Abstract
In the context of the FlyBase annotated gene models in Drosophila melanogaster, we describe the many exceptional cases we have curated from the literature or identified in the course of FlyBase analysis. These range from atypical but common examples such as dicistronic and polycistronic transcripts, noncanonical splices, trans-spliced transcripts, noncanonical translation starts, and stop-codon readthroughs, to single exceptional cases such as ribosomal frameshifting and HAC1-type intron processing. In FlyBase, exceptional genes and transcripts are flagged with Sequence Ontology terms and/or standardized comments. Because some of the rule-benders create problems for handlers of high-throughput data, we discuss plans for flagging these cases in bulk data downloads.
Keywords: bicistronic, stop-codon suppression, multiphasic exon, shared promoter, non-AUG translation start
The D. melanogaster genomic sequence assembly is of exceptionally high quality (Celniker et al. 2002; Hoskins et al. 2007, 2015) and is one of the few for which gene models have been manually annotated and assessed for all protein-coding and lncRNA genes (Matthews et al. 2015, which is the companion to this article). This has allowed FlyBase (dos Santos et al. 2015) to more easily identify and handle the rule-benders: gene models that incorporate exceptional or atypical transcription, splicing, or translation events. We summarize our current catalog of such exceptional gene models and events, including polycistronic transcripts, noncanonical splices, trans-spliced transcripts, noncanonical translation starts, stop-codon readthroughs, multiphasic coding exons, and ribosomal frameshifting. It is hoped that this extensively vetted compilation will support opportunities for further investigations into some of these rule-bending phenomena, including their biological impact, mechanistic bases, regulation, evolutionary development, and phylogenetic distribution.
Materials and Methods
The FlyBase gene model set consists entirely of manually annotated transcripts, with the exception of some classes of small noncoding RNAs. Gene model annotation guidelines and datasets informing the gene annotation process are described in Matthews et al. (2015). All data and gene models are available at FlyBase (http://flybase.org).
Results and Discussion
Exceptional cases are flagged at the gene and transcript levels
Atypical gene models, those that do not follow the canonical rules, can create confusion among biologists accustomed to better-behaved genes and can cause unexpected errors in bulk data assessments. Some categories, notably trans-spliced transcripts, are frequently assumed to be errors. Overlapping genes complicate the design of sequence-based reagents (Hu et al. 2013) and global assessments of levels of gene expression. To flag these exceptions for users, FlyBase identifies known cases of rule-bending gene models at several levels. Appropriate Sequence Ontology (SO) terms (Eilbeck et al. 2005) are associated with the gene records (Table 1). These may be found in the gene reports in the “Sequence Ontology: Class of Gene” subsection under the “Gene Model and Products” section. This is a controlled field with links from the terms to the FlyBase Vocabularies tool. A comment in the “Comments on Gene Models” field includes the relevant SO term and often additional information regarding the nature and attribution of the exception. Standardized comments have been added to individual rule-bending transcripts when appropriate (Supporting Information, Table S1); FlyBase is in the process of adding similar comments to the transcript headers in our bulk data files (Table 2). Flags of the type “translation exception” are particularly important because they break the rules by which a predicted protein is derived from an annotated transcript. Exceptional cases are also flagged in GenBank RefSeq transcript and protein entries (Table S2). Each of the following sections concludes with a description of the SO, transcript and GenBank flags used for that type of exceptional gene model.
Table 1. Gene-associated Sequence Ontology terms.
| SO Term | SO ID Number |
|---|---|
| gene_with_dicistronic_mRNA | SO:0000722 |
| gene_with_polycistronic_transcript | SO:0000690 |
| gene_with_trans_spliced_transcript | SO:0000459 |
| gene_with_unconventional_translation_start_codon | SO:0001739 |
| gene_with_translation_start_codon_CUG | SO:0001740 |
| gene_with_stop_codon_redefined_as_selenocysteine | SO:0000710 |
| gene_with_stop_codon_read_through | SO:0000697 |
| gene_with_transcript_with_translational_frameshift | SO:0000712 |
Table 2. Proposed transcript-associated flags to be included in FASTA files.
