Abstract
Extracellular cysteine cathepsins are known to drive cancer progression, but besides degradation of extracellular matrix proteins little is known about their physiological substrates and thus the molecular mechanisms they deploy. One of the major mechanisms used by other extracellular proteases to facilitate cancer progression is proteolytic release of the extracellular domains of transmembrane proteins or ectodomain shedding. Here we show using a mass spectrometry-based approach that cathepsins L and S act as sheddases and cleave extracellular domains of CAM adhesion proteins and transmembrane receptors from the surface of cancer cells. In cathepsin S-deficient mouse pancreatic cancers, processing of these cathepsin substrates is highly reduced, pointing to an essential role of cathepsins in extracellular shedding. In addition to influencing cell migration and invasion, shedding of surface proteins by extracellular cathepsins impacts intracellular signaling as demonstrated for regulation of Ras GTPase activity, thereby providing a putative mechanistic link between extracellular cathepsin activity and cancer progression. The MS data is available via ProteomeXchange with identifier PXD002192.
Cysteine cathepsins, a family of cysteine proteases normally confined to the endolysosomal system, emerged as major players in cancer progression (1–3). Genetic ablation of several cathepsins, including cathepsins B, L, and S, significantly slowed down cancer growth and metastatic spread in several mouse cancer models including mammary gland tumors and pancreatic islet cancer (3–6). Moreover, inhibition of cathepsins by broad-spectrum small molecule inhibitors significantly delayed cancer progression in vivo, consistent with cathepsin knockout data (7, 8). Such inhibition was also shown to significantly sensitize mammary gland tumors to standard chemotherapeutics including paclitaxel (9). Similarly, inhibition of cathepsin L secretion by expression of a recombinant anti-cathepsin L single chain variable fragment completely abolished melanoma invasion in vivo (10), whereas inhibition of extracellular cathepsin S by specific antibodies or by the recombinant propeptide significantly reduced cancer cell invasion and angiogenesis (11, 12). In addition, a significant synergistic effect on angiogenesis inhibition was observed when cathepsin S therapy was combined with anti-VEGF therapy (11). Collectively, these examples suggest that cathepsins may present valid therapeutic targets for cancer treatment.
In cancer, cathepsins S and L are secreted into the tumor microenvironment by tumor cells, fibroblasts, endothelial cells, and infiltrating immune cells (13). Among the immune cells, macrophages are a major source of tumor-associated cathepsins (14). Secreted cathepsins were found to be involved in several processes that contribute to carcinogenesis, including extracellular matrix (ECM)1 degradation, activation of proteases such as urokinase-type plasminogen activator (uPA) and matrix metalloproteinases (MMPs), and in E-cadherin cleavage (2). However, this evidence comes predominantly from in vitro studies and little is known about the in vivo substrates of these enzymes. Identification of the substrates of secreted cathepsins is therefore key to understanding their biological functions in cancer (15).
Membrane-anchored proteins, including receptors, growth factors, cytokines, and adhesion proteins, have a major role in cancer progression. A general mechanism for their functional regulation is the release of their extracellular domains through limited proteolysis, also known as ectodomain shedding (16–18). Most of the proteases involved in ectodomain shedding are members of the two zinc-dependent protease families, matrix metalloproteases (MMPs) and disintegrin-type metaloproteases (ADAMs), among which the best known is ADAM17 (reviewed in (19, 20)).
Here we show that extracellular cathepsins can act as sheddases and release protein ectodomains from the surface of cancer cells. Among the identified substrates are cell adhesion proteins and membrane receptors. We confirmed cathepsin-mediated shedding of these substrates in cell based models as well as in vivo in a mouse model of pancreatic cancer. Collectively, this work has identified possible molecular mechanisms by which cysteine cathepsins may regulate cancer progression.
EXPERIMENTAL PROCEDURES
Cathepsins
Human cathepsin B was expressed in E. coli and purified as described in (21). Human cathepsins S and L were expressed in the methylotrophic yeast P. pastoris and purified as described in (22).
Cell Culture
Cancer cell lines MDA-MB-231, MCF-7, PANC-1, HT-144, and T98-G were grown to confluence in Dulbecco's modified Eagles media supplemented with 10% fetal bovine serum (FBS), 1% glutamine and penicillin/streptomycin (Lonza, Verviers, Belgium). U937 cells were grown in RPMI (Roswell Park Memorial Institute, Buffalo, NY) media supplemented with 10% FBS, 1% glutamine and 1% penicillin/streptomycin (Lonza). U937 cells were plated in a 12-well culture plate (7 × 105 cells per well) and differentiated into macrophages with 30 nm phorbol 12-myristate 13-acetate (PMA) (Sigma, St. Louis, MO) for 48 h, followed by 24 h of recovery without PMA in the completed RPMI media. For a coculture experiment, 1.4 × 106 of detached MDA-MB-231 cells were resuspended in PBS buffer (Lonza) (pH 6.0, 0.5 mm dithiothreitol (DTT) (Fluka Biochemica)) and plated in 12-well cell culture dish containing differentiated U937 cells (0.7 × 106 cells per well).
Cell Treatment with Recombinant Cathepsins
Cells were detached using an enzyme-free cell dissociation solution (Millipore, Darmstadt, Germany). Per condition, thirty million cells were incubated in parallel in 500 μl of PBS (Lonza) (pH 6.0, containing 0.5 mm DTT (Fluka Biochemica, Steinheim, Germany)), with human recombinant cathepsin L, S, or B (1 μm and 0.2 μm) or with E-64-inhibited cathepsin (1 μm cathepsin L, S, or B incubated in PBS containing 20 μm broad spectrum cysteine cathepsin inhibitor E-64 (Peptide Institute, Osaka, Japan) for 1 h at 37 °C) serving as a negative control for 1 h at 37 °C, followed by collection of the supernatant (sample was centrifuged for 5 min at 500 × g, supernatant was removed and centrifuged again for 5 min at full speed). Residual cathepsin activity was blocked by the addition of E-64 to each sample (20 μm final concentration). In the microscopy experiments, MDA-MB-231 cells were first grown to confluence, thereby establishing contacts with neighboring cells, in contrast to most other experiments herein that were performed on detached cells. Similar protease concentrations were used in vitro in recent degradomic studies to identify putative substrates of various matrix metalloproteases, caspase-3, and aspartic cathepsins D and E (23–26).
Mass Spectrometry Sample Preparation
The obtained protein supernatants were separated on a 12.5% SDS-PAGE gel (Lonza). The gel was stained with Comassie Brilliant Blue and each protein lane was cut into six bands (six samples), which were destained using destaining solution (25 mm NH4HCO3 (Fluka Biochemica), 50% acetonitrile (JT Baker, Deventer, The Netherlands)). The gel pieces were washed with acetonitrile and vacuum dried before rehydrating the gel pieces in reducing solution (10 mm DTT (Fluka Biochemica), 25 mm NH4HCO3) followed by an incubation at 56 °C for 45 min before exchanging to an alkylating solution (55 mm iodoacetamide (Amersham Biosciences, Little Chalfont, UK), 25 mm NH4HCO3). The reaction was allowed to proceed in the dark for 30 min before washing the gel pieces with 25 mm NH4HCO3 using intense vortexing. Afterward, the gel pieces were washed with acetonitrile and vacuum dried before rehydrating in 80 μl of trypsinization buffer (25 mm NH4HCO3) containing 1 μg of sequencing-grade modified porcine trypsin (Promega, Madison, WI) per sample. The gel pieces were allowed to rehydrate on ice for 15 min before adding more trypsinization buffer to cover the gel pieces completely. Trypsin was then left to digest overnight at 37 °C. Next day, the trypsin solution was collected and remaining peptides were extracted from the gel pieces using extraction solution (50% acetonitrile, 5% formic acid (JT Baker)). Trypsin solution was added to extraction solution and concentrated by vacuum drying to a final volume of about 20 μl.
Analysis by Nano LC-MS/MS
Liquid chromatography tandem MS (LC-MS/MS) analyses were performed with an EASY-nanoLC II HPLC unit (Thermo Scientific, San Jose, CA) coupled to an Orbitrap LTQ Velos mass spectrometer (Thermo Scientific). The peptide sample was first loaded on a C18 trapping column (Proxeon EASY-ColumnTM, 2 cm (length), 100 μm internal diameter, 5 μm 120Å, C18-A1 beads) and then separated on a 10 cm long C18 PicoFritTM AQUASIL analytical column, (75 μm internal diameter, 5 μm 100 Å, C18 beads) (New Objective, Woburn, MA) using forward flushing. Peptides were eluted with a 90 min linear gradient of 5–50% solvent B (0.1% formic acid in acetonitrile) at a flow rate of 300 nl/min. MS spectra were acquired in the Orbitrap analyzer with a mass range of 300–2000 m/z and 30,000 resolution. MS/MS spectra were obtained by higher-energy collision dissociation fragmentation (normalized collision energy at 35) of the nine most intense precursor ions from the full MS scan. Dynamic exclusion was enabled with repeat count of 2 and 120 s exclusion time.
