Abstract
G protein-coupled receptors (GPCRs) constitute the largest family among mammalian membrane proteins and are capable of initiating numerous essential signaling cascades. Various GPCR-mediated pathways are organized into protein microdomains that can be orchestrated and regulated through scaffolding proteins, such as PSD-95/discs-large/ZO1 (PDZ) domain proteins. However, detailed binding characteristics of PDZ–GPCR interactions remain elusive because these interactions seem to be more complex than previously thought. To address this issue, we analyzed binding modalities using our established model system. This system includes the 13 individual PDZ domains of the multiple PDZ domain protein 1 (MUPP1; the largest PDZ protein), a broad range of murine olfactory receptors (a multifaceted gene cluster within the family of GPCRs), and associated olfactory signaling proteins. These proteins were analyzed in a large-scale peptide microarray approach and continuative interaction studies. As a result, we demonstrate that canonical binding motifs were not overrepresented among the interaction partners of MUPP1. Furthermore, C-terminal phosphorylation and distinct amino acid replacements abolished PDZ binding promiscuity. In addition to the described in vitro experiments, we identified new interaction partners within the murine olfactory epithelium using pull-down-based interactomics and could verify the partners through co-immunoprecipitation. In summary, the present study provides important insight into the complexity of the binding characteristics of PDZ–GPCR interactions based on olfactory signaling proteins, which could identify novel clinical targets for GPCR-associated diseases in the future.
PDZ domain proteins comprise one of the largest families among interaction domain scaffolding proteins and are highly abundant in various multicellular eukaryotic species. These proteins fulfill important physiological functions in a broad range of different tissues and cells as they orchestrate complex protein networks. Among putative PDZ interaction partners, one important protein family is the group of GPCRs1, constituting the largest family of membrane proteins in mammals (1). Here, signal efficiency, speed, desensitization, and internalization can be modulated by PDZ proteins (2–5). Olfactory receptors (ORs) represent a multigene family within this group of seven-transmembrane domain proteins and encompass 2% of the mammalian genome (6). Belonging to class I GPCRs, ORs share many general features of this receptor family, making them an interesting target for interactions involving PDZ proteins. Until recently, an organizing complex builder, such as the inactivation no afterpotential D (InaD) protein in the visual system of Drosophila melanogaster (7, 8), could not be clearly identified for olfactory signaling.
The multiple PDZ domain protein 1, with 13 individual PDZ domains, represents the largest of the described PDZ proteins to date (9) and interacts with different GPCRs (10–12). One well-described example is its interaction with GABAB receptors, leading to enhanced receptor stability at the plasma membrane and prolonged signaling duration (2). In previous studies, we demonstrated that PDZ domains 1 + 2 can interact with a selected subset of ORs (13). Furthermore, we showed that MUPP1 binds to a specific OR and that most of the described proteins are involved in mammalian olfactory signal transduction in the native system, making MUPP1 a promising candidate for orchestrating the olfactory system (14).
Many PDZ–ligand interactions depend on classical binding motifs at the ligand's C-terminal end, thereby building weak transient protein complexes (15, 16). However, an increasing number of PDZ interactions have emerged that seem to provide more complex binding modalities, differing from the canonical interactions (17, 18). Ligand binding seems not to be exclusively restricted to C-terminal sites, and PDZ domains cannot be distinctly classified but are evenly distributed throughout a selective space (17, 19–21). Therefore, it is of great interest to analyze OR–PDZ interactions to characterize the putative binding requirements and to further investigate the role of MUPP1 in olfactory signaling.
In the present study, we characterized the binding modalities between the 13 individual PDZ domains of MUPP1 and a broad range of murine olfactory receptors in a large-scale approach, indicating that classical binding motifs were not overrepresented among the evaluated binding partners. In addition, we identified new binding partners from the murine olfactory epithelium using pull-down-based interactomics.
EXPERIMENTAL PROCEDURES
DNA Constructs
GST-fusion constructs of the PDZ domains 1–13 of MUPP1 were generated by cloning the single domains into the pGEX-3X expression vector (Amersham Biosciences, Buckinghamshire, UK), and pCDNA3_MUPP1 served as the template. All of the constructs were verified by sequencing. For a detailed primer list, see the previous publication (14).
Antibodies
The primary antibodies were anti-MUPP1 rabbit polyclonal (provided by H. Luebbert, Ruhr-University Bochum), anti-MUPP1 mouse monoclonal (#1510; Santa Cruz, Dallas, TX), anti-HA mouse monoclonal (#H9658; Sigma, St. Louis, MO), anti-adenylyl cyclase III goat polyclonal (#32113; Santa Cruz), anti-adenylyl cyclase III rabbit polyclonal (#588; Santa Cruz), anti-calmodulin mouse monoclonal (#137079; Santa Cruz), anti-Gαolf goat polyclonal (#26764; Santa Cruz), and anti-Gαolf rabbit polyclonal (#1209; Santa Cruz). The secondary antibodies were horseradish peroxidase (HRP) coupled goat anti-mouse, donkey anti-goat and goat anti-rabbit IgGs (1:10,000) (Bio-Rad, Muenchen, Germany), and goat anti-rabbit/mouse Alexa 546 nm/660 nm (1:1,000) (Molecular Probes, Eugene, OR).
Immunohistochemistry
Heterozygous OMP–GFP transgenic mice (22) were raised and maintained according to institutional and governmental instructions. Horizontal cryosections were performed on P18 mice with a layer thickness of 14 μm. The sections were blocked for 1 h (PBS–, 1% gelatin) at room temperature and incubated with the corresponding antibody solution overnight at 4 °C. After several washing steps, the slices were coated with the appropriate fluorophore-coupled secondary antibody for 1 h at room temperature. After embedding the slices with Prolong Antifade Gold (Life Technologies, Carlsbad, CA), fluorescence images were obtained using a confocal microscope (Zeiss LSM 510 Meta) with a 40× oil immersion objective. Control experiments without primary antibodies showed a very low level of background staining (data not shown).
