Abstract
The goal of our study was to evaluate at the systems-level, the effect of sex hormones on thymic epithelial cells (TECs). To this end, we sequenced the transcriptome of cortical and medullary TECs (cTECs and mTECs) from three groups of 6 month-old mice: males, females and males castrated at four weeks of age. In parallel, we analyzed variations in the size of TEC subsets in those three groups between 1 and 12 months of age. We report that sex hormones have pervasive effects on the transcriptome of TECs. These effects were exquisitely TEC-subset specific. Sexual dimorphism was particularly conspicuous in cTECs. Male cTECs displayed low proliferation rates that correlated with low expression of Foxn1 and its main targets. Furthermore, male cTECs expressed relatively low levels of genes instrumental in thymocyte expansion (e.g., Dll4) and positive selection (Psmb11 and Ctsl). Nevertheless, cTECs were more abundant in males than females. Accumulation of cTECs in males correlated with differential expression of genes regulating cell survival in cTECs and cell differentiation in mTECs. The sexual dimorphism of TECs highlighted here may be mechanistically linked to the well-recognized sex differences in susceptibility to infections and autoimmune diseases.
The immune system of vertebrates shows a major sexual dimorphism. Thus, relative to males, females usually display stronger immune response to vaccination and infection but suffer a higher propensity to many autoimmune diseases1,2,3,4. Though we have limited insight into the mechanistic bases for these differences4, evidence suggests that they result from both direct and indirect effects of sex steroids on innate and adaptive immune responses. Thus, sex hormones can affect immune cells directly or via modulation of the gut microbiome which in turn influences the immune response5,6,7,8,9. Sex-based differences in thymus biology have been observed in many studies. Thymic involution, which affects all vertebrates10,11, accelerates at puberty and progresses at different rates in males and females12,13,14. Also, administration and ablation of androgens respectively accelerates and reverses, albeit transiently, thymic involution15,16,17,18,19. Nevertheless, it remains unclear to what degree sex steroids’ impact on thymic cellularity may reflect the influence of hormones on thymic epithelial cells (TECs)3 vs. other cell types (thymocytes, mesenchymal cells).
Recent studies have shown that sex steroids do affect TECs. In adult mice, cortical and medullary TECs (cTECs and mTECs) have slower proliferation rates in males than females20. Furthermore, following acute ablation in a transgenic mouse model, cTECs were found to regenerate in females and castrated (Cx) males, but not in males or androgen-treated females21. Finally, it was recently shown that, in TECs, androgens repress transcription of the Notch ligand Dll422 which is essential for specification, commitment, and development of thymocytes23. Nevertheless, studies in other cell types (e.g., liver, adipose tissue, muscle, and brain) have shown that the effects of sex hormones are pleiotropic and exquisitely cell-type dependent24,25. Hence, the impact of sex hormones on TECs cannot be inferred from data obtained in other cell types. The main goal of our study was therefore to evaluate at the systems-level, the effect of sex hormones on TEC subsets. To this end, we sequenced the transcriptome of cTECs and mTECs from three groups of 6 month-old mice: males, females and males castrated at four weeks of age. In parallel, we analyzed variations in the size of TEC subsets in those three groups between 1 and 12 months of age. We report that sex hormones have pervasive effects on TEC biology. In particular, adult males accumulate more cTECs than females, but male cTECs display low proliferation rates and show evidence of functional impairment. Furthermore, relative to castrated males, mTECs from males and females showed decreased expression of promiscuously expressed genes.
Results
Dynamics of TEC populations as a function of age
We expected that, in addition to divergent effects, sex hormones would have some redundant effects on TECs. Therefore, to get a global picture of these effects, we analyzed not only male and female mice but also Cx male mice. The onset of male puberty in inbred laboratory mice usually occurs between 34 and 38 days after birth26. Hence, in order to prevent any exposure to high levels of androgens, castration of male mice was performed before full sexual maturity, at one month of age (see Supplementary Figure S1a for experimental design). Also, knowing that castration induces a transient thymic hypertrophy16, Cx males were not studied in the early weeks following surgery; they were analyzed before surgery (1 month-old) and at 3–12 months of age.
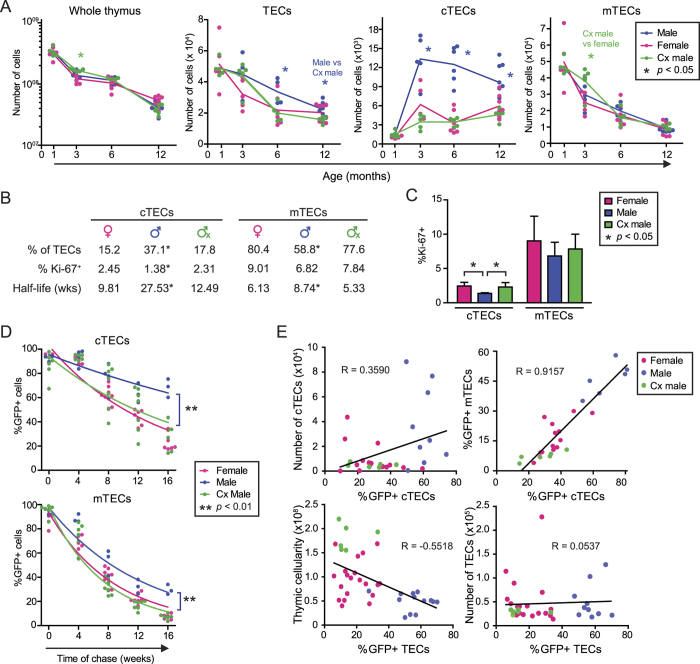
Total thymic cellularity, which essentially reflects thymocyte numbers, was similar in the three experimental groups over time (Fig. 1a, left). Only a minimal and transient hypercellularity was observed at three months in Cx males, probably reflective of the ephemeral thymic rebound induced by castration16. In adult mice, about 80% of TECs are mTECs and 20% cTECs 20,27. Notably, at 6 and 12 months, males possessed higher numbers of TECs than females and Cx males (Fig. 1a, middle left). This discrepancy was due solely to the greater abundance of cTECs in males, from 3 to 12 months of age (Fig. 1a middle right and 1b). In contrast, the loss of mTECs with age was similar in the three experimental groups (Fig. 1a, right). In order to understand the dramatic differences in cTEC numbers in males relative to other groups, we first analyzed TEC proliferation using two models. In the first model, the percentage of cycling cells was assessed using Ki-67 staining. As previously observed in adult mice20, mTECs proliferated more actively than cTECs in all groups (p < 0.05, Fig. 1c). Interestingly, males displayed a significantly lower percentage of Ki-67+ cTECs than females and Cx males (Fig. 1c). The mTEC compartment showed a similar tendency which, however, did not reach significance. In the second model, we use the H2B-GFP:rtTA transgenic mouse model in which the extent of cell proliferation can be inferred from H2B-GFP fluorescence intensity (see Supplementary Figure S1b)20,28. After administration of doxycycline during 6 weeks, TEC populations were analyzed during a chase period of 16 weeks (Fig. 1d). Nonlinear regression analysis of H2B-GFP dilution revealed that cTECs and mTECs in adult mice proliferated less actively in males than in females and Cx males (Fig. 1b,d). The half-life of GFP+ TECs was similar in females and Cx males (Fig. 1b). Therefore both methods yielded congruent results showing that cTECs proliferate less actively in males than in females and Cx males. Hence, the greater abundance of cTECs in males cannot be ascribed to a higher proliferation rate. Also, since the level of androgens is the sole feature that differentiates males from both Cx males and females (Supplementary Figure S1a), these results suggest that androgens have an anti-proliferative effect on TECs, and particularly on cTECs.