Data Analysis
The database search and quantification by spectral counting were performed using the MaxQuant proteomics software (version 2.0.18), with imbedded Andromeda search engine (27, 28). Search was performed against the human IPI protein database v.385 (89,952 sequences, 36,291,020 residues), using the trypsin cleavage specificity with maximum two missed cleavages. Carbamidomethylation of cysteines was set as static, whereas methionine oxidation and N-terminal acetylation were set as dynamic modifications. Precursor and fragment mass tolerances were set at 6 and 20 ppm. Reversed database search was performed and false discovery rate (FDR), was set at 1% for peptide and protein identifications. In MaxQuant, identified peptides are assigned to protein groups rather than proteins and groups with at least two identified peptides were considered as positive identifications. Relative quantification of identified proteins was performed by spectral counting of their razor and unique peptides. This approach is known to reliably detect differences in protein abundance if at least a twofold difference in spectral count is observed (29). To minimize the number of false positives especially in the case of proteins with lower spectral counts (5–10 counts per protein), only proteins with at least threefold spectral count change were considered to have significantly altered abundance (30). Protein spectral count ratios (SCRs) were computed by the division of spectral counts in cathepsin treated sample with spectral counts in negative control (sample treated with inhibited cathepsin). Spectral counts of all proteins identified in compared data sets were increased by 1 in order to avoid division by zero. Gene Ontology analysis was performed by g:Profiler web interface (31).
Flow Cytometry
MDA-MB-231 cells were grown to confluence and detached with enzyme-free cell dissociation solution (Milipore). A total of 7.2 million cells per parallel were incubated in 120 μl PBS (Lonza) (pH 6.0, with 0.5 mm DTT (Fluka Biochemica)), with added human recombinant cathepsin L (0.2 or 1 μm) or inhibited cathepsin (1 μm cathepsin 1 h preincubated with 20 μm E-64 (Peptide Institute)) as a negative control. After 1 h of incubation at 37 °C the cells were washed with PBS. Annexin V-PE and PI were used to determine the phosphatidylserine exposure and the loss of membrane integrity according to the manufacturer's instructions (BD PharmigenTM, Erembodegem, Belgium). Analysis was made with a FACSCalibur flow cytometer (Becton Dickinson) and the CellQuest software.
Western Blotting
The supernatants used in the sample preparation for mass spectrometry were also analyzed on 12% SDS-PAGE gels followed by immunoblotting. For the detection of the shed proteins in the supernatant, antibodies against neuropilin 1 (R&D Systems, Minneapolis, MN) (sheep polyclonal, dilution 1:500), transferrin receptor protein 1 (R&D Systems, goat polyclonal, dilution 1:500), CD44 (R&D Systems, mouse monoclonal, dilution 1:500), ALCAM (R&D Systems, goat polyclonal, dilution 1:500), L1CAM (R&D Systems, goat polyclonal, dilution 1:500), plexin B2 (R&D Systems, sheep polyclonal, dilution 1:200), MUC18 (R&D Systems, goat polyclonal, dilution 1:500), and ephrin type A receptor 2 (Novus Biologicals, Abingdon, UK) (rabbit polyclonal, dilution 1:5000) were used. Cathepsins B and S in the RIP1-Tag2 tumors were detected by antibodies against cathepsin B (R&D Systems, goat polyclonal, dilution 1:2000) and cathepsin S (R&D Systems, goat polyclonal, dilution 1:500). K-ras was detected using sheep polyclonal antibodies (R&D Systems, dilution 1:200). Secondary antibodies were used at 1:5000 dilutions and ECL kit was used for detection (GE Healthcare).
Cathepsin Activity Assay
The cleavage of the fluorogenic substrate Z-Phe-Arg-AMC (Bachem, Bubendorf, Switzerland) was used to determine cathepsin activity in the coculture supernatants. Fifty microliters of each sample were mixed in a 96-well plate with buffer (100 mm phosphate buffer, 1 mm EDTA, 1 mm dithiothreitol, and 0.1% (w/v) polyethyleneglycol, pH 6.0) to the final volume of 90 μl. After 15 min incubation at 37 °C, substrate was added to a final concentration of 10 μm and its hydrolysis continuously measured in a 96-well plate reader (Tecan Safire, Mannedorf, Switzerland) at excitation and emission wavelengths of 370 and 460 nm, respectively. In an additional experiment, the active concentration of the cathepsins in the supernatants was determined by active site titration with E-64 (32, 33). For activity probe labeling, differentiated U937 cells (7 × 105 cells per well) and MDA-MB-231 cells (1.4 × 106 cells per well) were grown separately and in a coculture on a 12-well culture plate. Cells were incubated for 2 h at 37 °C in PBS buffer (Lonza) (pH 6.0, 0.5 mm DTT (Fluka Biochemica)) in the presence of 10 μm DCG-04 probe. After the incubation, the PBS buffer was collected and centrifuged for 5 min at 500 × g and 30 min at maximum speed. The supernatant was then used for immunological detection of biotinylated cathepsins.
Substrate Shedding in a Cell Coculture
MDA-MB-231 and differentiated U937 cells were plated in a coculture as described in the cell culture section. After 2 h incubation at 37 °C, the PBS buffer was collected and centrifuged once for 5 min at 500 × g and once for 5 min at the maximum speed. The buffer was then used for imunodetection of the shed ectodomains by Western blotting. As a negative control, differentiated U937 and MDA-MB-231 cells were pretreated with E-64 (25 μm) (Peptide Institute), GM6001 (10 μm) (Calbiochem, San Diego, CA) or batimastat (10 μm) (Tocris Bioscience, Bristol, UK). Cells were pretreated with inhibitor for 2 h before the coculturing and same inhibitor concentration was also present during the coculture Additionally, differentiated U937 and MDA-MB-231 cells were also incubated with PBS buffer (Lonza) for 2 h (pH 6.0, 0.5 mm DTT (Fluka Biochemica)) in the presence and absence of 50 μm E-64, 10 μm GM6001, and batimastat.
The cathepsin activity in the coculture and macrophage supernatant was monitored as described in the Cathepsin activity assay.
Migration Assay
MDA-MB-231 cells were grown to confluence followed by starvation in FBS-free media for 48 h with 1% nutridoma (Roche, Basel, Switzerland) added. Cells were subsequently detached with a cell dissociation solution. A total of 4.2 × 104 cells per experimental setup were incubated in 100 μl of PBS buffer (Lonza) (pH 6.0 or 7.4, with 0.5 mm DTT (Fluka Biochemica)) with cathepsin B, S, or L (0.2 μm and 0.02 μm) or inhibited cathepsins (0.2 μm cathepsin, 1 h preincubated with 20 μm E-64 (Peptide Institute)) as a negative control. After 10 min at 37 °C the buffer with cathepsin was removed and the cells were put into a 24-well cell culture insert (BD Falcon Cell culture inserts, 8 μm pore size) to assay migration. 10% FBS (Lonza) was used as chemoattractant. Cells that migrated through the pores of the insert were counted after 40 h. To check whether the difference between the samples' arithmetic means were statistically significant, we used a homoscedastic (two samples with equal variance) Student's t-test with two-tailed distribution. If the probability associated with the Student's t-test (p value) was lower than 0.05, the sample was considered statistically significantly different from the control and was marked with an asterisk (*). The same statistical approach was applied also for the invasion assay below.
Invasion Assay
MDA-MB-231 cells were grown to confluence, followed by starvation in the FBS-free medium containing 1% nutridoma (Roche) for 48 h. The cells were detached with a cell dissociation solution. A total of 4.2 × 104 cells per experimental condition was incubated in 100 μl of PBS buffer (Lonza) (pH 6.0 or 7.4, with 0.5 mm DTT (Fluka Biochemica)) with cathepsin B, S, or L (0.2 μm and 0.02 μm) or inhibited cathepsin (0.2 μm cathepsin B, S, or L pre-incubated with 20 μm E-64 (Peptide Institute) for 1 h) as a negative control. After 10 min at 37 °C the buffer was removed and the cells were put into a 24-well cell culture insert with an 8 μm pore size polycarbonate membrane coated with ECMatrixTM (Milipore QCMTM cell invasion assay). Ten percent FBS (Lonza) was used as a chemoattractant. The invaded cells that migrated through the ECM layer were dissociated from the membrane and detected by CyQuant GR® dye using a fluorescence plate reader.
RIP1-Tag2 Soluble Tumor Extracts
Tumors were prepared from wild-type, cathepsin B-deficient and cathepsin S- deficient RIP1-Tag2 mice as described previously (4). Three wild-type tumors (28 mm3, 33 mm3, and 65 mm3) and three tumors from each cathepsin knockout mouse (1.2 mm3, 4.2 mm3, and 6.5 mm3, respectively, from cathepsin B-deficient mice, and 1.2 mm3, 1.8 mm3, and 2.4 mm3, respectively, from cathepsin S-deficient mice) were used in the study, each originating from a different mouse. In agreement with previous results, tumors from cathepsin-deficient mice were considerably smaller than wild-type tumors (4). Samples were weighed and dounce homogenized on ice in an ice-cold PBS buffer (Lonza) containing 0.5 mm EDTA (Serva). The buffer volume was adjusted according to the tumor mass (700 μl buffer per 1 mg tumor mass). The homogenate was centrifuged at 4 °C for 30 min at the maximum speed. The supernatant was used for the immuno-detection of substrate ectodomains by Western blotting. Actin was used as a loading control.
Light Microscopy
MDA-MB-231 cells were grown to confluence in a 24-well plate. They were washed twice with PBS. In each well the PBS was then replaced with 500 μl PBS (pH 7.4) with 0.5 mm DTT and active cathepsin B, L, or S at the final concentration of 1 μm. No cathepsins were added to the control wells. The cells were observed under the light microscope (Olympus IX81, 100× magnification). The pictures were taken immediately after addition of the cathepsins (t = 0), 3 (t = 3 min), and 10 min (t = 10 min) after the incubation.
