Abstract
This manuscript describes the enrichment and mass spectrometric analysis of intact glycopeptides from mouse liver, which yielded site-specific N- and O-glycosylation data for ∼130 proteins. Incorporation of different sialic acid variants in both N- and O-linked glycans was observed, and the importance of using both collisional activation and electron transfer dissociation for glycopeptide analysis was illustrated. The N-glycan structures of predicted lysosomal, endoplasmic reticulum (ER), secreted and transmembrane proteins were compared. The data suggest that protein N-glycosylation differs depending on cellular location. The glycosylation patterns of several mouse liver and mouse brain glycopeptides were compared. Tissue-specific differences in glycosylation were observed between sites within the same protein: Some sites displayed a similar spectrum of glycan structures in both tissues, whereas for others no overlap was observed. We present comparative brain/liver glycosylation data on 50 N-glycosylation sites from 34 proteins and 13 O-glycosylation sites from seven proteins.
The term protein glycosylation covers a wide variety of posttranslational modifications (PTMs). Protein glycosylation may occur within the cell, where a single GlcNAc is deposited on the side-chain of Ser and Thr residues to fulfill a regulatory/signaling function (1). However, the majority of glycosylation occurs on proteins traveling through the ER and Golgi, where protein domains on the lumenal side of a membrane, secreted proteins, and the extracellular domains of transmembrane proteins are modified on Trp, Asn, Ser, Thr, or Tyr side chains with simple or elaborately elongated oligosaccharide structures (2, 3). Numerous enzymes participate in this process, and the heterogeneity of the resulting structures is overwhelming. Traditionally, protein glycosylation studies have focused on the in-depth analysis of enzymatically or chemically released glycan pools (4–6). This approach is still the most reliable method for obtaining detailed structural information about the protein-modifying carbohydrates as the protein-level heterogeneity, both in terms of site occupancy and the number of site-specific structures, represent exceptional challenges for analysis. However, information about protein and site-specific glycosylation is lost by this approach, so there is a growing need for routine glycopeptide analysis, as glycosylation has been implicated as a key player in cell–cell interactions, host/pathogen interactions, enzymatic processing, and even intracellular signaling (7–13). Studies have shown that glycosylation is species-, and tissue-specific, and can be altered by disease or physiological changes (14–22). It has also been reported that cellular localization and protein structure influence/determine protein- and site-specific N-glycosylation (23, 24). It should be noted that intact glycopeptide studies usually only allow the determination of glycan compositions; the identity of the oligosaccharide units and their linkage may be obtained from released glycan studies (5, 6).
In this manuscript, we present data on the site-specific N- and O-glycosylation of mouse liver proteins. We report cellular compartment-dependent glycosylation based on glycopeptide data. We also compare the glycosylation pattern of several mouse liver and mouse brain glycopeptides (25). While individual proteins have been studied this way (15, 26), this is the first time that cellular-localization-specific and tissue-specific glycosylation have been compared on a larger scale at a glycosylation-site-specific level.
MATERIALS AND METHODS
The sample preparation has been published earlier (27). Here, we provide a brief description.
Mouse Liver Sample Preparation
Three livers, from 10-day-old mice, were homogenized, in 10 mm N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES-KOH, pH 7.9), 1.5 mm MgCl2, 10 mm KCl. The lysis buffer also contained O-GlcNAcase inhibitor PUGNAc (Sigma, St. Louis, MO; 50uM) and protease and phosphatase inhibitors (Roche, South San Francisco, CA, and Sigma, respectively). A previously published two-step differential solubilization and centrifugation protocol was followed to prepare a crude nuclear extract (28). Our goal was to reduce the complexity of the mixture primarily by eliminating the cytoplasmic proteins. The resulting protein mixture was denatured with 6 m guanidine hydrochloride in 50 mm ammonium bicarbonate buffer; disulfide bridges were reduced with tris (2-carboxyethyl)phosphine hydrochloride, and free sulfhydryls were alkylated with iodoacetamide. Tryptic digestion proceeded in 0.8 m guanidine hydrochloride for 16 h at 37 °C. The digest was desalted and lyophilized.
Lectin Weak Affinity Chromatography (LWAC)
Wheat germ agglutinin (Vector Labs, Burlingame, CA) covalently linked to POROS beads (Life Technologies, Grand Island, NY) (26) was used for the affinity-chromatography in a 100 mm Tris-HCl, 150 mm NaCl, 2 mm MgCl2, 2 mm CaCl2 buffer (pH ∼7.7) that also contained 5% acetonitrile. Three rounds of glycopeptide enrichment were performed as described previously (29). The final glycopeptide mixture was fractionated using high pH reverse phase chromatography.
