Abstract
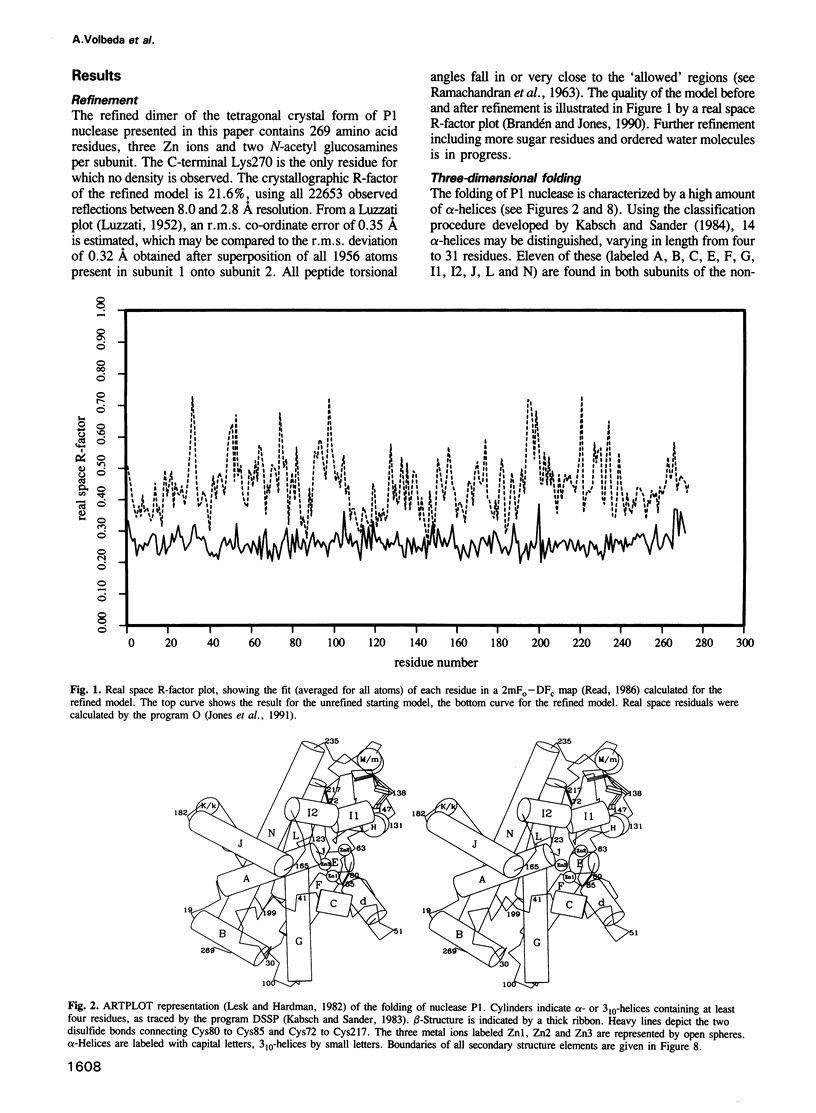
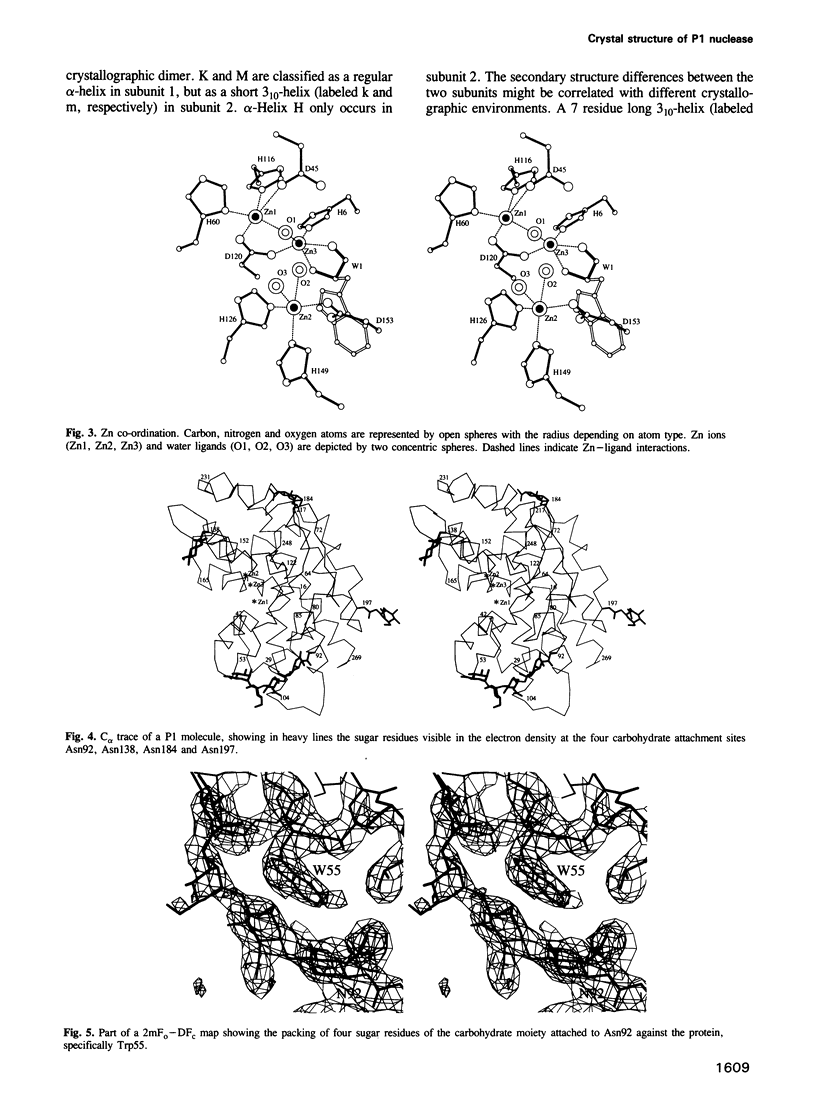
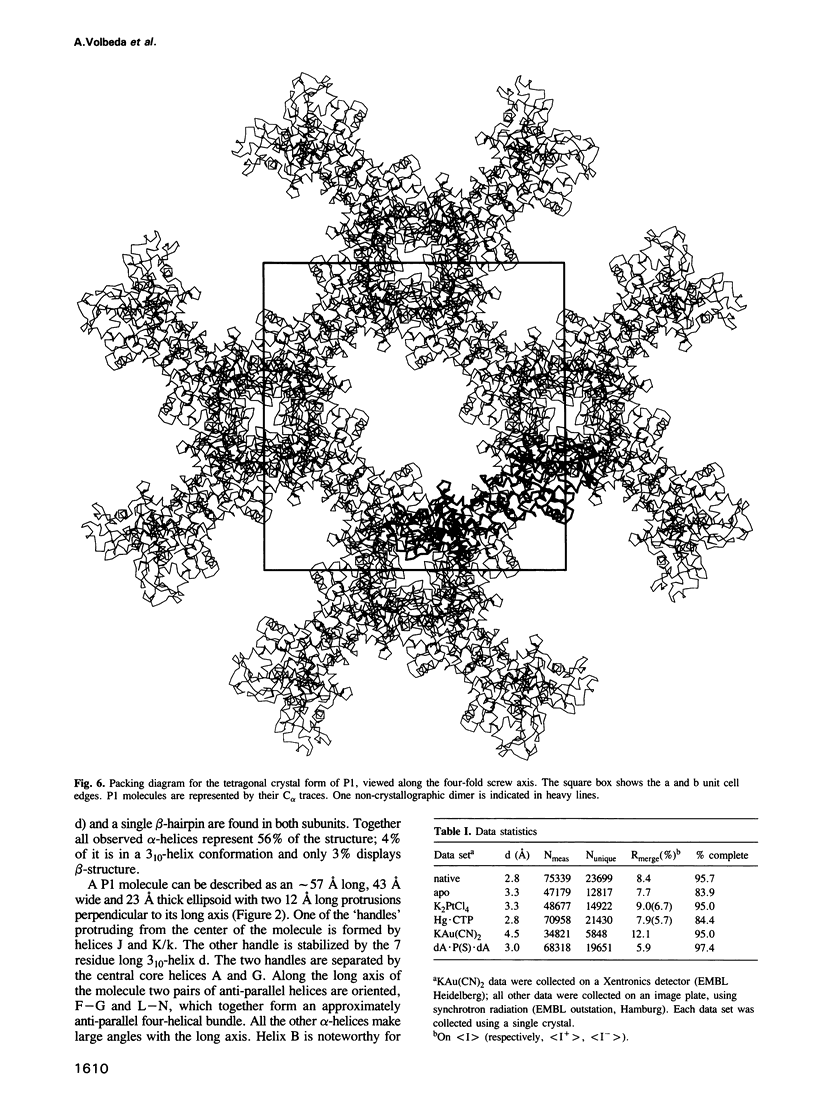
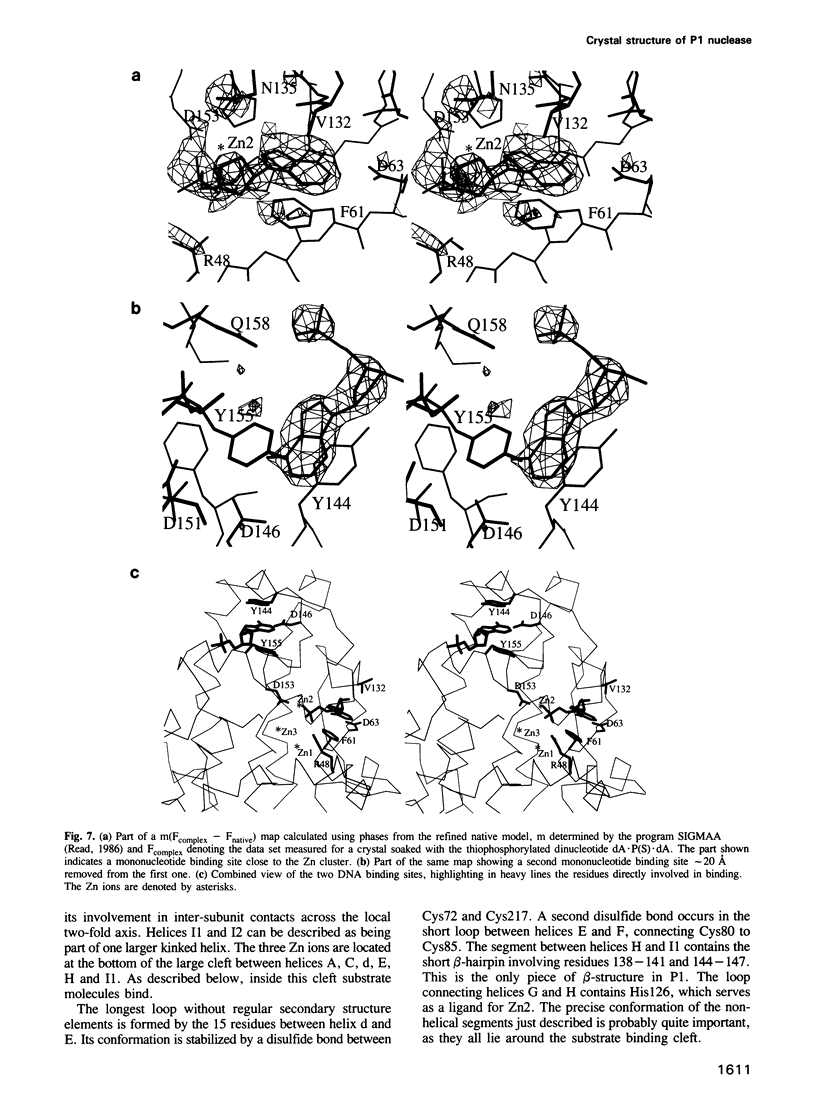
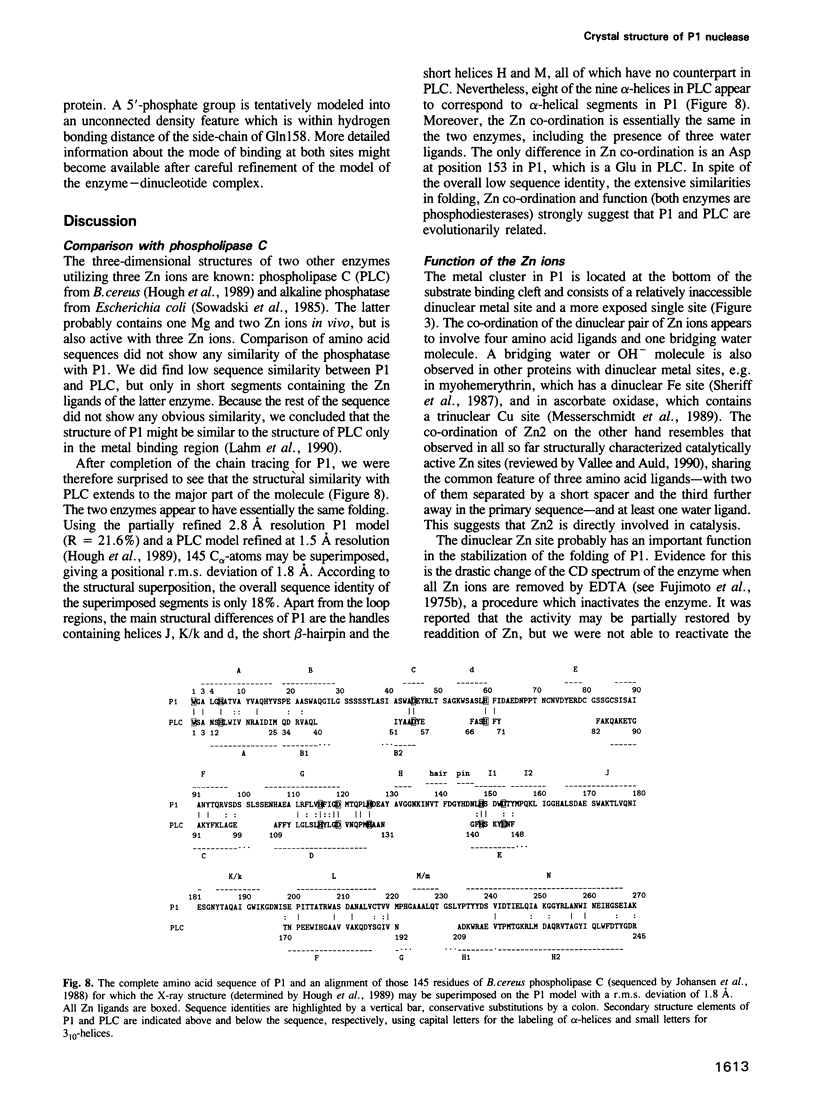
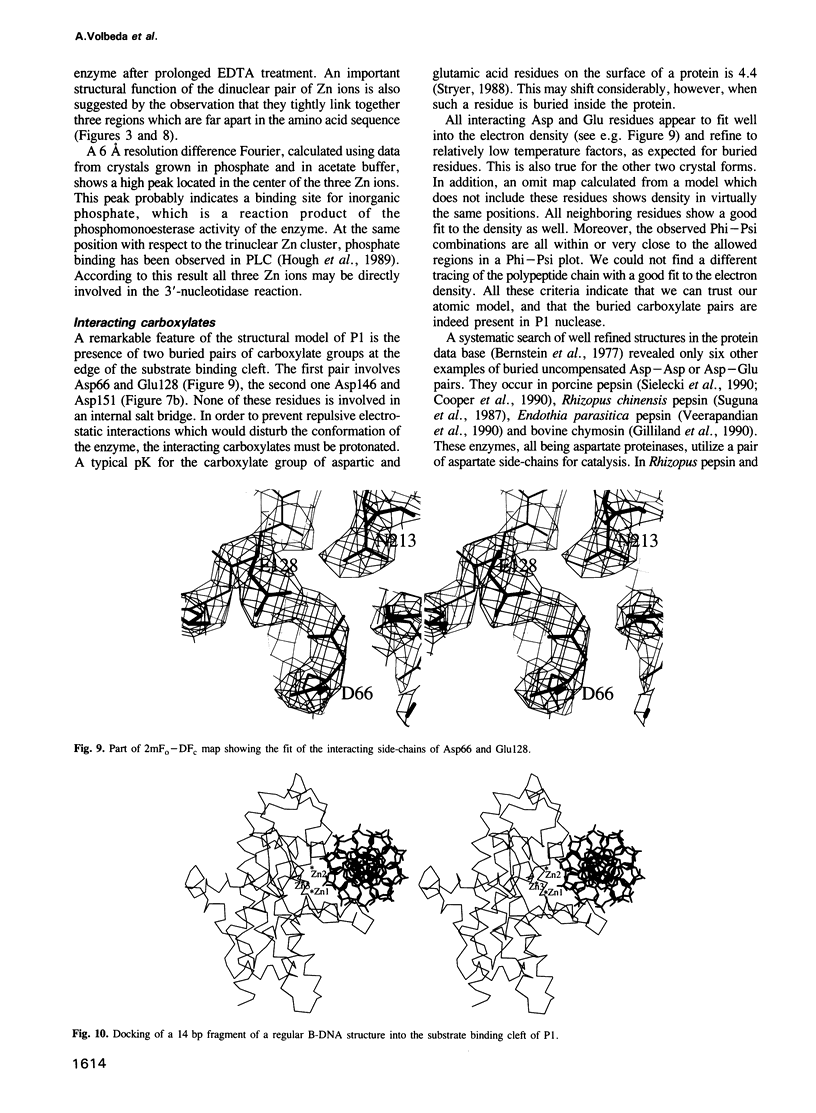
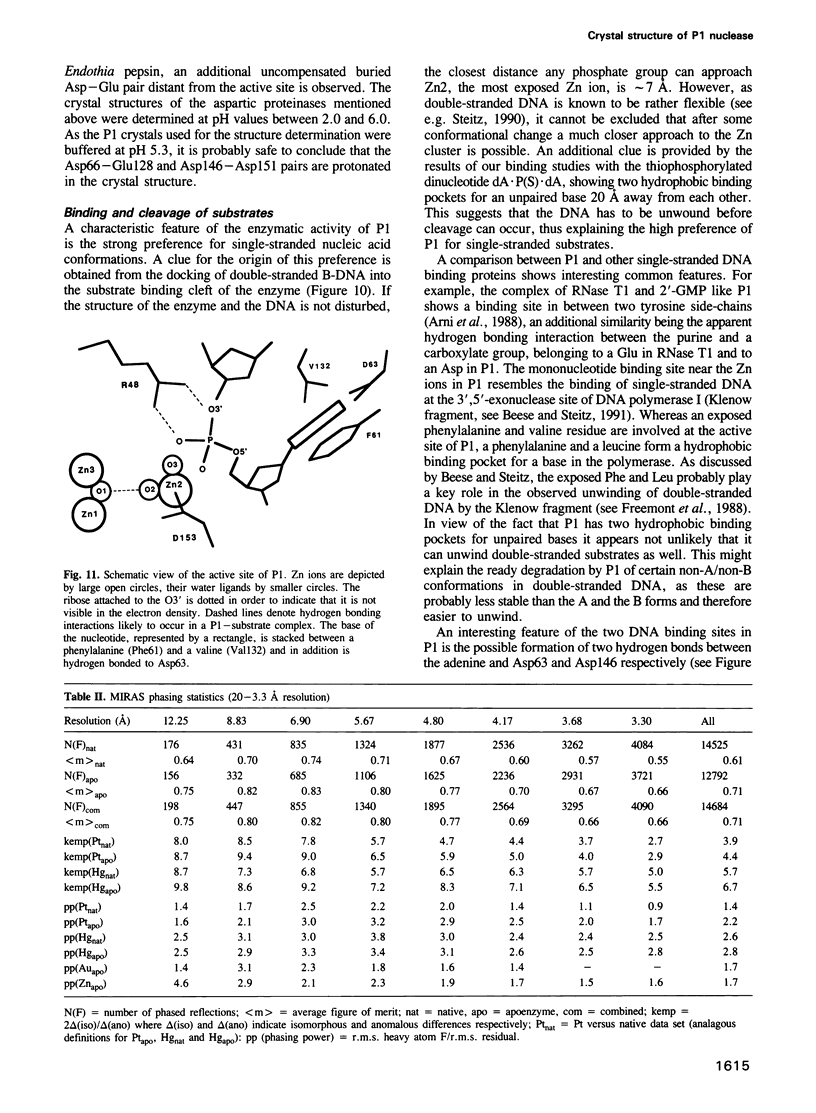
P1 nuclease from Penicillium citrinum is a zinc dependent glyco-enzyme consisting of 270 amino acid residues which cleaves single-stranded RNA and DNA into 5'-mononucleotides. The X-ray structure of a tetragonal crystal form of the enzyme with two molecules per asymmetric unit has been solved at 3.3 and refined at 2.8 A resolution to a crystallographic R-factor of 21.6%. The current model consists of 269 amino acid residues, three Zn ions and two N-acetyl glucosamines per subunit. The enzyme is folded very similarly to phospholipase C from Bacillus cereus, with 56% of the structure displaying an alpha-helical conformation. The three Zn ions are located at the bottom of a cleft and appear to be rather inaccessible for any phosphate group in double-stranded RNA or DNA substrates. A crystal soaking experiment with a dinucleotide