Abstract
Genetically susceptible bacteria become antibiotic tolerant during chronic infections, and the mechanisms responsible are poorly understood. One factor that may contribute to differential sensitivity in vitro and in vivo is differences in the time-dependent tobramycin concentration profile experienced by the bacteria. Here, we examine the proteome response induced by subinhibitory concentrations of tobramycin in Pseudomonas aeruginosa cells grown under planktonic conditions. These efforts revealed increased levels of heat shock proteins and proteases were present at higher dosage treatments (0.5 and 1 μg/ml), while less dramatic at 0.1 μg/ml dosage. In contrast, many metabolic enzymes were significantly induced by lower dosages (0.1 and 0.5 μg/ml) but not at 1 μg/ml dosage. Time course proteome analysis further revealed that the increase of heat shock proteins and proteases was most rapid from 15 min to 60 min, and the increased levels sustained till 6 h (last time point tested). Heat shock protein IbpA exhibited the greatest induction by tobramycin, up to 90-fold. Nevertheless, deletion of ibpA did not enhance sensitivity to tobramycin. It seemed possible that the absence of sensitization could be due to redundant functioning of IbpA with other proteins that protect cells from tobramycin. Indeed, inactivation of two heat shock chaperones/proteases in addition to ibpA in double mutants (ibpA/clpB, ibpA/PA0779 and ibpA/hslV) did increase tobramycin sensitivity. Collectively, these results demonstrate the time- and concentration-dependent nature of the P. aeruginosa proteome response to tobramycin and that proteome modulation and protein redundancy are protective mechanisms to help bacteria resist antibiotic treatments.
The opportunistic pathogen Pseudomonas aeruginosa is ubiquitous in the natural environment and causes human infections (1). P. aeruginosa can metabolize various carbon and nitrogen compounds and persists under nutrient-poor and hostile growth environments (2, 3). One example is P. aeruginosa pulmonary infection of cystic fibrosis (CF) patients. Despite stress induced by host defenses and high concentrations of antibiotics, P. aeruginosa cells are able to persistently colonize CF airways (4).
The aminoglycoside tobramycin is a front-line drug currently used in the treatment of P. aeruginosa in CF and other diseases. It is supplied in the forms of inhaled solution (TOBI) and intravenous injection. The tobramycin concentrations in airways after 300-mg dosage TOBI inhalation can reach 1,000 μg per g of sputum (5, 6). This concentration is in the range of 10 to 1,000 times of the minimal inhibitory concentration (MIC) for P. aeruginosa clinical isolates tested ex vivo (6). However, even with such high tobramycin concentrations, chronic P. aeruginosa infections are rarely eradicated (6). This is true even when the infecting bacteria are antibiotic sensitive, as is the case early in disease (7).
One possible reason for P. aeruginosa persistence in vivo could relate to the time dependence of local concentrations of tobramycin experienced by P. aeruginosa in CF patient airways. Many factors, including inflammatory responses, blood and lymphatic circulations, and air flow distribution (for inhaled antibiotics), can alter the local antibiotic concentrations. In addition, P. aeruginosa cells can form biofilms in CF lungs and other infection sites (8), and biofilm exopolysaccharide layers may slow the diffusion of tobramycin (9, 10). P. aeruginosa cells in the inner layers of biofilms may experience lower concentrations and more gradual increase of tobramycin levels than those in outer layers (10, 11). Furthermore, even if final tobramycin concentration levels inside the biofilm eventually grow to match the highest levels experienced elsewhere, bacteria in these inner regions have experienced a slower increase, during which time proteome levels could be altered to promote the “adapted resistant state” (12). Adaptive resistance can also be induced in planktonic (free-living) P. aeruginosa (13, 14), and conventional MIC assays are not designed to measure this.
Once induced, the adaptive resistance confers bacteria higher resistance to antibiotic treatments (13, 14) and is associated with decreased clinical antibiotic treatment efficacy (15). Interestingly, the adaptive resistance is time dependent and reversible. Typical adaptive resistance was observed starting 1 h after antibiotic exposure, and the drug susceptibility was regained after 36 h intervals (14, 15). Thus, adaptive resistance mechanisms may contribute in part to the disparity of in vivo persistence and ex vivo susceptibility to antibiotics in MIC tests.
As an initial step toward defining adaptive resistance mechanisms, we investigated the time- and concentration-dependence of P. aeruginosa proteome response to tobramycin in planktonic conditions. Since the most effective protective responses may operate before killing begins and the rate of change of drug levels is likely to depend on ambient conditions, we studied bacteria exposed to low, subinhibitory levels of tobramycin (0.1, 0.5, and 1.0 μg/ml) at a range of time points (15, 60, 120, and 360 min) after exposure. The candidate proteome marker of P. aeruginosa for tobramycin response, heat shock protein IbpA, was further investigated with genetic mutagenesis and MIC assays.
EXPERIMENTAL PROCEDURES
P. aeruginosa Strains
P. aeruginosa strain MPAO1 (16) was used for proteome analysis. Single transposon insertion mutants were obtained from the two-allele mutant library (17). The specific mutants examined were as follows: PA0779 (PW2411, PW2413, PW2414), clpB (PW8651, PW8652), hslV (PW9485, PW9486, PW9487), and the transposon-containing control PA3303 (18).
Two ibpA loss-of-function mutant alleles were used in this study. The first ibpA::luxCDABE was constructed in two recombination steps by replacing wild-type ibpA with an ibpA-lux gene fusion carried on plasmid pEX19Tc (19). The lux insertion site was at the N-terminal fifth amino acid of ibpA. The structure of the constructed allele was confirmed by PCR, and the absence of IbpA expression was confirmed by the selected reaction monitoring (SRM)1. The lux control strain PKH181 was constructed by insertion of pUC18-mini-Tn7T-Gm-lux vector (without a cloned promoter) into MPAO1, according to methods of Choi and Schweizer (20). The second ibpA mutant (ΔibpA) corresponded to an in-frame deletion that joined sequences coding for the N-terminal fifth amino acid to the fourth amino acid from the C terminus. The deletion was generated by recombination in MPAO1 with pEX19Tc plasmid carrying the deletion allele. The in-frame ΔftsH deletion mutant has been previously described (21).
To generate the double mutants (ibpA/PA0779, ibpA/clpB, and ibpA/hslV), chromosomal DNA isolated from the transposon-containing strains was transformed into ibpA-lux insertion inactivation mutant via lambda red recombination. Positive transformants were selected by tetracycline resistance as previously described (22).
Tobramycin Treatment for Proteomics Samples
MPAO1 cells were grown in salt-free LB containing 10 g tryptone, 5 g yeast extract per liter at 37 °C to optical density OD600 = 1.0, and aliquotted to 5 ml volumes. For the dosage-dependent treatments, tobramycin (Sigma-Aldrich, St. Louis, MO) at 0.1, 0.5, or 1.0 μg/ml (final concentration) was added to the cell cultures with shaking for 60 min at 37 °C. For the time course treatments, cells were treated with 1.0 μg/ml tobramycin (final concentration) were exposed for 15, 60, 120, and 360 min at 37 °C. Cells harvested at time zero (at the time that tobramycin treatment started) were used as controls. Three biological replicates for dosage treatments and two biological replicates for the time course were analyzed.