Mass Spectrometry
Fractions collected during the high pH reverse phase fractionation were analyzed by LC-MS/MS using a NanoAcquity (Waters, Milford, MA) and a LTQ-Orbitrap Velos (Thermo, San Jose, CA). Chromatography was performed using an EASY-Spray Nano-LC source with a 15 cm × 75 μm inner diameter column packed with 3 μm C18 particles. Solvent A was 0.1% formic acid, 5% DMSO in water; Solvent B was 0.1% formic acid, 5% DMSO in acetonitrile. The flow rate was 400 nl/min. The samples were injected directly on-column, then a 60-min linear gradient, 2–25% solvent B was developed, eventually the organic content was increased to 50% over 5 min. Data were acquired for 85 min. Mass measurements (m/z 350–1,400) were performed in the Orbitrap, and the three most abundant multiply charged ions were selected for sequential beam-type CID (HCD, measured in the Orbitrap) and ETD analyses (measured in the linear trap). Dynamic exclusion for precursor ion selection was enabled for 45 s. The trigger intensity was 2,000. The automatic gain control (AGC) settings were 2 × 106 for the mass measurements, 9 × 104 for the HCD acquisitions, and 104 for the ETD experiments. Supplemental activation for the ETD experiments was enabled.
MS/MS Data Analysis
Raw files were converted into peak list files using Proteome Discoverer v1.4.1.14. Database searches were performed with ETD data using Protein Prospector v5.13.2. An iterative searching strategy was employed to obtain information about the glycan pool, similar to as previously published (25). Briefly, ETD data were initially searched permitting mass modifications within a mass range of 200–2,000 on Asn, Ser, or Thr residues in the mouse protein subset of the SwissProt database downloaded on June 27, 2013 (16,622 entries searched). The results from this initial search were used to create an accession number list, a second search was performed considering only these proteins, and the upper limit of the mass modification was increased to 3,000 Da. A histogram was generated of the mass modifications detected (Fig. 1), and the most frequently detected masses were translated into glycan compositions. This yielded a candidate glycan list (Supplemental Table 1). All of these compositions were then added as potential variable modifications to Protein Prospector for a further search, along with a few additional structures, including some structures identified in our previous brain glycopeptide study (25). The complete glycan list used in this database search is presented in Supplemental Table 1. All of these glycans were included in a database search against all mouse proteins in the UniProt database downloaded on May 13, 2014 and concatenated with a random sequence for each entry (74,528 entries searched). Only one of each glycan per peptide was considered with the exception of HexNAc, and two variable modifications total per peptide were permitted. The other variable modifications considered included methionine oxidation, acetylation of protein N-termini, and cyclization of N-terminal Gln residues. Only tryptic cleavages were considered, with up to two missed cleavages permitted. The mass accuracy requirements were within 12 ppm and 0.6 Da for precursor ions and fragments, respectively. Only proteins (789) meeting acceptance criteria (minimum score = 22, max E = 0.1) and featuring at least one distinguishing sequence were included in a final search, performed with the same parameters, that yielded additional glycoforms. The best ETD spectrum representing each unique glycopeptide in the carefully curated results can be viewed in MS-Viewer (30) on the Protein Prospector public website (prospector.ucsf.edu), search key: qunwxqxpy4.
Fig. 1.
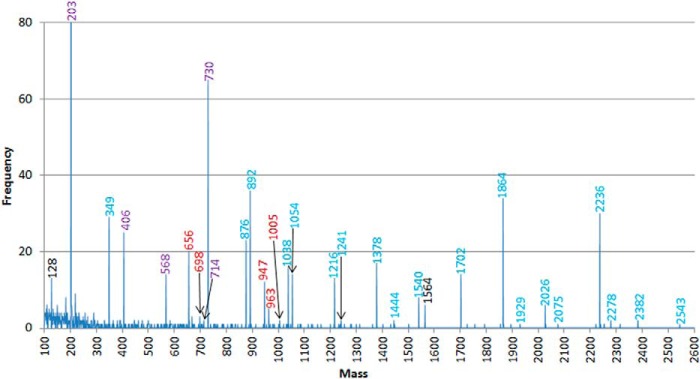
The histogram of mass modifications generated from liver glycopeptide ETD data permitting unspecified modifications on any Asn, Ser, or Thr residue of selected mouse proteins listed in the SwissProt database. Masses labeled in red correspond to O-linked glycan masses; those in blue correspond to N-linked glycan masses; those labeled in purple could be either N- or O-linked compositions. Masses in black do not correspond to glycan structures.
RESULTS
A tryptic digest of mouse liver proteins was subjected to LWAC chromatography using wheat germ agglutinin. The resulting glycopeptide mixture was further fractionated and analyzed by on-line LC/MS/MS using both beam-type collision-induced and electron transfer dissociation (HCD and ETD, respectively). The sample yielded numerous targeted GlcNAcylated peptides, mainly from nuclear proteins, but the presence of other glycan oxonium ions indicated the additional enrichment of N- or O-glycopeptides in the secretory pathway. To identify these, we applied an iterative searching strategy using Protein Prospector that has previously been successful in identifying other glycopeptides from LWAC data (25), where first a search is performed allowing unspecified modifications up to 2,000 Da on any Asn, Ser, or Thr residue. An identical search to this is then performed, except considering only the list of proteins identified in the initial search and increasing the permitted mass modification to 3,000 Da. This approach yielded the histogram shown in Fig. 1, where almost all frequently detected mass modifications correspond to potential oligosaccharide structures (see Supplemental Table 1).