Mass Spectrometry
To reduce unspecific binding events, 50 μl of glutathione-covered beads coupled with the GST-tagged PDZ domain were blocked in 100 μl of TBS+/+ (100 mm Tris-HCl, 150 mm NaCl) containing 1% BSA for 30 min under ice-cold conditions. The entire suspension was centrifuged for 3 min at 1,000 g, the supernatant was subsequently decanted, and the beads were resuspended in ice-cold interaction buffer (50 mm ammonium bicarbonate, 480 μm MgCl2, 100 mm KCl, 500 μm EDTA, 1 mm DTT, 20 mm HEPES, 1% BSA). The interaction was initiated by the incubation of GST-tagged PDZ domain with 4 μl of total protein lysate of murine olfactory epithelium overnight at 4 °C. As a background control, we performed the analysis three times using purified GST in place of the fusion proteins. After this incubation procedure, the protein-coupled beads were washed four times in 100 μl of ice-cold wash buffer (50 mm ammonium bicarbonate, 480 μm MgCl2, 100 mm KCl, 500 μm EDTA, 1 mm DTT, 20 mm HEPES) followed by centrifugation for 3 min at 1,000 g at 4 °C. The beads were resuspended in 100 of μl 50 mm ammonium bicarbonate buffer (Fluka, Buchs, Switzerland) and 20% methanol (Avantor, Center Valley, PA). Thereafter, 2 μg of sequencing-grade trypsin (Promega, Fitchburg, WI) was added for an overnight proteolytic digestion at 37 °C. Subsequently, the extracted peptides were diluted in 0.1% trifluoroacetic acid and 2% acetonitrile and were loaded directly onto a fused silica capillary (inner diameter: 100 μm, Polymicro, Lisle, IL) that was packed with a reversed phase (RP, C18 Luna, 3 μm, 100 Å, Phenomenex, Torrance, CA) and strong cation exchanger (SCX, PolySULPHOETHYL A, 5 μm, 200 Å, PolyLC, Columbia, SC) chromatography resin (10–12 cm RP/4 cm SCX/3 cm RP). A multidimensional protein identification technology analysis was performed using a quaternary U-HPLC pump that was directly connected by a polyether ether ketone (PEEK) microcross to a Thermo LTQ Orbitrap or an LTQ XL ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). For optimal peptide separation, an effective flow rate of 200 (±50) nl/min, a spray voltage of 1.8 kV, and a transfer capillary temperature of 180 °C were applied. For an eight-step multidimensional protein identification technology experiment, each step was represented by one instrument method file with identical settings for the HPLC gradient program. The mass spectrometer was set to detect a full MS spectrum between 400–2,000 m/z for the precursor ion followed by MS/MS scans of the top 10 precursor ions from the preceding MS scan to detect fragment ions by the ion trap. MS/MS data interpretation was accomplished with Proteome Discoverer software version 1.2.0.208. The data were searched against the UniProt murine database (release 10.0, 20,752 entries) with semi-tryptic peptides, a mass accuracy of 10 ppm or 2.5 Da, a fragment ion tolerance of 1 Da, and the oxidation of methionine as a variable modification, allowing two missed cleavage sites. All of the accepted results had an XCorr of 1.8 for singly charged peptides, 2.5 for doubly charged peptides, and 3.5 for triply charged peptides (23, 24). At least two different peptides must be identified per protein. The FDR value was greater than 2.7% based on a decoy database search. The analysis of the spectral count data was performed using the SAINTexpress software (25, 26) and an AvgP cutoff > 0.66 was used to determine positive hits for further testing.
Co-Immunoprecipitations and Western Blotting
For co-immunoprecipitations, olfactory epithelium lysate from BL6 mice (>P20) was ground in RIPA buffer (150 nm NaCl, 50 nm Tris-HCl, 1% Nonidet, 0.5% deoxycholate (w/v), 0.1% SDS (w/v), and proteinase inhibitor) and processed using the Catch & Release system (Catch & Release v 2.0, Millipore, Darmstadt, Germany). After centrifugation at 1,000 g for 10 min, the lysate (4 mg of total protein) was incubated with primary antibodies (each 0.8 μg) overnight at 4 °C in a spin column. After several washing steps, the proteins were eluted using a denaturing elution buffer. Additionally, an input control and a negative control using a normal rabbit IgG-antibody were prepared in the same manner. The samples were loaded onto an SDS gel to separate them according to size and immunoblotted on a nitrocellulose membrane (Porablot NCL, Machery-Nagel, Düren, Germany). The interactions were detected through specific antibodies (1:50–1:250) using the ECL Western blotting detection reagent (GE Healthcare, Little Chalfont, UK).
Phosphoinositide(4,5)-bisphosphate (PIP2)/Phosphatidylinositol Agarose Beads Interaction Assay
Phosphoinositide(4,5)-bisphosphate (PIP2) and phosphatidylinositol-coupled agarose beads (Molecular Probes) were added to cell/tissue lysates in PIP2-binding buffer and rocked at 4 °C for 3 h. The beads were washed four times in PIP2-binding buffer and then eluted using Laemmli buffer (1:20 DTT) and boiled for 5 min at 95 °C. Then, 15 μl of the supernatant was then loaded onto an SDS gel, along with a sample of the cell/tissue lysate as an input control and a negative control using uncoated agarose beads. Western blotting was performed as described, and anti-MUPP1 or anti-GFP antibodies were used to detect any interaction between MUPP1 and PIP2.
PIP2 StripTM Protein-Lipid Overlay Assay
PIP strips are nitrocellulose membranes containing 100 pmol samples of 15 different phospholipids and a blank control sample. Hana3A (27) cells were transfected with MUPP1-GFP and harvested 24 h after transfection. The cell pellet was homogenized in 800 μl of Ringer′s solution containing protease inhibitors. Then, the lysate was diluted in 3% fatty acid-free BSA in TBS-T and added to the PIP strips for 2.5 h at room temperature. After several washing steps, the blot was analyzed using anti-GFP and anti-rabbit secondary antibody coupled to horseradish peroxidase.
Peptide Microarrays
CelluSpotsTM Peptide Arrays (Intavis AG, Cologne, Germany) were blocked for 2 h at room temperature with 3% protein mixture (3K Eiweiss Shake, Layenberger, Rodenbach, Germany) in NaCl/Tris/Tween. The arrays were incubated with single PDZ domains (∼2 μg of protein) of MUPP1 (1–13) that were fused to HA overnight at 4 °C. Several washing steps were performed using NaCl/Tris/Tween. Then, the microarrays were incubated with anti-HA antibodies (1:250) for 4 h at room temperature and detected with HRP-coupled secondary antibodies using ECL plus a Western blotting detection reagent (GE Healthcare). Hits were considered positive when a signal was detectable in three out of three experiments.
GST Fusion Peptides
Single PDZ domains of MUPP1 (1–13) were produced in Escherichia coli BL21 DE3pLysS. The culture was induced using 0.5–1 mm isopropyl β-d-thiogalactoside (IPTG) and purified via glutathione-coupled Sepharose beads (Becton-Dickinson Biosciences, Franklin Lakes, NJ). Accurate protein sizes were verified using SDS gelelectrophoresis and Coomassie staining. The protein amount was measured using a Bradford protein assay (Coomassie Plus Protein Assay, Thermo Scientific).
Data Analysis
Microarray data were obtained using VilberLourmat Fusion Software. The spot intensities were analyzed with TIGR Spotfinder 3.2.1. The intensities (0–255) were calculated as mean values for spots where a positive signal was observed three out of three times. The individual highest intensity of one array was set as 100%, while all of the other intensity values were normalized to this intensity. To depict the data, heat maps were designed using the R Program (3.13.0, www.r-project.org). 3-D scatter plots (mass, isoelectric point, and hydropathicity (Grand average of hydropathicity (GRAVY))) were obtained with ProtoParam (www.expasy.org). The plots of bound and unbound interaction partners were prepared with Sigma Plot 8.02. Further mass spectrometry analysis was performed with SAINTexpress (25, 26) (http://saint-apms.sourceforge.net/). PSPP (0.8.4) was used for binary logistic regression analysis (http://www.gnu.org/software/pspp/).