Tobramycin MIC Assays
Minimum inhibitory concentration (MIC) assays were performed as described by Lee et al. (18). Single colonies were inoculated into 96-well plates containing 250 μl salt-free LB per well and grown for 18 h at 37 °C in a humidity chamber. 200 μl of these cultures were removed, added to a new plate, and 10-fold dilutions were made with salt-free LB. The salt was not added in LB for initial liquid culture because Δftsh mutant is salt sensitive (21), and salt-free conditions resulted in relatively uniform growth for strains. Diluted cells were allowed to grow for 90 min at 37 °C in a humidity chamber, at which point cells were spotted to tobramycin LB agar (including 137 mm NaCl) to deposit approximate 105 cells per spot. Plates were incubated at 37 °C for 18 h and photographed. MIC was defined as the lowest antibiotic concentration preventing the lawn growth of the spotted cells.
Protein Extraction of P. aeruginosa Cells
MPAO1 cells were harvested by centrifugation at 3,500 rpm for 10 min. Cell pellets were resuspended with lysis buffer (50 mm ammonium bicarbonate, pH 8.0; 8 m urea; and 10 mm DTT). Cells were lysed by incubation in lysis buffer on ice for 30 min, with vortexing for 30 s every 5 min. Cells in lysis buffer underwent one −80 °C freeze-thaw cycle to maximize cells lysis. Protein extracts were briefly sonicated to sheer DNA. Cell debris was removed by centrifugation at 20,000 g for 20 min. Extracted protein concentration was measured with Bradford assay (Thermo Fisher Scientific, Waltham, MA).
100 μg proteins from each extraction were diluted twofold with digestion buffer (40 mm ammonium bicarbonate, 5% acetonitrile) for alkylation by 10 mm iodoacetamide for 30 min in the dark. The mixture then was further diluted to urea concentration less than 1.5 m with digestion buffer for overnight trypsin digestion (Promega, Madison, WI). Peptides were purified with Sep-Pak cartridges (Waters Corporation, Milford, MA), dried, and resuspended with 0.1% formic acid. 1 μg of tryptic digest was loaded in each LC-MS/MS analysis.
LC-MS/MS Analysis
The proteomics samples were analyzed with NanoAcquity UPLC (Waters) coupled to a linear quadrupole ion trap mass spectrometer (LTQ-XL from Thermo Fisher Scientific). The column length of 3 cm trap column packed in house with 200 Å C18 magic beads (Bruker-Michrom Inc, Auburn, CA) and 30 cm analytical column packed with 100 Å C18 magic beads were used in all LC/MS-MS analysis to maintain the uniformity of peptide chromatography profiles. A 120 min linear gradient (5–35% acetonitrile) at a flow rate of 300 nl/min was used, and peptides were ionized by electrospray.
Data-dependent acquisition (DDA) mode was used in the spectral counting analysis. The top five most intense ions in the MS1 scan were selected for tandem mass spectrometry analysis.
For the selected reaction monitoring (SRM) analysis, scan-type SRM was used. Targeted peptides and transition ions were manually selected from DDA identification spectra. Ion intensity and preferentially larger fragment masses were the main selection criteria. Five to eight fragment ions were included per peptide. SRM instrument settings for this study were standardized, which included parent ions isolation width 3 m/z, transition ions scan width of 1 m/z, collision-induced disassociation (CID) collision energy 35, activation Q 0.25, activation time 30 ms, and automatic gain control (AGC) MSn value 1 × 104, maximum ion time 500 ms.
Data Analysis
The DDA raw files were converted to mgf peak list files using ReAdW (Ver. 4.3.1) (23). The data were searched in Mascot (version 2.3.02) against the Pseudomonas aeruginosa (PAO1) database (consisting of 5,680 genes or splicing fragments, annotation version 2012-Nov) (24). The search parameters included 3.0 Da precursor mass tolerance, 0.6 Da fragment mass tolerance, fixed modification cysteine carbamidomethylation, variable modification methionine oxidation, digestion enzyme trypsin, and maximum missed cleavages 2. The peptide expectation value 0.05 was used as the cutoff, and the false discovery rate (FDR) was determined by searching the same mgf file in the randomized decoy database in Mascot. The FDR of the presented dataset was ∼1%.
The Exponentially Modified Protein Abundance Index (emPAI) values derived from Mascot search were used for spectral quantitation analysis (25). The data of a total of seven LC-MS/MS analyses for each dosage treatment and five LC-MS/MS analyses for each time course treatment were used for spectral counting statistical analysis.
We required proteins for quantitation analysis to be quantified minimally by three unique peptides from at least two biological replicates. Proteins with significant fold changes (P ≤ 0.05) as well as proteins with no detection in one state but detected more than four times in another state were considered statistically altered proteins and were further subject to gene ontology analysis.
The SRM data in this experiment were processed in Skyline (version 1.30) (26). Selection of chromatogram peaks for quantitation was manually verified. Peak smoothing method Savitzky–Golay was used. Chromatogram extracted peak areas for transition ions were summed up to yield total peak areas. The ratio of the total peak areas represented the abundance ratio of the peptides. Peptide ratios of each protein were further averaged to estimate the protein fold changes. The quantitation normalization was based upon equivalent total injection amount (1 μg) in each SRM run. A quantitation linear range of four orders of magnitude was shown (down to femtomole) (Fig. S1).
RESULTS
Evaluation of P. aeruginosa Proteome Quantitation Methods
Spectral Counting and Selected Reaction Monitoring (SRM)
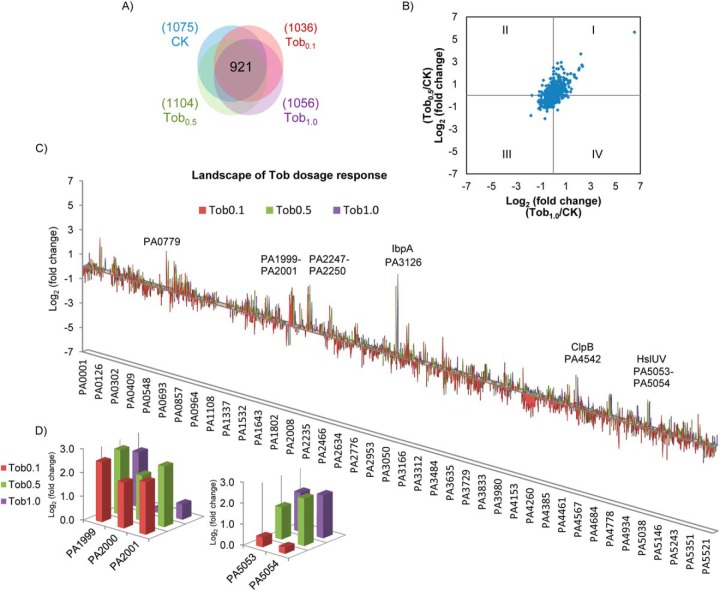
We first evaluated the reliability of our proteome quantitation methods. Spectral counting quantitation allows broad coverage of P. aeruginosa proteins and was used to identify potential protein targets. More than 1,000 P. aeruginosa proteins were examined in these assays (Fig. 1A). The Exponentially Modified Protein Abundance Index (emPAI) (25) values of 921 proteins commonly observed in all cell samples were used to calculate the fold change for each protein. The linear quantitation range of emPAI for the P. aeruginosa proteome was assessed with bovine serum albumin (BSA) spike-in assays, and a linear range over three orders of magnitude (down to 1:10,000 dilution w/w) was observed for BSA proteins (Fig. S1A). The detected coefficient of variation (CV) of emPAI for these proteins appeared protein-dependent as one might expect and scored a median CV of ∼40% (Fig. S2). The majority of protein emPAI ratios localized at the center of the scatter plot (log21 = 0), while the proteins with altered abundance appeared to be consistent between treatments, showing enrichment in Quadrant I and III of the Cartesian plane (Fig. 1B).