Ras GTPase Activity Assay
MDA-MB-231 cells were grown to confluence in Dulbecco's modified Eagles media. They were detached with enzyme-free cell dissociation solution (Milipore). Cells (30,000 cells per setup) were incubated in 300 μl PBS (Lonza) (pH 6.0, with 0.5 mm DTT (Fluka Biochemica)), with added human recombinant cathepsin L or S (0.05 μm) or inhibited cathepsin (0.05 μm cathepsin 1 h preincubated with 20 μm E-64 (Peptide Institute)) as a negative control. After 1 h of incubation at 37 °C whole-cell extracts were prepared and 100 μg of total protein was used for ELISA-based chemiluminescence assay according to manufacturer's instructions (Active Motif, La Hulpe, Belgium). Briefly, Ras proteins were bound to GST-Raf-RBD and loaded into glutathione coated 96-well plate. Immobilized Ras proteins were labeled with Ras antibody (specific against human H-Ras and K-Ras) and quantified by chemiluminescence measurement (Tecan Infinite M1000 Pro). The experiment was performed using three biological replicates.
Whole Cell Lysates
MDA-MB-231 and MCF-7 cells were grown to confluence, detached with enzyme-free cell dissociation solution (Millipore) and washed with PBS. 1 × 106 cells were lysed in 100 μl 1% SDS in PBS buffer. The whole cell lysates were analyzed on 12% SDS-PAGE gels followed by immunoblotting.
RESULTS
Cathepsins S and L Shed Several Membrane-anchored Proteins from the Cell Surface In Vitro
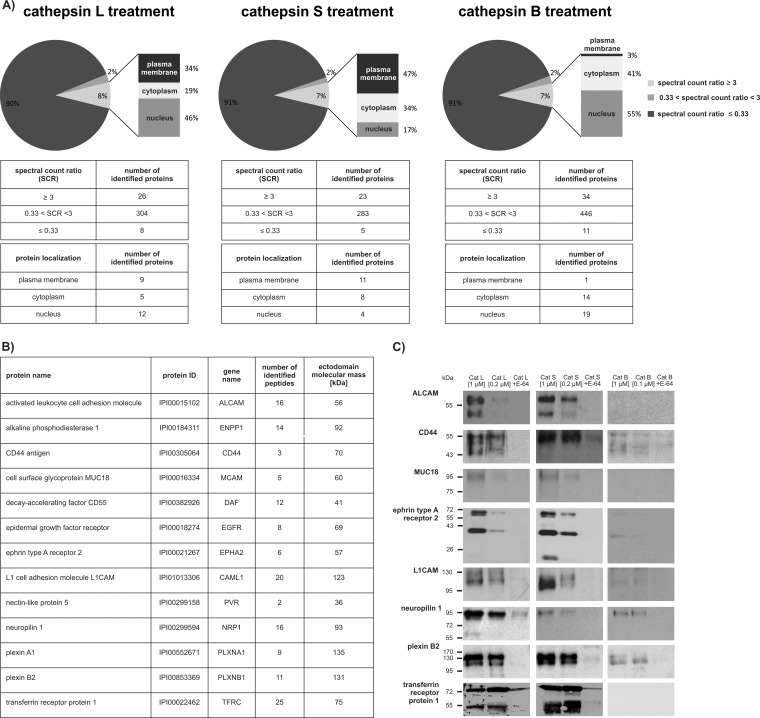
In order to identify putative cathepsin substrates on the surface of tumor cells we initially treated intact breast cancer cells MDA-MB-231 cells with recombinant cathepsins L, S, and B. Cells were treated in multiple biological replicates (three for cathepsins L and S and two for cathepsin B) and E-64 inhibited cathepsins were used as negative controls. Cathepsin treatment did not affect cell viability (∼ 95%; (supplemental Fig. S1). Next, mass spectrometry and spectral counting were used to identify and assign protein fragments released into the cellular supernatant, thereby reflecting differences in protein abundance (34) (supplemental Table S3). The majority of identified proteins (∼90%) were abundant intracellular proteins, which were not further considered as their abundance did not change significantly compared with the negative control (Fig. 1A). Such intracellular proteins largely originate from cancer cells that are known to secrete significant amounts of intracellular proteins. A recently reported secretome analysis of MDA-MB-231 cells thus showed that after 24 h intracellular proteins represent as much as 55% of all proteins present in the medium (35). In addition, intracellular proteins in the medium partially resulted from the small portion of damaged cells (< 5% total; see supplemental Fig. S1). Because of the short incubation time (1 h), these proteins were enriched in the medium as compared with the secreted proteins. However, the proteins that reproducibly showed at least a threefold increase in their SCR in the presence of active cathepsins were considered for further analysis. In the case of cathepsins L and S, over one third of these proteins were cell surface membrane proteins. Mass spectrometry analysis identified nine putative extracellular membrane substrates of cathepsin L and 11 substrates of cathepsin S, which were found in all three biological replicates (Fig. 1A). Moreover, data comparison showed that cathepsins L and S shed the same group of 13 proteins (Fig. 1B) and that four of cathepsin L substrates and two of cathepsin S substrates were not observed in one out of three biological replicates (supplemental Table S1). Furthermore, all identified peptides were located in the extracellular portion of these molecules, which confirms that cleavages occurred on the surface of the intact cells (supplemental Fig. S2).
Fig. 1.
Identification of cell surface substrates of cysteine cathepsins. A, Intact MDA-MB-231 cells were treated with active or E-64-inhibited cathepsins L, S, or B. The pie charts for the cathepsin L, S, and B treatment show the ratios between the proteins with unchanged abundance (0.3 < SCR < 3), proteins with decreased abundance (SCR ≤ 0.3) and proteins with increased abundance (spectral count ratio ≥ 3) upon cathepsin treatment. Proteins with SCR ≥ 3 were further subdivided according to their subcellular localization (membrane, cytoplasmic, or nuclear). B, List of potential cathepsin substrates that display plasma membrane localization with at least threefold increased spectral counts in the cathepsin-treated samples. For each substrate, the number of identified peptides and the theoretical mass of its ectodomain are shown. C, Intact cells were treated with active cathepsins L, S, or B or E-64-inhibited cathepsins as negative controls. Shed protein domains of ALCAM, CD44, ephrin type A receptor 2, L1CAM, MUC18, neuropilin 1, plexin B2, and transferrin receptor protein 1 were detected by immunoblotting the supernatant of cells.
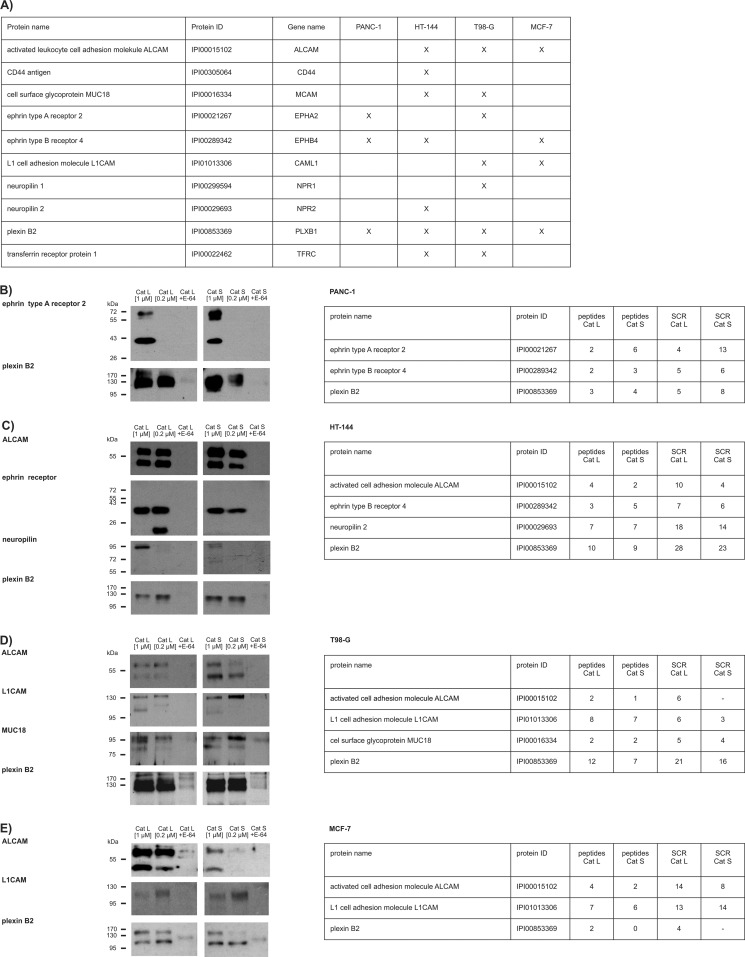
Among the identified substrates, there were several members of the immunoglobulin superfamily of cell adhesion molecules (CAMs) and several receptors. Although all the identified proteins were cleaved by cathepsins S and L, only neuropilin 1 was cleaved by cathepsin B. In the control experiment in the presence of the selective cysteine protease inhibitor E-64, processing was essentially abolished, indicating that cathepsins were responsible for the processing. Immunological detection of 8 selected substrate candidates (out of 13 identified) showed that cathepsins L and S, but not B, processed these proteins at one or more specific sites in a similar fashion, thereby generating stable protein fragments of the sizes that correlated well with the complete extracellular parts of the identified substrates (Fig. 1B and 1C), suggesting that cathepsins indeed shed these proteins from the cell surface. The same set of substrates was also identified using attached MDA-MB-231 cells, however, the yields of shed proteins were generally much lower and therefore this approach was not further pursued (data not shown). We next evaluated cathepsins L and S as potential sheddases in a number of other cancer cell lines, including the melanoma (HT-144), glioblastoma (T98-G), pancreatic carcinoma (PANC-1), and breast adenocarcinoma (MCF-7) cell lines. As shown in Fig. 2, both cathepsins S and L also acted as sheddases on these cell lines, indicating that cathepsin-mediated shedding is not limited to a single cancer cell line. Moreover, the majority of substrates identified in the MDA-MB-231 cell line was also identified in the other cell lines tested, suggesting that the process of cathepsin-mediated shedding is well-conserved.