gives clear evidence for two mononucleotide binding sites separated by approximately 20 A. One site shows binding of the phosphate group to one of the zinc ions. At both sites there is a hydrophobic binding pocket for the base, but no direct interaction between the protein and the deoxyribose. A cleavage mechanism is proposed involving nucleophilic attack by a Zn activated water molecule.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arni R., Heinemann U., Tokuoka R., Saenger W. Three-dimensional structure of the ribonuclease T1 2'-GMP complex at 1.9-A resolution. J Biol Chem. 1988 Oct 25;263(30):15358–15368. [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Breslow R., Huang D. L., Anslyn E. On the mechanism of action of ribonucleases: dinucleotide cleavage catalyzed by imidazole and Zn2+. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1746–1750. doi: 10.1073/pnas.86.6.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Chambers J. Crystal structure and stability of a DNA duplex containing A(anti).G(syn) base-pairs. J Mol Biol. 1989 May 20;207(2):455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- Cooper J. B., Khan G., Taylor G., Tickle I. J., Blundell T. L. X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 A resolution. J Mol Biol. 1990 Jul 5;214(1):199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Legg M. J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3',5'-bisphosphate-calcium ion complex at 1.5-A resolution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981 Apr 28;20(9):2361–2370. [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A. E., Kylsten P. M., Jones T. A., Liljas A. Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN- ion to the zinc at high pH. Proteins. 1988;4(4):283–293. doi: 10.1002/prot.340040407. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland G. L., Winborne E. L., Nachman J., Wlodawer A. The three-dimensional structure of recombinant bovine chymosin at 2.3 A resolution. Proteins. 1990;8(1):82–101. doi: 10.1002/prot.340080110. [DOI] [PubMed] [Google Scholar]
- Holmes M. A., Matthews B. W. Binding of hydroxamic acid inhibitors to crystalline thermolysin suggests a pentacoordinate zinc intermediate in catalysis. Biochemistry. 1981 Nov 24;20(24):6912–6920. doi: 10.1021/bi00527a026. [DOI] [PubMed] [Google Scholar]