Fig. 1.
Landscape of P. aeruginosa proteome detection and quantitation. (A) Venn diagram showing over 1,000 P. aeruginosa proteins were quantified in each cellular state, including 921 common proteins quantified in all four dosages. Tobramycin dosage level is indicated with colors. CK: no tobramycin treatment (T0 collected cells); Tob0.1, Tob0.5, and Tob1.0: tobramycin treatment of 0.1, 0.5, and 1.0 μg/ml. (B) The distribution of log2 (fold change) of the 921common proteins. Comparison of Tob1.0/CK is shown at x axis, and Tob0.5/CK at y axis. (C) Landscape of protein abundance changes in P. aeruginosa during tobramycin dosage treatment. X axis: P. aeruginosa gene locus. Y axis: log2 protein fold change of Tob0.1/CK, Tob0.5/CK or Tob1.0/CK. The bar colors illustrate the concetration of Tobramycin in experiment and bar heights indicate the change in each protein level. The genes or operons with the most dramatic increase in protein abundance were IbpA, ClpB, PA0779, HslU-V, PA1999-PA2001, and PA2247–2250. (D) Inset of proteome landscape showing concentration dependent changes of PA1999-PA2001 and PA5053–5054.
Selected reaction monitoring (SRM) achieves high specificity and accurate quantitation and was used for validation of targets identified by spectral counting. The linear quantitation range of SRM was also accessed by BSA spike-in assays, a linear quantitation range of four orders magnitude was achieved for BSA peptides (Fig. S1B), which is consistent with other reports (27, 28).
To determine if the protein-level changes measured by spectral counting or SRM were consistent, quantitation data of a set of 10 P. aeruginosa proteins (including 49 SRM peptide assays) at different tobramycin treatment conditions measured with both methods were compared. A general correlation (R2 = 0.61) was observed (Fig. S3). Therefore, emPAI spectral counting and SRM provide consistent measurements for P. aeruginosa proteins and allow us to systematically evaluate the proteome response of P. aeruginosa to tobramycin antibiotic treatment.
Concentration- and Time-Dependent Proteome Changes of P. aeruginosa in Response to Tobramycin Treatment
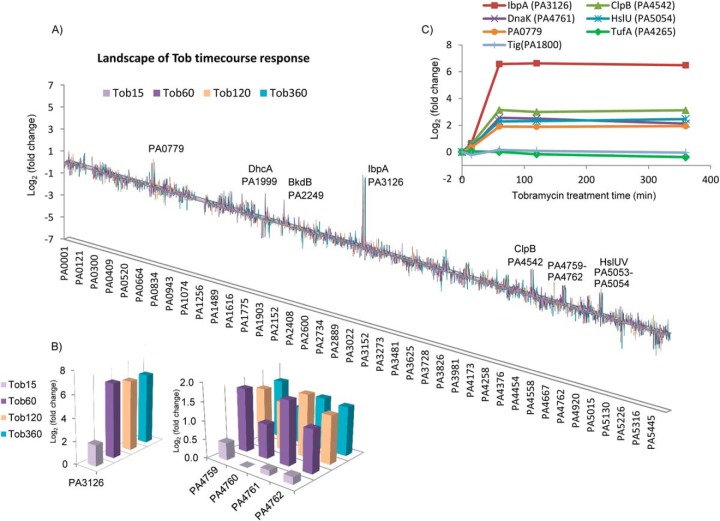
Since we are interested in the adaptive proteome response as a mechanism in protecting cells from antibiotic killing, P. aeruginosa cells grown under planktonic conditions were challenged with low and subinhibitory levels of tobramycin (0.1, 0.5, and 1.0 μg/ml) for periods of (15, 60, 120, and 360 min), and the proteome response was assessed. We constructed proteome landscape plots to visualize the global protein fold change as a function of gene locus and treatment dosage (Fig. 1C) or time (Fig. 2A). These plots show groups or clusters of genes that appear similarly increased or decreased with tobramycin treatment concentration or duration. Bar heights in the plot indicate the level of protein abundance change, and grouping of the bars implies the coregulation of protein changes among neighboring genes. The landscape readily distinguished heat shock protein IbpA (PA3126) as the highest fold change protein. It increased 50-fold at 0.5 μg/ml dosage and 90-fold at 1 μg/ml dosage and was observed with increased levels throughout the time course measurement from 60 min to 360 min. In addition to IbpA, the landscape also captured significant increases in PA0779, PA1999–2001 (DhcA-B, AtoB), PA2247–2250 (BkdA1-A2-B, LpdV), PA4542 (ClpB), PA4759–4762 (DapB, DnaJ-DnaK-GrpE), and PA5053–5054 (HslU-V) (Figs. 1C and 2A). Based on emPAI quantitation, many of these increases were fourfold or higher. Examination of their induction patterns revealed concentration- and time-dependent expression (Figs. 1D and 2B).
Fig. 2.
Time-dependent proteome response of P. aeruginosa to tobramycin. (A) Landscape of P. aeruginosa proteome response showing genes/operons that were most drastically changed in tobramycin time course treatment. X axis: P. aeruginosa gene locus. Y axis: log2 protein fold change of Tob15min/CK, Tob60min/CK, Tob120min/CK, and Tob360min/CK. Duration of tobramycin treatment is indicated with bar colors. PA0779, DhcA, BkdB, IbpA, ClpB, PA4759-PA4762, HslU-V were up-regulated, consistent with the results in the dosage treatment shown in Fig. 1C. (B) Inset of the proteome landscape showing time-dependent changes of IbpA and PA4759-PA4762. (C) Selected reaction monitoring (SRM) analysis for proteins that were dramatically increased in the tobramycin time course treatment. The SRM protein abundance ratio is shown, which represents the average of the peptide ratios from the protein. Interestingly, rapid increase of heat shock proteins and proteases occurred from 15 min to 60 min after tobramycin treatment, and the abundance level sustained till 360 min (last time point measured).