RESULTS
Proteomic Analysis of Interactions between Mouse Olfactory Receptor C Termini and MUPP1 PDZ Domains
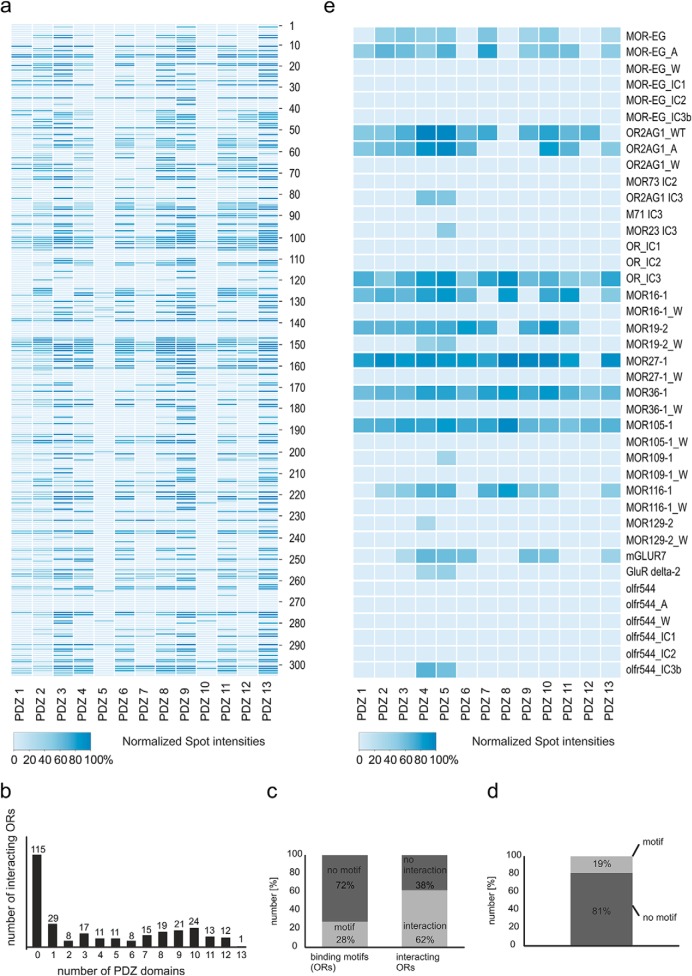
To unravel the interaction characteristics of GPCRs and PDZ domains more precisely, we selected the protein family of murine ORs and the largest described PDZ protein to date, MUPP1, as model systems. We analyzed the binding of the 13 single PDZ domains of MUPP1 to OR C termini using a large-scale in vitro approach. We designed peptide microarrays consisting of peptides corresponding to the 20 C-terminal amino acids of 304 murine ORs. Based on a phylogenetic tree of all of the mouse ORs, we randomly chose target sequences from every mouse OR subfamily and immobilized the corresponding C-terminal peptides onto a cellulose layer. We also produced the individual PDZ domains 1–13 of MUPP1 in E. coli and purified them via a GST-tag using glutathione-coupled beads. For interaction studies, we incubated the microarrays with 2 μg of purified HA-tagged PDZ domains; bound PDZ domains were detected by HA antibody incubation and chemiluminescence and purified GST was used as a negative control (Fig. 1A). Peptide interaction assays were performed three times for each PDZ domain, and the interactions were scored as positive when the signal was detected in all of the experiments. We analyzed the relative strength of the observed interactions between the 13 PDZ domains and their interacting receptors by normalizing the intensity values to the individual highest intensity of the spots on the microarrays and calculating the mean value of three replicates. The resulting intensity values are given in Fig. 1A as a color-coded heat map (for a detailed list of OR positions and intensity values, see Supplemental Table 1). With this approach, we could confirm known interactions for OR2AG1 (13) and mGluR7 (14, 28) with several PDZ domains, while published noninteractors (OR2AG1 tryptophan mutant, NMDA R2A, beta2 adrenergic R) failed to bind the tested PDZ domains (partly tested in Baumgart et al. 2014; Supplemental Table 2) (14). Most of the ORs bound to more than one of the domains that were used for the interaction analysis (Fig. 1B). A total of 62% of the selected OR subsets showed interaction with different MUPP1 PDZ domains (Fig. 1C), while only 28% of the ORs bore classical C-terminal PDZ interaction motifs (15). Remarkably, 81% of the ORs that bound to PDZ domains did not contain classical C-terminal PDZ interaction motifs (Fig. 1D). In addition, we quantified the relative binding intensities of these PDZ domains to mutant OR C termini peptides of 15 amino acids in length (Fig. 1E), showing that X-W-X-W mutations abolished PDZ-GPCR binding, while X-A-X-A mutations could even enhance the relative interaction strength. Immobilized intracellular loops 1 (IC1) and 2 (IC2) of the ORs showed no interactions with the PDZ domains compared with the C termini, while the third intracellular loop (IC3) seemed to be a promising candidate for PDZ interaction (Fig. 1E). Together, the 13 MUPP1 domains interacted with 189 of the 304 OR C-terminal peptides that were immobilized on the microarrays. Many ORs were attached to more than one specific domain, resulting in a total of 1,229 interactions.
Fig. 1.
Peptide microarray-based OR–PDZ interactions. (A) Heat map displaying the spot intensities between individual PDZ domains of MUPP1 (1–13) and interacting olfactory receptors (C-terminal peptides, 20 amino acids length). For a detailed list of microarray positions, see Supplemental Table 1. (B) Bar chart of the number of interacting peptides in relation to the number of bound PDZ domains. In total, 115 peptides did not bind to any PDZ domain, and one OR-peptide was able to interact with all 13 of the tested PDZ domains. (C) Bar chart depicting the predictions (in %) of OR–PDZ interactions using theoretical binding motifs versus the practical findings of the peptide interaction assays. Left panel: OR-peptides without a classical PDZ interaction motif are shown in dark gray and those with interaction motif in light gray. Right panel: Interacting OR-peptides are depicted in light gray and noninteracting in dark gray. (D) Bar chart showing all of the interacting peptides. Bound peptides with classical PDZ motif are shown in light gray and those without in dark gray. (E) Heat map of the spot intensities between the individual PDZ domains of MUPP1 (1–13) and C termini, intracellular loops, alanine- (A) and tryptophan-mutated C termini (W) of different interacting olfactory receptor peptides (peptides of 15 amino acids length). For a detailed list of microarray positions, see Supplemental Table 2.
Physicochemical Properties of MUPP1′s PDZ Domains
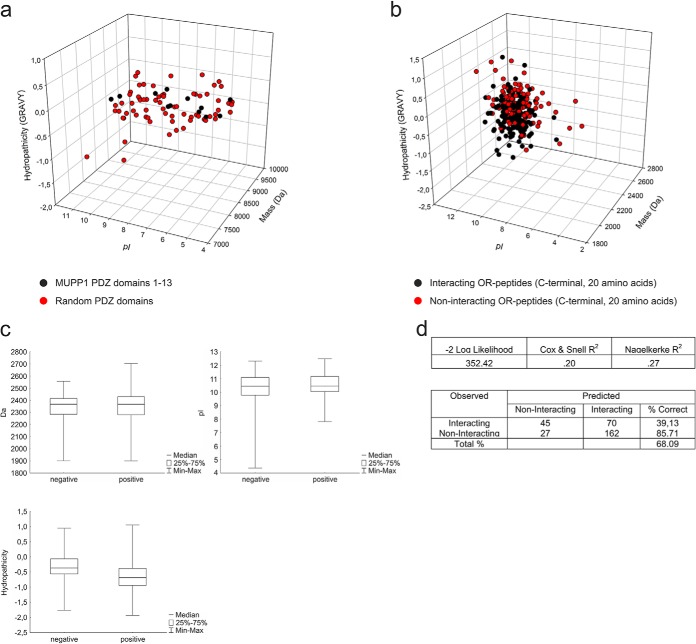
We further analyzed the physicochemical properties of each PDZ domain of MUPP1 compared with the PDZ domains of other scaffolding proteins (Supplemental Table 7) and calculated 3-D scatter plots displaying the hydropathicity (GRAVY), pI, and mass of the PDZ domains (Fig. 2A). The PDZ domains of MUPP1 are very similar regarding their size and their hydropathicity (-0.153 to 0.327) and cluster in relatively narrow ranges compared with PDZ domains in general (-1.499 to 0.651 for our selection).