Figure 1. Male mice show an accumulation of cTECs with age that is not due to enhanced cell proliferation.
(a) Thymic cell populations in mice aged from 1 to 12 months. The gating strategy is depicted in Supplementary Figure S1c. (b) Percentage of TEC populations in 6 month-old mice, mean percentage of Ki-67+ cells at 4–5 months of age, and half-life (in weeks) of H2B-GFP+ TECs calculated using one phase decay nonlinear regression. (c) Percentages of Ki-67+ TECs in 4 to 5 month-old mice. (d) H2B-GFP dilution in adult mice during a chase period of 16 weeks. (e) Correlation (Pearson) between different features of TEC populations in 6 month-old mice. Each dot represents onemouse.
Next, we evaluated the correlation between TEC proliferation and cellularity after 16 weeks of chase. Surprisingly, there was a modest positive correlation (R = 0.3590) between the percentage of GFP+ cTECs and the number of cTECs (Fig. 1e, upper left). This means that thymi in which cTECs proliferate more actively (and therefore have a low percentage of GFP + cells) actually have less cTECs. As a corollary, there must be a greater loss of cTECs in females and Cx males than in males. Two non-mutually exclusive mechanisms may be responsible for cTEC loss: cTEC apoptosis vs. differentiation of cTEC-phenotype cells into mTEC-phenotype cells. The latter possibility must not be discarded since the cTEC compartment (EpCAM+Ly51+UEA1–) contains bipotent TEC progenitors that can ultimately adopt a cTEC or an mTEC fate29,30. Besides, proliferation of cTECs was highly correlated with mTEC proliferation (R = 0.9157) (Fig. 1e, upper right). Furthermore, global TEC proliferation showed a significant correlation (R = 0.5518) with overall thymic cellularity (i.e., thymocyte numbers; Fig. 1e, lower left) but not with TEC numbers (Fig. 1e, lower right). The facts that cTEC proliferation was positively correlated with mTEC proliferation and thymocyte numbers suggests that cell proliferation in these three cell compartments may be co-regulated.
The impact of sex on the transcriptome of TEC subsets–An overview
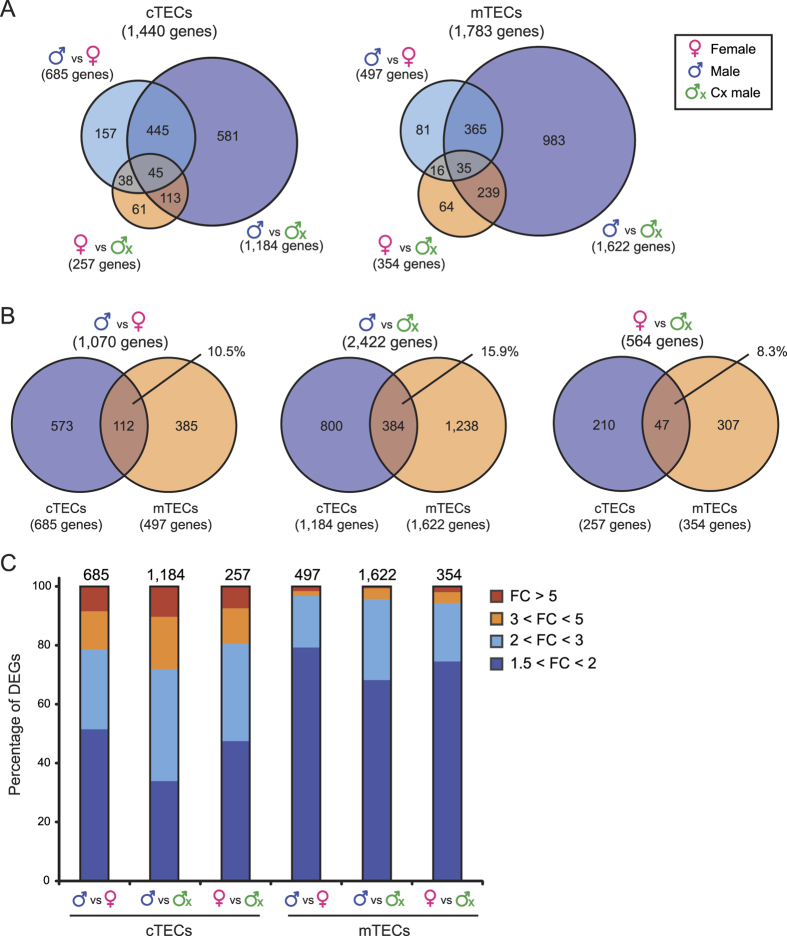
In order to assess the extent and molecular bases of sexual dimorphism in adult TECs, we performed RNA sequencing on cTECs and mTECs from 6 month-old mice from our three experimental groups (see gating strategy in Supplementary Figure S1c). DEGs were identified with the DESeq package31 using the following criteria: adjusted p value < 0.1, fold change ≥1.5 and minimal mean expression of 1 RPKM. A total of 1,440 and 1,783 DEGs were found in the three experimental groups in cTECs and mTECs, respectively (Fig. 2a). As observed in other tissues, sex-related differences in gene expression were generally modest, being <3-fold in most cases (Fig. 2c). Males and Cx males showed the greatest number of DEGs, with 1,184 (82%) and 1,622 (91%) DEGs in cTECs and mTECs, respectively. This result was striking since the sole physiological difference between these two groups is the level of androgens (Supplementary Figure S1a). Together with the relatively low number of DEGs in females vs. Cx males (Fig. 2a), these data indicate that androgens have a greater impact on TEC transcriptome than female sex hormones or the type of sex chromosomes. Previous studies on other types of somatic cells have shown that sexual dimorphism in gene expression is highly tissue-specific24,25. This was vividly illustrated by the fact that in various pairwise comparisons, we found that only 8 to 16% of DEGs were common to cTECs and mTECs (Fig. 2b).