The masses observed indicated the presence of numerous truncated N-linked glycan structures ranging from only the core GlcNAc to paucimannose structures. Of the oligomannose structures Man9 (mass 1,864) was the most abundant, and while several complex structures were observed, the disialo biantennary structure featuring N-glycolylneuraminic acids (mass 2,236) was the most abundant complex N-glycan observed. Mono- and disialo mucin-type core 1 structures represented the majority of the extracellular O-glycosylation, and different types and combinations of sialic acids decorated these structures: The masses showed that both N-acetyl- and N-glycolylneuraminic acids were present in the O-glycans, and O-acetylation of these further increased the glycan heterogeneity.
Based on the results of the mass modification search, 38 different N-linked glycans and 14 different O-glycans were included into a defined variable modification database search (Supplemental Table 1), where all mouse entries in the UniProt database were considered, two variable modifications were permitted per peptide, but with the exception of a single O-linked HexNAc, only one glycan was allowed per peptide. Analysis of the peak lists using MS-Filter (27) indicated 50.3% of spectra contained a HexNAc oxonium ion at m/z 204.086 ± 20 ppm within the 60 most abundant peaks, suggesting half of the mixture was glycopeptides. Approximately 2,000 of the 2,600 glycopeptide spectra assigned represented N-linked glycosylation. We present carefully curated lists for both N- and O-linked extracellular glycopeptides (Supplemental Tables 2 and 3, MS-Viewer). Only the most confident (based on E-value) ETD-based identifications were kept (replicates were removed), the N- and O-linked glycopeptides were separated, and the intracellular O-GlcNAc modified peptides are not presented.
N-glycosylation
The N-linked glycopeptide list contained 628 different glycopeptides, representing 106 gene products, 168 unique glycosylation sites, and 534 unique glycoforms. One doubly modified N-linked glycopeptide was identified bearing truncated structures at both sites and the dataset also featured nonconsensus N-glycosylation of apolipoprotein E (Supplemental Fig. 1). HexNAc2Hex10 structures were detected, suggesting the presence of immature N-glycans, i.e. Glc-capped Man9 structures (Supplemental Fig. 2). The detection of these glycoforms indicates that some proteins were isolated from the ER, enroute to their final destination. Glycoforms with trisialo biantennary structures were observed, indicating the presence of oligosaccharides with a Gal beta 1–3 GlcNAc linkage, as the sugars with the more common Gal beta 1–4 linkage cannot be sialylated on the subterminal GlcNAc (Supplemental Fig. 3).
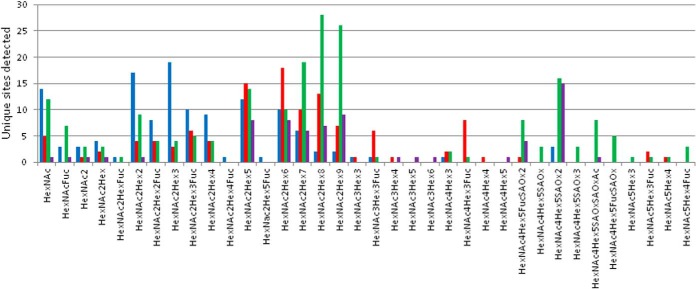
As glycopeptides rather than released glycans were analyzed, it was possible to break down the global glycan distribution into glycan distributions for proteins found in different cellular compartments, such as the lysosome and ER, and also secreted and transmembrane proteins. These results are shown in Fig. 2.
Fig. 2.
Comparison of the N-glycan distribution between lysosomal (blue), ER (red), secreted (green), and transmembrane (purple) proteins. Protein localization was assigned from UniProt, and is listed for each protein in Supplemental Table 2. Individual charts can be seen in Supplemental Table 4. Each glycoform was considered only once, even when it was identified from multiple different (for example, extended or oxidized) peptides.
These comparisons show that there are significant differences in the N-glycan pool depending on the cellular location of the glycoprotein. Lysosomal proteins mainly featured truncated sugars, paucimannose structures, and the two smallest members of the oligomannose family. Glycoproteins associated with the ER seem to be enriched in oligomannose structures and some core-fucosylated agalacto complex glycans. Secreted glycoproteins featured the complete list of truncated glycans, although the larger oligomannose structures were most common, along with disialo biantennary complex glycans with or without core fucosylation. Transmembrane proteins barely displayed truncated structures, the oligomannose structures were detected evenly distributed, and the disialo biantennary complex structure was represented at the highest level.