- Hough E., Hansen L. K., Birknes B., Jynge K., Hansen S., Hordvik A., Little C., Dodson E., Derewenda Z. High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus. Nature. 1989 Mar 23;338(6213):357–360. doi: 10.1038/338357a0. [DOI] [PubMed] [Google Scholar]
- Johansen T., Holm T., Guddal P. H., Sletten K., Haugli F. B., Little C. Cloning and sequencing of the gene encoding the phosphatidylcholine-preferring phospholipase C of Bacillus cereus. Gene. 1988 May 30;65(2):293–304. doi: 10.1016/0378-1119(88)90466-0. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kim H., Lipscomb W. N. Crystal structure of the complex of carboxypeptidase A with a strongly bound phosphonate in a new crystalline form: comparison with structures of other complexes. Biochemistry. 1990 Jun 12;29(23):5546–5555. doi: 10.1021/bi00475a019. [DOI] [PubMed] [Google Scholar]
- Koizumi K., Okada Y., Fujimoto E., Takagi Y., Ishigami H., Hara K., Hashimoto H. Separation and characterization of three positional isomers of dimaltosyl-cyclomaltoheptaose (dimaltosyl-beta-cyclodextrin). Chem Pharm Bull (Tokyo) 1991 Aug;39(8):2143–2145. doi: 10.1248/cpb.39.2143. [DOI] [PubMed] [Google Scholar]