Fig. 2.
Identification of shed membrane proteins in PANC-1, HT-144, T98-G, and MCF-7 cells and their immunological detection. Intact cells were treated with active cathepsins L, S, or B or E-64-inhibited cathepsins as negative controls. For each cell line, proteomic data and immunological detection of selected targets is shown. A, Three substrates identified using the MDA-MB-231 cell line were detected in the PANC-1 cell line, five in the HT-144 cell line, seven in the T98-G cell line and three in the MCF-7 cell line. Shedding of related receptor families was also observed in some cell lines (neuropilins 1 and 2, ephrin receptors A2 and B4), indicating cathepsin cleavage preference toward functionally related proteins. B, PANC-1 cells. Shed protein domains of ephrin type A receptor 2 and plexin B2 were detected by immunobloting using the culture media of treated cells. C, HT-144 cells. Shed protein domains of ALCAM, ephrin receptor, neuropilin, and plexin B2 were detected by immunobloting using the culture media of treated cells. In this cell line, neuropilin 2 and ephrin type B receptor 4, which are close homologs of neuropilin 1 and ephrin type A receptor 2, were identified by mass spectrometry. D, T98-G cells. Shed protein domains of ALCAM, L1CAM, MUC18 and plexin B2 were detected by immunobloting in the culture media. The same group of proteins was detected by mass spectrometry. ALCAM was identified only in one proteomic experiment, hinting to its lower abundance. E, MCF-7 cells. ALCAM, L1CAM, and plexin B2 were detected by immunobloting in the culture media.
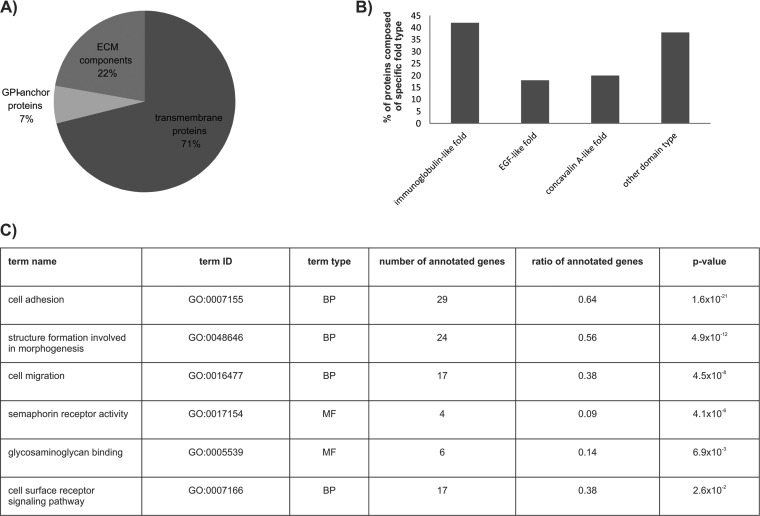
However, in other cell lines, additional substrates were identified, including other members of the neuropilin, ephrin receptor, and CAM families, accounting for 45 putative membrane substrates in total in the five cell lines tested (supplemental Table S2, supplemental Table S4). Among these, 32 are transmembrane proteins, three are GPI-anchored proteins, and the remaining 10 proteins are present on the cell surface as components of the ECM (Fig. 3A). The majority of identified substrates were not previously reported to be cleaved by cathepsins. However, several of the identified ECM components were previously shown to be cathepsin substrates (fibronectin, laminin, tenascin, collagen, nidogen, and perlecan (1, 36, 37)), validating our approach as a means to study cathepsin-mediated substrate shedding. A number of the identified substrates share a high degree of similarity and closely related family members of substrates were identified in different cell lines (neuropilin 2, ephrin type B receptor 4). In addition, protein domain annotation using the InterPro database (38) showed that 60% of the shed proteins have immunoglobulin, EGF, and/or concavalin A lectin-like folds (Fig. 3B), suggesting that cathepsins may have a preference to shed distinct and structurally related groups of proteins from the cell surface. A functional analysis of the identified targets using Gene Ontology annotation showed enrichment for proteins involved in cell adhesion, migration, morphogenesis, and receptor signaling. In the molecular function category, semaphorin receptor activity showed the most prominent enrichment (Fig. 3C), suggesting that cathepsin-mediated shedding plays an important role in these processes.
Fig. 3.
Structural and functional annotation of membrane substrates identified in all tested cell lines. A, Substrates were divided according to their attachment to the membrane surface. The majority of them are classical transmembrane proteins whereas only three of them are attached to the membrane using a GPI anchor. Components of the extracellular matrix (ECM) are secreted from the cell. B, Structural diversity of the identified substrates. About 60% of the cathepsin substrates have immunoglobulin, concavalin A or EGF-like fold, or a combination of the three folds. Only 40% of them displayed a different domain architecture. C, Functional enrichment of the identified membrane substrates determined by Gene Ontology. Highly enriched biological processes (BP) and molecular functions (MF) are listed according to their p value.
Cathepsins Secreted by Macrophages Induce Ectodomain Shedding
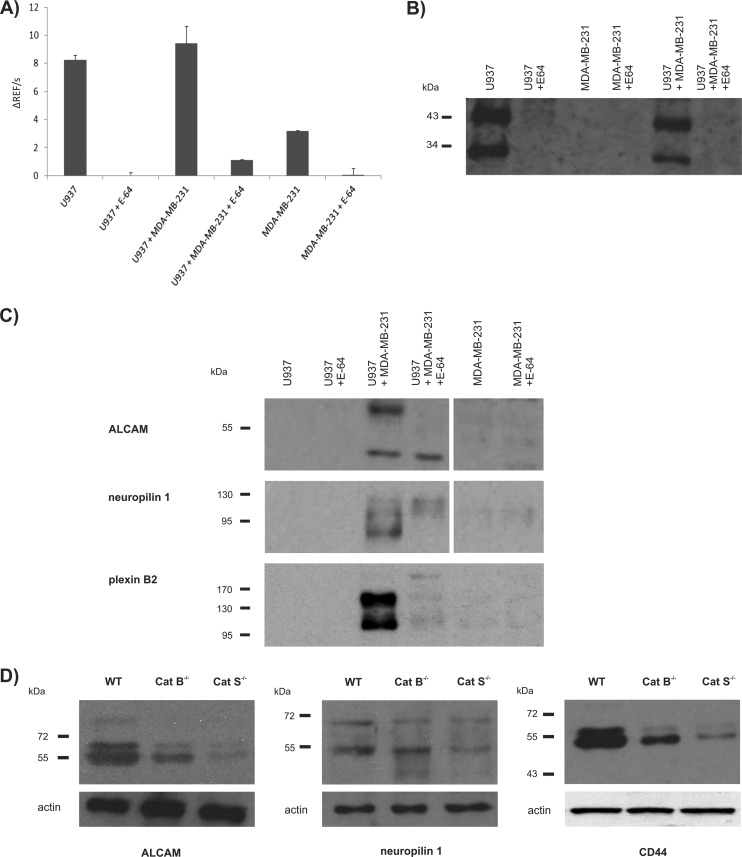
We next tested whether cathepsins secreted from macrophages, which are the main source of extracellular cathepsins in the tumor microenvironment (14, 39, 40), can induce shedding of the same membrane-anchored proteins from tumor cells. We therefore used a well-established cellular model of macrophage-differentiated U937 cells, and performed coculture experiments with MDA-MB-231 breast cancer cells. Cathepsin activity measurements verified macrophages as the main source of extracellular cathepsin activity in coculture both by hydrolysis of a small general cathepsin substrate Z-Phe-Arg-AMC (Fig. 4A) and by the broad spectrum cathepsin activity-based probe DCG-04 (41) (Fig. 4B). Moreover, by active site titration we found out that the concentration of secreted cysteine cathepsins in macrophage secretome was ∼65 nm (supplemental Fig. S3). Because a much higher cell density was used in the same reaction volume in the initial in vitro experiments (∼45-fold higher), this further suggests that 1 μm cathepsin concentration, which was applied in these experiments, was a good estimate. In addition, cathepsins are known to reach millimolar concentrations in the lysosomal compartments of cancer cells and macrophages (42) and their secretion in the tumor microenvironment was reported to be highly localized (43), which further implies that cathepsin concentrations in the low micromolar range can be expected in the specific regions of the tumor microenvironment.
Fig. 4.
Cathepsins mediate shedding in macrophage/tumor cell cocultures and in solid tumors in vivo. A, Cathepsin activity in the cell culture medium was based on continuous monitoring the turnover of the fluorogenic substrate Z-Phe-Arg-AMC. Error bars show standard deviations based on duplicate values for each data set. B, Activity-based profiling of cathepsin activity in the cell culture medium using the fluorescent broad spectrum cathepsin activity-based probe DCG-04. C, Immunoblot analysis of shedding of ALCAM, neuropilin 1 and plexin B2 in the culture media of the coculture of U937 cells differentiated into macrophages and MDA-MB-231 cells. No shedding was detected in individual cell lines or in inhibitor-treated cells (typical result from three biological replicates is shown). D, Shedding of ALCAM, CD44 and neuropilin 1 in soluble RIP1-Tag2 tumor extracts. Actin was used as a loading control.