Figure 2. Transcriptomic signatures of cTECs and mTECs in 6 month-old male, female and Cx male mice.
(a) Venn diagram representation of DEGs in cTECs and mTECs from the three experimental groups. (b) Overlap between DEGs in cTECs and mTECs. (c) Percentages of DEGs for selected thresholds of mRNA expression fold-change (FC). Numbers of DEGs are indicated above bars.
Androgens inhibit cell death and cell differentiation in TECs
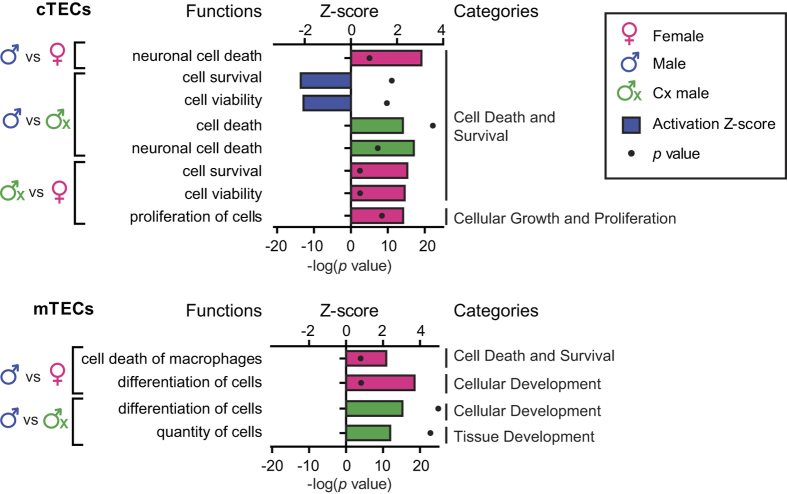
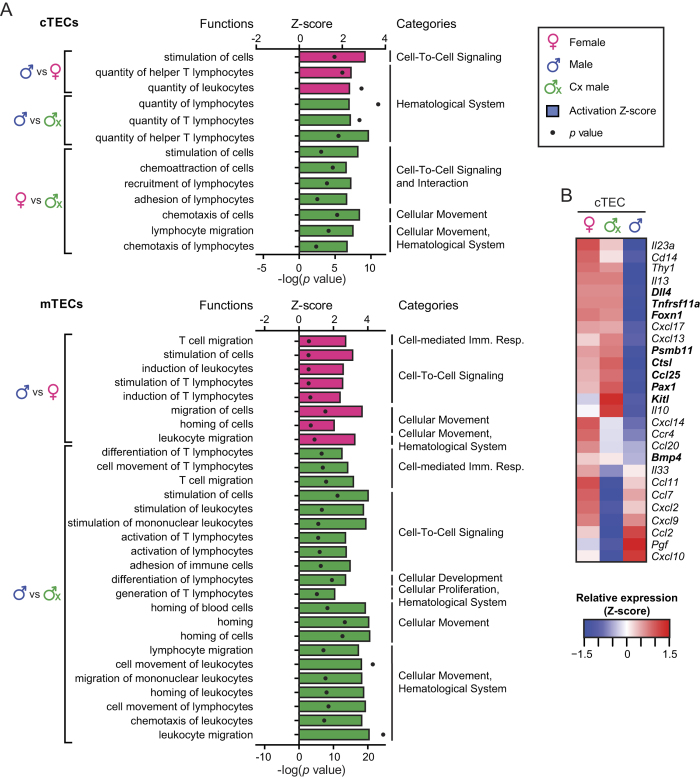
To better understand of the impact of sex hormones on TEC function, we performed a downstream effect analysis on our lists of DEGs using the Ingenuity Pathway Analysis software (IPA, Ingenuity® Systems, www.ingenuity.com). We used two metrics to identify the most important downstream effects of these DEGs: activation Z-score and p-value. Positive and negative Z-scores indicate increased and decreased functional activity, respectively. The p-value, calculated with the Fischer’s exact test, indicates the likelihood that the association between a set of DEGs and a biological function is significant. The complete list of biological processes regulated by DEGs can be found in Supplementary Figures S2 and S3. Of particular relevance in view of differences in TEC subsets depicted in Fig. 1, the following functional categories were highlighted as being differentially activated in the three experimental groups: cell death and survival and cell differentiation (Fig. 3). More specifically, IPA analysis predicted that, relative to the two other groups, male TECs would show i) less cell death (cTECs and mTECs) and ii) less cell differentiation (mTECs). Superior cTEC survival coupled to lessened differentiation of cTEC-phenotype progenitors into mTECs offer a plausible explanation for the higher numbers of cTECs observed in males (Fig. 1a).