Comparison to Glycopeptides in Mouse Brain
Having previously generated a large mouse synaptosomal glycopeptide dataset using the same LWAC enrichment protocol (25, 29), we thought it interesting to compare the mouse liver glycosylation to mouse brain tissue. Glycosylation data published from the synaptosome study (25), with some additional glycoforms identified manually or with improved database searches, were compared with the liver glycoforms reported in this manuscript. Although tissue-specific glycosylation is well documented, these studies have nearly all been of released glycans, meaning it has not been possible to compare glycosylation differences at the modification site level. The two tissue preparations obviously delivered a different protein population; e.g. the liver glycopeptides contained many more extracellular matrix proteins such as collagens and fibronectin and also serum proteins, such as haptoglobin and complement proteins. However, there is overlap in the glycoprotein content of these samples, albeit these proteins generally contributed different relative amounts in the two samples. Certain proteins, such as Prolow-density lipoprotein receptor-related protein 1 and glucosylceramidase were present at about the same level in both mixtures, while the relative amounts of practically all other components seem to have been higher in the liver samples based on their relative contribution to all peptides identified in the samples (Supplemental Table 5). Obviously, one has to be aware that not all glycoforms present were identified from either mixture, but the most abundant forms were detected for each site, and as samples were LWAC enriched and analyzed in the same way, glycopeptide biases due to enrichment and acquisition should be the same for each dataset. We were able to compare the site-specific N-glycosylation in brain and liver for 50 glycosylation sites within 34 proteins (Supplemental Table 6).
A few general differences in sugar composition were observed. Whereas the brain glycans almost exclusively used NeuAc, most sialic acids in the liver were NeuGc. Many acetylated sialic acids were observed in liver glycopeptides, whereas none were detected in the brain sample. A final general difference was that with the exception of Cathepsin D, no fucosylated high mannose glycans were observed in the liver, whereas many were observed in the brain (fucosylated paucimannose and complex glycans were observed in both tissues).
Numerous truncated structures, starting with the core GlcNAc either alone or fucosylated were observed in glycopeptides isolated from both sources. Some glycosites consistently only featured these short glycans; for example, Asn-192 of Cathepsin B or Asn-69 of Galectin-3-binding protein. There were a few examples when a series of identical or very similar glycoforms were detected in both samples. For example, Asn-236 of Multiple inositol polyphosphate phosphatase 1 was detected either bearing a Man6 glycan or a GlcNAc4Man3Fuc agalacto biantennary structure, or Asn-47 of reticulocalbin-1 displayed a similar set of oligomannose and complex glycans in each sample. In general, little difference was observed for oligomannose-bearing sites, but complex glycans clearly showed tissue-specific differences, the most striking being on transmembrane proteins. Brain glycopeptides usually featured more glycoforms, including large branched, antenna-fucosylated structures. In contrast, the detected complex liver glycans were mostly biantennary complex oligosaccharides, with or without core fucosylation, and the capping residues were mostly N-glycolylneuraminic acids, which were also detected O-acetylated in a few instances.
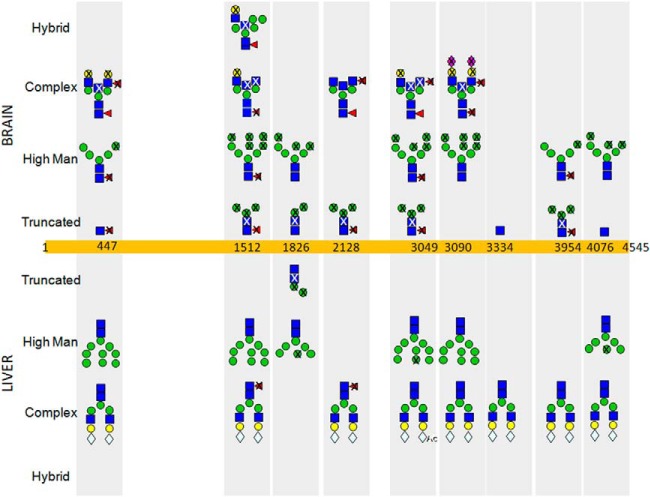
In some cases, tissue-specific differences were only observed for a subset of glycosylation sites within the same protein. The glycosylation of nine N-linked glycosylation sites in Prolow-density lipoprotein receptor-related protein 1 were observed in both samples, and these results are summarized in Fig. 3. In both tissues, Asn-1826 was only observed with oligomannose structures without fucosylation. Asn-3954 also only bore oligomannose structures in the brain (although in this case core fucosylation was also observed). However, in the liver, only the dominant disialo biantennary complex glycan was found attached to this site. Asn-447, -1512, -3049, and -3090 displayed a mixture of oligomannose and complex glycans in both tissues. However, far greater heterogeneity in structures was observed in the brain, both in terms of sugar length and fucosylation patterns; e.g. 19 glycoforms were detected on Asn-3090 in the brain, whereas only two were identified in the liver: Man9 and the common disialo biantennary structure.