Next, we evaluated substrate shedding in the culture medium by Western blot. Three of the identified substrates, ALCAM, neuropilin 1, and plexin B2, were shed into the medium only when cells were grown in coculture, suggesting that macrophage-secreted cathepsins are responsible for the shedding (Fig. 4C). This was confirmed by the addition of E-64, which significantly reduced the release of their ectodomains. Moreover, the broad-spectrum metalloprotease inhibitors GM6001 and batimastat did not prevent shedding (supplemental Fig. S4), indicating that cysteine cathepsins are required for the shedding of these proteins.
Cathepsins are Involved in Ectodomain Processing of Several Substrates In Vivo
The in vivo relevance of substrate shedding by cathepsins was evaluated in the tumor microenvironment in a murine pancreatic islet cancer model (RIP1-Tag2) (44). Initially, we found evidence of ectodomain processing in the soluble extracts of RIP1-Tag2 tumors from wild-type mice for three proteins; neuropilin 1, ALCAM, and CD44. Processing of other targets identified from the cell-based screen was not found, possibly reflecting the difference between the situation in vitro and in vivo, as well as between human and mouse. Next, a comparison was made with tumors from RIP1-Tag2 mice with deletion of cathepsins B or S (supplemental Fig. S5), which in this model significantly impaired tumor invasion, with effects also observed on tumor growth, cell proliferation, tumor formation, and angiogenesis (4). The genetic loss of cathepsin S expression largely abolished the processing of ALCAM and CD44, whereas processing of neuropilin 1 was diminished. In the cathepsin B knockouts, however, a much smaller effect on ectodomain processing was observed, in agreement with its generally poor shedding efficiency observed in in vitro experiments (Fig. 4D). Addition of high concentration of E-64 (20 μm) in the buffer before homogenization of RIP1-Tag2 tumors did not affect the observed cleavage pattern, indicating that the cathepsins eventually released during preparation of tumor extracts were not involved in the processing (supplemental Fig. S6). These results further suggest that cysteine cathepsins, in particular cathepsin S, are responsible for processing of a subset of membrane proteins in vivo and that in some cases, they cannot be substituted by other proteases, such as metalloproteases. Interestingly, ALCAM, CD44, and neuropilin 1 were not identified among the substrates, which were in vitro shed from the human pancreatic carcinoma cell line PANC1 (supplemental Table S2). This is likely because of the different origin of the two cell types as the PANC1 cell line is derived from the pancreatic ductal carcinoma, whereas RIP1-Tag2 tumors are formed in the islets of Langerhans.
Cathepsin Treatment Increases Migration and Invasion Rate of Cancer Cells
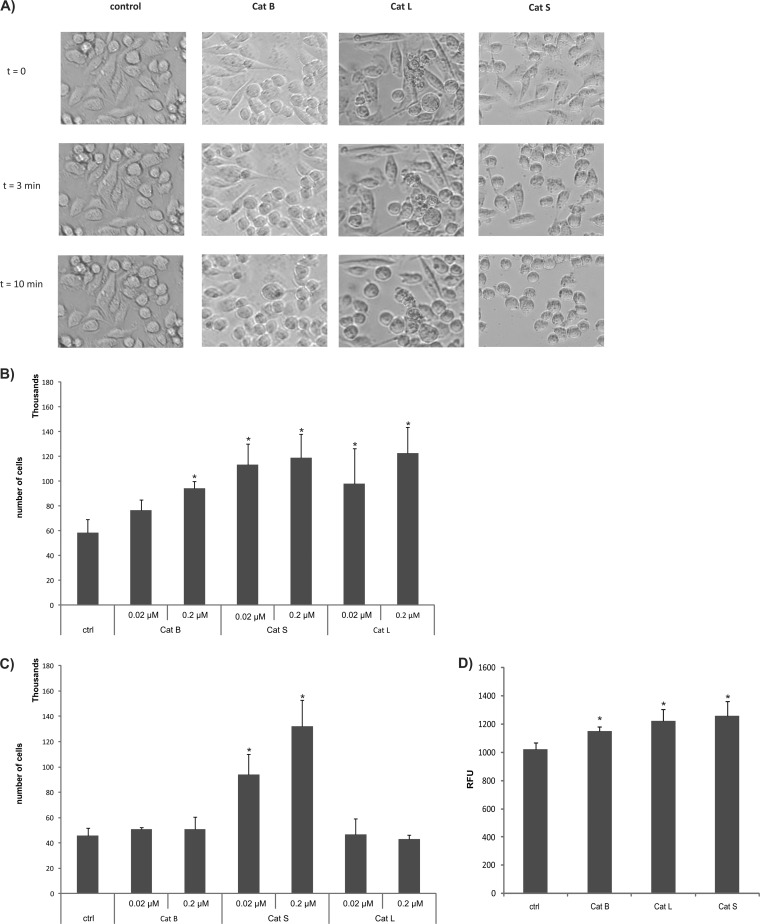
As there were a number of CAM molecules among the identified cathepsin targets, which have a major role in cell adhesion and establishment of cell–cell contacts (45), we next investigated whether their shedding influenced cell migration and invasion capabilities. We first evaluated whether cathepsins can disrupt cell–cell contacts and influence cell surface attachment using MDA-MB-231 cells grown to confluence. Following exposure to exogenously added cathepsins S, L, and B, an extremely rapid rounding and detachment of the cells was observed upon cathepsin S treatment even at neutral pH. Cathepsin L was slightly less potent, possibly because of its lower stability under these conditions (46), whereas cathepsin B was the least efficient (Fig. 5A).This supported the idea that cell–cell contacts are lost at least in part because of cathepsin-mediated shedding of CAM proteins.
Fig. 5.
Cysteine cathepsin shedding promotes migration and invasion of MDA-MB-231 cells. A, Detachment of MDA-MB-231 cells by cathepsins B, L, and S. Cells were monitored under the light microscope and pictures were taken immediately after adding PBS (t = 0), after 3 min and after10 min of incubation with active cathepsins (t = 3 min or t = 10 min). B, Migration of cathepsin-treated MDA-MB-231 cells through the PET membrane after cathepsin treatment (10 min at pH 6.0). After 40 h, the cells that had migrated through the membrane were counted in a counting chamber. *p < 0.05, compared with the control setup in the absence of cathepsins. C, Migration of cathepsin-treated MDA-MB-231 cells through the PET membrane after cathepsin treatment at pH 7.4. *p < 0.05, compared with the control setup in the absence of cathepsins. Error bars show the standard deviations based on triplicate values of each data set. D, Invasion of cathepsin treated MDA-MB-231 cells into the EC matrix at pH 6.0. After 6 h the cells that had invaded the matrix were quantified by the CyQuant GR dye using a plate reader. *p < 0.05, compared with the control setup in the absence of cathepsins. Error bars show the standard deviations based on triplicate values of each dataset.
The rapid detachment of cells further suggests that such individual cells could have higher migration properties that could facilitate metastatic spread. At slightly acidic pH, cathepsins S and L statistically significantly increased the migration potential of tumor cells compared with the negative control at both concentrations tested, whereas cathepsin B increased the migration potential of the cells to a smaller extent and only at a higher concentration (Fig. 5B). At neutral pH, only cathepsin S increased cell mobility (Fig. 5C), in agreement with the high stability and activity of this enzyme under these conditions (47).
Because elevated cell migration could be linked to increased metastatic dissemination, we next investigated the effects of cathepsins L, S, and B on invasion in cell-based assays. All three cathepsins statistically significantly increased the invasion of MDA-MD-231 cells compared with the untreated cells. Cathepsin S was the most potent, followed by cathepsins L and B (Fig. 5D).
Extracellular Cathepsins Decrease the Activity of Intracellular Ras GTPases
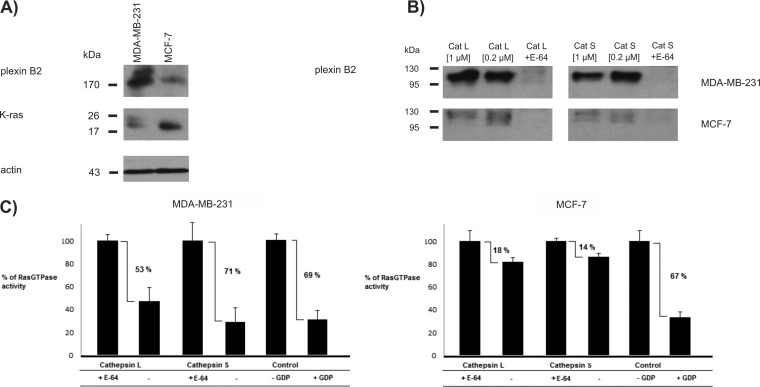
Finally, we evaluated whether shedding affected intracellular signaling pathways that are known to have an important role in cancer signaling. One such pathway is the Ras signaling pathway, which is known to affect a large variety of cancer-related processes and which is regulated by several receptors identified as cathepsin substrates. Among these are plexins and their coreceptors neuropilins, which have a vital role in the semaphorin signaling pathway (48). Here, semaphorin binding activates the cytosolic GTPase-activating (GAP) domain of plexins, which in turn inactivates signaling proteins from the Ras GTPase family (49, 50). Because genetic removal of the extracellular domain of plexins results in their constitutive activation (51), we evaluated whether proteolytic removal of this domain would have a similar effect. In addition, EGFR is another receptor known to strongly influence Ras activity (52), and ablation of EGFR activity was shown to strongly decrease K-Ras activity in pancreatic ductal adenocarcinoma (53). To test whether extracellular cathepsins can influence intracellular Ras activity through such receptor shedding we used MDA-MB-231 cells that were found to express high levels of receptors (plexin A1, plexin B2, EGFR, and neuropilin), and MCF-7 cells, which express these at substantially lower levels (e.g. plexin B2 was the only semaphorin receptor identified in the shed fraction of MCF-7 cells by mass spectrometry). Interestingly, levels of K-Ras, the downstream target in this receptor signaling, were higher in MCF-7 cells than in MDA-MB-231 cells (Fig. 6A). Shedding of plexin B2 by cathepsins S and L was compared between the two cell lines and a much higher amount of plexin B2 was shed from MDA-MB-231 as compared with MCF-7 cells (Fig. 6B), consistent with the difference in the expression levels of the proteins. Receptor shedding by cathepsins was therefore expected to either largely suppress the Ras activity (MDA-MB-231 cells) or to have a more moderate effect (MCF-7 cells). We focused on K-Ras and H-Ras, which are known for their role in cancer (54), using a selective ELISA-based chemiluminescence assay. A major suppression of K-Ras and H-Ras activity was observed in the MDA-MB-231 cells, essentially reaching the basal level as observed in the presence of GDP, which is known to abolish signaling, whereas only a marginal effect was observed in MCF-7 cells, thereby demonstrating an important role of cathepsin-mediated plexin shedding in the regulation of Ras activity in cancer (Fig. 6C).