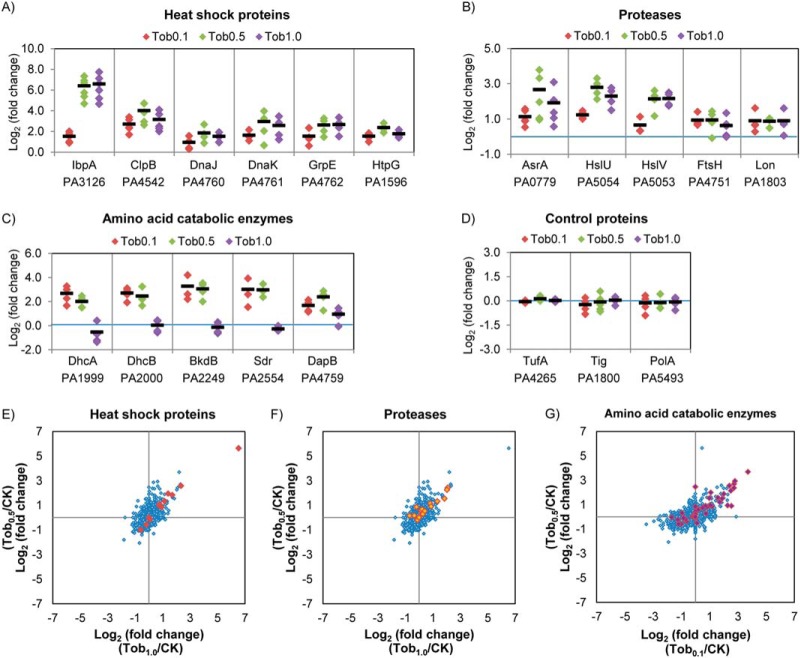
To validate the tobramycin dosage effects on the observed proteome induction, we used SRM to analyze the dosage-dependent levels of a number of these proteins (Fig. 3A–3C). The proteins can be categorized into three groups based on their biological functions, heat shock proteins, proteases, and amino acid catabolic enzymes (29). Heat shock proteins (IbpA, ClpB, DnaJ, DnaK, GrpE, and HtpG) were significantly increased by tobramycin at all three dosage levels, with the increases at the higher dosage levels (0.5 and 1.0 μg/ml) most marked. IbpA was again observed with largest fold change, with 84-fold induction at 0.5 μg/ml and 95-fold at 1.0 μg/ml tobramycin treatment, consistent with the quantitation from spectral counting. For the proteases (PA0779, HslU, and HslV), larger increases were also observed at the higher dosage treatment (0.5 and 1.0 μg/ml), while FtsH and PA1803 (Lon protease) exhibited similar increases at all three dosage levels. For the amino acid catabolic enzymes, only DapB was significantly increased at all three tobramycin concentrations, while DhcA, DhcB, BkdB, and Sdr were significantly increased only at 0.1 and 0.5 μg/ml treatments. For the control proteins (TufA, Tig, and PolA), no significant abundance level change was observed at any dosage treatment (Fig. 3D).
Fig. 3.
Confirmation of the up-regulation of heat shock proteins, proteases and amino acid catabolic enzymes in P. aeruginosa during tobramycin treatments with selected reaction monitoring (SRM). (A–D) SRM analysis confirmed heat shock proteins (IbpA, ClpB, DnaJ-DnaK-GrpE, and HtpG) and proteases (PA0779, HslU-V, FtsH, and PA1803) were significantly increased in tobramycin treatment of all three dosages, and the extent of increase was more dramatic at the higher dosages 0.5 and 1.0 μg/ml. Amino acid catabolic enzymes DhcA, DhcB, BkdB, and Sdr were significantly increased at 0.1 and 0.5 μg/ml dosages, and enzyme DapB was significantly increased in all three dosages. Control proteins TufA, Tig, and PolA did not show significant fold changes in tobramycin treatment. Three to five SRM peptides (each containing 5–8 transition ion quantitation) were used to quantify each protein. The diamond represents the measured fold change for each peptide. The black line indicates the fold change of the protein by averaging the fold change of all the SRM peptides. (E)-(G) Scatter plot showing the induction for heat shock proteins, proteases, and amino acid catabolic enzymes. The spectral counting data log2 (fold change) of Tob1.0/CK (x axis) and Tob0.5/CK (y axis) are shown in (E) and (F), and log2 (fold change) of Tob0.1/CK (x axis) and Tob0.5/CK (y axis) are shown in (G). The whole quantified proteome is provided in blue diamond as background. The heat shock proteins are highlighted in red, proteases in orange, amino acid catabolic enzymes in purple. Proteins in these three pathways contributed most of the largest fold changes in the proteome.
To interrogate the time dependence of inducible proteome changes, we used SRM to measure the tobramycin-induced proteins (IbpA, ClpB, DnaK, PA0779, and HslU) at the time points of 0, 15, 60, 120, and 360 min after 1.0 μg/ml treatment (Fig. 2C). Interestingly, for all five proteins, the rate of increase was most marked from 15 min to 60 min after tobramycin treatment and leveled off at 120 and 360 min. The temporal characteristics of IbpA increase resembles that seen after heat treatment (30). Again, the control proteins TufA and Tig did not significantly change.
We compared the proteome changes with published transcriptome results (18) in order to further comprehend the temporal regulation of proteome response. The transcriptome analysis by Lee et al. (18) was performed with similar tobramycin treatment conditions (1 μg/ml tobramycin 15 min treatment, OD600 = 1.0 MPAO1 cells) as in the present study so provided a useful reference fold change of RNAs. As shown in Fig. S4, 11 genes were found to be tobramycin induced in both transcriptome and proteome analyses, including seven of the genes that corresponded to proteins showing the greatest increase in proteome changes (Figs. 1C and 2A): PA0779, ibpA, clpB, dnaJ, grpE, hslU, and hslV.
Other genes that were observed to increase at the RNA level were not detected at the protein level, which may be due to low protein abundance levels, proteome sample collection, or regulatory changes that dissociate mRNA and protein expression. For genes identified as induced using both approaches, smaller changes were generally observed at the protein level than RNA level at the 15 min time point. This may reflect the longer time required to increase protein than RNA levels after transcriptional induction. However, at 60 min, the levels of protein fold changes became more pronounced. In particular, the increase of five genes (ibpA, clpB, hslU-V, and PA0779) was higher at the protein level than RNA level. The additional increase suggests the importance of protein-level regulation of tobramycin response in P. aeruginosa.
Functional Enrichment of Tobramycin-Induced Proteins in P. aeruginosa
To further define the proteome response induced by tobramycin, we used a one-way unpaired t test (also known as a one-way ANOVA test) to distinguish statistically significant changes of P. aeruginosa protein levels in different treatment states. In addition to the statistical cutoff (P value ≤ 0.05), proteins with significantly altered levels were also required to be repetitively quantified minimally by three unique peptides from at least two of the seven replicates in dosage treatments or two of the five replicates in time course treatments. Proteins that were found by spectral counting with statistically significant level changes but lower than twofold changes were further validated by other quantitation methods (Fig. S5).