| Proposed Flag | Type |
|---|---|
| dicistronic_mRNA | Transcript exception |
| polycistronic_transcript | Transcript exception |
| non_canonical_splice_site | Transcript exception |
| endonuclease_spliced_intron | Transcript exception |
| trans_spliced_transcript | Transcript exception |
| non-canonical_start_codon | Translation exception |
| stop_codon_redefined_as_selenocysteine | Translation exception |
| stop_codon_read_through | Translation exception |
| transcript_with_translational_frameshift | Translation exception |
| mitochondrial_genetic_code | Translation exception |
| mitochondrial_incomplete_stop_codon | Translation exception |
| start_codon_not_determined | Translation exception |
| mutation in strain | Sequence alteration |
| genomic sequence error or gap | Sequence alteration |
Exceptional transcript structure (1): polycistronic (primarily dicistronic) transcripts are not uncommon
One of the surprising results of the first manual gene model annotation sweep of D. melanogaster performed by FlyBase (Misra et al. 2002) was the number of new dicistronic transcripts. At that time nine dicistronic loci had been previously described (Pauli et al. 1988; Schulz et al. 1990; Andrews et al. 1996; Brogna and Ashburner 1997; Ibnsouda et al. 1998; Niimi et al. 1999; Gray and Nicholls 2000; Liu et al. 2000; Walker et al. 2000). Misra et al. (2002) expanded the number to 31 dicistronic gene pairs that were confidently identified and another 17 that were tentatively identified. Dicistronic loci are no longer a surprise: there are 159 dicistronic gene pairs in annotation release 6.04. In addition, there are nine sets with transcripts encoding more than two genes (seven tricistronic and two tetracistronic) and one complex case involving overlapping dicistronics (of seq, Kdm4B, and CG17724). In total, 350 genes are annotated as sharing one or more transcripts with a neighboring protein-coding gene. Thus, 2.5% of protein-coding gene models include at least one polycistronic transcript. Note that FlyBase annotates each member of a dicistronic or polycistronic locus as a separate gene; this allows unambiguous association of gene-specific information, such as protein domains, molecular function, and mutant phenotypes. A listing of all polycistronic genes with additional information is found in File S1. [The mod(mdg4) polycistronic trans-splicing precursors are excluded from these totals and from this discussion; see description of trans-splicing below.]
In 2002, annotation of dicistronic loci was dependent on isolation of cDNA clones that spanned both genes. Now, RNA-Seq coverage data (Graveley et al. 2011; Brown et al. 2014) and whole genome assessments for transcription start sites (Hoskins et al. 2011; Batut et al. 2013) provide independent evidence of transcript extent and structure. Originally, some polycistronic genes were missed because it was difficult to determine which of many small open reading frames (ORFs) might correspond to functional protein-coding genes. Since then, the availability of genomic sequence information for multiple Drosophila species has allowed identification of some conserved ORFs as small as 20–30 amino acids (Lin et al. 2007, 2011; Hayden and Bosco 2008).
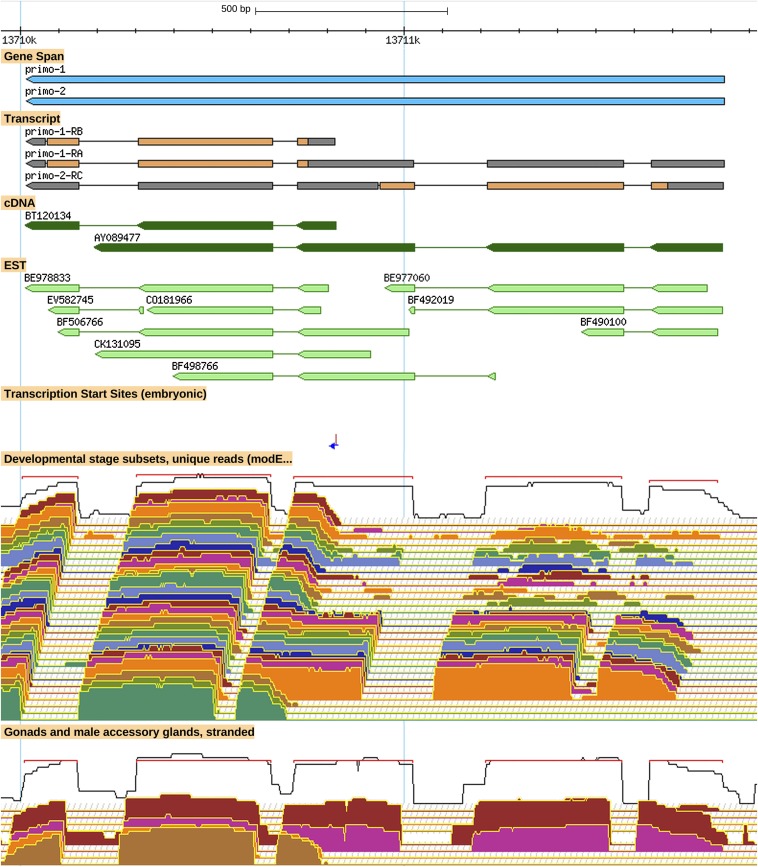
Polycistronic loci often encode alternative transcripts that are monocistronic. Of the 159 currently annotated dicistronic loci, there are only 45 for which polypeptides from both genes of the pair appear to be produced exclusively from dicistronic transcripts. There are also 45 loci for the opposite case, with support for alternative monocistronic transcripts for both genes. In some of these cases, the dicistronic transcript appears to be produced at a relatively low frequency or only in specific circumstances. For the remaining 69 loci, one gene has both monocistronic and dicistronic transcripts and the other is encoded only by dicistronic transcripts (example in Figure 1). The nine loci that include tricistronic and tetracistronic transcripts may encode a mix of polycistronic types and several also include an alternative monocistronic transcript (example in Figure S1). Although we cannot be certain that we have identified all alternative monocistronic transcripts, currently 181 gene models include only polycistronic transcript isoforms. Even if this is a significant overestimate, it is likely that more than 1% of protein-coding genes in D. melanogaster are encoded exclusively by polycistronic transcripts.
