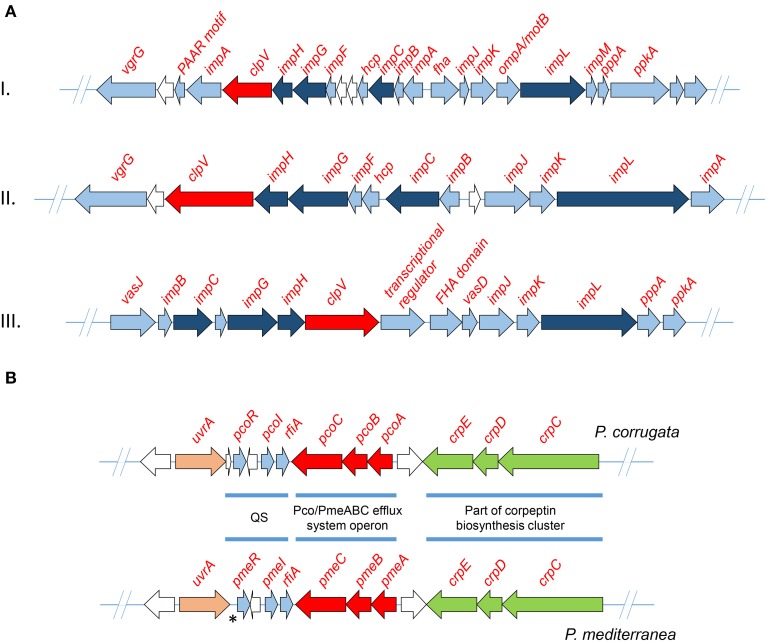
Three phylogenetically distinct clusters coding for T6SSs have been characterized in the genome of various P. syringae pathovars (Sarris et al., 2010, 2012). In order to identify gene clusters coding for T6SS components of the assembled genomes, all the Pcor and Pmed predicted proteomes were screened using the T6SS core components ImpB, ImpC and the ClpV1 ATPase of P. syringae as sequence queries. BLASTP analysis of the nine predicted proteomes was used to reveal the presence of three independent gene clusters that were extracted from each genome (Table 5). Several genes coding for VgrG (Valine-Glycine Repeat Protein G) proteins were found randomly distributed in the genome of both species. This is a common feature of Pseudomonas species (Sarris et al., 2010, 2012; Sarris and Scoulica, 2011). Nucleotide alignments of the three T6SS clusters revealed a high degree of similarity between the nine sequenced strains of Pcor and Pmed, and the P. brassicacearum strains. Interestingly, the T6SS-III locus was not identified in Pmed strains TEIC1022 and CFBP5447 or in P. brassicacearum NFM421, but the cluster is present in the genome of P. fluorescens F113. The three phylogenetically-distinct consensus T6SS loci are schematically presented in Figure 5A.
Figure 5.
(A) Maps of the three detected T6SS clusters of Pseudomonas corrugata and P. mediterranea strains. Arrows indicate detected ORFs and their direction shows the direction of transcription. All ORFs annotated as type VI secretion system associated proteins are colored light blue. Those which designate the T6SS phylogeny are colored dark blue. The ClpV protein, which was used to mine the T6SS from each genome, is colored red. White arrows indicate ORFs encoding hypothetical, uncharacterized proteins. (B) The map of P. corrugata and P. mediterranea consensus quorum sensing gene clusters. The asterisk indicates the locus of P. mediterranea that lacks an ORF when compared to the genome of P. corrugata. QS, quorum sensing.
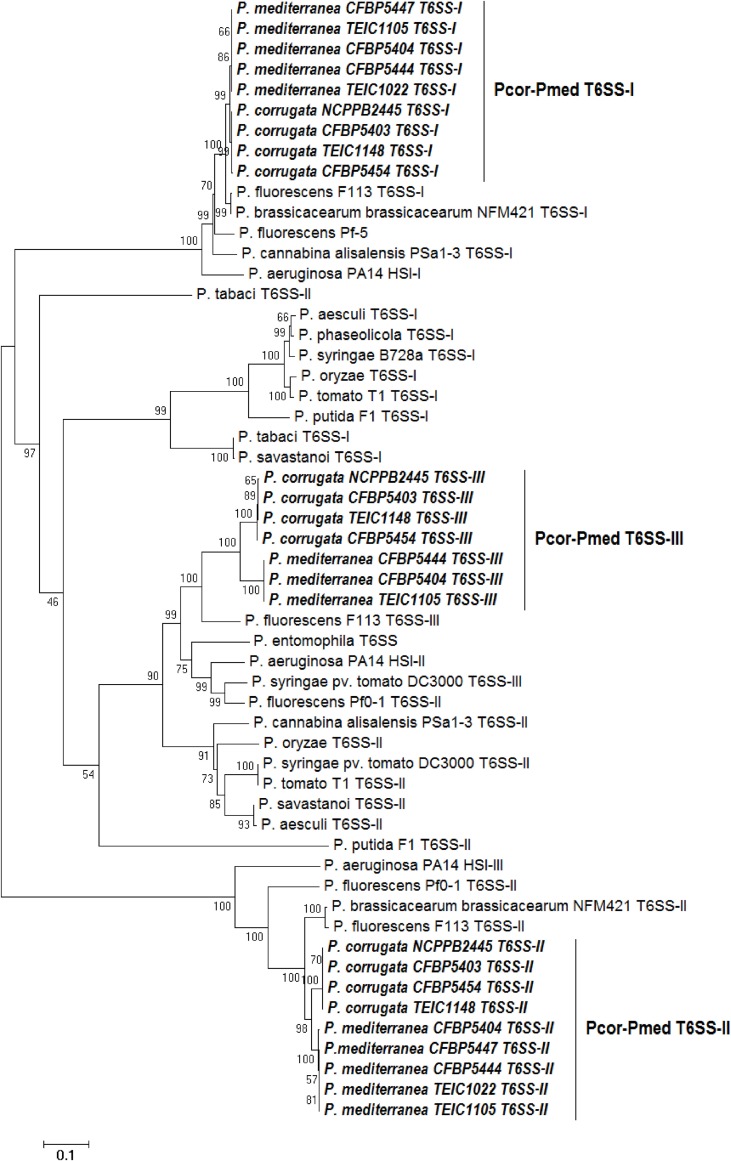
Phylogenetic analysis of various Pseudomonas species was performed using concatenated protein sequences of four highly conserved T6SS core proteins (ImpC, ImpG, ImpH, ImpL) (Figure 6). The consensus phylogenetic tree obtained indicates that pseudomonad T6SSs are scattered into three main clusters (Figure 6), and this matches what has been previously shown (Sarris et al., 2012).