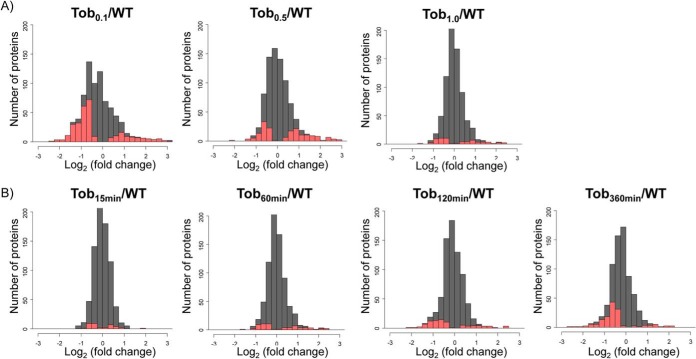
Using this approach, as many as 200 proteins appeared with statistically significant altered levels in different tobramycin treatment states. Consistent with SRM quantitation that showed tobramycin dosage- and time-dependent effects on individual protein level changes (Figs. 2C and 3A, 3B, and 3C), the global proteome induction also showed systematic tobramycin dosage and duration effects (Fig. 4). For instance, a higher number of proteins (and higher percentage of proteome) was observed with significant altered levels with 0.1 μg/ml tobramycin compared with the higher dosage 1.0 μg/ml, while 0.5 μg/ml treatment exhibited intermediate changes (Fig. 4A). Extending 1.0 μg/m tobramycin treatment to 360 min also gradually increased the number of proteins that were observed with statistically significant decreased levels (Fig. 4B).
Fig. 4.
Global protein fold change showing the dosage- and time-dependent effects of tobramycin treatments. The log2 protein fold change by spectral counting was shown. The statistically altered proteins (P ≤ 0.05) are highlighted in red, and the entire quantified proteome is shown in gray. (A) Dosage-dependent effects. The lower dosage tobramycin treatment 0.1 and 0.5 μg/ml for 60 min induced wider proteome changes compared with 1.0 μg/ml. (B) Time-dependent effects. Extending 1.0 μg/ml tobramycin treatment to 360 min gradually increased the number of proteins that were statistically significantly altered.
Interestingly, examination of the top 30 proteins with greatest fold changes from the three tobramycin dosage treatments indicated that very few proteins were common among the three dosage levels, and the proteins that were common in two dosage levels were more likely to be shared between more similar doses (i.e. 0.1 and 0.5 μg/ml or 0.5 and 1.0 μg/ml) (Fig. 5A). These results suggest that bacterial responses become less similar as the differences in the tobramycin dose increase. Because significant proteome induction was observed at the very low dosage levels (0.1 μg/ml, 214 nm, 1/10 MIC), it also illustrates that P. aeruginosa cells can sense a very low amount of tobramycin molecules, supporting the notion that cells first exposed to lower-level tobramycin treatment could yield acclimation phenotypes.
Fig. 5.
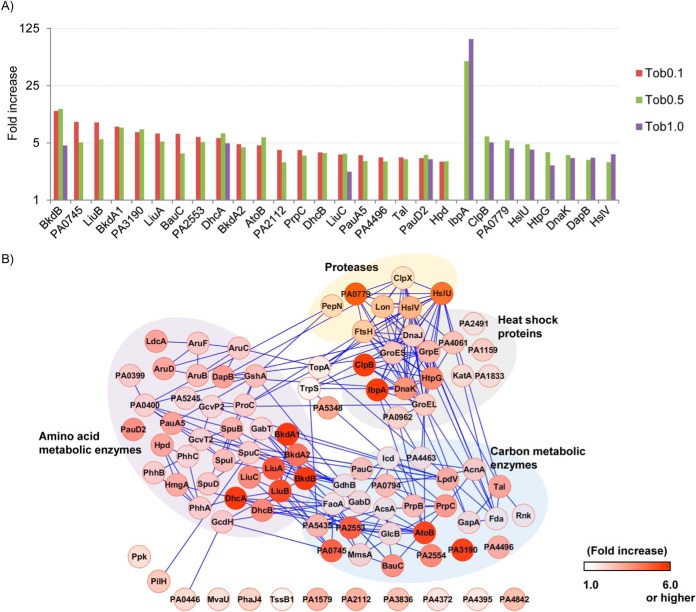
Dosage-dependent induction of proteome subsets and quantitative proteome interaction network. (A) The top 30 proteins with greatest fold changes from the three tobramycin dosage treatments were compared. The emPAI fold changes of those common in at least two dosages were plotted. Interestingly, very few proteins were common to be the top 30 in all three dosages, and the proteins that were common in two dosages were more likely to be shared between 0.1 and 0.5 μg/ml or 0.5 and 1.0 μg/ml. (B) Quantitative proteome interaction network for proteins significantly increased (P ≤ 0.05) in 0.5 μg/ml tobramycin treatment. The network was visualized with Cytoscape (version 2.8) (47). Each node indicates the protein identity, and the extent of increase is shown in red. The protein fold change from spectral counting was used. The edges indicate the protein co-regulation evidence of the connected nodes referenced from STRING database (33). Four functional clusters are observed in the network.
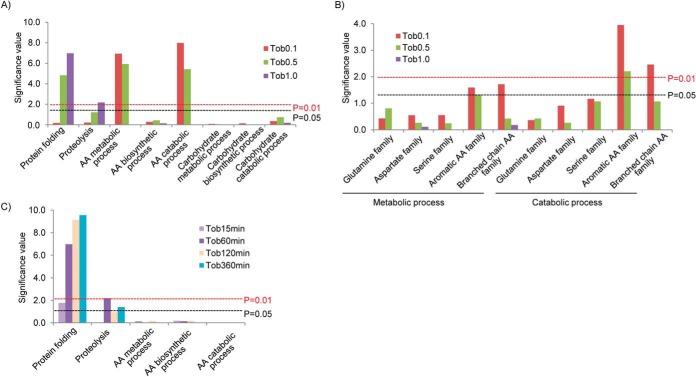
We next characterized the gene ontology of proteins that were observed to increase with tobramycin (Fig. 6A-6C). At higher tobramycin dosages of 0.5 and 1.0 μg/ml, protein folding function (heat shock proteins) and proteolysis function (proteases) were significantly overrepresented as compared with other gene ontology categories. These functions were temporally up-regulated at 60, 120, and 360 min with 1.0 μg/ml tobramycin treatments. The up-regulation of protein folding and proteolysis functions highlights the importance of protein quality control after higher tobramycin exposures (21, 31). At lower dosages of 0.1 and 0.5 μg/ml, amino acid metabolic/catabolic (but not biosynthetic) pathways were statistically overrepresented. Thus, different functional activation was observed dependent on tobramycin treatment dosages, and one would anticipate differences in cell phenotypes when specific subsets of proteins were up-regulated (32).
Fig. 6.
Gene ontology analysis of the significantly up-regulated proteins in P. aeruginosa by tobramycin. (A) Up-regulated proteins are enriched in the pathways of protein folding, proteolysis, and amino acid metabolic (catabolic) process. Significance test was performed at BiNGO (version 2.44) (48) with protein annotations from Gene Onology (49). Significance level was determined by hypergeometric test with Benjamini–Hochberg FDR correction. The resultant values were converted to y axis significance value by -log10, and the corresponding 0.01 and 0.05 significance levels are indicated in the figure. (B) The amino acid catabolic pathways rather than biosynthetic pathways were significantly up-regulated by tobramycin. The catabolism of all amino acid families was up-regulated, but the increase in aromatic and branch chain amino acid family was more significant. (C) Protein folding and proteolysis pathways were also up-regulated in tobramycin time course treatment.