Figure 3. IPA analysis of DEGs in cTECs and mTECs.
IPA analysis of DEGs predicts decreased cell death and cell differentiation in males. Activation Z-scores are depicted with bars, whereas p values are shown with black dots. The color of bars shows in which group a given process is activated (e.g., blue = males). All functions represented on the graph were significantly enriched (p < 0.05).
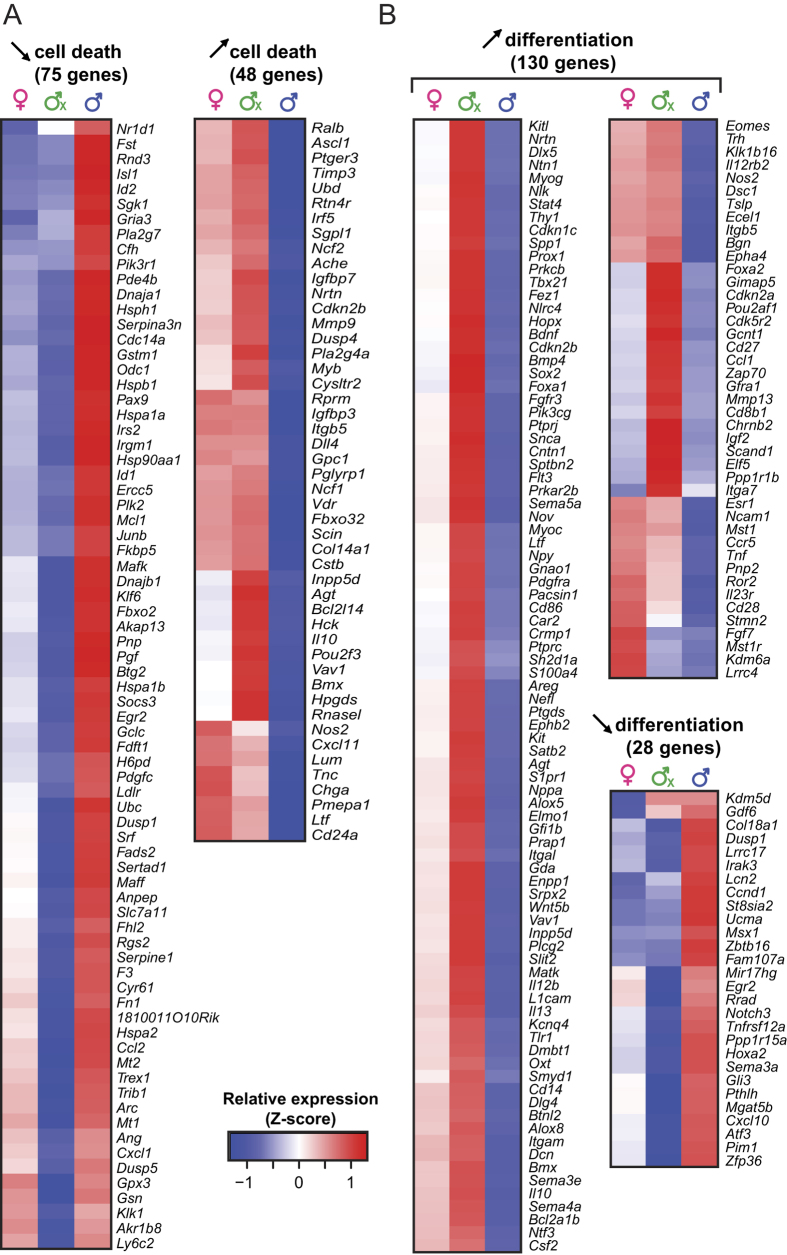
We then sought to gain further mechanistic insights into processes regulated by sex hormones by analyzing relevant DEGs, i.e. genes whose differential expression was consistent with the predicted activation status in the three experimental groups (Fig. 4). Genes were separated into activators or inhibitors of cell death or cell differentiation. The dataset related to cell death included DEGs associated with the following functional categories: cell death and survival, cellular growth and proliferation, and tissue development. Notably, Pax9 and Sgpl1, which are known to regulate TEC survival and apoptosis32,33, were among DEGs linked to inhibition of cell death in male TECs (Fig. 4a). Furthermore, many genes known to regulate survival of different epithelial cell types are present in that list, including Sgk1, Id1, Vdr, Bcl2l14, Dusp1, Bmx and Igfbp3 (mammary gland), Pgf, Irf5, Pla2g4a and Pmepa1 (intestine), and Hspa1a (ovary)34,35,36. Finally, several members of key pathways regulating cell survival/death were among sexually dimorphic genes: heat shock proteins (Hspa1a, Hspa1b, Hspa2, Hspb1) and members of the p53 (Rnd3, Plk2, Sgpl1, Igfbp3, Pmepa1), Pi3k/Akt (Pik3r1, Pdgfc, Fn1) and Fas/caspases pathways (Mcl1, Hspa1b, Fn1, Bcl2l14, Lum).
Figure 4. Relative expression of genes contributing to the inhibition of cell death and cell differentiation in male TECs.
(a) Relative expression of genes that affect cell death in cTECs. (b) Relative expression of genes that affect cell differentiation in mTECs. Relative gene expression is depicted as a Z-score, calculated separately for cTECs and mTECs. Red corresponds to higher expression, whereas blue corresponds to lower expression.
On the other hand, many genes which are instrumental in differentiation of ectoderm-and endoderm-derived epithelia were differentially expressed and may therefore contribute to the inhibition of cell differentiation in male mTECs: Elf5, Oxt, Dmbt1, Areg, Spp1 (mammary gland), Kitl, Alox8, Dsc1, Atf3 (skin), Notch3, Mir17hg and Foxa2 (lung epithelial cells), Ccnd1, Cdkn2a and Dmbt1 (colon)37,38,39,40. In conclusion, IPA analysis provides a molecular framework that can explain why cell death and cell differentiation are inhibited in male TECs relative to female and Cx male TECs.