Fig. 2.
3-D scatter plots of PDZ domains and binding partners. (A) 3-D scatter plot depicting the pI, mass (Da) and hydropathicity (GRAVY) of all 13 of the PDZ domains of MUPP1 (black) and randomly chosen PDZ domains of other PDZ proteins (red) showed the clustering of MUPP1 PDZ domains in a certain region. (B) 3-D scatter plot of the pI, mass (Da) and hydropathicity (GRAVY) of olfactory receptors (C-terminal peptides, 20 amino acids length) that interacted with one or more PDZ domains of MUPP1 (black) or showed no interaction (red). (C) Box and whisker plot of the three different physiochemical properties of the interacting (positive) and noninteracting (negative) olfactory receptor peptides. The plots show the median, the central 50% region as well as the minimum and maximum whiskers. (D) Binary logistic regression analysis showed a prediction efficiency of 68.09% for a model with the three chemical properties.
In addition, we analyzed the physicochemical properties of GPCR interactors and noninteractors and designed 3-D scatter plots representing the results. We did not observe any prominent differences between interacting and noninteracting OR C termini (Fig. 2B) concerning their chemical properties; both had relatively high pI values and medium hydropathicity. Box whisker plots show a nearly equal distribution of all of the properties between interacting and noninteracting peptides (Fig 2C). Furthermore, a binary logistic regression analysis showed only a 68.09% correct prediction based on the three factors (Fig 2D). A general binding affinity regarding these chemical properties could therefore not be determined for GPCR–PDZ interactions.
Phosphorylation of PDZ Binding Partners Alters PDZ Interactions
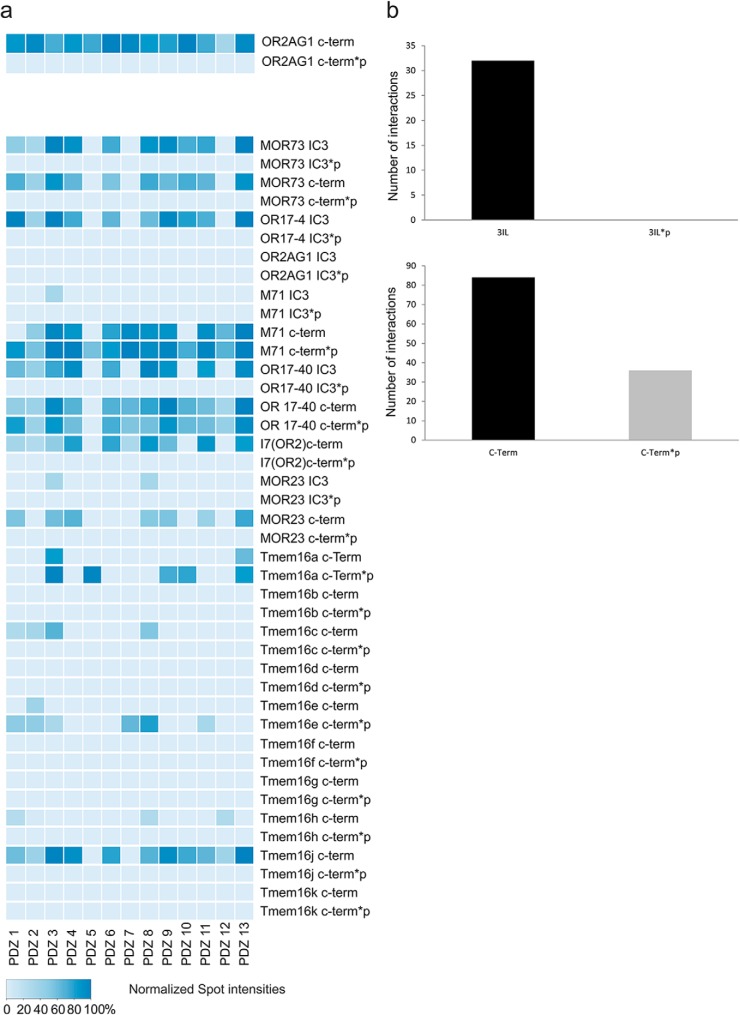
To account for the significance of phosphorylated PDZ binding partners regarding the modulation of interactions, we designed peptide microarrays of 20 amino acids in length with unmodified and phosphorylated C-terminal and IC3 (intracellular loop 3) peptides of deorphanized ORs and the olfactory-relevant Cl− channel family TMEM16 (29–32) and performed interaction assays with single 13 PDZ domains of MUPP1 as described above (Fig. 3A). Remarkably, the phosphorylation of both the C-terminal region as well as IC3 seems to strongly influence PDZ binding. All of the observed interactions (32 in total) between the 13 PDZ domains of MUPP1 and the tested IC3 regions of different ORs and ion channels were completely abolished after phosphorylation (Fig. 3A). The interactions of the tested PDZ domains with the C-terminal regions of diverse ORs were altered to both higher (8) and lower (2) relative binding affinities after phosphorylation. However, specifying a general tendency, the number of interactions was reduced to nearly half (71/36) after phosphorylation (Fig. 3B).
Fig. 3.
Phosphorylation modulates the MUPP1 interaction. (A) Heat map showing the spot intensities of the interactions between the individual PDZ domains of MUPP1 (1–13) and the C termini or intracellular loops of several olfactory receptors as well as their phosphorylated versions (peptides of 20 amino acids in length). For a detailed list of microarray positions, see Supplemental Table 4. (B) Bar charts comparing the number of interactions between the native and phosphorylated versions of the intracellular loops or the C termini of all of the spotted olfactory receptor peptides with PDZ domains. Phosphorylated intracellular loops completely lose the ability to interact with MUPP1, while C termini show a 49% decrease in the number interactions.