Fig. 3.
Glycoforms detected in mouse brain and liver for 9 N-glycosylation sites of Prolow-density lipoprotein receptor-related protein 1. The nomenclature proposed by the Functional Glycomics Consortium is used. “X”s through residues indicate that the glycan was also detected lacking that particular building block. From the mass spectrometry data, the linkages usually cannot be determined, only the composition of the oligosaccharide.
O-glycosylation
The number of O-glycosites observed was much lower: 85 glycopeptides representing 26 proteins (Supplemental Table 3), of which five were also detected N-glycosylated. Site assignment represents a bigger challenge in O-glycosylation than in N-glycosylation. All site localizations in these results were automatically assessed for reliability using the built-in SLIP (site localization in peptide) scoring in Protein Prospector (31). Results with SLIP scores below 6 (95% confidence of correct site localization) are reported as ambiguous with all potential site localizations in supplemental Table 3. However, for the glycopeptides discussed in detail below, the ETD data were inspected manually, and the site assignments are reported accordingly. The presented list contains 36 confidently assigned O-glycosylation sites. In contrast to N-glycosylation, we identified nine doubly O-glycosylated peptides. Some feature different glycans on each site; others were identified in a follow-up search where for the already identified O-glycoproteins the occurrences of two identical sugar compositions within the same glycopeptide were permitted.
Thirteen O-glycosylation sites (or regions) in seven glycoproteins were detected in both the synaptosome and liver samples (Supplemental Table 7). The majority of the structures are sialylated mucin-type core 1 glycans, with the liver displaying a higher variety of these due to the presence of sialic acid variants. Interestingly, if the glycan contained NeuGc, the HCD data indicated in most cases that it was attached to the core GalNAc. The brain glycopeptide data were revisited in order to investigate whether any O-glycans featuring sialic acid variants were detected. NeuGc and NeuAc2 were identified once each. The former was detected in a tetrasaccharide on a Glypican 4 peptide that featured the same structure in the liver sample (Supplemental Table 7). There may be some difference in the frequency of O-glycopeptides with structures other than mucin-type core 1. A GlcNAcGalNAc, which could originate from mucin-type core-2 or core-3 oligosaccharides, was detected at four glycosites in the brain sample but was only observed once in the liver dataset.
To illustrate the heterogeneity one may encounter when characterizing O-glycopeptides, we discuss the results from one protein further. Nucleobindin-1 is a glycoprotein residing in the Golgi, to which three O-glycosylation sites were identified in liver samples: Thr-41, Ser-406, and Ser-410; the latter was only observed glycosylated in doubly modified sequences. The same residues were reported glycosylated in the synaptosome study. Thr-41 displayed mono or disialo mucin-type core 1 structures in the brain, but in the liver, the sialic acid variants employed produced a much bigger assortment of structures, partly due to presence of acetylated NeuGc and NeuAc2: residues that were only observed in the liver. We reported recently that Thr-41 was detected with eight different glycans, and relying on diagnostic beam-type CID fragments, one could distinguish the isomeric Neu(Ac)2Gal(NeuGc)GalNAc and NeuAcGal(NeuGc,Ac)GalNAc structures (27). In the brain samples, Ser-406 was detected with the mucin-type tri- and tetrasaccharides and also observed displaying just the core GalNAc. In the liver, four additional glycoforms were observed, containing NeuGc as well as O-acetyl sialic acid variants (Supplemental Table 3). The glycan distributions for the two sites were dramatically different (Supplemental Figs. 4A and 4B), with trisaccharide structures more prominent on Ser-406, whereas NeuGc-containing structures were more abundant on Thr-41. Sequences containing Ser-406 were also detected doubly modified with identical glycans (GalNAc or the tetrasaccharide) on both Ser-406 and -410.
DISCUSSION
It has been reported that wheat germ agglutinin preferentially binds GlcNAc or neuraminic acid (32). Taking advantage of this binding specificity, LWAC chromatography with WGA has been used for the enrichment of GlcNAcylated peptides in several studies (33–35). However, in a more recent study, the wider binding specificity of WGA has been documented, attesting that the presence of nonreducing end GlcNAc or neuraminic acid is not required (36). Thus, while WGA is efficient for the enrichment of GlcNAcylated sequences, it also enriches glycopeptides carrying glycans added in the ER and the Golgi, as demonstrated by our previous LWAC study of mouse brain (25).
We utilized the same LWAC enrichment approach in this study and gathered data about the N- and O-glycosylation of liver proteins at the glycopeptide level. Intact glycopeptide analysis offers advantages over analysis of released glycans, primarily the ability to obtain protein- and site-specific glycosylation information. For example, there is growing evidence for the occurrence of nonconsensus motif glycosylation (25, 37–40), as observed on apolipoprotein E in this study. This site was previously identified as formerly glycosylated in a large-scale proteomic study after PNGase F digestion (39).