To further interrogate the response of amino acid metabolic enzymes, the metabolic subpathways for different amino acid families were analyzed (Fig. 6B). Interestingly, overrepresentation for the amino acid metabolic pathway was largely due to the catabolic pathways rather than the synthesis pathways. In terms of the amino acid family, the induction was common to enzymes in all five families, but the increases in the aromatic and branched chain amino acid families were more pronounced than the other families and achieved level changes with P ≤ 0.01. Such increases were most apparent with the low level of tobramycin treatment 0.1 μg/ml.
To characterize the functional relationship of tobramycin-induced proteins, we analyzed the gene coregulation referenced from the STRING database (33). The induced proteins observed with 0.5 μg/ml tobramycin dosage are shown in Fig. 5B. Fourteen proteins related to heat shock and proteases functions were represented, and they were tightly connected in the network. Amino acid metabolic enzymes PA1999–2001 (DhcAB, AtoB) in carnitine catabolism, PA2013–2015 (LiuABC) in leucine degradation, and PA2247–2250 (BkdA1A2B, LpdV) in valine degradation (29) were also significantly induced. Although many carbon metabolic enzymes were also increased by tobramycin, the enrichment of the pathway was not statistically significant (Fig. 5B), considering the large base number of carbon metabolic enzymes in P. aeruginosa genome.
To verify the widespread increase for heat shock proteins, proteases, and amino acid catabolic enzymes, we examined the protein fold change of all the proteins quantified in the three pathways. As shown in Figs. 3E-3G, proteins in the three pathways contributed most of the highest fold changes in P. aeruginosa proteome, indicating that the three pathways are critical to P. aeruginosa tobramycin response.
As a complementary analysis, we also analyzed the tobramycin repressed proteome in P. aeruginosa. Proteins with levels that decreased during tobramycin treatment also showed dosage and time dependent effects (Fig. 4). Those proteins were functionally enriched in protein synthesis, nucleotide metabolism, tricarboxylic acid (TCA) carbon metabolism and energy derivation, and electron transport activities (Fig. S6). Gene coregulation analysis demonstrated that the down-regulated proteins were functionally connected (Fig. S7).
Tobramycin Sensitivity Phenotypes of ibpA Single and Double Mutants
Because IbpA levels were increased by the greatest amount in the observed proteome response to tobramycin, we constructed ibpA gene-inactivation mutants to determine if the absence of IbpA proteins may result in changes of P. aeruginosa tobramycin sensitivity. Surprisingly, neither ibpA-lux inactivation mutant nor ΔibpA in-frame deletion mutant exhibited significant changes in tobramycin sensitivity compared with MPAO1 WT (Table I and Fig. S8). This observation suggested that either IbpA is not directly involved in tobramycin resistance or the function of IbpA is compensated by other heat shock chaperones (34), leading to no obvious changes in tobramycin sensitivity. To examine these possibilities, we further constructed the double mutants of ibpA with clpB, (another heat shock protein) or PA0779 (lon protease) or hslV (ATP-dependent protease subunit), all of which showed significantly increased protein levels with tobramycin treatments (Figs. 3A and 3B) and exposure time (Figs. 2A and 2C).
Table I. Summary of tobramycin MIC assays for P. aeruginosa mutantsa.
| Genotype and alleles | Strains | Tob MIC (μg/ml) |
|---|---|---|
| WT | MPAO1 | 0.5–1 |
| ftsH gene deletion | ΔftsH | 0.03–0.06 |
| ibpA in-frame deletion | ΔibpA | 0.5–1 |
| ibpA gene inactivation with lux insert | ibpA-lux | 0.5–1 |
| Control containing lux insert | PKH181 | 0.5–1 |
| Control for Tn insertion | PA3303 | 0.5–1 |
| clpB single mutant, Tn 1076(2565) | clpB (PW8651) | 1 |
| clpB single mutant, Tn 1076(2565) | clpB (PW8652) | 1 |
| PA0779 single mutant, Tn 2245(2400) | PA0779 (PW2411) | 1 |
| PA0779 single mutant, Tn 1351(2400) | PA0779 (PW2414) | 1 |
| PA0779 single mutant, Tn 1345(2400) | PA0779 (PW2413) | 1 |
| hslV single mutant, Tn 223(534) | hslV (PW9487) | 0.25 |
| hslV single mutant, Tn 168(534) | hslV (PW9485) | 0.5 |
| hslV single mutant, Tn 244(534) | hslV (PW9486) | 0.5–1 |
| double mutant ibpA/clpB | ibpA/PW8651 (1,2,3) | 0.5, 0.5, 0.5 |
| double mutant ibpA/clpB | ibpA/PW8652 (1,2,3) | 0.5, 0.5, 0.5 |
| double mutant ibpA/PA0779 | ibpA/PW2411 (1,2,3) | 0.5, 0.5, 0.5 |
| double mutant ibpA/PA0779 | ibpA/PW2414 (1,2,3) | 0.5, 0.5, 0.5 |
| double mutant ibpA/PA0779 | ibpA/PW2413 (1,2,3) | 0.5–1, 0.5, 0.5 |
| double mutant ibpA/hslV | ibpA/PW9487 (1,2,3) | 0.25, 0.25, 0.25 |
| double mutant ibpA/hslV | ibpA/PW9485 (1,2,3) | 0.25, 0.25, 0.25 |
| double mutant ibpA/hslV | ibpA/PW9486 (1,2,3) | 0.25, 0.25, 0.25 |
a The sequence-verified transposon insertion mutants (Tn) are as reported in Held et al. (17), and information of mutant-specific insertion sites (gene lengths), e.g. 1,076(2,565) and library names, e.g. clpB (PW8651) are indicated in the table. Two lines of ibpA/clpB mutants, three lines of ibpA/PA0779, and ibpA/hslV were examined. For each double mutant line, three independent transformants (1,2,3) were assayed. Tobramycin MIC was defined as the lowest antibiotic concentration preventing the lawn growth of the spotted cells. A listing of a range of concentration levels indicates that a very thin lawn was observed at the lower concentration and this lawn was entirely abolished at the higher concentration in the range. See Figure S8 for additional details.