Figure 1.
A dicistronic transcript isoform for primo-1 and primo-2 is produced from a stage- and tissue-specific promoter. A GBrowse view showing (top to bottom): the gene extents and the gene models; cDNAs and ESTs; transcription start site(s); unstranded RNA-Seq coverage data corresponding to a developmental series (early embryos, top, to adults, bottom); and stranded RNA-Seq coverage data (plus strand top, minus strand bottom) corresponding to testis (red), male accessory gland (magenta), ovary from virgin females (orange), and ovaries from mated females (tan). More information on data presented in GBrowse may be found at http://flybase.org/wiki/FlyBase:GBrowse_Tracks#General.
Genes that encode small polypeptides are over-represented among polycistronic genes. There are 10 genes for which all annotated polypeptides are less than 25 amino acids; all are polycistronic, although one is also annotated with an alternative monocistronic transcript. Of the 65 genes for which all annotated polypeptides are between 25 and 45 amino acids, 15 (23%) are polycistronic. A number of genes in this category correspond to small conserved ORFs found in the 5′ or 3′ untranslated regions (UTRs) of longer coding genes (Hayden and Bosco 2008); these are defined as separate genes in FlyBase. There are undoubtedly additional translated ORFs less than 50 amino acids that will be identified by new techniques, such as ribosomal profiling combined with proteomic validation (see Gawron et al. 2014 for discussion of emerging approaches), so the list of polycistronic genes encoding small polypeptides is likely to grow.
Polycistronic loci are not a homogenous group (File S1). The distance between the upstream stop codon and the downstream start codon ranges from negative (overlapping in different ORFs) to over a kilobase. For loci that produce only polycistronic transcripts (no alternative monocistronics), the range between upstream stop and downstream start is −34 nt to 319 nt. Alternative monocistronic transcripts for the 3′ coding region are usually transcribed from an alternative downstream (or external) promoter; monocistronic transcripts for the 5′ coding region usually terminate at an upstream polyadenylation site. However, a number of alternative monocistronic transcripts are the result of alternative splices that disrupt one of the two coding regions. Approximately 18% of polycistronic loci include similar genes; many of these may have been created by tandem duplication. Among the other 82%, a few are known to be functionally related (Liu et al. 2000; Phillips et al. 2000), including the highly conserved example of the large and small subunits of Molybdopterin synthase 2, Mocs2 and CG42503 (Hayden and Bosco 2008).
Ben-Shahar et al. (2007) present preliminary evidence that the CheB42a and ppk25 genes are transcribed as a dicistronic pre-mRNA; they postulate that this pre-mRNA undergoes a unique cleavage event to produce two monocistronic mRNAs. This proposed mechanism differs from the polycistronic loci we define, for which polycistronic processed mRNAs are supported, and for which each cistron’s product is postulated to be translated from a single mRNA isoform. The gene models we describe as polycistronic necessitate internal initiation of translation for the downstream gene by use of an internal ribosome entry site (IRES) (reviewed in Hellen and Sarnow 2001), resumption of ribosomal scanning (Wall et al. 2005), or some other mechanism. Use of an IRES may be common, because although some dicistronic pairs appear to meet the requirements of a ribosomal scanning mechanism, most do not [see the discussion in Misra et al. (2002) and additional information tabulated in File S1].