Interaction Analysis of the PDZ Domains of MUPP1 and GPCR-Associated Signaling Components
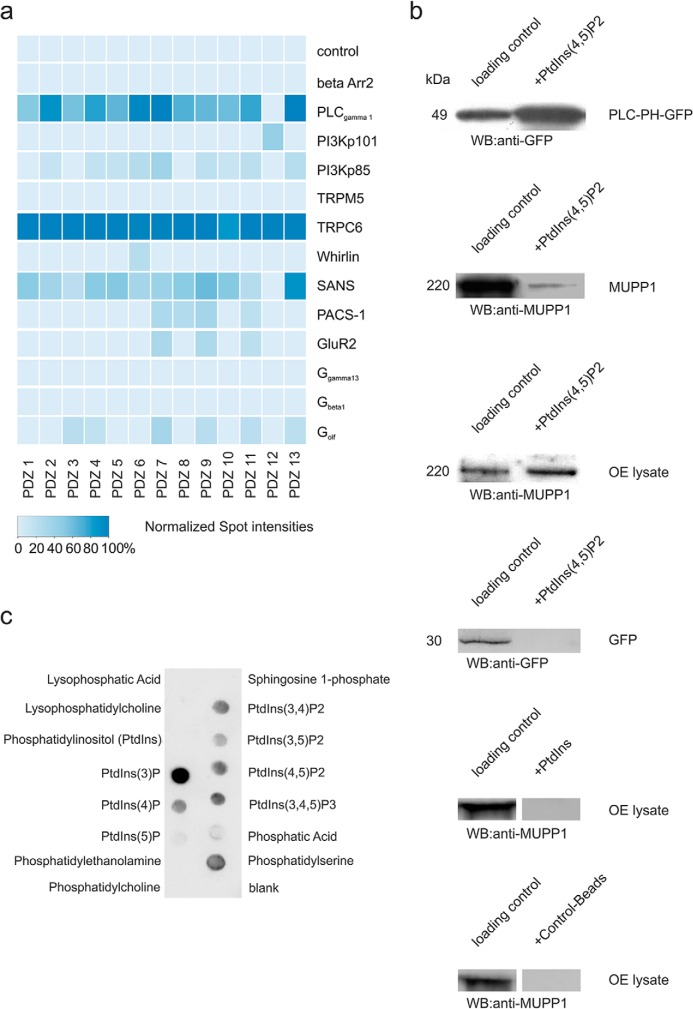
In addition to the interactions of PDZ domains with GPCRs, we were also interested in binding events to GPCR-related and other signaling proteins. Therefore, we probed a signaling peptide-containing microarray with purified 13 PDZ domains of MUPP1 and detected putative interactions as described above (Fig. 4A). The C-terminal peptide sequences of these microarrays were of 20 amino acids in length. We detected interactions for MUPP1 PDZ domains with phospholipase C γ1 (PLC-γ1), phosphoinositide-3-kinase, TRPC6, Whirlin, SANS, PACS-1, GluR2, and Golf.
Fig. 4.
Interactions between signaling proteins and MUPP1. (A) Heat map displaying the spot intensities of the interactions between the individual PDZ domains of MUPP1 (1–13) and the different binding partners (C-terminal peptides, 20 amino acids length). For a detailed list of microarray positions, see Supplemental Table 3. (B) First panel: Control interaction showing PLC-PH-GFP binding to PIP2-agarose-coated beads. Second panel: Interaction of overexpressed MUPP1 (Hana3A cells (27)) with PIP2-agarose beads. Third panel: Specific interaction of MUPP1 (olfactory epithelial lysate) with PIP2-agarose beads. Fourth panel: Control showing no interaction of overexpressed GFP (Hana3A cells (27)) with PIP2-agarose beads. Fifth panel: No detectable interaction of MUPP1 (olfactory epithelial lysate) with PIP-agarose beads. Sixth panel: Control showing no interaction of MUPP1 (olfactory epithelial lysate) with control-agarose beads. Lane 1 = loading control, Lane 2 = protein pulled down by PIP-agarose beads. (C) PIP StripsTM (Molecular Probes) spotted with 15 different phospholipids. The spots show interactions between MUPP1-GFP from transfected Hana3A cells and various phospholipids.
Because phosphoinositide signaling is often cross-linked to GPCR-mediated signal transduction cascades, we were interested in the importance of the PDZ domain binding for these interactions. We found that recombinantly expressed MUPP1 as well as MUPP1 isolated from murine olfactory epithelium can be precipitated with PIP2-coated beads, while a pull down with unphosphorylated phosphatidylinositol beads did not show any interaction (Fig. 4B). Additionally, we detected interactions of recombinantly expressed MUPP1 with various phosphoinositides using a protein–lipid overlay assay (Fig. 4C). Here, we could observe interactions of MUPP1 with phosphatidylinositol monophosphates (PtdIns(3)P, PtdIns(4)P, PtdIns(5)P), phosphatidylinositol bisphophosphates (PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2), and phosphatidylinositol trisphophosphate (PtdIns (3,4,5)P3), with PtdIns(3)P being the spot with the highest and PtdIns(5)P the spot with the lowest intensity. According to the previously described Inositol monophosphate bead experiment, no interaction with the unphosphorylated phosphatidylinositol could be detected.
Pull-Down-Based Mass Spectrometry of PDZ (MUPP1) Interaction Partners
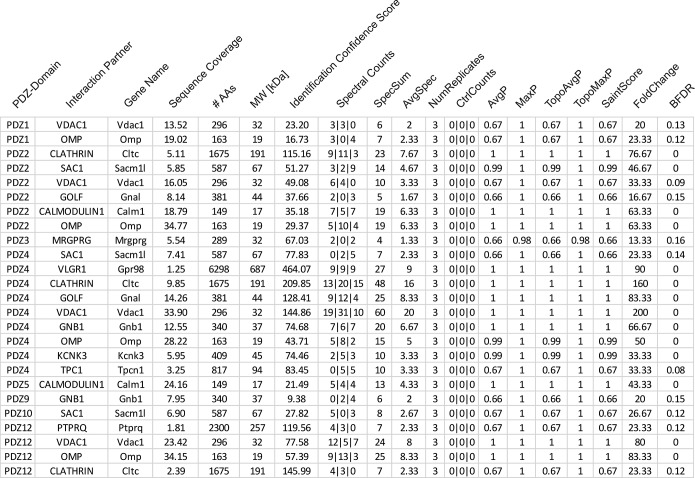
In addition to the characterization of PDZ-GPCR interactions in the heterologous system, we wanted to examine the detailed binding events of our PDZ domains and proteins. Therefore, we performed pull-down experiments with nine single PDZ domains of MUPP1 and proteins from murine OE lysate and analyzed PDZ interaction partners by mass spectrometry. We used the PDZ domains that yielded the highest and purest peptide amounts for further examination. Analyses were performed three times using individual OE and PDZ samples. The background estimation was performed three times by pulling down OE proteins with purified GST protein and acted as a control for the mass spectrometry data. For the probabilistical analysis of these protein–protein interaction datasets, we used the SAINTexpress Software (25, 26) to evaluate the quality of the identified interactions. The raw data from the mass spectrometry analysis are listed in Supplemental Table 5, while the SAINTexpress evaluation of these data is shown in Supplemental Table 6. A list of the most interesting interactions regarding the olfactory system with an AvgP cutoff at 0.66 is shown in Fig. 5. The tested PDZ domains of MUPP1 interacted with various olfactory signaling proteins, such as Golf, Gs β-subunit, calmodulin (CaM), olfactory marker protein (OMP), and clathrin as well as with other promising signaling proteins and ion channels, e.g. KCNK3 or VLGR1.
Fig. 5.
Mass spectrometry analysis of novel binding partners for MUPP1. List of binding partners of the 13 PDZ domains of MUPP1 as determined through mass spectrometry with an AvgP > 0. 66 and a relevance to the olfactory system. For a detailed list of the mass spectrometry results, see Supplemental Table 5.