Detectable increases in tobramycin sensitivity were observed for most of the ibpA double mutants. The double mutants ibpA/hslV exhibited tobramycin MIC of 0.25 μg/ml, which was twofold more sensitive as compared with single mutant ibpA and MPAO1 WT (Table I and Fig. S8). Two of the ibpA/hslV strains also showed twofold enhancement in tobramycin sensitivity compared with hslV single mutant. For double mutants ibpA/clpB, ibpA/PA0079, tobramycin MIC of 0.5 μg/ml was identified. Although the extent of tobramycin sensitivity change was relatively small, close examination of colony lawn density showed that ibpA/clpB and ibpA/PA0079 strains were almost absent at tobramycin MIC of 0.5 μg/ml, whereas a clear thin lawn was still observed for single mutants ibpA, clpB, and PA0079 and MPAO1 WT at the same plates (Table I and Fig. S8). These sensitivity differences were reproducible for different transformants of double mutants tested. Thus, ibpA/clpB and ibpA/PA0079 double mutants were also more tobramycin sensitive compared with the single mutants. The MIC results support the hypothesis that IbpA, as well as ClpB, HslV, and PA0779 proteins, indeed play a role in tobramycin resistance in P. aeruginosa cells; however, some of these chaperones and proteases may share overlapped function, which preclude the observation of strong tobramycin sensitivity changes for single-gene inactivation mutants.
DISCUSSION
In this study, we investigated the dynamic concentration- and time-dependent proteome response of P. aeruginosa to the aminoglycoside antibiotic tobramycin. Marked proteome changes were observed after 60 min of tobramycin exposure and in response to the dosage as low as 0.1 μg/ml (1/10th MIC). The concerted proteome changes led to the functional induction of heat shock proteins and proteases at higher dosages (1.0 and 0.5 μg/ml) and amino acid catabolic enzymes at lower dosages (0.1 and 0.5 μg/ml). We further showed that inactivation of proteome markers ibpA/clpB, ibpA/PA0779, or ibpA/hslV was observed with increased tobramycin sensitivity changes in P. aerugionsa, which supports the notion that proteome response indeed has effects on antibiotic resistance in P. aeruginosa.
Our proteomics approach complements previous genetic analyses from the P. aeruginosa transposon mutant library (16, 18, 35, 36) and reveals the dynamic changes of many essential candidate genes during tobramycin treatment. For example, protein levels for essential candidate genes DnaK, GrpE, GroEL, and GroES heat shock proteins were significantly increased at dosages 0.5 and 1.0 μg/ml and time points 60, 120, and 360 min. Likewise, essential metabolic enzymes DhcA, DhcB, and DapB were significantly up-regulated by 0.1 and 0.5 μg/ml dosages. The observed protein-level changes support the hypothesis that many of these essential genes are not only indispensable for P. aeruginosa normal growth but also play important roles in tobramycin response.
Adaptive resistance of P. aeruginosa to aminoglycoside antibiotics has been described both in vivo and in vitro (13–15). It is associated with enhanced bacterial drug resistance and reduced clinical treatment efficacy (15). Some previously described mechanisms of adaptive response involved membrane impermeability (37) and the up-regulation of efflux pump MexXY (38). We hypothesize that such adaptive resistance may also involve other changes at the proteome level. Results presented in this manuscript demonstrate that the P. aeruginosa proteome response is dependent on the tobramycin levels experienced by cells, with induction of different groups of proteins being observed with exposure to various subinhibitory levels (0.1–1 μg/ml) of tobramycin. Thus, P. aeruginosa sense and respond differently to tobramycin, depending on the antibiotic concentration, supporting the adaptive resistance concept (12).
The observed time scale of proteome response is likely sufficient to produce functional impact in antibiotic defense. For example, time-lapse confocal microscopy analyses indicate that exopolysaccharide layers of biofilms can delay the biocide penetration to the central cell clusters up to 60 min (and in some cases, even longer) (10, 11, 39). In addition, CF patients that inhaled a second dose tobramycin 1 h after an initial dose also experienced the time-dependent decrease in drug efficacy (15). This effect was hypothesized to be resultant from induced adaptive resistance of P. aeruginosa in vivo. Thus, this time dependence of bacterial adaptive resistance is consistent with the observed time dependence in proteome changes in the present study. We also show that prolonged exposure of P. aeruginosa to tobramycin up to 6 h is associated with sustained proteome acclimation, during which time adaptive resistance also occurs (13–15).
Distinct proteome subsets were induced when P. aeruginosa cells were exposed to increasing concentrations of tobramycin. Such concentration-dependent changes underscore the diverse mechanisms in P. aeruginosa adaptive response. Widespread induction of heat shock proteins and proteases were observed with 0.5 and 1.0 μg/ml tobramycin treatments, consistent with previous transcriptome analysis (31, 40). Recent Escherichia coli proteome analysis of streptomycin treatments also highlights the roles of heat shock chaperones and proteases (41, 42), suggesting the response may be conserved among bacterial species and aminoglycoside drugs.
In particular, heat shock protein IbpA was the highest fold increased protein in the present study. Inactivation of ibpA did not yield significant tobramycin MIC changes. However, inactivation of two heat shock proteins/proteases ibpA/clpB, ibpA/PA0779, or ibpA/hslV led to increased tobramycin sensitivity changes in P. aeruginosa. The lack of strong changes in tobramycin sensitivity for ΔibpA may be owing to the overlapped function of IbpA with other heat shock chaperones and proteases in P. aeruginosa. In E. coli, IbpA was found to perform concerted actions with ClpB and DnaK-DnaJ-GrpE to reverse protein aggregation induced by high temperatures (34, 43).
In addition to the induction of heat shock chaperones and proteases, changes in amino acid metabolic and biosynthesis functions were observed with tobramycin treatments at the lower dosages 0.1 and 0.5 μg/ml and longer time points 120 and 360 min. These include the up-regulation of essential candidate genes of dhcA and dhcB, which are dehydrocarnitine CoA transferase subunits involved in metabolism of the amino acid carnitine, and dapB, which is a 4-hydroxy-tetrahydrodipicolinate reductase involved in lysine biosynthesis. Consistent with our findings that show amino acid metabolic enzyme levels were altered during tobramycin exposure, the interaction effects of amino acids and aminoglycoside antibiotics have been reported. For instance, supplementing 0.4% l-arginine in tryptic soy agar plate enhances the tobramycin killing of P. aeruginosa biofilm colonies over 10-fold (44). Conversely, the presence of 20 mm cadaverine (a decarboxylation product of lysine amino acid) promotes P. aeruginosa resistance to aminoglycoside kanamycin and gentamycin by fourfold (45), while suppressing its resistance to carboxypenicillins (46). Although much still needs to be understood, the results demonstrated here show that bacterial cells exposed to low levels of tobramycin exhibit increased levels of enzymes that metabolize and synthesize amino acids that could alter drug sensitivity. If so, manipulation of amino acid homeostasis pathways may provide new ways to modulate proteome response in P. aeruginosa and improve antibiotic treatment effects.
Supplementary Material
Acknowledgments
We thank Dr. Priska D. von Haller for helpful discussions, and Samuel Lee for experimental contributions.
Footnotes
Author contributions: X.W., P.K.S., C.M., and J.E.B. designed the research; X.W., K.H., B.J.S., and J.D.C. performed the research; C.R.W. contributed new reagents or analytic tools; X.W., K.H., C.Z., J.K.E., and J.E.B. analyzed the data; and X.W., P.K.S., C.M., and J.E.B. wrote the paper.
* This research was supported in part by National Institutes of Health grants 5R01HL110879, 5R01AI101307, 5R01GM086688, and 7S10RR025107, and the University of Washington's Proteomics Resource (UWPR95794).