FlyBase and GenBank flags:
Genes with dicistronic or polycistronic transcripts are identified by specific SO terms (Table 1). At the transcript level, comments are included in FlyBase transcript reports (Table S1), and a “dicistronic gene” exception is included in the GenBank RefSeq transcript entry (Table S2). The proposed bulk data flag is of the “transcript exception” type (Table 2).
Exceptional transcript structure (2): adjacent genes may share exons, noncoding and coding
There are many cases in which genes on the same strand overlap each other. The most extreme cases are the polycistronic gene models, as described above. Two categories we have not viewed as sufficiently unruly that they merit special treatment or comments: a gene that is nested in the intron of a gene on the same strand or a gene with a 3′ UTR that overlaps the 5′ UTR of the downstream gene. Intermediate cases in which two genes share an exon, usually including a shared splice site, are described below. See File S2 for a complete listing of the genes discussed in this section.
The first example in the intermediate category is that of shared exons, but not shared coding DNA sequence (162 gene models flagged). Most common are cases in which two genes share a promoter and 5′ noncoding exons (130 genes); this includes 13 pairs for which one gene is coding and the other is noncoding. In some cases, the shared promoter is used by all transcripts of both genes, for example, Ip259 and RpS27A. In other cases, for example, CG2911 and Spec2, one or both gene models include transcripts derived from unshared promoters. Genes may also share 3′ UTRs (14 genes); an extreme example is the set comprising inaF-A, inaF-B, inaF-C, and inaF-D (Cheng and Nash 2007). There are eight sets (18 genes) described as complex or atypical cases, usually in which both 5′ and 3′ UTRs overlap.
The second example in the intermediate category consists of genes that share exons including a short extent of the coding sequence (in the same open reading frame), usually at the amino terminus. Historically, FlyBase categorized any transcripts that shared any amount of coding sequence as belonging to one gene. This policy has been changed for a small number of cases in which the genes in question are clearly functionally and evolutionarily distinct. Of the cases currently annotated (10 pairs and one triplet), most of the genes share a promoter, the first exon, and a translation start for at least one pair of transcript isoforms; some of these genes also have alternative transcript isoforms with different promoters that do not share coding sequences with the second gene.
A well-studied case of such coding sequence (CDS) overlap is that of Su(var)3-9 and eIF-2γ (Krauss et al. 2006), which may have resulted from the transposition of Su(var)3-9 into an intron of eIF-2γ. The two genes share a promoter, translation start, and 80 N-terminal residues that are similar to the N-terminus of eIF-2γ-like proteins in other species. The two genes, however, encode polypeptides with very different functions: histone methylation and translation initiation. This gene fusion encodes the only D. melanogaster orthologs for two different highly conserved eukaryotic genes. There is no transcript isoform for either gene that includes the characteristic domains of both Su(var)3-9 and eIF-2γ. The gene fusion appears to be insect-specific, with instances of re-fission in aphids (Krauss et al. 2006).
FlyBase flags:
Several comments are used to flag FlyBase gene models with shared noncoding exons: “Shares 5′ UTR,” “Shares 3′ UTR,” “Shares 5′ exon(s),” and “Complex/atypical overlap,” followed by additional explanatory information. Genes that contain overlapping coding extents are flagged with gene model comments that begin “Genes with CDS overlap,” followed by additional specific information. The affected transcripts are not flagged; an exceptional translation flag is not necessary.
Exceptional transcript structure (3): overlapping genes or alternative transcripts may share multiphasic or bidirectional regions of coding sequence
Coding regions for which more than one overlapping ORF on the same strand appears to be used are described as “multiphasic.” There are 269 gene models flagged with a comment indicating that the current annotation includes transcripts that share a multiphasic region; a complete listing is available in File S3. These annotations are based on data supporting different transcript isoforms; in most cases, there are no data addressing whether both protein isoforms are biologically relevant. The majority of cases correspond to transcripts that include a variably spliced intron that results in a frameshift in the next exon, a short multiphasic extent, and an alternative stop codon. If the multiphasic extent is less than 40 nucleotides (N = 133), we flag the gene model with the comment “Alternative translation stop created by use of multiphasic reading frames within coding region.” Because the nucleotide extent includes the stop codon, this corresponds to less than 13 amino acids. In some instances, the frameshift produces a significantly truncated protein, for example, Ucp4B and Start1; in others, the resulting change in carboxy sequence appears to be minor, for example, CG15278. Transcripts with a trunctated CDS are flagged with an additional comment that concludes “results in premature stop codon and/or downstream start; may or may not produce functional polypeptide” (see Matthews et al. 2015). The multiphasic extent may be longer than 40 nucleotides (N = 118) or involve the initial coding exon (N = 18); and examples include NetA and CG31948. These are flagged with the comments “Multiphase exon postulated: exon reading frame differs in alternative transcripts” or “Multiphase exon postulated: reading frame of first coding exon differs in alternative transcripts.” For 83 genes, the annotated multiphasic overlap exceeds 63 nucleotides (>20 amino acids); the longest is the 704-nucleotide multiphasic overlap found for transcripts of the moody gene (Bainton et al. 2005). For several genes, production and function of both alternative protein isoforms derived from a multiphasic exon have been demonstrated (including Ada2b, Pankotai et al. 2013; moody, Bainton et al. 2005; and Xbp1, a unique case described below).