Figure 6.
Distance tree of T6SSs of various pseudomonads based on the sequence of four T6SS core proteins (ImpC, ImpG, ImpH, and ImpL). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1500 replicates is taken to represent the evolutionary history of the proteins analyzed (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in theunits of the number of amino acid substitutions per site. The analysis involved 59 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 829 positions in the final dataset. Evolutionary analyses were conducted using MEGA6 (Tamura et al., 2013). “Pcor” stands for Pseudomonas corrugata while “Pmed” stands for P. mediterranea. “TEIC” strains came from the Technological Educational Institute of Crete Collection. CFBP strains came from the French Collection of Plant-associated Bacteria, part of the International Center for Microbial Resources.
The T6SS clades from Pcor and Pmed tend to cluster with sequences from P. fluorescens F113 and P. brassicacearum NFM421. Our analysis revealed that the T6SS-II clusters of Pcor and Pmed are highly related to the T6SS-III of the opportunistic human pathogen P. aeruginosa (Figure 6). In addition, the grouping of Pcor and Pmed indicates minor divergence of the T6SS between the two species, since no mixing among the Pcor and Pmed strains was observed. The divergence was higher for the T6SS-II and T6SS-III clusters than for the T6SS-I cluster.
Other virulence factors
Quorum sensing
Highly similar AHL-QS systems were identified in Pcor strain CFBP5454 and Pmed strain CFBP5447; these LuxI/R QS systems were designated PcoI/R and PmeI/R, respectively (Licciardello et al., 2007, 2012). As described earlier, the AHL-QS system and more specifically the PcoR and PmeR, as well as the RfiA regulators of both species, are required for full virulence in tomato (Licciardello et al., 2009, 2012; Strano et al., 2014). Analysis of all examined genomes revealed that all strains have genes of only one canonical paired LuxI/R system (Figure 5B). The QS gene topology was conserved in both Pcor and Pmed strains, with the luxI/luxR genes oriented in the same direction and separated by a divergently oriented ORF, which codes for a putative homoserine/threonine efflux protein. All strains have the rfiA gene coding for a transcriptional regulator downstream of, and in the same orientation as, luxI. Licciardello et al. demonstrated that the two genes are co-transcribed in both species (Licciardello et al., 2009, 2012).
The LuxI and LuxR protein sequences are highly conserved within Pcor (between 99 and 100%) and Pmed species (100%) and also between the two species (similarities >84% and >95%, respectively). A significant difference was observed between the promoter regions of pcoR and pmeR; the promoter region of Pmed strains is 31 bp longer than that of Pcor. It is not known whether this property may have any effects on the regulation of pmeR compared to pcoR, but it could account for some of the differences observed between the two species.
The regulatory gene, rfiA, was located at the right border of the PcoI/PmeI gene in all strains. RfiA contains a C-terminal helix-turn-helix (HTH) DNA-binding motif, which is characteristic of the LuxR family of bacterial regulatory proteins, but lacks the auto-inducer-binding domain found in the quorum-sensing-associated LuxR regulators (Licciardello et al., 2009). RfiA showed a similarity of 100% within species and >94% between species. These kinds of LuxR-type regulators have been demonstrated to be involved in the biosynthesis of a number of cyclic lipopeptides in several pseudomonads (reviewed in Raaijmakers et al., 2010).
P. corrugata and P. mediterranea harbor a large arsenal of secondary metabolites
Many plant associated Pseudomonas produce secondary metabolite cyclic lipopeptides (CLPs) that are composed of a fatty acid tail, linked to a short oligo-peptide which is cyclized to form a lactone ring between two amino acids in the peptide chain. They possess surfactant, antimicrobial, anti-predation, and cytotoxic properties (Raaijmakers et al., 2006, 2010). Based on the length and composition of the fatty acid tail as well as the number, type, and configuration of the amino acids in the peptide moiety, CLPs have been classified into six groups. Biosynthesis of CLPs is directed by non-ribosomal peptide synthetases (NRPS), which are multi-enzyme complexes with a modular arrangement. Each module consist of several domains for condensation (C), thiolation (T) and aminoacyl adenylation (A), responsible for incorporation, recognition and activation of each amino acid unit into the growing peptide (Raaijmakers et al., 2006). Genes that code for these large multi-modular enzymes can span up to 75 kb of DNA. This repetitive structure is difficult to assemble with next generation sequencing approaches; such large enzymes may be split and span several contigs. This is the case for Pcor and Pmed genomes, for which the software pipeline antiSMASH (Blin et al., 2013) detected corpeptin NRPS spanning several contigs.