 This article contains supplemental material Figs. S1-S8.
This article contains supplemental material Figs. S1-S8.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- DDA
- data dependent acquisition
- emPAI
- exponentially modified protein abundance index
- CF
- cystic fibrosis
- Tob
- tobramycin
- MIC
- minimum inhibitory concentrations
- BSA
- bovine serum albumin.
REFERENCES
- 1. Trautmann M., Lepper P. M., Haller M. (2005) Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am. J. Infection Control 33, S41–49 [DOI] [PubMed] [Google Scholar]
- 2. Williams H. D., Zlosnik J. E., Ryall B. (2007) Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microbial Physiol .52, 1–71 [DOI] [PubMed] [Google Scholar]
- 3. Frimmersdorf E., Horatzek S., Pelnikevich A., Wiehlmann L., Schomburg D. (2010) How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Env. Microbiol. 12, 1734–1747 [DOI] [PubMed] [Google Scholar]
- 4. Goss C. H., Burns J. L. (2007) Exacerbations in cystic fibrosis. 1: Epidemiology and pathogenesis. Thorax 62, 360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geller D. E., Pitlick W. H., Nardella P. A., Tracewell W. G., Ramsey B. W. (2002) Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest 122, 219–226 [DOI] [PubMed] [Google Scholar]
- 6. Cheer S. M., Waugh J., Noble S. (2003) Inhaled tobramycin (TOBI): a review of its use in the management of Pseudomonas aeruginosa infections in patients with cystic fibrosis. Drugs 63, 2501–2520 [DOI] [PubMed] [Google Scholar]
- 7. Mayer-Hamblett N., Kronmal R. A., Gibson R. L., Rosenfeld M., Retsch-Bogart G., Treggiari M. M., Burns J. L., Khan U., Ramsey B. W. (2012) Initial Pseudomonas aeruginosa treatment failure is associated with exacerbations in cystic fibrosis. Ped. Pulmonol. 47, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., Greenberg E. P. (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764 [DOI] [PubMed] [Google Scholar]
- 9. Slack M. P., Nichols W. W. (1981) The penetration of antibiotics through sodium alginate and through the exopolysaccharide of a mucoid strain of Pseudomonas aeruginosa. Lancet 2, 502–503 [DOI] [PubMed] [Google Scholar]
- 10. Tseng B. S., Zhang W., Harrison J. J., Quach T. P., Song J. L., Penterman J., Singh P. K., Chopp D. L., Packman A. I., Parsek M. R. (2013) The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Env. Microbiol. 15, 2865–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davison W. M., Pitts B., Stewart P. S. (2010) Spatial and temporal patterns of biocide action against Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 54, 2920–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szomolay B., Klapper I., Dockery J., Stewart P. S. (2005) Adaptive responses to antimicrobial agents in biofilms. Env. Microbiol. 7, 1186–1191 [DOI] [PubMed] [Google Scholar]
- 13. Daikos G. L., Jackson G. G., Lolans V. T., Livermore D. M. (1990) Adaptive resistance to aminoglycoside antibiotics from first-exposure down-regulation. J. Infect. Diseases 162, 414–420 [DOI] [PubMed] [Google Scholar]
- 14. Barclay M. L., Begg E. J., Chambers S. T. (1992) Adaptive resistance following single doses of gentamicin in a dynamic in vitro model. Antimicrob. Agents Chemother. 36, 1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barclay M. L., Begg E. J., Chambers S. T., Thornley P. E., Pattemore P. K., Grimwood K. (1996) Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J. Antimicrob. Chemother. 37, 1155–1164 [DOI] [PubMed] [Google Scholar]
- 16. Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Held K., Ramage E., Jacobs M., Gallagher L., Manoil C. (2012) Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee S., Hinz A., Bauerle E., Angermeyer A., Juhaszova K., Kaneko Y., Singh P. K., Manoil C. (2009) Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U.S.A. 106, 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]
- 20. Choi K. H., Schweizer H. P. (2006) Mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nature protocols 1, 153–161 [DOI] [PubMed] [Google Scholar]
- 21. Hinz A., Lee S., Jacoby K., Manoil C. (2011) Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol. 193, 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallagher L. A., McKnight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. (2002) Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184, 6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the trans-proteomic pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winsor G. L., Lam D. K. W., Fleming L., Lo R., Whiteside M. D., Yu N. Y., Hancock R. E., Brinkman F. S. L. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596-D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 26. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weisbrod C. R., Eng J. K., Hoopmann M. R., Baker T., Bruce J. E. (2012) Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 11, 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Romero P., Karp P. (2003) PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J. Mol. Microb. Biotech. 5, 230–239 [DOI] [PubMed] [Google Scholar]
- 30. Krajewski S. S., Nagel M., Narberhaus F. (2013) Short ROSE-like RNA thermometers control IbpA synthesis in Pseudomonas species. PloS One 8, e65168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kindrachuk K. N., Fernández L., Bains M., Hancock R. E. (2011) Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1874–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whiteley M., Bangera M. G., Bumgarner R. E., Parsek M. R., Teitzel G. M., Lory S., Greenberg E. P. (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 [DOI] [PubMed] [Google Scholar]
- 33. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561-D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mogk A., Deuerling E., Vorderwülbecke S., Vierling E., Bukau B. (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50, 585–595 [DOI] [PubMed] [Google Scholar]
- 35. Gallagher L. A., Shendure J., Manoil C. (2011) Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio, 2, e00315–10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fajardo A., Martinez-Martín N., Mercadillo M., Galán J. C., Ghysels B., Matthijs S., Cornelis P., Wiehlmann L., Tümmler B., Baquero F., Martínez J. L. (2008) The neglected intrinsic resistome of bacterial pathogens. PloS One 3, e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilleland L. B., Gilleland H. E., Gibson J. A., Champlin F. R. (1989) Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J. Med. Microbiol. 29, 41–50 [DOI] [PubMed] [Google Scholar]
- 38. Hocquet D., Vogne C., El Garch F., Vejux A., Gotoh N., Lee A., Lomovskaya O., Plésiat P. (2003) MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47, 1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jefferson K. K., Goldmann D. A., Pier G. B. (2005) Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 49, 2467–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anderson G. G., Moreau-Marquis S., Stanton B. A., O'Toole G. A. (2008) In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ling J., Cho C., Guo L. T., Aerni H. R., Rinehart J., Söll D. (2012) Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goltermann L., Good L., Bentin T. (2013) Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in Escherichia coli. J. Biol. Chem. 288, 10483–10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thomas J. G., Baneyx F. (2000) ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 36, 1360–1370 [DOI] [PubMed] [Google Scholar]
- 44. Borriello G., Richards L., Ehrlich G. D., Stewart P. S. (2006) Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50, 382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwon D. H., Lu C. D. (2006) Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 50, 1615–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Manuel J., Zhanel G. G., de Kievit T. (2010) Cadaverine suppresses persistence to carboxypenicillins in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 54, 5173–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maere S., Heymans K., Kuiper M. (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 49. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Consortium G. O. (2000) Gene Ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.