Other groups have surveyed the glycan pool in mouse liver, including a recent study that analyzed released glycans from FFPE (formalin fixed paraffin embedded) and frozen tissue slices (18). Comparing their results to the histogram of modifications observed in the present study (Fig. 1), both datasets feature biantennary disialo complex glycans and Man9 oligosaccharides as abundant, but the LWAC results display much higher abundance of shorter oligomannose structures, and the released glycan results completely missed all of the truncated glycans. There are probably multiple factors contributing to these differences. The results from studying the glycan pool bear the strongest similarity to the glycan distribution we observed for transmembrane proteins, which is consistent with the fact that in the Turiak-study protein digestions were performed on the surface of intact tissue prior to N-glycan release (18). Hence, one would expect the results to be biased toward cell surface glycans. Secondly, there are questions over how efficiently PNGase F can digest paucimannose and even more truncated sugar structures, and it has been reported that it will not remove the core GlcNAc from the modified peptide chain (41). Finally, even if some of the small sugar structures were released, the sample clean-up and mass spectrometry analysis methods were optimized for larger glycans. We report the presence of trisialo biantennary complex glycans and NeuGc,Ac-containing structures. These oligosaccharides were mostly observed on secreted proteins, which could explain their absence in the glycan-pool study. It should also be mentioned that the efficiency of ETD is dependent on the charge-density of the precursor ion. Thus, the method is biased against larger and negatively charged glycoforms (25), offering an explanation for the lack of triantennary or bigger structures among the glycopeptide results.
Our results illustrate the importance of the acquisition of both CID/HCD and ETD data in glycopeptide research. ETD data alone may not provide sufficient information for glycan composition assignments. The difference between a fucose and a hexose or NeuAc and NeuGc is a single oxygen, and covalent modifications of some of the building blocks, such as O-acetylation of sialic acids, also should be considered. Thus, unfortunately, different combinations of sugar units may yield the same mass. In the present study, the mass increment of 2,278 Da could have corresponded to three different oligosaccharide structures: GlcNAc2Man3(GlcNAcGal)3NeuAc, a monosialo triantennary complex glycan; FucGlcNAc2Man3GlcNAc(GlcNAcGal)2NeuGc, a core fucosylated bisecting biantennary glycan with a single NeuGc; or GlcNAc2Man3 (GlcNAcGal)2NeuGc,NeuGc,Ac, a biantennary complex glycan capped with a NeuGc and an O-acetyl NeuGc. The characteristic oxonium ions in the HCD spectra (27) showed the last structure to be the correct assignment in the glycopeptides listed. Side-reactions modifying the peptide also may interfere: Met, Trp, and Cys residues readily undergo oxidation. Considering this and the frequently limited peptide backbone fragmentation in ETD experiments, CID/HCD data may provide the decisive information, revealing whether the peptide bore the additional oxygen or it was present in the glycan. Supplemental Table 2 lists Met-containing peptides from Collagen alpha-2(VI) chain, integrin beta-2, and GPI transamidase component PIG-T where the assignments made use of supporting HCD data.
A few years ago, we published a study on the site-specific N-and O-glycosylation of proteins found in the murine synaptosome (25). In the present study, we compared these results with our glycosylation findings in mouse liver. Tissue-specific N-glycosylation data at the glycan level have been reported in great detail for rat brain (16, 17) and less comprehensively for mouse liver (18). However, as far as we know, this is the first time that tissue-specific glycosylation has been compared between these tissues, at a protein- as well as site-specific manner, by reporting the analysis of intact glycopeptides. Our results revealed some expected differences, namely the replacement of N-acetyl neuraminic acids with N-glycolyl derivatives in practically all N-linked structures. This is a well-documented difference, with brain known to use NeuGc sparingly (42, 43). O-linked glycans also showed the same shift in sialic acid use as the N-linked glycans but not completely. In the O-linked glycopeptides identified, the sialic acid modifying the Gal in the mucin-type core 1 structure was always NeuAc, while that linked to the GalNAc in ∼40% of the cases was NeuGc. This is consistent with previous studies that reported lower levels of 2–3 linked NeuGc attached to Gal than 2–6 linked to GalNAc in mouse submaxillary mucins (44). We detected O-acetyl sialic acids in both the N- and O-linked glycans in the liver samples but not in the brain. Previous studies indicated a higher level of Neu5,9Ac2 in the liver than in the brain: ∼1.5% of the protein-bound sialic acids versus 0.7% (43). We have not found any previous reports about O-acetyl NeuGc distribution.
In summary, our data reveal protein- and site-specific similarities and striking differences in both N- and O-glycosylation when comparing mouse brain and liver tissues. These observations emphasize that glycosylation must be studied at the glycopeptide level in order to further our understanding of its biological role.