Pcor and Pmed strains produce phytotoxic and antimicrobial cationic CLPs (Emanuele et al., 1998; Scaloni et al., 2004; Licciardello et al., 2012). Corpeptin A and corpeptin B, two isoforms consisting of 22 amino acid residues, were identified in the culture of the Pcor type strain. Both are phytotoxic against tobacco leaves and possess antimicrobial activity against Bacillus megaterium (Emanuele et al., 1998). A strain-dependent production of cormycin, a lipodepsinonapeptide, has been earlier described (Scaloni et al., 2004). This molecule produced a strong in vitro inhibition of B. megaterium and Rhodotorula pilimanae that was higher than that of nonapeptides from P. syringae, and also induced chlorosis in tobacco leaves (Scaloni et al., 2004).
Pcor and Pmed are able to inhibit the in vitro growth of various Gram positive and Gram negative bacteria, and phytopathogenic fungi (Catara, 2007). The antimicrobial activity of strains described in this study was tested against two microorganisms: the Gram positive bacterium Bacillus megaterium ITM100 and the yeast Rhodotorula pilimanae ATTC26432, which are both sensitive to CLPs, and against the Gram negative phytopathogens P. syringae pv. tomato PVCT28.3.1 and X. campestris pv. campestris PVCT 62.4. All Pcor and Pmed strains displayed activity that inhibited the growth of R. pilimanae. Only the five R-phenotype strains were able to inhibit the growth of B. megaterium and P. syringae pv. tomato (Figure 1D and Table 2). Cormycin and corpeptins are both produced in cultures which displayed antimicrobial activity, so we therefore decided to test the culture filtrates of four strains grown in CLP-inducing conditions; two for Pcor (CFBP5454, CFBP5403) and two for Pmed strains respectively (CFBP5447, CFBP5444) (Surico et al., 1988). The culture filtrates of three strains sharing the R-phenotype showed antimicrobial activity against both R. pilimanae and B. megaterium, whereas Pmed CFBP5444 S-type strain culture filtrate did not display antagonistic activity (Figure 1D). This correlates with the previous observation that culture filtrates of Pcor strain CFBP5454 and Pmed CFBP5447 mutants unable to produce these CLPs lose the ability to inhibit the in vitro growth of the two bio-indicator microorganisms (Licciardello et al., 2009, 2012). Moreover, a null mutant strain for the QS transcriptional regulator PcoR, which is impaired in CLP production, is still able to antagonize fungal and bacterial plant pathogens in dual culture assays (Licciardello et al., 2007). Thus, we propose that the residual antifungal activity against R. pilimanae is due at least partially to another secreted antifungal metabolite. These results support the importance of further in-depth studies for the identification of antimicrobial components of both Pcor and Pmed species for their application against important plant pathogens.
Antimicrobial cluster mining
We used the software pipeline antiSMASH (Blin et al., 2013) for the automated identification of secondary metabolite biosynthesis clusters in each genome. The analysis identified, in all genomes, a number of non-ribosomal peptide synthetases (NRPS) clusters, a gene cluster related to bacteriocin biosynthesis, a type I PKS cluster a siderophore and an arylpolyene gene cluster. A lantipeptide gene cluster was exclusively detected in Pmed genomes (Supplementary File 2).
In the Pcor CFBP5454 genome, which has a relatively low number of scaffolds, we identified at least 10 scaffolds with genes similar to those for syringomycin and syringopeptin biosynthesis, secretion and regulation in P. syringae pv. syringae strains. In particular, we found NRPSs putatively involved in cormycin and corpeptin production and their secretion. One of the antiSMASH clusters matches with the GenBank sequence KF192265. This cluster also includes genes for the PcoI/PcoR AHL-QS system, the transcriptional regulator gene RfiA and CrpCDE corpeptin biosynthetic cluster coding for an approximately 6000 bp DNA segment that encodes two modules of a NRPS (crpC) and an ABC transporter responsible for corpeptin secretion (crpDE) (Strano et al., 2014). The CrpDE efflux system is similar to the PseEF efflux system of P. syringae pv. syringae strain B301D, which is involved in the export of syringomycin and syringopeptin (Cho and Kang, 2012). The scaffolds containing the QS system have the same topology in all Pcor and Pmed strains but the sequence of the corpeptin synthetase gene C (crpC) is always prematurely interrupted by the end of a contig. The analysis of NRPS clusters in the other strains supported the presence of the same cormycin and corpeptin biosynthesis, secretion and regulatory apparatus for all strains.
BLAST searches of the other NRPS genes mined by antiSMASH suggest that three other different NRPS clusters are present in all strains. One of them is putatively involved in the production of the lipopeptide siderophore corrugatin (see Siderophores paragraph below). Thus, more bioactive metabolites can be produced by the strains by means of the other two NRPS clusters and by a conserved type I PKS cluster which was not detected in other Pseudomonas species deposited in GenBank and IMG databases. Best similarities (approximately 40%) were detected by BLAST within the genomes of the symbiotic bacterium Rhizobium sullae wsm1592 and Mesorhizobium loti MAFF303099 plasmid pMLa.