Multiphasic regions may involve two genes. There are 36 genes (18 pairs) flagged with the comment “Multiphase exon postulated: this gene shares a region of coding sequence with an overlapping gene, but different reading frames are utilized in the overlapping coding region.” Of these, 26 are encoded on polycistronic transcripts. Most overlap by less than 40 nucleotides; however, for five pairs the multiphasic overlap exceeds 63 nucleotides. The most dramatic example is the att-ORFA and att-ORFB pair, which overlaps for 553 nucleotides across two exons and for which there is evidence that both protein products are produced (using an in vitro translation assay) (Madigan et al. 1996). Currently, there is a single pair of genes annotated with overlapping coding extents on opposite strands: CG34148 and P5CDh2. We describe such a shared region as “bidirectional” and flag these genes with the comment “Bidirectional region of coding sequence postulated: a portion of the CDS of this gene overlaps a portion of the CDS of a gene on opposite strand.” Criteria for annotating bidirectional genes are discussed in Matthews et al. (2015).
FlyBase flags:
Genes with multiphasic exons or bidirectional regions are flagged with the comments described above. The affected transcripts are not flagged; an exceptional translation flag is not necessary.
Exceptional splicing (1): noncanonical splices are rare
For 99% of the ∼60,000 annotated introns, the primary canonical splice donor-acceptor pair GT-AG is used; for most of the remaining 1%, the secondary canonical splice donor-acceptor pair GC-AG is used (563 GC-AG out of 60,223 total in release 6.03). Thus, the number of introns for which other splice donor-acceptor pairs are used is very low: only 79 are annotated in current gene models (a complete listing is available in File S4). Considering the high levels of cDNA and RNA-Seq junction data that have been incorporated into the current gene model annotations, it is likely that most typically used noncanonical splices have been identified. Introns supported only by very low-frequency data are not usually included in FlyBase gene models (Matthews et al. 2015); there are some additional noncanonical splices in this category, primarily representing potential alternatively spliced introns within 5′ UTRs. Introns processed by the U12 spliceosome have been well characterized (Schneider et al. 2004; Alioto 2007; Lin et al. 2010); remarkably, many are conserved between flies and humans (Lin et al. 2010). Most of the AT-AC introns in the current annotation set are of the U12 type (Table 3) and had been previously identified.
Table 3. Introns with noncanonical splice sites and/or U12-type 5′ consensus sequence.
| Splice Donor-Acceptor Pair | Number in Release 6.04 | Number with RNA-Seq Junction Support | Number with Similar Alternative Splice | Within Coding | Within 5′ UTR | Within lncRNA |
|---|---|---|---|---|---|---|
| AT-AC (U12) | 9 | 9 | 1 | 9 | 0 | 0 |
| AT-AC (U2) | 4 | 4 | 2 | 4 | 0 | 0 |
| GT-TG | 23 | 19 | 22 | 13 | 9 | 1 |
| GT-GG | 6 | 5 | 5 | 3 | 2 | 1 |
| GT-CG | 8 | 8 | 8 | 6 | 2 | 0 |
| GT-AT | 14 | 11 | 14 | 11 | 3 | 0 |
| GT-AA | 3 | 3 | 3 | 2 | 1 | 0 |
| GA-AG | 12 | 12 | 5 | 8 | 3 | 1 |
| GG-AG | 0 | — | — | — | — | — |
| GT-AC | 0 | — | — | — | — | — |
| Total | 79 | 71 | 60 | 56 | 20 | 3 |
| GT-AG (U12) | 10 | 9 | 9 | 1 | 0 | |
| GC-AG (U12) | 1 | 1 | 1 | 0 | 0 |
With the exception of AT-AC (and the mechanistically different HAC1-type intron splice sites, see below), the noncanonical splice sites currently annotated in D. melanogaster differ from GT-AG by only one base, with the G of the donor site being invariant. Of the eight possible pairs (excluding GT-AG and GC-AG) that fit these criteria, six are observed (Table 3). The distribution among genes is nonrandom: 10 genes are annotated with more than one intron defined by a noncanonical splice pair, despite the very low frequency of such introns (File S4). For the majority of cases there is a similar alternative canonical splice (Table 3); this has also been observed in humans (Szafranski et al. 2007; Parada et al. 2014). Although in some cases this may be a mechanism to increase protein variation (example in Figure 2A), similar alternative splices are also observed for splices that occur in 5′ UTRs. The key exceptions to this pattern of similar alternative splices are U12 spliceosome introns and GA-AG introns.