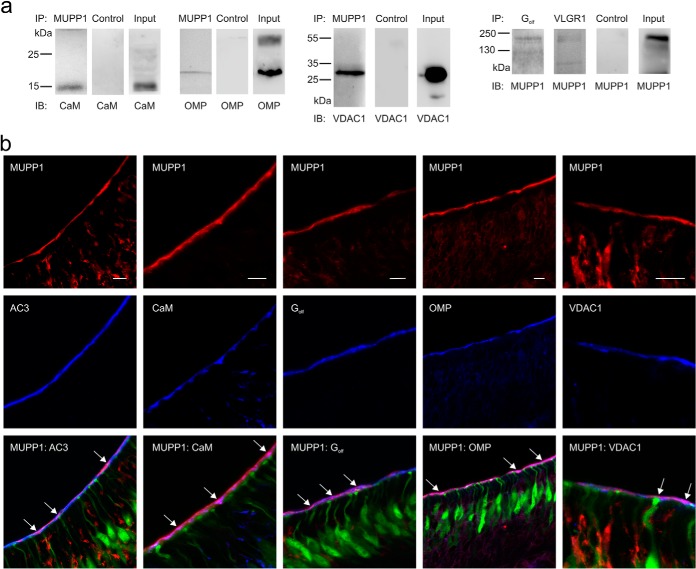
To validate the mass spectrometry data, we performed co-immunoprecipitations using OE lysate and antibodies against MUPP1 and other signaling proteins to pull out protein complexes and detect specific interaction partners (Fig. 6A). We used a normal rabbit IgG-antibody for control pull downs and OE lysate as an input control. Thereby, we could validate the interactions for MUPP1 with CaM, OMP, VDAC1, Golf, and VLGR1, indicating the accuracy of the mass-spectrometry-based approach. To observe the detailed localization of MUPP1 interaction partners, we performed immunohistological stainings using a transgenic heterozygous OMP-GFP mouse strain (22) and specific antibodies (Fig. 6B). Thereby, we could detect the colocalization of MUPP1 with Golf, CaM, AC3, OMP, and VDAC1 within the apical region of the murine OE, indicating a close proximity of these proteins within this tissue.
Fig. 6.
MUPP1 interacts and colocalizes with different signaling proteins. (A) Co-immunoprecipitations of MUPP1 with different potential binding partners as identified through mass spectrometric analysis. Rows 1–3: Co-immunoprecipitations of endogenous MUPP1 with calmodulin, olfactory marker protein (OMP) and voltage-dependent anion channel 1 (VDAC1), including normal rabbit IgG-antibody controls and input controls. Row 4: Co-Immunoprecipitations of endogenous Golf and VLGR1 with MUPP1, including normal rabbit IgG-antibody control and input control. Murine OE lysate was immunoprecipitated with the protein mentioned above and detected with antibodies against the interacting protein. (B) Confocal images of horizontal cryosections using heterozygous OMP-GFP mice stained with specific antibodies against MUPP1 (red) and Golf, calmodulin, AC3, OMP, VDAC1 (blue). Scale bar = 10 μm. Arrows in overlay images indicate colocalization of MUPP1 and Golf, calmodulin, AC3, OMP, VDAC1 (pink/purple).
DISCUSSION
PDZ domains are one of the most frequently encountered domains that are specifically responsible for protein–protein interactions. These domains consist of ∼90 amino acids and can occur in multiple copies inside a protein (33, 34). PDZ proteins can fulfill various functions, build large protein networks, and are involved in the modulation of many different cellular processes. Especially in neuronal signal transduction processes, the spatial and temporal organization of its components is of utter importance. Therefore, PDZ domains are key players in these systems, regulating the interactions of all of the involved signaling proteins and enhancing the efficiency and speed of signal processing (35, 36). Among the multitude of PDZ interactions partners, one important group is the diverse family of GPCRs (37). These membrane proteins are essential for various cellular processes and can be involved in different diseases, such as cancer, obesity, diabetes, or central nervous system disorders (38–40), and therefore make excellent therapeutic targets. Thus far, the detailed binding characteristics and regulatory mechanisms of PDZ–GPCR interactions are still under debate and must be further investigated. Here, we carried out a comprehensive interaction study of the 13 single PDZ domains of MUPP1 and 304 murine olfactory receptors, representing the protein family of GPCRs, using peptide microarrays. The analyzed PDZ domains of MUPP1 show very conserved chemical characteristics compared with the PDZ domains of other proteins, suggesting a specialized scaffolding complex. Interestingly, the chemical properties of PDZ-interacting and noninteracting peptides seem to be relatively similar to each other, demonstrating the complexity of PDZ interactions. In contrast to studies in which classical hieratic binding motifs were presented (15), we demonstrated a disagreement between those motifs and our practical findings. The proportion of interacting and noninteracting peptides to classical binding motifs was nearly vice versa. However, PDZ domains do not fall in discrete classes but are distributed throughout selectivity space, thereby avoiding cross reactivity (17, 41). Furthermore, extensive interaction studies in Caenorhabditis elegans revealed frequent noncanonical interactions between PDZ domains and their binding partners, indicating the extreme fine-tuning of PDZ–ligand interactions (18). The individual domains bind with different affinities to various immobilized ORs, suggesting the presence of a highly dynamic and flexible interactome (42–44).
Additionally, we analyzed the interactions of PDZ domains and various signaling peptides. Here, we could validate a published MUPP1 interactor (Golf) (14) but also identify new PDZ binding partners, such as PLC-γ1, PIP2, TRPC6, SANS, or PACS-1. This identification indicates that the putative interactions of PDZ proteins are not exclusively with peptides but also with phospholipids. Different studies have demonstrated the relevance of PDZ–phosphoinositide interactions and have claimed the necessity to further characterize such binding events (45). Interestingly, only phosphorylated versions of phosphatidylinositol were able to interact with MUPP1 in either of our approaches, further indicating the importance of such modifications for PDZ binding. PDZ–lipid interactions are important for cell signaling, receptor trafficking, and receptor cargo recycling (46). Furthermore, an interaction of MUPP1 and tandem-PH-domain-containing protein-1 (TAPP1) is important for PIP2-mediated signaling (47), which further strengthens the idea of an interactome consisting of PDZ proteins, GPCRs, signaling peptides, and lipids. Interestingly, associated enzymes, such as PLC-γ1, were also able to interact with the PDZ domains of MUPP1. In this context, different PLC subtypes bear classical PDZ binding motifs, but detailed binding modalities must be further determined (48).
We were interested in the mechanisms regulating and influencing interactions of PDZ proteins with GPCRs. Because posttranslational modifications, such as phosphorylation, are important regulatory instruments, we focused on PDZ binding to GPCR peptides with phosphorylated C termini and IC3. We found that PDZ binding promiscuity was diminished for most interactions after introducing a phospho-group to both IC3 and C termini. This finding agrees with other studies demonstrating that the phosphorylation of PDZ ligands modifies binding properties (49). The phosphorylation of AMPA receptor subunit GluR2 decreases binding to the PDZ protein GRIP1 but not to PICK1, indicating the differential dynamics of such a regulatory mechanism (50). Similar to our results, in some cases, the binding affinities of PDZ–ligand interactions are even enhanced after phosphorylation (51). Additionally, distinct C-terminal mutations (X-A-X-A and X-W-X-W) of the PDZ ligands led to altered relative binding affinities. Alanine substitutions caused even higher relative intensity values, whereas sterically complex tryptophan mutations abolished PDZ–ligand binding in all of the tested scenarios. Tryptophans at C-terminal positions 0 and -2 can rarely be found within PDZ interactors, and these mutations are capable of inhibiting interactions (13, 14, 17). These observations indicate the importance of C-terminal positions 0 and -2 and suggest that underlying amino acids can be variable within a certain range.