In addition, an aryl-polyene (APE) gene cluster was detected in all strains Recently APE gene clusters were found widely but discontinuously distributed among Gram-negative bacteria (Cimermancic et al., 2014). Aryl polyenes are responsible for yellow pigmentation such as the brominated aryl- polyenes xanthomonadins of Xanthomonas species (Cimermancic et al., 2014). Lantipeptides, which are potentially bioactive metabolites, are estimated to be produced only by Pmed strains. Lantipeptides are ribosomal synthesized and post-translational modified peptides, formerly called lantibiotics, characterized by thio-ether cross-links (Knerr and van der Donk, 2012).
We mined each genome by BLAST using the biosynthetic loci of compounds contributing to biological control in other Pseudomonas strains such as phenazines, hydrogen cyanide (HCN), pyrrolnitrin, 2,4-diacetylphloroglucinol (DAPG), and pyoluteorin as queries (Loper et al., 2012). Only the hydrogen cyanide gene cluster was found in all Pcor and Pmed genomes. However, using the Cyantesmo detection card only, six out of the nine strains produced detectable levels of HCN (Table 2). The HCN and the DAPG gene clusters have been reported in strain Q8r1-96 of the closely related P. brassicacearum and in P. fluorescens strain F113 (Loper et al., 2012; Redondo-Nieto et al., 2013).
Mining for bacteriocins
The assembled genomes were queried for genes encoding bacteriocins (Riley and Wertz, 2002), which are antimicrobial peptides produced by bacteria to inhibit the growth of closely related bacterial strains (Hassan et al., 2012). The translated ORFs of the nine genomes under study and the P. brassicacearum NFM421 were blasted with antimicrobial peptides from the BAGEL database (Van Heel et al., 2013) and protein hits were recorded (Supplementary File 1). In all Pmed strains, a gene for the synthesis of bacteriocin Colicin E was detected. However, no Colicin E biosynthesis genes were detected in the genomes of the Pcor strains, although genes for the peptides Carocin D and Zoocin A were detected. In the related P. brassicacearum NFM421, no bacteriocin biosynthesis genes were detected, while in the P. fluorescens F113 a Carocin D biosynthesis gene was detected.
Siderophores
Typically, Pseudomonas sensu stricto produces yellow-green fluorescent pigments when grown under iron-limiting conditions. These fluorescent compounds are pyoverdines that act as siderophores (Cornelis and Matthijs, 2002), which are iron chelating compounds secreted to survive during conditions of iron deprivation. As expected, since Pcor and Pmed are members of the non-fluorescent group of the species, no pyoverdine biosynthesis cluster was found.
However, Pcor was found to produce corrugatin, a CLP siderophore (Meyer et al., 2002). The antiSMASH analysis of Pcor and Pmed genomes revealed the presence of two different gene clusters putatively coding for proteins involved in siderophore biosynthesis or utilization. The first cluster was characterized by the presence of NRPS genes and was split in all strains into two different contigs (Supplemental File 1). The second cluster revealed similarity to the achromobactin biosynthesis cluster. BLASTX analysis with the predicted NRPS sequences showed similarity to a putative siderophore gene cluster in P. fluorescens strain SBW25 (Cheng et al., 2013). The SBW25 gene cluster contains five NRPS genes (PFLU3220, 3222–3225) that are predicted to encode an eight-amino acid peptide that resembles ornicorrugatin (similar to corrugatin, but the amino acid 2,4-diaminobutanoic acid is replaced by the amino acid ornithine), a siderophore produced by P. fluorescens AF76 (Matthijs et al., 2008). This cluster is also present in P. brassicacearum subsp. brassicaceraum NFM421 and in P. fluorescens F113, the genomes of which are closely related to the Pcor and Pmed strains.
We discovered a cluster that has high similarity to the achromobactin biosynthesis and utilization cluster of P. syringae pv. syringae B728a (Psyr2582-2593) and that has been detected in six different P. syringae pathovars (Berti and Thomas, 2009). All Pcor and Pmed strains in this study possess highly conserved genes of this cluster, but lack the entire ABC transporter system encoded by cbrA, cbrB, cbrC (Psyr2590-2592). A comparative analysis with the genetically related bacterial strains P. brassicacearum subsp. brassicaceraum strain NFM421 and P. fluorescens strain F113 showed the same cluster without the transporter system.
Conclusion
We sequenced the genomes of nine P. corrugata and P. mediterranea strains in order to undertake an in-depth genome comparison study. The comparison of nine genomes with each other and with other closely related genomes revealed the absence of virulence genes corresponding to the T3SS from the genome of Pcor and Pmed. These findings raise the question regarding the absence of a T3SS from Pcor and Pmed. Was this system deleted specifically in these lineages? This may have happened at a much earlier point in the lineage, or the P. fluorescens lineages that have a T3SS may have acquired it via horizontal gene transfer (HGT); this second conclusion would mean that the ancestor of Pcor and Pmed lineages did not have this cluster. Our whole genome phylogeny provides a potential answer to these questions. According to our analysis, the closer relatives to Pcor and Pmed species (the P. brassicacearum strains NFM421, F113 and the P. fluorescens strains SBW25, A506), possess a T3SS indicating that the loss of the T3SS from Pcor and Pmed strains occurred specifically in these species. However, further studies are required for the elucidation of the mechanisms of virulence (e.g., through the exploitation of toxins) of these species.