Androgens impact on genes involved in thymopoiesis
IPA analysis of DEGs in the three experimental groups predicted that several functions linked to T-cell production were inhibited in male TECs. These functions included stimulation, homing and chemoattraction of lymphoid cells (Fig. 5a). DEGs involved in these functional categories included several cytokines, transcription factors and costimulatory molecules of prime relevance in thymopoiesis (Fig. 5b). Foxn1, the key regulator of thymus development and maintenance41,42, was downregulated by ~2 fold in cTECs from males relative to females and Cx males. In accordance with this, expression of Foxn1 targets Ccl25, Dll4, Pax1 and Kitl43,44 was also lower in male cTECs. Other genes having a crucial role in thymocyte development45,46,47 were also downregulated in male cTECs: i) Psmb11 (AKA β5t), a cTEC-specific proteasome subunit required for positive selection of CD8 thymocytes48, ii) Ctsl, a peptidase instrumental in the positive selection of CD4 thymocytes49 and iii) Tnfrsf11a (AKA RANK), the TNF family receptor necessary for mTEC maturation50. Overall, these data suggest that androgens have a negative impact on TEC-mediated processes that are central to normal thymopoiesis.
Figure 5. Several TEC genes involved in thymopoiesis are downregulated in male TECs.
(a) IPA analysis of DEGs predicts differential activation of biological functions related to homing, stimulation of cells, chemotaxis and production of T lymphocytes. Activation Z-score is depicted with bars, whereas p values are shown with black dots. The color of bars shows in which group a given process is activated (e.g., green = Cx males). All functions represented on the graph are significantly enriched (p < 0.05). (b) Heatmaps of relative cTEC expression of IPA-discovered genes that may affect thymopoiesis. Genes that have well-characterized functions in the thymus are highlighted in bold. The gene expression is depicted as a Z-score, calculated separately for each cell type. Red corresponds to higher expression, whereas blue corresponds to lower expression.
Cx males show increased promiscuous gene expression in mTECs
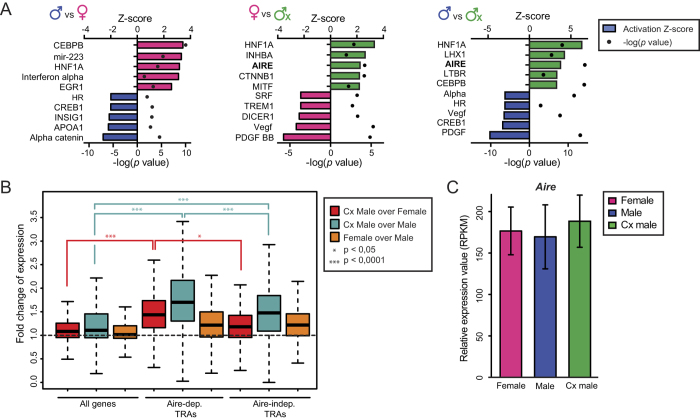
Induction of so-called “central tolerance” in the thymic medulla depends on ectopic expression of proteins otherwise restricted to differentiated organs in the periphery51. Collectively, mTECs express almost all protein-coding genes52,53 and can therefore induce tolerance to a wide array of tissue-restricted antigens (TRAs). Promiscuous gene expression by mTECs is therefore essential to induce tolerance to “extrathymic proteins”, and is regulated in part by the autoimmune regulator (AIRE). According to IPA analysis, several upstream regulators were differentially activated in mTECs from our three experimental groups (Fig. 6a). One of the top regulators was Aire, which was activated in mTECs from Cx males compared to males and females (Fig. 6a). This led us to evaluate expression of TRAs in mTECs from our three groups. We adopted the definition stipulating that TRA-coding genes are those that are tissue-enriched (i.e. expressed at relatively high levels) in at most five tissues54. Our definition of AIRE-dependent and –independent TRAs was based on RNA-seq analysis of mTECs from wild-type and Aire−/− littermates55. We found that Aire-dependent TRAs were significantly upregulated in Cx males compared to females (red) and males (turquoise), whereas males and females showed no difference (orange, Fig. 6b). Furthermore, Cx males expressed higher levels of Aire-independent TRAs than males, although the difference was less than for Aire-dependent TRAs (Fig. 6b). However, Aire expression was similar in the three experimental groups (Fig. 6c). These data indicate that the differential abundance of TRAs was not due to differential expression of Aire. In addition, they argue against the possibility that Cx males might have a higher proportion of mature mTECs than other groups, since mTEC maturity correlates with Aire expression. We therefore conclude that sex hormones directly or indirectly repress the promiscuous gene expression of Aire-dependent and –independent TRAs, without affecting the expression of Aire itself.
Figure 6. Cx males show higher expression of TRAs.
(a) Ten most activated upstream regulators predicted by IPA analysis of mTECs DEGs in female vs male (left), in Cx male vs female (center) and in Cx male vs male (right, all predicted activators shown are significant p < 0.05). The color of bars shows in which group a given upstream regulator is activated (e.g., blue = males). (b) Fold-difference in expression of all genes, Aire-dependent TRAs and Aire-independent TRAs in mTECs. The gene expression ratio (log2 RPKM) of Cx male over female is depicted in red, of Cx male over male in turquoise and of female over male in orange. (c) Relative expression of Aire in mTECs (RPKM).
Discussion
The present work shows that sex hormones have pervasive effects on the transcriptome of TECs: the number of DEGs was 1,440 in cTECs and 1,783 in mTECs (Fig. 2a). Studies on adipocytes as well as brain, liver and muscle cells revealed that sex-related changes in gene expression were highly tissue-specific24,25. It is remarkable that the same holds true in cells that are as closely related, ontogenetically, as cTECs and mTECs. Indeed, the overlap between cTEC DEGs and mTEC DEGs in various pairwise comparisons was only about 12% (Fig. 2b). This means that the sex differences are exquisitely context dependent.