- Lahm A., Volbeda A., Suck D. Crystallisation and preliminary crystallographic analysis of P1 nuclease from Penicillium citrinum. J Mol Biol. 1990 Sep 20;215(2):207–210. doi: 10.1016/S0022-2836(05)80337-7. [DOI] [PubMed] [Google Scholar]
- Lesk A. M., Hardman K. D. Computer-generated schematic diagrams of protein structures. Science. 1982 Apr 30;216(4545):539–540. doi: 10.1126/science.7071602. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A., Rossi A., Ladenstein R., Huber R., Bolognesi M., Gatti G., Marchesini A., Petruzzelli R., Finazzi-Agró A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini. Analysis of the polypeptide fold and a model of the copper sites and ligands. J Mol Biol. 1989 Apr 5;206(3):513–529. doi: 10.1016/0022-2836(89)90498-1. [DOI] [PubMed] [Google Scholar]
- Potter B. V., Connolly B. A., Eckstein F. Synthesis and configurational analysis of a dinucleoside phosphate isotopically chiral at phosphorus. Stereochemical course of Penicillium citrum nuclease P1 reaction. Biochemistry. 1983 Mar 15;22(6):1369–1377. doi: 10.1021/bi00275a008. [DOI] [PubMed] [Google Scholar]
- Potter B. V., Romaniuk P. J., Eckstein F. Stereochemical course of DNA hydrolysis by nuclease S1. J Biol Chem. 1983 Feb 10;258(3):1758–1760. [PubMed] [Google Scholar]
- RAMACHANDRAN G. N., RAMAKRISHNAN C., SASISEKHARAN V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963 Jul;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Sheriff S., Hendrickson W. A., Smith J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987 Sep 20;197(2):273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- Shishido K., Habuka N. Purification of S1 nuclease to homogeneity and its chemical, physical and catalytic properties. Biochim Biophys Acta. 1986 Oct 29;884(1):215–218. doi: 10.1016/0304-4165(86)90247-3. [DOI] [PubMed] [Google Scholar]
- Sielecki A. R., Fedorov A. A., Boodhoo A., Andreeva N. S., James M. N. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution. J Mol Biol. 1990 Jul 5;214(1):143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]