Although the examined Pcor and Pmed strains were isolated from different geographic areas and hosts, no host or geographical differentiation in their genomes could be identified. Based on these findings, we hypothesize that the two species may have separated by factors other than the selection pressures of host range and geography. Likewise, differentiation of colony morphology, experimental antimicrobial activity and induction of an HR-like response in N. benthamiana plants, resulted in intra- instead of inter-species differentiation for all Pcor and Pmed strains used in this study. Furthermore, analysis of Pcor and Pmed genomes showed three genomic clusters coding for T6SSs (T6SS-I, T6SS-II and T6SS-III). The number of T6SS clusters per genome reflects the importance of this secretion system to the life cycle of Pcor and Pmed species. Interestingly, the first two T6SS clusters are syntenically and phylogenetically related to the P. aeruginosa HSI-I and HSI-III. However, the potential contribution of the T6SS to pathogenicity of Pcor and Pmed species remains to be investigated; this will be of great interest in light of the absence of a functional T3SS in both species.
Pcor and Pmed are particularly interesting pathogens because they can cause disease on a large number of phylogenetically divergent hosts, including several economically important crop species. This indicates that both Pcor and Pmed are able to manipulate host immunity without using any known virulence effector protein (e.g., type III effectors). The phylogenetic relationship with saprophyte species P. fluorescens and P. brassicacearum places Pcor and Pmed as intermediates between saprophytic and plant pathogenic lifestyles. To date, Pcor and Pmed have displayed peculiar and unique features. Now that we have analyzed and compared their genomes, future investigations have resources with the exciting potential to reveal the presence of additional virulence mechanisms in the genomes of Pcor and Pmed and the elucidation of the nature and role of these mechanisms during host colonization. Our study provides the basis for such investigations, and not only gives mechanistic insights into the molecular interactions underlying alternative virulence mechanisms.
Author contributions
PS and VC conceived the study, and contributed to its design and coordination and helped to draft the manuscript. DG contributed to genome sequencing. ET, GL, NA, KW, DG, VC, and PS contributed to genome assembly and annotation of the genomes. NA contributed to core genome analyses. FV, DG, and JJ contributed materials or analytic tools. VC, CS and DG contributed to antimicrobial and pathogenicity tests. All authors contributed to the writing and editing of the manuscript and approved the final version of it.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GL and VC were supported by MIUR by means of the national Program PON R&C 2007-2013, co-funded by EU, project “PolyBioPlast – Technologies and processes for the production of diversely functionalised sheets based on microbial biopolymers and biosurfactants (PON01_1377).” NFA: Fundect (grant TO0096/2012), CNPq (grant 305857/2013-4).
Funding
DEG was supported by Research Actions in Phytopathology lab, Education and Research Committee, Technological Educational Institute of Crete.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00811
References
- Achouak W., Thiéry J. M., Roubaud P., Heulin T. (2000). Impact of crop management on intraspecific diversity of Pseudomonas corrugata in bulk soil. FEMS Microbiol. Ecol. 31, 11–19. 10.1111/j.1574-6941.2000.tb00666.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai Y., Kim H., Park J. Y., Wakabayashi H., Oyaizu H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50(Pt 4), 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Ball G., Durand E., Lazdunski A., Filloux A. (2002). A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43, 475–485. 10.1046/j.1365-2958.2002.02759.x [DOI] [PubMed] [Google Scholar]
- Barnett S. J., Alami Y., Singleton I., Ryder M. H. (1999). Diversification of Pseudomonas corrugata 2140 produces new phenotypes altered in GC-FAME, BIOLOG, and in vitro inhibition profiles and taxonomic identification. Can. J. Microbiol. 45, 287–298. 10.1139/w99-006 [DOI] [PubMed] [Google Scholar]
- Berti A. D., Thomas M. G. (2009). Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J. Bacteriol. 191, 4594–4604. 10.1128/JB.00457-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Medema M. H., Kazempour D., Fischbach M. A., Breitling R., Takano E., et al. (2013). antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212. 10.1093/nar/gkt449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- Boetzer M., Pirovano W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. 10.1186/gb-2012-13-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Catara V. (2007). Pseudomonas corrugata: plant pathogen and/or biological resource? Mol. Plant Pathol. 8, 233–244. 10.1111/j.1364-3703.2007.00391.x [DOI] [PubMed] [Google Scholar]
- Catara V., Arnold D., Cirvilleri G., Vivian A. (2000). Specific oligonucleotide primers for the rapid identification and detection of the agent of Tomato Pith Necrosis, Pseudomonas corrugata, by PCR amplification: evidence for two distinct genomic groups. Eur. J. Plant Pathol. 106, 753–762. 10.1023/A:1026507423848 [DOI] [Google Scholar]
- Catara V., Gardan L., Lopez M. (1997). Phenotypic heterogeneity of Pseudomonas corrugata strains from southern Italy. J. Appl. Microbiol. 83, 576–586. 10.1046/j.1365-2672.1997.00268.x [DOI] [Google Scholar]