In addition to in vitro interaction analyses, we focused on PDZ–ligand binding using murine olfactory epithelium and pull-down-associated mass spectrometry. We could thereby validate the published interactions of MUPP1′s PDZ domains with Golf and CaM (14) and identify unknown binding partners. Here, OMP was able to bind to several PDZ domains and has been reported to play important roles in olfactory signaling and development of the olfactory system (52–54). VLGR1, the largest known GPCR, which is part of a PDZ-organized protein network that is associated with a type of sensineuronal hearing loss called Usher syndrome (55, 56) is another new, interesting interaction partner of MUPP1. To validate the accuracy of our mass-spectrometry-based interaction analyses, we performed additional co-immunoprecipitations and colocalization studies that confirmed our findings.
In conclusion, we provide here a comprehensive overview of the binding characteristics of diverse PDZ–GPCR interactions based on the murine olfactory system, indicating the complexity und variability of these binding events, and found that phosphorylation of both the receptor′s C terminus and IC3 alters PDZ binding promiscuity. Thus, we anticipate that our study will greatly enhance the understanding of these important protein–protein and protein–lipid interactions and might identify novel clinical targets for GPCR-associated diseases.
Supplementary Material
Acknowledgments
We want to thank the Intavis Bioanalytical Instruments AG for supplying the “CelluSpots™ - Custom Peptide Arrays.” We also thank Simon Pyschny for his valuable technical support in all of the biochemical and immunohistochemical techniques. Without him, this study would not have been possible.
Footnotes
Author contributions: F.J., H.H., E.M.N., and S.O. designed the research; F.J., B.K., B.F., M.O., and S.O. performed research; D.W. contributed new reagents or analytic tools; F.J., B.K., P.S., B.F., and S.O. analyzed the data; F.J., B.F., and S.B. wrote the paper; and B.K., P.S., D.W., H.H., and E.M.N. provided proofreading.
* The research for this paper was financially supported by the Deutsche Forschungsgemeinschaft grant numbers SFB958, SFB642, and Exc257; the Studienstiftung des deutschen Volkes; the International Max-Planck Research School in Chemical Biology (IMPRS-CB); the Ruhr-University Research School; and the Wilhelm und Guenter Esser-Stiftung.
 This article contains supplemental material Supplement Tables 1 to 7.
This article contains supplemental material Supplement Tables 1 to 7.
1 The abbreviations used are:
- AC3
- adenylyl cyclase III
- CaM
- calmodulin
- CNS
- central nervous system
- C-term
- carboxy-terminus
- GABA
- gamma-aminobutyric acid
- Golf
- olfactory G protein
- GPCR
- G protein-coupled receptors
- GST
- glutathione S-transferase
- IC
- intracellular loop
- InaD
- inactivation no afterpotential
- MUPP1
- multiple PDZ domain protein 1
- OE
- olfactory epithelium
- OMP
- olfactory marker protein
- OR
- olfactory receptor
- PDZ
- PSD-95/discs-large/ZO1
- PIP
- phosphatidylinositol
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PLC
- phospholipase C
- TAPP1
- tandem-PH-domain-containing protein-1
- TRP
- transient receptor potential.
REFERENCES
- 1. Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 [DOI] [PubMed] [Google Scholar]
- 2. Balasubramanian S., Fam S. R., Hall R. A. (2007) GABAB receptor association with the PDZ scaffold MUPP1 alters receptor stability and function. J. Biol. Chem. 282, 4162–4171 [DOI] [PubMed] [Google Scholar]
- 3. Magalhaes A. C., Dunn H., Ferguson S. S. G. (2011) Regulation of G protein-coupled receptor activity, trafficking and localization By GPCR-interacting proteins. Br. J. Pharmacol. 165(6), 1717–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Romero G., von Zastrow M., Friedman P. a. (2011) Role of PDZ Proteins in Regulating Trafficking, Signaling, and Function of GPCRs: Means, Motif, and Opportunity. 1st ed Elsevier, Inc; Adv Pharmacol. 62, 279–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bockaert J., Fagni L., Dumuis A., Marin P. (2004) GPCR interacting proteins (GIP). Pharmacol. Ther. 103, 203–221 [DOI] [PubMed] [Google Scholar]
- 6. Glusman G., Yanai I., Rubin I., Lancet D. (2001) The complete human olfactory subgenome. Genome Res. 11, 685–702 [DOI] [PubMed] [Google Scholar]
- 7. Xu X. Z., Choudhury A., Li X., Montell C. (1998) Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J. Cell Biol. 142, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsunoda S., Sierralta J., Sun Y., Bodner R., Suzuki E., Becker A., Socolich M., Zuker C. S. (1997) A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature 388, 243–249 [DOI] [PubMed] [Google Scholar]
- 9. Ullmer C., Schmuck K., Figge A., Lübbert H. (1998) Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 424, 63–68 [DOI] [PubMed] [Google Scholar]
- 10. Becamel C., Figge A., Poliak S., Dumuis A., Peles E., Bockaert J., Lubbert H., Ullmer C. (2001) Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J. Biol. Chem. 276, 12974–12982 [DOI] [PubMed] [Google Scholar]
- 11. Parker L. L., Backstrom J. R., Sanders-Bush E., Shieh B.-H. (2003) Agonist-induced phosphorylation of the serotonin 5-HT2C receptor regulates its interaction with multiple PDZ protein 1. J. Biol. Chem. 278, 21576–21583 [DOI] [PubMed] [Google Scholar]
- 12. Guillaume J.-L., Daulat A. M., Maurice P., Levoye A., Migaud M., Brydon L., Malpaux B., Borg-Capra C., Jockers R. (2008) The PDZ protein MUPP1 promotes Gi coupling and signaling of the Mt1 melatonin receptor. J. Biol. Chem. 283, 16762–16771 [DOI] [PubMed] [Google Scholar]
- 13. Dooley R., Baumgart S., Rasche S., Hatt H., Neuhaus E. M. (2009) Olfactory receptor signaling is regulated by the post-synaptic density 95, Drosophila discs large, zona-occludens 1 (PDZ) scaffold multi-PDZ domain protein 1. FEBS J. 276, 7279–7290 [DOI] [PubMed] [Google Scholar]
- 14. Baumgart S., Jansen F., Bintig W., Kalbe B., Herrmann C., Klumpers F., Köster S. D., Scholz P., Rasche S., Dooley R., Metzler-Nolte N., Spehr M., Hatt H., Neuhaus E. M. (2014) The scaffold protein MUPP1 regulates odorant-mediated signaling in olfactory sensory neurons. J. Cell Sci. 127, 2518–2527 [DOI] [PubMed] [Google Scholar]
- 15. Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 16. Jemth P., Gianni S. (2007) PDZ domains: Folding and binding. Biochemistry 46, 8701–8708 [DOI] [PubMed] [Google Scholar]
- 17. Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenfant N., Polanowska J., Bamps S., Omi S., Borg J.-P., Reboul J. (2010) A genome-wide study of PDZ-domain interactions in C. elegans reveals a high frequency of non-canonical binding. BMC Genomics 11, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hillier B. J., Christopherson K. S., Prehoda K. E., Bredt D. S., Lim W. A. (1999) Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284, 812–815 [PubMed] [Google Scholar]
- 20. Birrane G., Chung J., Ladias J. A. (2003) Novel mode of ligand recognition by the Erbin PDZ domain. J. Biol. Chem. 278, 1399–1402 [DOI] [PubMed] [Google Scholar]
- 21. Kozlov G., Banville D., Gehring K., Ekiel I. (2002) Solution structure of the PDZ2 domain from cytosolic human phosphatase hPTP1E complexed with a peptide reveals contribution of the β2–β3 loop to PDZ domain–ligand interactions. J. Mol. Biol. 320, 813–820 [DOI] [PubMed] [Google Scholar]