Supplementary Material
Footnotes
Author contributions: K.K. and R.J.C. designed the research; K.K. performed the research; K.F.M. and R.J.C. analyzed data; and K.F.M., K.K., and R.J.C. wrote the paper.
* This work was supported by NIH grant NIGMS 8P41GM103481 and by the Howard Hughes Medical Institute (to the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF, Director: A.L. Burlingame).
 This article contains supplemental material Supplemental Tables 1–7 and Supplemental Figs. 1–4.
This article contains supplemental material Supplemental Tables 1–7 and Supplemental Figs. 1–4.
1 The abbreviations used are:
- CID
- collision-induced dissociation
- ER
- endoplasmic reticulum
- ETD
- electron transfer dissociation
- HCD
- higher energy C-trap dissociation
- LWAC
- Lectin weak affinity chromatography
- WGA
- wheat germ agglutinin.
REFERENCES
- 1. Hart G. W. (2014) Three decades of research on O-GlcNAcylation - A major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front Endocrinol. 5, 183 doi: 10.3389/fendo.2014.00183. eCollection 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Varki A, Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds. (2009) Essentials of Glycobiology. 2nd edition Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 3. Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dwek R. A., Edge C. J., Harvey D. J., Wormald M. R., Parekh R. B. (1993) Analysis of glycoprotein-associated oligosaccharides. Annu. Rev. Biochem. 62, 65–100 [DOI] [PubMed] [Google Scholar]
- 5. Tharmalingam T., Adamczyk B., Doherty M. A., Royle L., Rudd P. M. (2013) Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides. Glycoconj. J. 30, 137–146 [DOI] [PubMed] [Google Scholar]
- 6. Alley W. R., Jr., Mann B. F., Novotny M. V. (2013) High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem. Rev. 113, 2668–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kroslak T., Laforge K. S., Gianotti R. J., Ho A., Nielsen D. A., Kreek M. J. (2007) The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J. Neurochem. 103, 77–87 [DOI] [PubMed] [Google Scholar]
- 8. Springer S. A., Gagneux P. (2013) Glycan evolution in response to collaboration, conflict, and constraint. J. Biol. Chem. 288, 6904–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venkatakrishnan V., Packer N. H., Thaysen-Andersen M. (2013) Host mucin glycosylation plays a role in bacterial adhesion in lungs of individuals with cystic fibrosis. Expert Rev. Respir. Med. 7, 553–576 [DOI] [PubMed] [Google Scholar]
- 10. Srinivasan S., Romagnoli M., Bohm A., Sonenshein G. E. (2014) N-glycosylation regulates ADAM8 processing and activation. J. Biol. Chem. 289, 33676–33688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott H., Panin V. M. (2014) The role of protein N-glycosylation in neural transmission. Glycobiology 24, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bochner B. S., Zimmermann N. (2015) Role of siglecs and related glycan-binding proteins in immune responses and immunoregulation. J. Allergy Clin. Immunol. 135, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glavey S. V., Huynh D., Reagan M. R., Manier S., Moschetta M., Kawano Y., Roccaro A. M., Ghobrial I. M., Joshi L., O'Dwyer M. E. (2015) The cancer glycome: Carbohydrates as mediators of metastasis. Blood Rev. (in press) [DOI] [PubMed] [Google Scholar]
- 14. Méflah K., Bernard S., Massoulié J. (1984) Interactions with lectins indicate differences in the carbohydrate composition of the membrane-bound enzymes acetylcholinesterase and 5′-nucleotidase in different cell types. Biochimie 66, 59–69 [DOI] [PubMed] [Google Scholar]
- 15. Parekh R. B., Tse A. G., Dwek R. A., Williams A. F., Rademacher T. W. (1987) Tissue-specific N-glycosylation, site-specific oligosaccharide patterns and lentil lectin recognition of rat Thy-1. EMBO J. 6, 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y. J., Wing D. R., Guile G. R., Dwek R. A., Harvey D. J., Zamze S. (1998) Neutral N-glycans in adult rat brain tissue—Complete characterisation reveals fucosylated hybrid and complex structures. Eur. J. Biochem. 251, 691–703 [DOI] [PubMed] [Google Scholar]
- 17. Zamze S., Harvey D. J., Chen Y. J., Guile G. R., Dwek R. A., Wing D. R. (1998) Sialylated N-glycans in adult rat brain tissue—A widespread distribution of disialylated antennae in complex and hybrid structures. Eur. J. Biochem. 258, 243–270 [DOI] [PubMed] [Google Scholar]
- 18. Turiák L., Shao C., Meng L., Khatri K., Leymarie N., Wang Q., Pantazopoulos H., Leon D.R., Zaia J. (2014) Workflow for combined proteomics and glycomics profiling from histological tissues. Anal. Chem. 86, 9670–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruhaak L. R., Uh H. W., Deelder A. M., Dolhain R. E., Wuhrer M. (2014) Total plasma N-glycome changes during pregnancy. J. Proteome Res. 13, 1657–1668 [DOI] [PubMed] [Google Scholar]