Fig. 6.
Cathepsin treatment abrogates Ras GTPase activity in MDA-MB-231, but not in MCF-7 cells. A, Immunological detection of plexin B2 and K-ras in the whole cell lysates of MDA-MB-231 and MCF-7 cells. B, Immunological detection of plexin B2 in the supernatant of MDA-MB-231 and MCF-7 cells, after treatment with cathepsins L or S. Presence of plexin B2 is barely detectable in MCF-7 cells, which is consistent with the proteomic data. C, Ras GTPase activity (H-Ras and K-Ras) in cell lysates after treatment with cathepsins L and S. In control experiment cell lysates were incubated with GDP, which inhibits the GTPase activity. A major decrease in Ras GTPase activity was observed (53% with cathepsin L, 71% with cathepsin S, 65% with GDP) in MDA-MB-231, but not in MCF-7 cells (< 20%).
DISCUSSION
Although cysteine cathepsins are established promoters of tumor growth and cancer invasion, little is known about their molecular mechanisms of action. In order to get better insights into these mechanisms, we applied proteomics for the identification of extracellular substrates. Cysteine cathepsins L and S were found to shed a distinct group of membrane proteins from the cell surface, and the majority of these proteins are known mediators of cancer progression and are involved in regulation of cell adhesion and signaling. Their shedding could explain many tumorigenic processes related to extracellular cathepsin activity such as angiogenesis, invasion, and metastasis formation (55).
Among the substrates identified in the MDA-MB-231 cell line, ALCAM, MUC18, L1CAM, and nectin-like protein 5 are members of the immunoglobulin superfamily of CAM proteins. They influence cell migration, motility, and invasion either by direct cell–cell adhesion or through intracellular signaling (reviewed in (56–59). CAM proteins are not the only regulators of cell migration among the identified substrates. CD44 is an important regulator of cell binding to the ECM, because its major ligand hyaluronan (HA) is one of the most abundant components of extracellular matrix (60). Plexin A1, plexin B2, and neuropilin 1 are involved in the regulation of semaphorin signaling. Neuropilin 1 was originally discovered as an adhesion molecule, but it also interacts with plexins in order to mediate semaphorin signaling (61). Although B type plexins interact with semaphorins directly, A type plexins strictly need neuropilin 1 as a coreceptor for this interaction. This pathway regulates angiogenesis, cell migration, and other signaling pathways within the tumor microenvironment (48, 62). A second larger group of identified substrates were proteins primarily involved in cell signaling, including EGFR, ephrin type A receptor 2, and the decay accelerating factor (CD55). EGFR is a well-known oncogene involved in a variety of cancer processes such as tumor cell motility, adhesion, angiogenesis, proliferation, and metastasis (63, 64), and antibodies against EGFR have already been approved for colorectal cancer treatment (65). Another promising new therapeutic target is the ephrin receptor family. Up-regulation of their activity significantly decreases migration and invasion of many types of cancer cells and tumor growth in mouse cancer models (reviewed in (66)). Inactivation of the ephrin receptor activity by proteolytic processing could therefore enhance oncogenic signaling pathways. Finally, CD55 is the major inhibitor of the complement system. It is broadly expressed in malignant tumors, where it promotes tumorigenesis by decreasing complement-mediated tumor cell lysis. This protein can also promote invasion and metastasis formation through activation of the tyrosine kinase pathway (67). Several additional cathepsin substrates were identified in other cell lines, the most common being ECM components and components of the basement membrane (supplemental Table S2). One example is perlecan, a ubiquitous pro-angiogenic proteoglycan here found to be shed by cathepsins L and S in all tested nonbreast cancer cell lines.
Some of the substrates identified here have already been reported as substrates of metalloproteases, including ADAM 17, which was found to shed ectodomains of CD44, ALCAM, L1CAM, and MUC18 (reviewed in (68)). This shedding influences cell migration and invasion. In a similar manner, metalloproteases have been proposed to shed transferrin receptor protein 1 and EGFR although it remains unclear which metalloproteases are involved (69, 70). However, our coculture experiments in the presence of the broad-spectrum metalloprotease inhibitors GM6001 and batimastat, and the in vivo results from the cathepsin S-deficient RIP1-Tag2 tumors suggest that cathepsins are indispensable for the shedding of at least some of these substrates. This further suggests that cathepsins could have been involved in the generation of the soluble forms of ALCAM, CD44, and EGFR, which were detected in the serum of cancer patients and in mouse cancer models, with increased serum levels found to correlate with poor prognosis (71, 72), thereby having a potential as cathepsin-dependent biomarkers. Moreover, several of the identified substrates, plexin A1, ephrin type A receptor 2, nectin-like protein 5, and E-NPP 1, were not yet reported to be proteolytically shed. It can be therefore suggested that cathepsins influence cellular processes in a unique way.
One of the major signaling pathways affected through cathepsin-mediated protein shedding seems to be semaphorin signaling. Semaphorin binding to plexins leads to the activation of the latter and facilitates their interaction with either guanine nucleotide exchange factors (GEFs) or GTPase activating proteins (GAPs) (73), thereby critically regulating the activity of Ras proteins and, indirectly, their downstream targets Raf and ERK (74). Plexin activation thus leads to inactivation of Ras GTPases, as demonstrated for plexin B1, which was found to inactivate R-Ras and M-Ras (49, 75). However, interaction of plexins with other Ras GTPases such as K-Ras, H-Ras, and N-Ras, which have major roles in cancer (76), has not been established as yet. Because genetic ablation of the semaphorin binding domain or the whole extracellular plexin domain leads to constitutive activation of plexin A1 (51), it can be suggested that cathepsin-mediated removal of the plexins' extracellular domain(s) also results in constitutive plexin activation and thereby sustained inactivation of Ras-GTPases as observed for the K-Ras and H-Ras activity in MDA-MB-231 cells (Fig. 6). EGFR, here identified as a cathepsin substrate, is another receptor involved in Ras signaling as its activation is known to lead to Ras activation. Ablation of its activity decreased the activity of K-Ras in pancreatic ductal adenocarcinoma (53), consistent with the idea that cathepsin-mediated shedding of EGFR would additionally impair Ras activation. However, this is a very complex process and the role of individual receptor shedding remains to be elucidated.
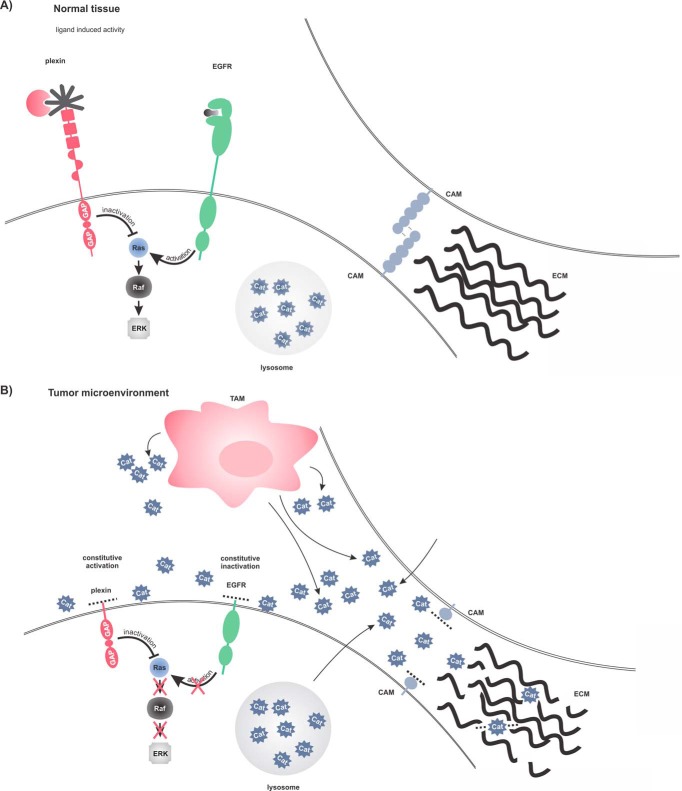
Based on these findings, we propose a model for how cathepsins that are secreted into the tumor microenvironment by various cell types could promote cancer progression at the molecular level (Fig. 7). Secreted cathepsins could originate from various cell types including cancer cells and tumor associated macrophages (TAMs) (1). Once in the extracellular milieu, these cathepsins, in addition to degrading ECM, shed a number of CAMs, thereby destroying cell–cell contacts and increasing cell mobility. Such detached tumor cells can easily enter the circulation and thus contribute to metastasis spread. In addition, the cathepsins might have partially a tumor suppressing role as demonstrated by the inactivation of small Ras GTPases through the disruption of semaphorin or EGFR signaling. However, cancer cells use numerous mechanisms to circumvent the organism's immune system, including numerous mutations in the critical regulatory genes such as Ras, resulting in inability of the organism to remove the tumor. This leads to a further recruitment of inflammatory cells to the tumor system and sustained inflammation, partially through the cathepsin-mediated chemokine processing (77), which then promotes tumorigenesis. Increased macrophage infiltration is often observed at the invasive front of tumors (78), which suggests that TAMs could directly promote tumor invasion, in agreement with our idea. However, this is still a simplified model and does not fully explain the complex role(s) of cathepsins in tumor progression. It is namely very likely that cathepsins do not affect tumorigenesis through a single pathway or by shedding of a single substrate but rather through a combination of multiple pathways, which in vivo is likely tissue- and cancer type-specific.
