Abstract
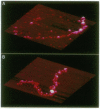
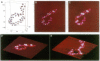
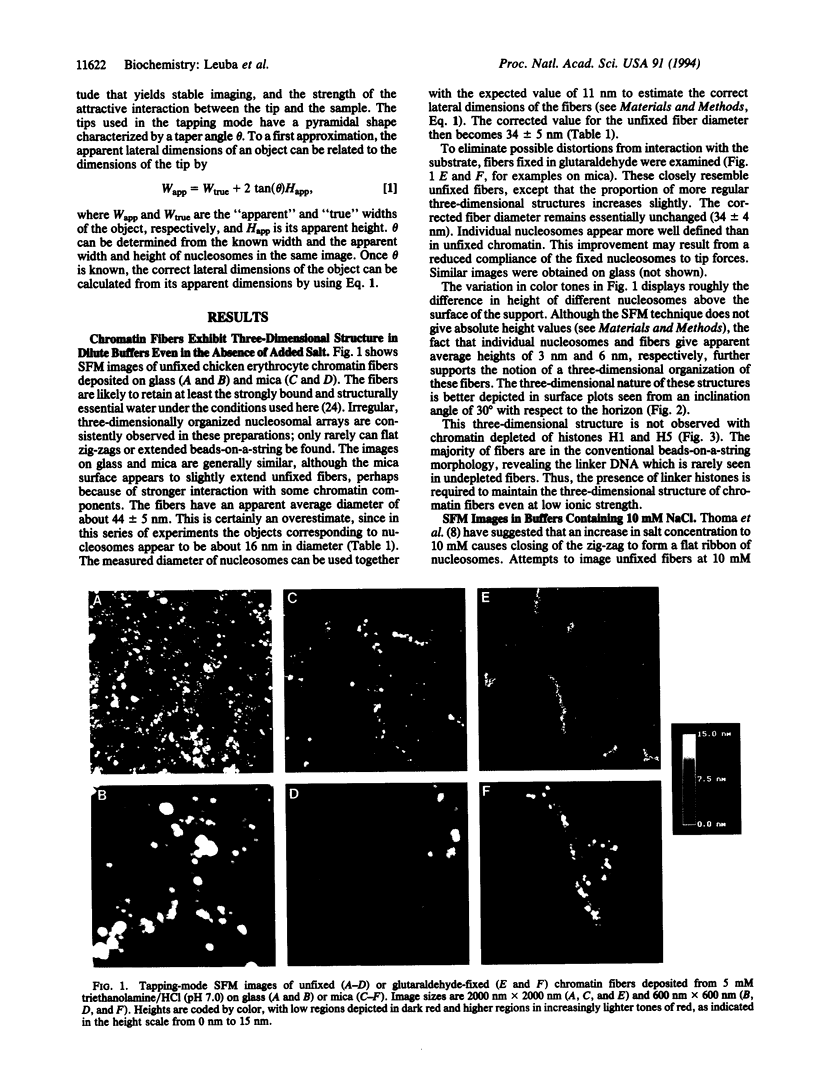
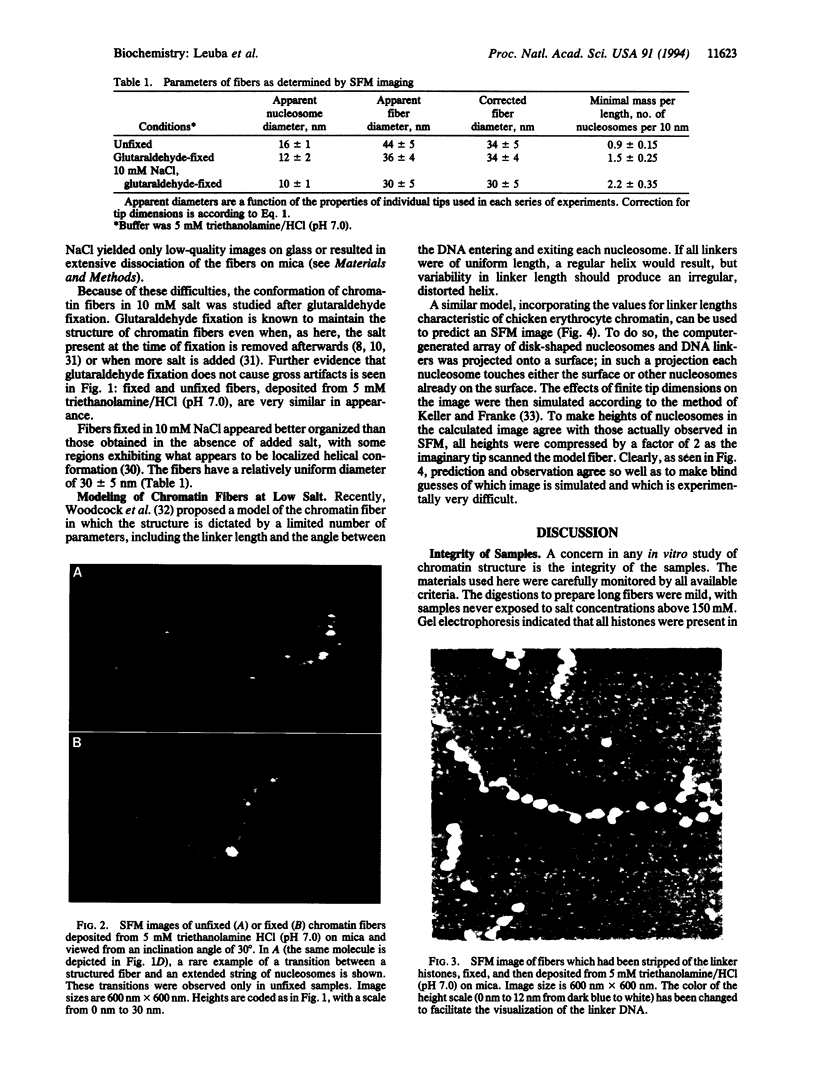
Unfixed chicken erythrocyte chromatin fibers in very low salt have been imaged with a scanning force microscope operating in the tapping mode in air at ambient humidity. These images reveal a three-dimensional organization of the fibers. The planar "zig-zag" conformation is rare, and extended "beads-on-a-string" fibers are seen only in chromatin depleted of histones H1 and H5. Glutaraldehyde fixation reveals very similar structures. Fibers fixed in 10 mM salt appear somewhat more compacted. These results, when compared with modeling studies, suggest that chromatin fibers may exist as irregular three-dimensional arrays of nucleosomes even at low ionic strength.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen M. J., Dong X. F., O'Neill T. E., Yau P., Kowalczykowski S. C., Gatewood J., Balhorn R., Bradbury E. M. Atomic force microscope measurements of nucleosome cores assembled along defined DNA sequences. Biochemistry. 1993 Aug 24;32(33):8390–8396. doi: 10.1021/bi00084a002. [DOI] [PubMed] [Google Scholar]
- Bavykin S. G., Usachenko S. I., Zalensky A. O., Mirzabekov A. D. Structure of nucleosomes and organization of internucleosomal DNA in chromatin. J Mol Biol. 1990 Apr 5;212(3):495–511. doi: 10.1016/0022-2836(90)90328-J. [DOI] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986 Mar 3;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. I. Synchrotron radiation X-ray scattering results. Eur Biophys J. 1986;13(3):157–173. doi: 10.1007/BF00542560. [DOI] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. II. Theoretical analysis of the X-ray scattering patterns and model calculations. Eur Biophys J. 1986;13(3):175–185. doi: 10.1007/BF00542561. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Cotter R. I., Pardon J. F. Light scattering measurements supporting helical structures for chromatin in solution. Nucleic Acids Res. 1978 May;5(5):1571–1580. doi: 10.1093/nar/5.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerchman S. E., Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Zlatanova J., van Holde K. On the location of histones H1 and H5 in the chromatin fiber. Studies with immobilized trypsin and chymotrypsin. J Mol Biol. 1993 Feb 20;229(4):917–929. doi: 10.1006/jmbi.1993.1096. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Zlatanova J., van Holde K. On the location of linker DNA in the chromatin fiber. Studies with immobilized and soluble micrococcal nuclease. J Mol Biol. 1994 Jan 21;235(3):871–880. doi: 10.1006/jmbi.1994.1045. [DOI] [PubMed] [Google Scholar]