- 22. Potter S. M., Zheng C., Koos D. S., Feinstein P., Fraser S. E., Mombaerts P. (2001) Structure and emergence of specific olfactory glomeruli in the mouse. J. Neurosci. 21, 9713–9723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolters D. A., Washburn M. P., Yates J. R., 3rd (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 24. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 25. Teo G., Liu G., Zhang J., Nesvizhskii A. I., Gingras A.-C., Choi H. (2014) SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi H., Larsen B., Lin Z.-Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A.-C., Nesvizhskii A. I. (2011) SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saito H., Kubota M., Roberts R. W., Chi Q., Matsunami H. (2004) RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 [DOI] [PubMed] [Google Scholar]
- 28. Cui H., Hayashi A., Sun H.-S., Belmares M. P., Cobey C., Phan T., Schweizer J., Salter M. W., Wang Y. T., Tasker R. A., Garman D., Rabinowitz J., Lu P. S., Tymianski M. (2007) PDZ protein interactions underlying NMDA receptor-mediated excitotoxicity and neuroprotection by PSD-95 inhibitors. J. Neurosci. 27, 9901–9915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasche S., Toetter B., Adler J., Tschapek A., Doerner J. F., Kurtenbach S., Hatt H., Meyer H., Warscheid B., Neuhaus E. M. (2010) Tmem16b is specifically expressed in the cilia of olfactory sensory neurons. Chem. Senses 35, 239–245 [DOI] [PubMed] [Google Scholar]
- 30. Stephan A. B., Shum E. Y., Hirsh S., Cygnar K. D., Reisert J., Zhao H. (2009) ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc. Natl. Acad. Sci. U.S.A. 106, 11776–11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pifferi S., Dibattista M., Menini A. (2009) TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch. 458, 1023–1038 [DOI] [PubMed] [Google Scholar]
- 32. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. J. V. (2008) TMEM16A, A membrane protein associated with calcium-dependent chloride channel activity. Science 322(5901), 590–594 [DOI] [PubMed] [Google Scholar]
- 33. Fanning A. S., Anderson J. M. (1996) Protein–protein interactions: PDZ domain networks. Curr. Biol. 6, 1385–1388 [DOI] [PubMed] [Google Scholar]
- 34. Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell 85, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 35. Tsunoda S., Zuker C. S. (1999) The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell Calcium 26, 165–171 [DOI] [PubMed] [Google Scholar]
- 36. Huber A. (2001) Scaffolding proteins organize multimolecular protein complexes for sensory signal transduction. Eur. J. Neurosci. 14, 769–776 [DOI] [PubMed] [Google Scholar]
- 37. Tovo-Rodrigues L., Roux A., Hutz M. H., Rohde L. a, Woods A. S. (2014) Functional characterization of G-Protein Coupled receptors: A bioinformatics approach. Neuroscience 277, 764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leyva-Illades D., Demorrow S. (2013) Orphan G protein receptor GPR55 as an emerging target in cancer therapy and management. Cancer Manag. Res. 5, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kimple M. E., Neuman J. C., Linnemann A. K., Casey P. J. (2014) Inhibitory G proteins and their receptors: Emerging therapeutic targets for obesity and diabetes. Exp. Mol. Med. 46, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nickols H. H., Conn P. J. (2014) Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol. Dis. 61, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gianni S., Haq S. R., Montemiglio L. C., Jurgens M. C., Engström Å., Chi C. N., Brunori M., Jemth P. (2011) Sequence specific long-range networks in PDZ domains tune their binding selectivity. J. Biol. Chem. 286, 27167–27175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mishra P., Socolich M., Wall M. A., Graves J., Wang Z., Ranganathan R. (2007) Dynamic scaffolding in a G protein-coupled signaling system. Cell 131, 80–92 [DOI] [PubMed] [Google Scholar]
- 43. Montell C. (2007) Dynamic regulation of the INAD signaling scaffold becomes crystal clear. Cell 131, 19–21 [DOI] [PubMed] [Google Scholar]
- 44. Liu W., Wen W., Wei Z., Yu J., Ye F., Liu C.-H., Hardie R. C., Zhang M. (2011) The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye. Cell 145, 1088–1101 [DOI] [PubMed] [Google Scholar]
- 45. Ivarsson Y., Wawrzyniak A. M., Kashyap R., Polanowska J., Betzi S., Lembo F., Vermeiren E., Chiheb D., Lenfant N., Morelli X., Borg J.-P., Reboul J., Zimmermann P. (2013) Prevalence, specificity and determinants of lipid-interacting PDZ domains from an in-cell screen and in vitro binding experiments. PLoS One 8, e54581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimmermann P. (2006) The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim. Biophys. Acta 1761, 947–956 [DOI] [PubMed] [Google Scholar]
- 47. Kimber W. A., Trinkle-Mulcahy L., Cheung P. C. F., Deak M., Marsden L. J., Kieloch A., Watt S., Javier R. T., Gray A., Downes C. P., Lucocq J. M., Alessi D. R. (2002) Protein MUPP1 in vivo. Society 536, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim J. K., Lim S., Kim J., Kim S., Kim J. H., Ryu S. H., Suh P.-G. (2011) Subtype-specific roles of phospholipase C-β via differential interactions with PDZ domain proteins. Adv. Enzyme Regul. 51, 138–151 [DOI] [PubMed] [Google Scholar]
- 49. Choi J., Ko J., Park E., Lee J.-R., Yoon J., Lim S., Kim E. (2002) Phosphorylation of stargazin by protein kinase A regulates its interaction with PSD-95. J. Biol. Chem. 277, 12359–12363 [DOI] [PubMed] [Google Scholar]
- 50. Chung H. J., Xia J., Scannevin R. H., Zhang X., Huganir R. L. (2000) Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J. Neurosci. 20, 7258–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adey N. B., Huang L., Ormonde P. A., Baumgard M. L., Pero R., Byreddy D. V., Tavtigian S. V., Bartel P. L. (2000) Threonine phosphorylation of the MMAC1/PTEN PDZ binding domain both inhibits and stimulates PDZ binding advances. Cancer Res. 60, 35–37 [PubMed] [Google Scholar]
- 52. Lee A. C., He J., Ma M. (2011) Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J. Neurosci. 31, 2974–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kass M. D., Moberly A. H., McGann J. P. (2013) Spatiotemporal alterations in primary odorant representations in olfactory marker protein knockout mice. PLoS One 8, e61431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kass M. D., Moberly A. H., Rosenthal M. C., Guang S. A., McGann J. P. (2013) Odor-specific, olfactory marker protein-mediated sparsening of primary olfactory input to the brain after odor exposure. J. Neurosci. 33, 6594–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McMillan D. R., Kayes-Wandover K. M., Richardson J. A., White P. C. (2002) Very large G protein-coupled receptor-1, the largest known cell surface protein, is highly expressed in the developing central nervous system. J. Biol. Chem. 277, 785–792 [DOI] [PubMed] [Google Scholar]
- 56. Weston M. D., Luijendijk M. W., Humphrey K. D., Möller C., Kimberling W. J. (2004) Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am. J. Hum. Genet. 74, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.