- Catara V., Sutra L., Morineau A., Achouak W., Christen R., Gardan L. (2002). Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1749–1758. 10.1099/ijs.0.02174-0 [DOI] [PubMed] [Google Scholar]
- Cheng X., de Bruijn I., Van Der Voort M., Loper J. E., Raaijmakers J. M. (2013). The Gac regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 5, 608–619. 10.1111/1758-2229.12061 [DOI] [PubMed] [Google Scholar]
- Cho H., Kang H. (2012). The PseEF efflux system is a virulence factor of Pseudomonas syringae pv. syringae. J. Microbiol. 50, 79–90. 10.1007/s12275-012-1353-9 [DOI] [PubMed] [Google Scholar]
- Cimermancic P., Medema M. H., Claesen J., Kurita K., Wieland Brown L. C., Mavrommatis K., et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421. 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P., Matthijs S. (2002). Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4, 787–798. 10.1046/j.1462-2920.2002.00369.x [DOI] [PubMed] [Google Scholar]
- Czechowska K., Mckeithen-Mead S., Al Moussawi K., Kazmierczak B. I. (2014). Cheating by type 3 secretion system-negative Pseudomonas aeruginosa during pulmonary infection. Proc. Natl. Acad. Sci. U.S.A. 111, 7801–7806. 10.1073/pnas.1400782111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos P., Goor M., Gillis M., De Ley J. (1985). Ribosomal ribonucleic acid cistron similarities of phytopathogenic Pseudomonas species. Int. J. Syst. Bacteriol. 35, 169–184. 10.1099/00207713-35-2-169 [DOI] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub R., Gartemann K. H. (2011). The Clavibacter michiganensis subspecies: molecular investigation of Gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 49, 445–464. 10.1146/annurev-phyto-072910-095258 [DOI] [PubMed] [Google Scholar]
- Emanuele M. C., Scaloni A., Lavermicocca P., Jacobellis N. S., Camoni L., Di Giorgio D., et al. (1998). Corpeptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata. FEBS Lett. 433, 317–320. 10.1016/S0014-5793(98)00933-8 [DOI] [PubMed] [Google Scholar]
- Farias N. C., Almeida N. F. (2013). Orthologsorter: Inferring Genotyping and Functionality from Ortholog Protein Families. Campo Grande: Federal University of Mato Grosso do Sul. [Google Scholar]
- Felsenstein J. (1985). Confidence-Limits on phylogenies - an approach using the Bootstrap. Evolution 39, 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Fiori M. (1992). A new bacterial disease of chrysanthemum: a stem rot by Pseudomonas corrugata Roberts et Scarlett. Phytopathol. Mediterr. 31, 110–114. [Google Scholar]
- Gazi A. D., Sarris P. F., Fadouloglou V. E., Charova S. N., Mathioudakis N., Panopoulos N. J., et al. (2012). Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiol. 12:188. 10.1186/1471-2180-12-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustine D. L., Sherwood R. T., Lukezic F. L., Moyer B. G. (1990). Metabolites from Pseudomonas corrugata elicit phytoalexin biosynthesis in white clover. Phytopathology 80, 1427–1432. 10.1094/Phyto-80-1427 [DOI] [Google Scholar]
- Gustine D. L., Sherwood R. T., Moyer B. G. (1995). Evidence for a new class of peptide elicitor of the hypersensitive reaction from the tomato pathogen Pseudomonas corrugata. Phytopathology 85, 848–853. 10.1094/Phyto-85-848 [DOI] [Google Scholar]
- Hassan M., Kjos M., Nes I. F., Diep D. B., Lotfipour F. (2012). Natural antimicrobial peptides from bacteria: characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 113, 723–736. 10.1111/j.1365-2672.2012.05338.x [DOI] [PubMed] [Google Scholar]
- Hosni T., Moretti C., Devescovi G., Suarez-Moreno Z. R., Fatmi M. B., Guarnaccia C., et al. (2011). Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 5, 1857–1870. 10.1038/ismej.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersters K., Ludwig W., Vancanneyt M., De Vos P., Gillis M., Schleifer K.-H. (1996). Recent changes in the classification of the pseudomonads: an overview. Syst. Appl. Microbiol. 19, 465–477. 10.1016/S0723-2020(96)80020-8 [DOI] [Google Scholar]
- Knerr P. J., van der Donk W. A. (2012). Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505. 10.1146/annurev-biochem-060110-113521 [DOI] [PubMed] [Google Scholar]
- Kovacevich P., Ryder M. (1991). Biocontrol performance characterization of Pseudomonas corrugata isolates PS 2140 and PS 2161. Phytopathology 81, 1178. [Google Scholar]
- Lelliott R. A., Stead D. E. (1987). Methods for the Diagnosis of Bacterial Diseases of Plants. Oxford: Blackwell; Science Inc. [Google Scholar]
- Li L., Stoeckert C. J., Jr., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licciardello G., Bella P., Catara V. (2011). Quantitative detection of Pseudomonas corrugata and P. mediterranea in tomato plants by duplex real-time PCR. J. Plant Pathol. 93, 595–602. 10.4454/jpp.v93i3.1227 [DOI] [Google Scholar]
- Licciardello G., Bella P., Devescovi G., Strano C. P., Sarris P. F., Catara A. F., et al. (2014a). Draft genome sequence of Pseudomonas mediterranea strain CFBP 5447T, a producer of filmable medium-chain-length polyhydroxyalkanoates. Genome Announc. 2:e01260-14. 10.1128/genomeA.01260-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licciardello G., Bertani I., Steindler L., Bella P., Venturi V., Catara V. (2007). Pseudomonas corrugata contains a conserved N-acyl homoserine lactone quorum sensing system; its role in tomato pathogenicity and tobacco hypersensitivity response. FEMS Microbiol. Ecol. 61, 222–234. 10.1111/j.1574-6941.2007.00338.x [DOI] [PubMed] [Google Scholar]