- Libertini L. J., Small E. W. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 1980 Aug 25;8(16):3517–3534. doi: 10.1093/nar/8.16.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C., Bezot P., Hesse-Bezot C., Roux B., Bernengo J. C. Conformation of chromatin oligomers. A new argument for a change with the hexanucleosome. Eur J Biochem. 1981 Nov;120(1):169–176. doi: 10.1111/j.1432-1033.1981.tb05685.x. [DOI] [PubMed] [Google Scholar]
- Marion C. The structural organization of oligonucleosomes. J Biomol Struct Dyn. 1984 Oct;2(2):303–317. doi: 10.1080/07391102.1984.10507569. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Bakken A. H. Morphological studies of transcription. Acta Endocrinol Suppl (Copenh) 1972;168:155–177. doi: 10.1530/acta.0.071s155. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Perez-Grau L., Bordas J., Koch M. H. Chromatin superstructure: synchrotron radiation X-ray scattering study on solutions and gels. Nucleic Acids Res. 1984 Mar 26;12(6):2987–2996. doi: 10.1093/nar/12.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russanova V. R., Dimitrov S. I., Makarov V. L., Pashev I. G. Accessibility of the globular domain of histones H1 and H5 to antibodies upon folding of chromatin. Eur J Biochem. 1987 Sep 1;167(2):321–326. doi: 10.1111/j.1432-1033.1987.tb13339.x. [DOI] [PubMed] [Google Scholar]
- Sperling L. The mass per unit length of chromatin by low-angle x-ray scattering. FEBS Lett. 1976 Apr 15;64(1):89–91. doi: 10.1016/0014-5793(76)80256-6. [DOI] [PubMed] [Google Scholar]
- Suau P., Bradbury E. M., Baldwin J. P. Higher-order structures of chromatin in solution. Eur J Biochem. 1979 Jul;97(2):593–602. doi: 10.1111/j.1432-1033.1979.tb13148.x. [DOI] [PubMed] [Google Scholar]
- Swedlow J. R., Agard D. A., Sedat J. W. Chromosome structure inside the nucleus. Curr Opin Cell Biol. 1993 Jun;5(3):412–416. doi: 10.1016/0955-0674(93)90005-b. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T. Unravelled nucleosomes, nucleosome beads and higher order structures of chromatin: influence of non-histone components and histone H1. J Mol Biol. 1981 Jul 15;149(4):709–733. doi: 10.1016/0022-2836(81)90354-5. [DOI] [PubMed] [Google Scholar]
- Williams S. P., Athey B. D., Muglia L. J., Schappe R. S., Gough A. H., Langmore J. P. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986 Jan;49(1):233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L., Rattner J. B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984 Jul;99(1 Pt 1):42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager T. D., McMurray C. T., van Holde K. E. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 1989 Mar 7;28(5):2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Leuba S. H., Yang G., Bustamante C., van Holde K. Linker DNA accessibility in chromatin fibers of different conformations: a reevaluation. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5277–5280. doi: 10.1073/pnas.91.12.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]