Sexual dimorphism was particularly conspicuous in cTECs where it was mainly regulated by the levels of testicular androgens. Androgens induced an accumulation of cTECs that were hypoproliferative and presented features suggestive of functional impairment. The low proliferation rate of male TECs can be explained by a low expression of Foxn1 and its main targets, which are required for both fetal and adult thymopoiesis41. In adult mice, minor variations in the expression or function of Foxn1 directly impact on thymic cellularity42,56. Interestingly, thymic cellularity (i.e., number of thymocytes) correlated with TEC proliferation (higher in females) (Fig. 1e) but not with cTEC or total TEC numbers (higher in males) (Fig. 1a,e). This observation is consistent with the fact that thymic output in human adults (number of signal-joint T cell receptor excision circles/T cell) is higher in females than males13. In addition, numerous genes that are instrumental in thymocyte development were repressed in male cTECs (Fig. 5). These DEGs included genes driving thymocyte expansion (e.g., Dll4) and positive selection (Psmb11 and Ctsl) and whose repression can certainly impinge on thymopoietic activity22.
But if cTECs proliferate more in females than males, why are they less abundant in females? Our IPA analyses suggest that cTEC accumulation in males results from enhanced survival coupled to impairment of cell differentiation (Fig. 4). Given the large numbers of relevant DEGs, direct evaluation of the role of individual genes would require numerous gain-and loss-of-function experiments. Finally, is there any rationale for the superior proliferative potential of female over male cTECs? Our favorite hypothesis is that evolution has favored high cTEC renewal potential in females because of the constraints imposed by pregnancy. Pregnancy causes a major, though transient, TEC-dependent thymic involution which is instrumental in enhancing reproductive fitness17,57. This process is progesterone-dependent, and transplantation of a progesterone receptor null thymus in wild-type female mice impairs fertility. Relative to recipients of a wild-type thymus, mice grafted with a progesterone receptor null thymus have smaller litters and higher numbers of resorbing embryos57. We therefore speculate that a high cTEC renewal potential allows females to withstand pregnancy-associated thymic involution and regain adequate thymic function rapidly in the postpartum period.
Unexpectedly, we found that both female and male sex hormones repressed promiscuous gene expression in mTECs. Both AIRE-dependent and –independent TRAs were expressed at higher levels in Cx males than in females and males. Hence, while sex hormones did not affect the expression of Aire per se, they must impinge on the activity of AIRE cofactors or unidentified regulator(s) of promiscuous gene expression in mTECs. Is this effect biologically significant? We speculate that, at least for the Aire-dependent TRAs, lower TRA expression in adult is probably not biologically important under physiological conditions. Indeed, Aire-controlled mechanisms of central tolerance are largely dispensable in the adult58. However, we hypothesize that the effect of sex hormones on TRA expression may be highly relevant in the context of allogeneic hematopoietic cell transplantation (AHCT) in adults. Indeed, following AHCT, donor hematopoietic progenitors must undergo a second round of ontogeny in the recipient’s thymus, in the presence of sex hormones if the recipient is an adult. During this period, the occurrence of graft-versus-host disease in many recipients leads to loss of AIRE+ mTECs and thereby to generation of autoreactive T cells59. By extension, our data would suggest that sex hormones might have a negative impact on the establishment of graft-host tolerance and might explain why post-AHCT autoimmunity [i.e., chronic graft-versus-host disease59] is much more common in adults than children60. If this were the case, pharmacological antagonists of sex hormones could mitigate the risk of chronic graft-versus-host disease, which remains the unrelenting nemesis of patients and physicians involved in AHCT60,61.
The sexual dimorphism of TECs highlighted here may be mechanistically linked to the well-recognized sex differences in susceptibility to infections and tissue-specific and systemic autoimmune diseases4. While these complex issues may be tackled from different perspectives, our next objective will be to investigate whether the sexual dimorphism of cTECs impacts on the diversity of the T-cell repertoire.
Methods
Mice
C57BL/6 mice, B6.Cg-Gt(ROSA)26Sortm1(rtTA*M2)Jae/J and STOCK Tg(tetO-HIST1H2BJ/GFP)47Efu/J mice purchased from The Jackson Laboratory (Bar Harbor) were bred and housed under specific-pathogen-free conditions in sterile ventilated racks at the Institute for Research in Immunology and Cancer. For H2B-GFP pulse-chase experiments, doxycyline was incorporated in food (2 g/kg) (Hartlan Laboratories) or in drinking water (2 mg/ml of doxycycline supplemented with 5% sucrose) (Sigma Aldrich)20. The pulse period lasted for six weeks and was initiated at 4–6 weeks of age. All procedures were in accordance with the Canadian Council on Animal Care guidelines and approved by the Comité de Déontologie et Expérimentation Animale de l’Université de Montréal.
Castration
Four week-old male mice were anesthetized and a small incision in the scrotal region was made to expose the testes. Testes were tied off with suture thread and removed.
Flow cytometry analysis and sorting
Enrichment of thymic stromal cells was performed as previously described52,62. Thymic stromal cells were stained with biotinylated Ulex Europaeus Lectin 1 (UEA1; Vector Laboratories) and PE-Cy7 or PE-TexasRed conjugated streptavidin (BD Biosciences) and the following antibodies: i) AlexaFluor-700 anti-CD45 from BD Biosciences, ii) AlexaFluor-647 anti-Ly51 and APC-Cy7 anti-EpCAM from BioLegend. Staining with 7-AAD and PE conjugated anti-Ki-67 (BD Biosciences) was used to assess cell viability and the proportion of cycling cells. TECs were selected as CD45−EpCAM+, and mTECs and cTECs were defined as UEA-1+Ly51− and UEA-1−Ly51+, respectively. Intranuclear staining of Ki-67 was performed using the Foxp3/Transcription Factor Staining Buffer Set (Affymetrix eBiosciences). For flow cytometry analyses, each group contained 4–8 mice per time point. Live cTECs and mTECs were sorted on a three laser FACSAria or analyzed on a three laser LSR II using FACSDiva (BD Biosciences) and FlowJo VX.0.7(FlowJo Enterprise) softwares.