- Licciardello G., Bertani I., Steindler L., Bella P., Venturi V., Catara V. (2009). The transcriptional activator rfiA is quorum-sensing regulated by cotranscription with the luxI homolog pcoI and is essential for plant virulence in Pseudomonas corrugata. Mol. Plant Microbe Interact. 22, 1514–1522. 10.1094/MPMI-22-12-1514 [DOI] [PubMed] [Google Scholar]
- Licciardello G., Jackson R. W., Bella P., Strano C. P., Catara A. F., Arnold D. L., et al. (2014b). Draft genome sequence of Pseudomonas corrugata, a phytopathogenic bacterium with potential industrial applications. J. Biotechnol. 175, 65–66. 10.1016/j.jbiotec.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Licciardello G., Strano C. P., Bertani I., Bella P., Fiore A., Fogliano V., et al. (2012). N-acyl-homoserine-lactone quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in the regulation of lipodepsipeptide production. J. Biotechnol. 159, 274–282. 10.1016/j.jbiotec.2011.07.036 [DOI] [PubMed] [Google Scholar]
- Loper J. E., Hassan K. A., Mavrodi D. V., Davis E. W., II., Lim C. K., Shaffer B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M., Rodriguez F., Montojo A., Salcedo R., Marti R. (1988). Pepper, a new host of Pseudomonas corrugata, in 5th International Congress of Plant Pathology (Kioto: ISPP 98. Abstracts book; ), 98. [Google Scholar]
- Magyarosy A., Buchanan B. (1995). First report of Pseudomonas corrugata causing pith necrosis on geraniums. Phytopathology 85, 1040. [Google Scholar]
- Markowitz V. M., Chen I.-M. A., Palaniappan K., Chu K., Szeto E., Pillay M., et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567. 10.1093/nar/gkt963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs S., Budzikiewicz H., Schäfer M., Wathelet B., Cornelis P. (2008). Ornicorrugatin, a new siderophore from Pseudomonas fluorescens AF76. Z. Naturforsch. Sect. C. 63, 8–12. 10.1515/znc-2008-1-202 [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M., Bertani I., Ferluga S., Cabrio L., Bigirimana J., Guarnaccia C., et al. (2011). The plant pathogen Pseudomonas fuscovaginae contains two conserved quorum sensing systems involved in virulence and negatively regulated by RsaL and the novel regulator RsaM. Environ. Microbiol. 13, 145–162. 10.1111/j.1462-2920.2010.02316.x [DOI] [PubMed] [Google Scholar]
- Mavromatis K., Ivanova N. N., Chen I. M., Szeto E., Markowitz V. M., Kyrpides N. C. (2009). The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand. Genomic Sci. 1, 63–67. 10.4056/sigs.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. M., Geoffroy V. A., Baysse C., Cornelis P., Barelmann I., Taraz K., et al. (2002). Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines. Arch. Biochem. Biophys. 397, 179–183. 10.1006/abbi.2001.2667 [DOI] [PubMed] [Google Scholar]
- Moore E. R., Mau M., Arnscheidt A., Böttger E. C., Hutson R. A., Collins M. D., et al. (1996). The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19, 478–492. 10.1016/S0723-2020(96)80021-X [DOI] [Google Scholar]
- Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet M., Lalucat J., García-Valdés E. (2010). DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12, 1513–1530. 10.1111/j.1462-2920.2010.02181.x [DOI] [PubMed] [Google Scholar]
- Ortet P., Barakat M., Lalaouna D., Fochesato S., Barbe V., Vacherie B., et al. (2011). Complete genome sequence of a beneficial plant root-associated bacterium, Pseudomonas brassicacearum. J. Bacteriol. 193, 3146. 10.1128/JB.00411-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen M. J., Beatson S. A., Bailey C. M. (2005). Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. 10.1186/1471-2180-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Palni L. M., Hebbar K. P. (2001). Suppression of damping-off in maize seedlings by Pseudomonas corrugata. Microbiol. Res. 156, 191–194. 10.1078/0944-5013-00102 [DOI] [PubMed] [Google Scholar]
- Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones B., Dulla G., Lindow S. E. (2005). Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18, 682–693. 10.1094/MPMI-18-0682 [DOI] [PubMed] [Google Scholar]
- Quiñones B., Pujol C. J., Lindow S. E. (2004). Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17, 521–531. 10.1094/MPMI.2004.17.5.521 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., de Bruijn I., de Kock M. J. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant Microbe Interact. 19, 699–710. 10.1094/MPMI-19-0699 [DOI] [PubMed] [Google Scholar]
- Raaijmakers J. M., De Bruijn I., Nybroe O., Ongena M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. 10.1111/j.1574-6976.2010.00221.x [DOI] [PubMed] [Google Scholar]
- Redondo-Nieto M., Barret M., Morrissey J., Germaine K., Martinez-Granero F., Barahona E., et al. (2013). Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics 14:54. 10.1186/1471-2164-14-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. A., Wertz J. E. (2002). Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56, 117–137. 10.1146/annurev.micro.56.012302.161024 [DOI] [PubMed] [Google Scholar]
- Ryder M., Rovira A. (1993). Biological control of take-all of glasshouse-grown wheat using strains of Pseudomonas corrugata isolated from wheat field soil. Soil Biol. Biochem. 25, 311–320. 10.1016/0038-0717(93)90129-Y [DOI] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sarris P. F., Scoulica E. V. (2011). Pseudomonas entomophila and Pseudomonas mendocina: potential models for studying the bacterial type VI secretion system. Infect. Genet. Evol. 11, 1352–1360. 10.1016/j.meegid.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Sarris P. F., Trantas E. A., Skandalis N., Tampakaki A. P., Kapanidou M., Kokkinidis M., et al. (2012). Phytobacterial type VI secretion system - gene distribution, phylogeny, structure and biological functions, in Plant Pathology, ed Cumagun C. J. (InTech; ). [Google Scholar]