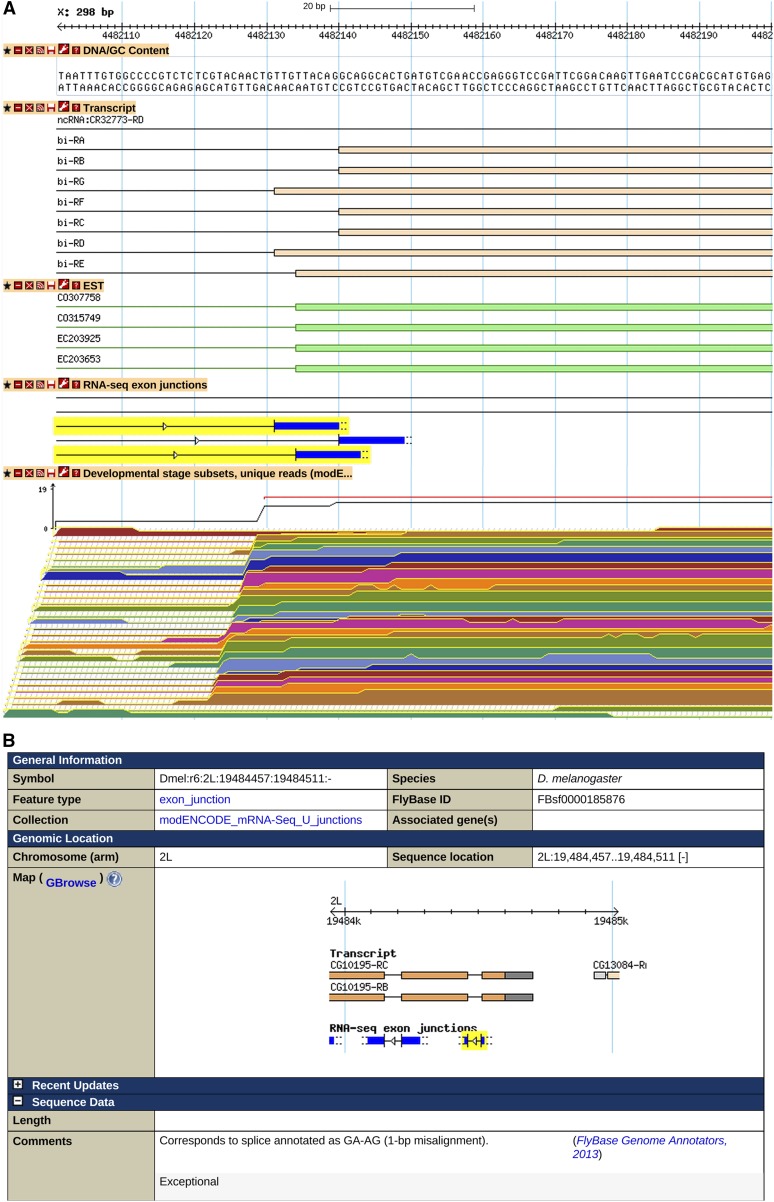
Figure 2.
Noncanonical splices supported by RNA-Seq junction data. (A) Of three alternative splice acceptors for intron 6 of the bifid (bi) gene, two are noncanonical TGs, including the splice acceptor used at the highest frequency (first highlighted junction). A GBrowse view showing (top to bottom): nucleotide sequence; region of the gene model showing one intron/exon boundary; EST data; RNA-Seq junction data; and unstranded RNA-Seq coverage data corresponding to a developmental series (early embryos, top, to adults, bottom). More information on data presented in GBrowse may be found at http://flybase.org/wiki/FlyBase:GBrowse_Tracks#General. (B) Report for an RNA-Seq junction that corresponds to a noncanonical splice but is aligned to incorrect noncanonical sites, one of several cases that were slightly misaligned.