Fig. 7.
Molecular mechanism of extracellular cathepsin involvement in tumorigenesis. A, Normal tissue. Plexins are inactive and only semaphorin ligand binding can trigger their GAP activity. EGFR can increase Ras activity upon ligand stimulation. Cell–cell contacts formed by CAM proteins are intact, as well as extracellular matrix. Cysteine cathepsins are mainly localized within the endo/lysosomal compartments. B, Tumor microenvironment. Infiltrated immune cells and tumor cells secrete cathepsins, which cleave receptor ectodomains. Plexin cytosolic GAP activity is constitutively activated and inactivates Ras proteins. Simultaneously, removal of the EGFR ectodomain inactivates the receptor, which loses capability of Ras activation. Shedding of CAM proteins and degradation ECM destroys cell–cell contacts and increases migration and invasion of cancer cells.
In summary, this study provides novel insights into the molecular pathways that govern the processes of tumorigenesis. Here we show that cathepsins can shed ectodomains of a group of membrane proteins involved in cancer progression, thereby providing a link between extracellular cathepsins and their regulation of cancer cell migration and invasion. Moreover, cathepsins were found to be indispensable for the processing of some of the substrates in vivo in a pancreatic islet mouse tumor model. Finally, through shedding of substrates, cathepsins were found to regulate intracellular signaling as demonstrated for the regulation of Ras protein activity.
Supplementary Material
Acknowledgments
We thank M. Prebanda and D. Caglič (Jožef Stefan Institute, Ljubljana) for preparing recombinant cathepsins L and B, M. Bogyo (Stanford University) for his comments on the manuscript and a generous gift of DCG-04, and Z. Werb (UCSF, USA) for critical reading of the manuscript. P. Van Damme is a Post-doctoral Fellow of the Research Foundation - Flanders (FWO-Vlaanderen).
Footnotes
Author contributions: B.S., B.T., and M.F. designed research; B.S., M.V., R.V., and V.T. performed research; P.V., V.G., J.A.J., and K.G. contributed new reagents or analytic tools; P.V., K.G., and M.F. analyzed data; B.S., B.T., and M.F. wrote the paper.
* This work was supported by grants from the Slovenian Research Agency (P1-0140 and J1-3602 to B.T.; J1-0185 and J1-5449 to M.F.) and by the FP7 project MICROENVIMET to B.T., K.G. acknowledges support from the Research Foundation - Flanders (FWO-Vlaanderen, project number G.0048.08), and J.A.J. received support from NIH R01 CA125162. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (79) via the PRIDE partner repository with the dataset identifier PXD002192.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
1 The abbreviations used are:
- ECM
- extracellular matrix
- ADAM
- disintegrin-type metaloprotease
- CAM
- cell adhesion molecule
- EGF
- epidermal growth factor
- GPI
- glycophosphatidylinositol
- PMA
- phorbol 12-myristate 13-acetate
- SCR
- spectral count ratio
- TAM
- tumor associated macrophages
- uPA
- urokinase-type plasminogen activator
- MMPs
- matrix metalloproteinases
- VEGF
- vascular endothelial growth factor.
REFERENCES
- 1. Mohamed M. M., Sloane B. F. (2006) Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer 6, 764–775 [DOI] [PubMed] [Google Scholar]
- 2. Palermo C., Joyce J. A. (2008) Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol. Sci. 29, 22–28 [DOI] [PubMed] [Google Scholar]
- 3. Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. (2007) Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 13, 387–403 [DOI] [PubMed] [Google Scholar]
- 4. Gocheva V., Zeng W., Ke D., Klimstra D., Reinheckel T., Peters C., Hanahan D., Joyce J. A. (2006) Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Develop. 20, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sevenich L., Schurigt U., Sachse K., Gajda M., Werner F., Muller S., Vasiljeva O., Schwinde A., Klemm N., Deussing J., Peters C., Reinheckel T. (2010) Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc. Natl. Acad. Sci. U.S.A. 107, 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasiljeva O., Turk B. (2008) Dual contrasting roles of cysteine cathepsins in cancer progression: apoptosis versus tumour invasion. Biochimie 90, 380–386 [DOI] [PubMed] [Google Scholar]
- 7. Joyce J. A., Baruch A., Chehade K., Meyer-Morse N., Giraudo E., Tsai F. Y., Greenbaum D. C., Hager J. H., Bogyo M., Hanahan D. (2004) Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell 5, 443–453 [DOI] [PubMed] [Google Scholar]
- 8. Mikhaylov G., Mikac U., Magaeva A. A., Itin V. I., Naiden E. P., Psakhye I., Babes L., Reinheckel T., Peters C., Zeiser R., Bogyo M., Turk V., Psakhye S. G., Turk B., Vasiljeva O. (2011) Ferri-liposomes as an MRI-visible drug-delivery system for targeting tumours and their microenvironment. Nat. Nanotechnol. 6, 594–602 [DOI] [PubMed] [Google Scholar]
- 9. Shree T., Olson O. C., Elie B. T., Kester J. C., Garfall A. L., Simpson K., Bell-McGuinn K. M., Zabor E. C., Brogi E., Joyce J. A. (2011) Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 25, 2465–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rousselet N., Mills L., Jean D., Tellez C., Bar-Eli M., Frade R. (2004) Inhibition of tumorigenicity and metastasis of human melanoma cells by anti-cathepsin L single chain variable fragment. Cancer Res. 64, 146–151 [DOI] [PubMed] [Google Scholar]
- 11. Ward C., Kuehn D., Burden R. E., Gormley J. A., Jaquin T. J., Gazdoiu M., Small D., Bicknell R., Johnston J. A., Scott C. J., Olwill S. A. (2010) Antibody targeting of cathepsin S inhibits angiogenesis and synergistically enhances anti-VEGF. PLoS ONE 5, e12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burden R. E., Snoddy P., Buick R. J., Johnston J. A., Walker B., Scott C. J. (2008) Recombinant cathepsin S propeptide attenuates cell invasion by inhibition of cathepsin L-like proteases in tumor microenvironment. Mol. Cancer Ther. 7, 538–547 [DOI] [PubMed] [Google Scholar]
- 13. Mason S. D., Joyce J. A. (2011) Proteolytic networks in cancer. Trends Cell Biol. 21, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gocheva V., Wang H. W., Gadea B. B., Shree T., Hunter K. E., Garfall A. L., Berman T., Joyce J. A. (2010) IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Develop. 24, 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turk B., Turk D., Turk V. (2012) Protease signalling: the cutting edge. EMBO J. 31, 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garton K. J., Gough P. J., Raines E. W. (2006) Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J. Leukoc. Biol. 79, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 17. Massague J., Pandiella A. (1993) Membrane-anchored growth factors. Annu. Rev. Biochem. 62, 515–541 [DOI] [PubMed] [Google Scholar]
- 18. Murphy G. (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 19. Arribas J., Borroto A. (2002) Protein ectodomain shedding. Chem. Rev. 102, 4627–4638 [DOI] [PubMed] [Google Scholar]
- 20. Reiss K., Saftig P. (2009) The “a disintegrin and metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 21. Rozman J., Stojan J., Kuhelj R., Turk V., Turk B. (1999) Autocatalytic processing of recombinant human procathepsin B is a bimolecular process. FEBS Lett. 459, 358–362 [DOI] [PubMed] [Google Scholar]
- 22. Mihelic M., Dobersek A., Guncar G., Turk D. (2008) Inhibitory fragment from the p41 form of invariant chain can regulate activity of cysteine cathepsins in antigen presentation. J. Biol. Chem. 283, 14453–14460 [DOI] [PubMed] [Google Scholar]
- 23. Impens F., Colaert N., Helsens K., Ghesquiere B., Timmerman E., De Bock P. J., Chain B. M., Vandekerckhove J., Gevaert K. (2010) A quantitative proteomics design for systematic identification of protease cleavage events. Mol. Cell. Proteomics 9, 2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Plasman K., Van Damme P., Kaiserman D., Impens F., Demeyer K., Helsens K., Goethals M., Bird P. I., Vandekerckhove J., Gevaert K. (2011) Probing the efficiency of proteolytic events by positional proteomics. Mol. Cell. Proteomics 10, M110 003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Starr A. E., Bellac C. L., Dufour A., Goebeler V., Overall C. M. (2012) Biochemical characterization and N-terminomics analysis of leukolysin, the membrane-type 6 matrix metalloprotease (MMP25): chemokine and vimentin cleavages enhance cell migration and macrophage phagocytic activities. J. Biol. Chem. 287, 13382–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 29. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 30. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 31. Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007) g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rozman-Pungercar J., Kopitar-Jerala N., Bogyo M., Turk D., Vasiljeva O., Stefe I., Vandenabeele P., Bromme D., Puizdar V., Fonovic M., Trstenjak-Prebanda M., Dolenc I., Turk V., Turk B. (2003) Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: when reaction mechanism is more important than specificity. Cell Death Differ. 10, 881–888 [DOI] [PubMed] [Google Scholar]
- 33. Turk B., Krizaj I., Kralj B., Dolenc I., Popovic T., Bieth J. G., Turk V. (1993) Bovine stefin C, a new member of the stefin family. J. Biol. Chem. 268, 7323–7329 [PubMed] [Google Scholar]