- Sarris P. F., Skandalis N., Kokkinidis M., Panopoulos N. J. (2010). In silico analysis reveals multiple putative type VI secretion systems and effector proteins in Pseudomonas syringae pathovars. Mol. Plant Pathol. 11, 795–804. 10.1111/j.1364-3703.2010.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaloni A., Dalla Serra M., Amodeo P., Mannina L., Vitale R., Segre A., et al. (2004). Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: cormycin A1. Biochem. J. 384, 25–36. 10.1042/BJ20040422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett C. M., Fletcher J. T., Roberts P., Lelliott R. A. (1978). Tomato pith necrosis caused by Pseudomonas corrugata n. sp. Ann. Appl. Biol. 88, 105–114. 10.1111/j.1744-7348.1978.tb00684.x [DOI] [Google Scholar]
- Schaad N. W., Jones J. B., Chun W. (1988). Laboratory Guide for Identification of Plant Pathogenic Bacteria. St. Paul, MN: APS press. [Google Scholar]
- Schisler D., Slininger P. (1994). Selection and performance of bacterial strains for biologically controlling Fusarium dry rot of potatoes incited by Gibberella pulicaris. Plant Dis. 78, 251–255. 10.1094/PD-78-0251 [DOI] [Google Scholar]
- Siverio F., Cambra M., Gorris M., Corzo J., Lopez M. M. (1993). Lipopolysaccharides as determinants of serological variability in Pseudomonas corrugata. Appl. Environ. Microbiol. 59, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandalis N., Sarris P., Panopoulos N. J. (2012). Targeting injectisomes of virulence: benefi ts of thirty years of research on bacterial secretion systems, to crop protection Hell. Plant Prot. J. 5, 31–47. [Google Scholar]
- Solaiman D. K., Catara V., Greco S. (2005). Poly(hydroxyalkanoate) synthase genotype and PHA production of Pseudomonas corrugata and P. mediterranea. J. Ind. Microbiol. Biotechnol. 32, 75–82. 10.1007/s10295-005-0205-6 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Strano C. P., Bella P., Licciardello G., Fiore A., Lo Piero A. R., Fogliano V., et al. (2014). Pseudomonas corrugata crpCDE is part of the cylic lipopeptide corpeptin biosynthetic gene cluster and is involved in bacterial virulence in tomato and in hypersensitive response in Nicotiana benthamiana. Mol. Plant Pathol. 16, 495–506. 10.1111/mpp.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surico G., Lavermicocca P., Iacobellis N. (1988). Production of syringomycin and syringotoxin in culture of Pseudomonas syringae pv. syringae. Phytopathol. Mediterr. 27, 163–168. 3980301 [Google Scholar]
- Sutra L., Siverio F., Lopez M. M., Hunault G., Bollet C., Gardan L. (1997). Taxonomy of Pseudomonas strains isolated from tomato pith necrosis: emended description of Pseudomonas corrugata and proposal of three unnamed fluorescent Pseudomonas Genomospecies. Int. J. Syst. Bacteriol. 47, 1020–1033. 10.1099/00207713-47-4-1020 [DOI] [PubMed] [Google Scholar]
- Tampakaki A. P., Fadouloglou V. E., Gazi A. D., Panopoulos N. J., Kokkinidis M. (2004). Conserved features of type III secretion. Cell. Microbiol. 6, 805–816. 10.1111/j.1462-5822.2004.00432.x [DOI] [PubMed] [Google Scholar]
- Tampakaki A. P., Skandalis N., Gazi A. D., Bastaki M. N., Sarris P. F., Charova S. N., et al. (2010). Playing the Harp: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370. 10.1146/annurev-phyto-073009-114407 [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). Clustal-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trantas E. A., Sarris P. F., Pentari M. G., Mpalantinaki E. E., Ververidis F. N., Goumas D. E. (2015). Diversity among Pseudomonas corrugata and Pseudomonas mediterranea isolated from tomato and pepper showing symptoms of pith necrosis in Greece. Plant Pathol. 64, 307–318. 10.1111/ppa.12261 [DOI] [Google Scholar]
- Troisfontaines P., Cornelis G. R. (2005). Type III secretion: more systems than you think. Physiology 20, 326–339. 10.1152/physiol.00011.2005 [DOI] [PubMed] [Google Scholar]
- Ussery D. W., Wassenaar T. M., Borini S. (2009). Computing for Comparative Microbial Genomics: Bioinformatics for Microbiologists. London: Springer Science & Business Media. [Google Scholar]
- Van Heel A. J., De Jong A., Montalbán-López M., Kok J., Kuipers O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453. 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30, 274–291. 10.1111/j.1574-6976.2005.00012.x [DOI] [PubMed] [Google Scholar]
- Walker C., Goodyear C., Anderson D., Titball R. (2000). Identification of arsenic-resistant bacteria in the soil of a former munitions factory at Löcknitz, Germany. Land Contam. Reclamation 8, 13–18. [Google Scholar]
- Wei H. L., Zhang L. Q. (2006). Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89, 267–280. 10.1007/s10482-005-9028-8 [DOI] [PubMed] [Google Scholar]
- Whitchurch C. B., Leech A. J., Young M. D., Kennedy D., Sargent J. L., Bertrand J. J., et al. (2004). Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52, 873–893. 10.1111/j.1365-2958.2004.04026.x [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Kasai H., Arnold D. L., Jackson R. W., Vivian A., Harayama S. (2000). Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146, 2385–2394. 10.1099/00221287-146-10-2385 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins, in Evolving Genes and Proteins, eds Bryson V., Vogel H. J. (New York, NY: Academic Press; ), 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.