RNA sequencing experiments
We analyzed the transcriptome of six populations of TECs: cTECs and mTECs from males, females and Cx males. We obtained three biological replicates from each TEC population, except for Cx male mTECs where we had 2 biological replicates. Hence, in toto, we analyzed 17 biological replicates (Supplementary Table S1). Each biological replicate contained pooled FACs-sorted cTECs or mTECS from four to six mice. Total RNA was isolated using TrizolTM as recommended by the manufacturer (Invitrogen), and then further purified using RNeasy Micro columns (Qiagen). Sample quality was assessed using Bioanalyzer RNA Pico chips (Agilent). Transcriptome libraries were made using the TruSeq RNA Sample Prep Kit (v2) (Illumina) following the manufacturer’s protocols. Library generation was then assessed using a Bioanalyzer platform (Agilent) and Illumina MiSeq-QC run. Then, sequencing was done using an Illumina HiSeq2000 using TruSeq SBS v3 chemistry at the Institute for Research in Immunology and Cancer’s Genomics Platform. Cluster density was targeted at around 800 k clusters/mm2. Data was mapped to the Mus musculus (mm10) reference genome using the ELANDv2 alignment tool from the CASAVA 1.8.2 software (Illumina). RNA-seq data have been deposited in GEO archives (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE66873 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66873) and are displayed in Supplementary Table S1. Analyses of RNA sequencing data were performed using the publicly available statistical software package “R” (http://www.r-project.org/). Differentially expressed genes (DEGs) were determined with the DESeq package from Bioconductor (http://bioconductor.org/), using thresholds of adjusted p-value of 0.1 and a minimal fold change of 1.5. To remove genes that were lowly expressed in our analysis, we further filtered the DEGs to only keep genes that had at least one sample with a relative expression higher than 1 read per kilobase of exon model per million reads mapped (RPKM). Enrichment of biological functions and predicted upstream regulators were assessed using the IPA software (Ingenuity Systems, http://www.ingenuity.com).
Statistical analyses
Unless stated otherwise, results are expressed as means ± SD, statistical significance was tested using Student unpaired two-tailed t test, and differences with a p value < 0.05 were considered significant. Significant differences in the percentages of Ki-67+ cells were assessed using a one-way ANOVA and a Newman-Keuls post-hoc test. The half-life of GFP+ cells were calculated using a one-phase decay nonlinear regression analysis and significance was assessed using an extra sum-of-squares F test. Correlations were calculated using Pearson’s correlation test. All statistical analyses were performed with GraphPad Prism software V5.01.
Additional Information
How to cite this article: Dumont-Lagacé, M. et al. Sex hormones have pervasive effects on thymic epithelial cells. Sci. Rep. 5, 12895; doi: 10.1038/srep12895 (2015).
Supplementary Material
Acknowledgments
We are grateful to Danièle Gagné and Gaël Dulude (flow cytometry and cell sorting), Marianne Arteau (RNA sequencing experiments), to Isabelle Caron for the surgical procedures and to the staff of IRIC animal care facility for their assistance. Special thanks to Sébastien Lemieux for advice on statistical analyses. This work was supported by grant MOP 42384 from the Canadian Institute of Health Research (CIHR). MDL is supported by a CIHR studentship and CP holds a Canada Research Chair in Immunobiology. The Institute for Research in Immunology and Cancer is supported in part by the Canada Foundation for Innovation and the Fonds de la Recherche en Santé du Québec.
Footnotes
Author Contributions M.D.L. and C.S.P. performed experiments and analyzed data. M.D.L. and C.P. designed the study, and wrote the first draft of the manuscript. All authors discussed results and reviewed the manuscript.
References
- Whitacre C. C. Sex differences in autoimmune disease. Nat Immunol 2, 777–780 (2001). [DOI] [PubMed] [Google Scholar]
- Fish E. N. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol 8, 737–744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J. G. & Fish E. N. SeXX matters in immunity. Trends Immunol. 35, 97–104 (2014). [DOI] [PubMed] [Google Scholar]
- Danska J. S. Sex matters for mechanism. Sci Transl Med 6, 258fs40 (2014). [DOI] [PubMed] [Google Scholar]
- Markle J. G. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- Markle J. G., Frank D. N., Adeli K., von Bergen M. & Danska J. S. Microbiome manipulation modifies sex-specific risk for autoimmunity. Gut Microbes. 5, 485–493 (2014). [DOI] [PubMed] [Google Scholar]
- Furman D. et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA 111, 869–874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. Female resistance to pneumonia identifies lung macrophage nitric oxide Synthase-3 as a therapeutic target. Elife. 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino-Rodriguez E., Berent-Maoz B. & Dorshkind K. Causes, consequences, and reversal of immune system aging. J Clin. Invest 123, 958–965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventevogel M. S. & Sempowski G. D. Thymic rejuvenation and aging. Curr. Opin. Immunol. 25, 516–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen N. J., Olson G., Viselli S. M., Gu X. & Kovacs W. J. Androgen receptors in thymic epithelium modulate thymus size and thymocyte development. Endocrinology 142, 1278–1283 (2001). [DOI] [PubMed] [Google Scholar]
- Mitchell W. A., Lang P. O. & Aspinall R. Tracing thymic output in older individuals. Clin. Exp Immunol 161, 497–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen N. J. & Kovacs W. J. Evidence that androgens modulate human thymic T cell output. J. Investig. Med. 59, 32–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland J. S. et al. Activation of thymic regeneration in mice and humans following androgen blockade. J. Immunol 175, 2741–2055 (2005). [DOI] [PubMed] [Google Scholar]
- Griffith A. V., Fallahi M., Venables T. & Petrie H. T. Persistent degenerative changes in thymic organ function revealed by an inducible model of organ regrowth. Aging Cell 11, 169–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley J. & Liston A. Molecular control over thymic involution: From cytokines and microRNA to aging and adipose tissue. Eur. J Immunol. 42, 1073–1079 (2012). [DOI] [PubMed] [Google Scholar]
- Ardiani A. et al. Combination therapy with a second-generation androgen receptor antagonist and a metastasis vaccine improves survival in a spontaneous prostate cancer model. Clin. Cancer Res. 19, 6205–6218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K. P. et al. Targeting thymic epithelia AR enhances T-cell reconstitution and bone marrow transplant grafting efficacy. Mol. Endocrinol. 27, 25–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont-Lagacé M., Brochu S., St-Pierre C. & Perreault C. Adult thymic epithelium contains nonsenescent label-retaining cells. J. Immunol. 192, 2219–2226 (2014). [DOI] [PubMed] [Google Scholar]