- 20. Gustafsson O. J., Briggs M. T., Condina M. R., Winderbaum L. J., Pelzing M., McColl S. R., Everest-Dass A. V., Packer N. H., Hoffmann P. (2015) MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney. Anal. Bioanal. Chem. 407, 2127–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dall'Olio F., Malagolini N., Trinchera M., Chiricolo M. (2014) Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 1840, 2752–2764 [DOI] [PubMed] [Google Scholar]
- 22. Maverakis E., Kim K., Shimoda M., Gershwin M. E., Patel F., Wilken R., Raychaudhuri S., Ruhaak L. R., Lebrilla C. B. (2015) Glycans in the immune system and the altered glycan rheory of autoimmunity: A critical review. J. Autoimmun. 57, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee L. Y., Lin C. H., Fanayan S., Packer N. H., Thaysen-Andersen M. (2014) Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thaysen-Andersen M., Packer N. H. (2012) Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 25. Trinidad J. C., Schoepfer R., Burlingame A. L., Medzihradszky K. F. (2013) N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics. 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. West M. B., Segu Z. M., Feasley C. L., Kang P., Klouckova I., Li C., Novotny M. V., West C. M., Mechref Y., Hanigan M. H. (2010) Analysis of site-specific glycosylation of renal and hepatic γ-glutamyl transpeptidase from normal human tissue. J. Biol. Chem. 285, 29511–29124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Medzihradszky K. F., Kaasik K., Chalkley R. J. (2015) Characterizing sialic Acid variants at the glycopeptide level. Anal. Chem. 87, 3064–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akashi M., Okamoto A., Tsuchiya Y., Todo T., Nishida E., Node K. (2014) A positive role for PERIOD in mammalian circadian gene expression. Cell Rep. 7, 1056–1064 [DOI] [PubMed] [Google Scholar]
- 29. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics. 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baker P. R., Chalkley R. J. (2014) MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics. 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baker P. R., Trinidad J. C., Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics. 10, M111.008078 doi: 10.1074/mcp.M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gallagher J. T., Morris A., Dexter T. M. (1985) Identification of two binding sites for wheat-germ agglutinin on polylactosamine-type oligosaccharides. Biochem. J. 231, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vosseller K., Trinidad J. C., Chalkley R. J., Specht. C. G., Thalhammer. A., Lynn A. J., Snedecor J. ., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 34. Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. . (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Myers S. A., Panning B., Burlingame A. L. (2011) Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9490–9495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang C. F., Pan J. F., Lin C. N., Wu I. L., Wong C. H., Lin C. H. (2011) Rapid characterization of sugar-binding specificity by in-solution proximity binding with photosensitizers. Glycobiology 21, 895–902 [DOI] [PubMed] [Google Scholar]
- 37. Valliere-Douglass J. F., Eakin C. M., Wallace A., Ketchem R. R., Wang W., Treuheit M. J., Balland A. (2010) Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 285, 16012–16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang H., Huang R. Y., Jalili P. R., Irungu J. W., Nicol G. R., Ray K. B., Rohrs H. W., Gross M. L. (2010) Improved mass spectrometric characterization of protein glycosylation reveals unusual glycosylation of maize-derived bovine trypsin. Anal. Chem. 82, 10095–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 40. Chandler K. B., Brnakova Z., Sanda M., Wang S., Stalnaker S. H., Bridger R., Zhao P., Wells L., Edwards N. J., Goldman R. (2014) Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J. Proteome Res. 13, 3314–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu F. K. (1986) Requirements of cleavage of high mannose oligosaccharides in glycoproteins by peptide N-glycosidase F. J. Biol. Chem. 261, 172–177 [PubMed] [Google Scholar]
- 42. Schauer R., Stoll S., Reuter G. (1991) Differences in the amount of N-acetyl- and N-glycoloyl-neuraminic acid, as well as O-acylated sialic acids, of fetal and adult bovine tissues. Carbohydr. Res. 213, 353–359 [DOI] [PubMed] [Google Scholar]
- 43. Rinninger A., Richet C., Pons A., Kohla G., Schauer R., Bauer H. C., Zanetta J. P., Vlasak R. (2006) Localisation and distribution of O-acetylated N-acetylneuraminic acids, the endogenous substrates of the hemagglutinin-esterases of murine coronaviruses, in mouse tissue. Glycoconj. J. 23, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savage A. V., Koppen P. L., Schiphorst W. E., Trippelvitz L. A., Van Halbeek H., Vliegenthart J. F., Van den Eijnden D. H. (1986) Porcine submaxillary mucin contains alpha 2-3- and alpha 2-6-linked N-acetyl- and N-glycolylneuraminic acid. Eur J Biochem. 160, 123–129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.