Noncanonical splices frequently foil gene model prediction programs and complicate cDNA alignments and algorithms for deriving RNA-Seq junction data. Currently, 71 of the 79 annotated noncanonical splices have a supporting RNA-Seq junction (Table 3). For most (57 of 71), the alignment is impressively accurate; however, a number of the junctions are slightly misaligned and thus do not appear to map precisely to the annotated intron. A particular problem for unstranded RNA-Seq data are GT-AT splices, which are usually called as AT-AC splices on the opposite strand (eight of 10 cases). For exceptional junctions that correspond to annotated introns, the noncanonical acceptor/donor and any alignment inconsistencies are identified in explanatory comments in the FlyBase RNA-Seq junction reports (Figure 2B).
A more significant problem for identification of noncanonical splices by RNA-Seq junction analysis is false positives; including AT-AC calls, there are more than 500 exceptional junctions from the combined Baylor (Daines et al. 2011) and modENCODE datasets (Graveley et al. 2011). Some of these can be culled by eliminating low-frequency junctions and those with weak confidence scores; however, exceptional junctions within the middle ranges of frequency or confidence scores need to be manually assessed. Common problems (shared with nonexceptional junctions) include spurious calls within coding sequences comprising repeated motifs (for example, CG10953 and Eig71Ee), mismatch of acceptor and donor calls across related tandem genes (for example, the Jon99C cluster and the trypsin cluster at 47F), atypical calls within very highly expressed genes (for example, RpS13 and Cyt-b5-r), and RNA-Seq data mapping to unmasked repeat elements. A number of exceptional junction calls are one nucleotide off from a well-supported canonical splice; these are assumed to be artifactual.
Currently, there are approximately 40 transcripts in FlyBase that are annotated with noncanonical splices as a matter of convenience; most of these splices are unlikely to occur in vivo and are not included in the description of supported noncanonical splices above. They are used to allow representation of an intact gene model when the gene extent is interrupted by a transposable element insertion or a genomic sequence gap or is within a heterochromatic region that may vary from strain to strain. These cases are flagged with specific explanatory comments (Table S2).
FlyBase and GenBank flags:
Genes with transcripts annotated with noncanonical splices are not flagged; there is no appropriate gene-level SO term. Comments at the transcript level are included in FlyBase transcript reports (Table S1) and as a “nonconsensus splice site” exception in the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “transcript exception” type (Table 2).
Exceptional splicing (2): a single HAC1-type intron splice is highly conserved
Several rule-defying exceptional gene models are, in fact, examples of uncommon, yet very highly conserved, phenomena. One notable example is that of Xbp1 (Ryoo et al. 2007), a gene with an alternatively spliced isoform that has a very short intron (23 nt) flanked by noncanonical splice junctions (CA-TG). This alternative isoform is translated into a protein with the carboxy terminus in a different reading frame with respect to the unspliced isoform. Although this looks like an especially egregious annotation error, it is actually an example of Ire1-mediated unconventional splicing, a process conserved from yeast to mammals as part of the unfolded protein response (UPR). In this response to ER degradation stress, the protein Ire1 acts as a sensor and mediates a signaling cascade; it also acts as an endonuclease with a single target: the HAC1 ortholog Xbp1. The unconventionally spliced Xbp1 transcript is translated into a bzip transcription factor that upregulates genes responsive to ER stress. Like most organisms, D. melanogaster has exactly one example of the HAC1-type intron splice mechanism (Sidrauski and Walter 1997; Plongthongkum et al. 2007; Hooks and Griffiths-Jones 2011).
FlyBase and GenBank flags:
The Xbp1 gene report includes an explanatory comment in the gene model comment section. The report for the transcript that results from Ire1-mediated splicing includes a similar explanatory comment (Table S1) plus the flag “endonuclease_spliced_intron” (Table 2). A “nonconsensus splice site” exception appears in the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “transcript exception” type (Table 2).
Exceptional splicing (3): trans-splicing is well supported for two genes
At least two genes in D. melanogaster undergo trans-splicing, a process by which a mature mRNA is created by a bimolecular splice between two independently transcribed pre-mRNAs. In both known cases, the gene encodes multiple DNA-binding proteins with a common amino BTB/POZ domain and variable carboxy zinc-finger domains. The initial and more dramatic example is mod(mdg4) (Labrador et al. 2001; Dorn et al. 2001), which encodes more than 30 protein isoforms, at least 18 of which appear to be trans-spliced. The second example is lola, a gene that encodes at least 20 protein isoforms, one of which shows evidence of being trans-spliced (Horiuchi et al. 2003). In recent work, Gao et al. (2015) have begun to elucidate the molecular mechanisms of trans-splicing in the mod(mdg4) and lola systems.