- Rode I. & Boehm T. Regenerative capacity of adult cortical thymic epithelial cells. Proc. Natl. Acad. Sci. USA 109, 3463–3468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velardi E. et al. Sex steroid blockade enhances thymopoiesis by modulating Notch signaling. J Exp. Med 211, 2341–2349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. K. & Zuniga-Pflucker J. C. An overview of the intrathymic intricacies of T cell development. J Immunol 192, 4017–4023 (2014). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16, 995–1004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais-Jarvis F. Estrogen and androgen receptors: regulators of fuel homeostasis and emerging targets for diabetes and obesity. Trends Endocrinol. Metab 22, 24–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig J. T., Blake J. A., Bult C. J., Kadin J. A. & Richardson J. E. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 43, D726–D736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. H. et al. Developmental kinetics, turnover and stimulatory capacity of thymic epithelial cells. Blood 108, 3777–3785 (2006). [DOI] [PubMed] [Google Scholar]
- Tumbar T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves N. L. et al. Serial progression of cortical and medullary thymic epithelial microenvironments. Eur. J. Immunol 44, 16–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. et al. Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep. 8, 1198–1209 (2014). [DOI] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer-Egger C. et al. Thymopoiesis requires Pax9 function in thymic epithelial cells. Eur. J. Immunol 32, 1175–1181 (2002). [DOI] [PubMed] [Google Scholar]
- Borowsky A. D. et al. Sphingosine-1-phosphate lyase expression in embryonic and adult murine tissues. J. Lipid Res. 53, 1920–1931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. C., Tsao P. N., Weng M. T., Cao Z. & Wong J. M. Flt-1 in colorectal cancer cells is required for the tumor invasive effect of placental growth factor through a p38-MMP9 pathway. J. Biomed. Sci. 20, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. L., Park J. H., Nishidate T., Nakamura Y. & Katagiri T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 9, R17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink M. et al. The interaction of HspA1A with TLR2 and TLR4 in the response of neutrophils induced by ovarian cancer cells in vitro. Cell Stress. Chaperones. 17, 661–674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh J. I. et al. 2-methoxyestradiol induces mammary gland differentiation through amphiregulin-epithelial growth factor receptor-mediated signaling: molecular distinctions from the mammary gland of pregnant mice. Endocrinology 148, 1266–1277 (2007). [DOI] [PubMed] [Google Scholar]
- Collard J. F., Mertens B. & Hinsenkamp M. In vitro study of the effects of ELF electric fields on gene expression in human epidermal cells. Bioelectromagnetics 32, 28–36 (2011). [DOI] [PubMed] [Google Scholar]
- Bonauer A. & Dimmeler S. The microRNA-17-92 cluster: still a miRacle? Cell Cycle 8, 3866–3873 (2009). [DOI] [PubMed] [Google Scholar]
- Moussalli M. J. et al. Mechanistic contribution of ubiquitous 15-lipoxygenase-1 expression loss in cancer cells to terminal cell differentiation evasion. Cancer Prev. Res. (Phila) 4, 1961–1972 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeaux T. et al. Thymopoiesis in mice depends on a Foxn1-positive thymic epithelial cell lineage. Proc. Natl. Acad. Sci. USA 107, 16613–16618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenkamp N., Nowell C. S. & Blackburn C. C. Regeneration of the aged thymus by a single transcription factor. Development 141, 1627–1637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell C. S. et al. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS. Genet. 7, e1002348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon L. & Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell 149, 159–172 (2012). [DOI] [PubMed] [Google Scholar]
- Desanti G. E. et al. Developmentally regulated availability of RANKL and CD40 ligand reveals distinct mechanisms of fetal and adult cross-talk in the thymus medulla. J. Immunol 189, 5519–5526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. & Takahama Y. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol 33, 256–263 (2012). [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P. M. & Hogquist K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev. Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T. et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32, 29–40 (2010). [DOI] [PubMed] [Google Scholar]
- Honey K., Nakagawa T., Peters C. & Rudensky A. Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J. Exp. Med. 195, 1349–1358 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S. W. et al. RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp. Med 204, 1267–1272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., Wirnsberger G. & Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9, 833–844 (2009). [DOI] [PubMed] [Google Scholar]
- St-Pierre C. et al. Transcriptome sequencing of neonatal thymic epithelial cells. Sci. Rep. 3, 1860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom S. N. et al. Population and single cell genomics reveal the Aire-dependency, relief from Polycomb silencing and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S. et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc. Natl. Acad. Sci. USA 110, E3497–E3505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre C., Trofimov A., Brochu S., Lemieux S. & Perreault C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol 195, 10.4049/jimmunol.1500558 (2015). [DOI] [PubMed] [Google Scholar]
- Chen L., Xiao S. & Manley N. R. Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood 113, 567–574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts T. A., DeMayo F., Rich S., Conneely O. M. & O’Malley B. W. Progesterone receptors in the thymus are required for thymic involution during pregnancy and for normal fertility. Proc. Natl. Acad. Sci USA 96, 12021–12026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerau-de-Arellano M., Martinic M., Benoist C. & Mathis D. Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J. Exp. Med. 206, 1245–1252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertschnig S., Hauri-Hohl M. M., Vollmer M., Hollander G. A. & Krenger W. Impaired thymic expression of tissue-restricted antigens licenses the de novo generation of autoreactive CD4+ T cells during murine acute GVHD. Blood 125, 2720–2723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara J. L., Levine J. E., Reddy P. & Holler E. Graft-versus-host disease. Lancet 373, 1550–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent K., Roy D. C., & Perreault C. Next-generation leukemia immunotherapy. Blood 118, 2951–2959 (2011). [DOI] [PubMed] [Google Scholar]
- Jain R. & Gray D. H. Isolation of thymic epithelial cells and analysis by flow cytometry. Curr. Protoc. Immunol 107, 3.26.1–3.26.15 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.