- Sowadski J. M., Handschumacher M. D., Murthy H. M., Foster B. A., Wyckoff H. W. Refined structure of alkaline phosphatase from Escherichia coli at 2.8 A resolution. J Mol Biol. 1985 Nov 20;186(2):417–433. doi: 10.1016/0022-2836(85)90115-9. [DOI] [PubMed] [Google Scholar]
- Spitzer S., Eckstein F. Inhibition of deoxyribonucleases by phosphorothioate groups in oligodeoxyribonucleotides. Nucleic Acids Res. 1988 Dec 23;16(24):11691–11704. doi: 10.1093/nar/16.24.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Suguna K., Bott R. R., Padlan E. A., Subramanian E., Sheriff S., Cohen G. H., Davies D. R. Structure and refinement at 1.8 A resolution of the aspartic proteinase from Rhizopus chinensis. J Mol Biol. 1987 Aug 20;196(4):877–900. doi: 10.1016/0022-2836(87)90411-6. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Active-site zinc ligands and activated H2O of zinc enzymes. Proc Natl Acad Sci U S A. 1990 Jan;87(1):220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerapandian B., Cooper J. B., Sali A., Blundell T. L. X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution. J Mol Biol. 1990 Dec 20;216(4):1017–1029. doi: 10.1016/S0022-2836(99)80017-5. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]