- 34. Li Z., Adams R. M., Chourey K., Hurst G. B., Hettich R. L., Pan C. (2012) Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J. Proteome Res. 11, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 35. Villarreal L., Mendez O., Salvans C., Gregori J., Baselga J., Villanueva J. (2013) Unconventional secretion is a major contributor of cancer cell line secretomes. Mol. Cell. Proteomics 12, 1046–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sage J., Leblanc-Noblesse E., Nizard C., Sasaki T., Schnebert S., Perrier E., Kurfurst R., Bromme D., Lalmanach G., Lecaille F. (2012) Cleavage of nidogen-1 by cathepsin S impairs its binding to basement membrane partners. PLoS ONE 7, e43494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fonovic M., Turk B. (2014) Cysteine cathepsins and extracellular matrix degradation. Biochim. Biophys. Acta 1840, 2560–2570 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., Punta M., Attwood T. K., Sigrist C. J., Redaschi N., Rivoire C., Xenarios I., Kahn D., Guyot D., Bork P., Letunic I., Gough J., Oates M., Haft D., Huang H., Natale D. A., Wu C. H., Orengo C., Sillitoe I., Mi H., Thomas P. D., Finn R. D. (2014) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43, D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reddy V. Y., Zhang Q. Y., Weiss S. J. (1995) Pericellular mobilization of the tissue-destructive cysteine proteinases, cathepsins B, L, and S, by human monocyte-derived macrophages. Proc. Natl. Acad. Sci. U.S.A. 92, 3849–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mikhaylov G., Klimpel D., Schaschke N., Mikac U., Vizovisek M., Fonovic M., Turk V., Turk B., Vasiljeva O. (2014) Selective targeting of tumor and stromal cells by a nanocarrier system displaying lipidated cathepsin b inhibitor. Angew Chem Int Ed Engl 53, 10077–10081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Greenbaum D., Medzihradszky K. F., Burlingame A., Bogyo M. (2000) Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 7, 569–581 [DOI] [PubMed] [Google Scholar]
- 42. Xing R., Addington A. K., Mason R. W. (1998) Quantification of cathepsins B and L in cells. Biochem. J. 332, 499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sevenich L., Joyce J. A. (2014) Pericellular proteolysis in cancer. Genes Dev. 28, 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanahan D. (1985) Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 315, 115–122 [DOI] [PubMed] [Google Scholar]
- 45. Wai Wong C., Dye D. E., Coombe D. R. (2012) The role of immunoglobulin superfamily cell adhesion molecules in cancer metastasis. Int. J. Cell Biol. 2012, 340296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turk B., Dolenc I., Turk V., Bieth J. G. (1993) Kinetics of the pH-induced inactivation of human cathepsin L. Biochemistry 32, 375–380 [DOI] [PubMed] [Google Scholar]
- 47. Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamagnone L. (2012) Emerging role of semaphorins as major regulatory signals and potential therapeutic targets in cancer. Cancer Cell 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 49. Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004) The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science 305, 862–865 [DOI] [PubMed] [Google Scholar]
- 50. Tong Y., Hota P. K., Penachioni J. Y., Hamaneh M. B., Kim S., Alviani R. S., Shen L., He H., Tempel W., Tamagnone L., Park H. W., Buck M. (2009) Structure and function of the intracellular region of the plexin-b1 transmembrane receptor. J. Biol. Chem. 284, 35962–35972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takahashi T., Strittmatter S. M. (2001) Plexina1 autoinhibition by the plexin sema domain. Neuron 29, 429–439 [DOI] [PubMed] [Google Scholar]
- 52. Wheeler D. L., Dunn E. F., Harari P. M. (2010) Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat. Rev. Clin. Oncol. 7, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ardito C. M., Gruner B. M., Takeuchi K. K., Lubeseder-Martellato C., Teichmann N., Mazur P. K., Delgiorno K. E., Carpenter E. S., Halbrook C. J., Hall J. C., Pal D., Briel T., Herner A., Trajkovic-Arsic M., Sipos B., Liou G. Y., Storz P., Murray N. R., Threadgill D. W., Sibilia M., Washington M. K., Wilson C. L., Schmid R. M., Raines E. W., Crawford H. C., Siveke J. T. (2012) EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karnoub A. E., Weinberg R. A. (2008) Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gocheva V., Joyce J. A. (2007) Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 6, 60–64 [DOI] [PubMed] [Google Scholar]
- 56. Ofori-Acquah S. F., King J. A. (2008) Activated leukocyte cell adhesion molecule: a new paradox in cancer. Transl. Res. 151, 122–128 [DOI] [PubMed] [Google Scholar]
- 57. Raveh S., Gavert N., Ben-Ze'ev A. (2009) L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 282, 137–145 [DOI] [PubMed] [Google Scholar]
- 58. Schwarz Q., Ruhrberg C. (2010) Neuropilin, you gotta let me know: should I stay or should I go? Cell Adh. Migr. 4, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 9, 603–615 [DOI] [PubMed] [Google Scholar]
- 60. Zoller M. (2011) CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 11, 254–267 [DOI] [PubMed] [Google Scholar]
- 61. Staton C. A., Kumar I., Reed M. W., Brown N. J. (2007) Neuropilins in physiological and pathological angiogenesis. J. Pathol. 212, 237–248 [DOI] [PubMed] [Google Scholar]
- 62. Capparuccia L., Tamagnone L. (2009) Semaphorin signaling in cancer cells and in cells of the tumor microenvironment–two sides of a coin. J. Cell Sci. 122, 1723–1736 [DOI] [PubMed] [Google Scholar]
- 63. Engebraaten O., Bjerkvig R., Pedersen P. H., Laerum O. D. (1993) Effects of EGF, bFGF, NGF and PDGF(bb) on cell proliferative, migratory and invasive capacities of human brain-tumour biopsies in vitro. Int. J. Cancer 53, 209–214 [DOI] [PubMed] [Google Scholar]
- 64. Petit A. M., Rak J., Hung M. C., Rockwell P., Goldstein N., Fendly B., Kerbel R. S. (1997) Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: angiogenic implications for signal transduction therapy of solid tumors. Am. J. Pathol. 151, 1523–1530 [PMC free article] [PubMed] [Google Scholar]
- 65. Jonker D. J., O'Callaghan C. J., Karapetis C. S., Zalcberg J. R., Tu D., Au H. J., Berry S. R., Krahn M., Price T., Simes R. J., Tebbutt N. C., van Hazel G., Wierzbicki R., Langer C., Moore M. J. (2007) Cetuximab for the treatment of colorectal cancer. New Engl. J. Med. 357, 2040–2048 [DOI] [PubMed] [Google Scholar]
- 66. Pasquale E. B. (2010) Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mikesch J. H., Schier K., Roetger A., Simon R., Buerger H., Brandt B. (2006) The expression and action of decay-accelerating factor (CD55) in human malignancies and cancer therapy. Cell Oncol. 28, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Scheller J., Chalaris A., Garbers C., Rose-John S. (2011) ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 32, 380–387 [DOI] [PubMed] [Google Scholar]
- 69. Kaup M., Dassler K., Reineke U., Weise C., Tauber R., Fuchs H. (2002) Processing of the human transferrin receptor at distinct positions within the stalk region by neutrophil elastase and cathepsin G. Biol. Chem. 383, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 70. Wilken J. A., Perez-Torres M., Nieves-Alicea R., Cora E. M., Christensen T. A., Baron A. T., Maihle N. J. (2013) Shedding of soluble epidermal growth factor receptor (sEGFR) is mediated by a metalloprotease/fibronectin/integrin axis and inhibited by cetuximab. Biochemistry 52, 4531–4540 [DOI] [PubMed] [Google Scholar]
- 71. Taguchi A., Politi K., Pitteri S. J., Lockwood W. W., Faca V. M., Kelly-Spratt K., Wong C. H., Zhang Q., Chin A., Park K. S., Goodman G., Gazdar A. F., Sage J., Dinulescu D. M., Kucherlapati R., Depinho R. A., Kemp C. J., Varmus H. E., Hanash S. M. (2011) Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell 20, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hansen A. G., Freeman T. J., Arnold S. A., Starchenko A., Jones-Paris C. R., Gilger M. A., Washington M. K., Fan K. H., Shyr Y., Beauchamp R. D., Zijlstra A. (2013) Elevated ALCAM shedding in colorectal cancer correlates with poor patient outcome. Cancer Res. 73, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Puschel A. W. (2007) GTPases in semaphorin signaling. Adv. Exp. Med. Biol. 600, 12–23 [DOI] [PubMed] [Google Scholar]
- 74. Bar-Sagi D., Hall A. (2000) Ras and Rho GTPases: a family reunion. Cell 103, 227–238 [DOI] [PubMed] [Google Scholar]
- 75. Saito Y., Oinuma I., Fujimoto S., Negishi M. (2009) Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO Rep. 10, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gysin S., Salt M., Young A., McCormick F. (2011) Therapeutic strategies for targeting ras proteins. Genes Cancer 2, 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Repnik U., Starr A. E., Overall C. M., Turk B. (2015) Cysteine cathepsins activate ELR chemokines and inactivate non-ELR chemokines. J. Biol. Chem. 290, 13800–13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Condeelis J., Pollard J. W. (2006) Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124, 263–266 [DOI] [PubMed] [Google Scholar]
- 79. Vizcaíno J. A., Deutch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated protoemics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.