These two examples of trans-splicing are in the category of intragenic trans-splicing, in contrast to the better-characterized category of spliced leader (SL) trans-splicing events originally observed in trypanosomes and nematodes (reviewed in Lasda and Blumenthal 2011) and in contrast to intergenic trans-splicing, which generates chimeric mRNAs. Both intragenic and intergenic trans-splicing have been observed in mammalian cells, but these events appear to be rare and in some cases are associated with neoplastic cells (reviewed in Horiuchi and Aigaki 2006).
In the case of mod(mdg4), five clusters of polycistronic transcripts encoding multiple 3′ alternative termini are supported by cDNA/EST and transcriptional start site data (Nechaev et al. 2010; Batut et al. 2013). These have been annotated as separate genes by FlyBase, with symbols such as pre-mod(mdg4)-T for the precursor of mod(mdg4)-RT. Two of these clusters are transcribed from the strand opposite to the rest of the mod(mdg4) exons, and are what precipitated the discovery of trans-splicing in D. melanogaster. It appears likely that this locus has undergone multiple small inversions since the Drosophilidae diverged from other dipterans: the genomic pattern of trans-encoded exons is observed in other Drosophila species (Gabler et al. 2005), but in mosquitoes it appears that all the 3′ alternative exons are located on the same strand (http://vectorbase.org, AGAP003439) (Krauss and Dorn 2004; Megy et al. 2012).
For lola there is clear support for at least one trans-spliced precursor, including EST and transcription start site data. It appears to be monocistronic and corresponds to the 3′-most alternative exon; it has been annotated as a separate gene in FlyBase, pre-lola-G.
Additional candidate genes subject to trans-splicing have been suggested but await definitive evidence (McManus et al. 2010). For broad, another gene encoding multiple isoforms with a constant BTB/POZ domain and variable zinc-finger domains, there are data supporting a transcription start site 5′ of the last two variable exons (Batut et al. 2013), suggesting that this gene may undergo trans-splicing. A number of complex gene model annotations include short 3′ isoforms with supported alternative downstream transcription starts; in some cases, there are also 5′ isoforms that do not overlap the 3′ isoforms (for example, vir-1, CG43427, dlg1). It is interesting to speculate that some of these loci may also be subject to trans-splicing.
FlyBase and GenBank flags:
Genes with trans-spliced transcripts, including the 3′ trans-splicing precursors, are identified by a SO term (Table 1). For the nine mod(mdg4) spliced transcripts derived from exons on opposite strands, a comment is included in FlyBase transcript reports (Table S1) and a “trans_splicing” comment appears in the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “transcript exception” type (Table 2).
Exceptional transcript modification: A-to-I RNA editing is noted at the genome level only
Post-transcriptional modifications of pre-mRNAs that result in changes to the nucleotide sequence are described as RNA editing. A-to-I RNA editing results from modification of an adenosine to inosine, which subsequently acts as would a guanine in terms of translation, splicing, and in the formation of secondary structures (reviewed in Mallela and Nishikura 2012). In D. melanogaster, 2005 putative A-to-I RNA editing sites have been identified and mapped on the genome, overlapping the transcripts of 1307 genes (Graveley et al. 2011; Rodriguez et al. 2012). A more recent study (St. Laurent et al. 2013) has not yet been incorporated into FlyBase. Most genes subject to A-to-I editing have at least three associated editing sites, and two genes have more than 20 overlapping editing sites (para is associated with 23, NaCP60E with 21). Because editing is developmentally regulated, and because the efficiency of each editing site varies, different combinations of editing are also possible for a given transcript, further adding to the vast number of possible permutations.
FlyBase does not attempt to represent alternative transcript or polypeptide sequences that may result from RNA editing. Identified A-to-I editing sites are annotated on the genome and are viewable in relation to gene model annotations on GBrowse. A Sequence Feature Report for each editing site details the observed editing frequency at various developmental stages, as well as the potential impact of a given editing site on the coding sequence of the related transcript (see, for example, A-I_edit_000244, FBsf0000383608). Each of these identified genomic sites is associated with the SO term “modified_RNA_base_feature” (SO:0000250). The SO term “gene_with_edited_transcript” (SO:0000548) is used to flag genes subject to A-to-I RNA editing.
Exceptional translation (1): noncanonical translation starts have been difficult to identify
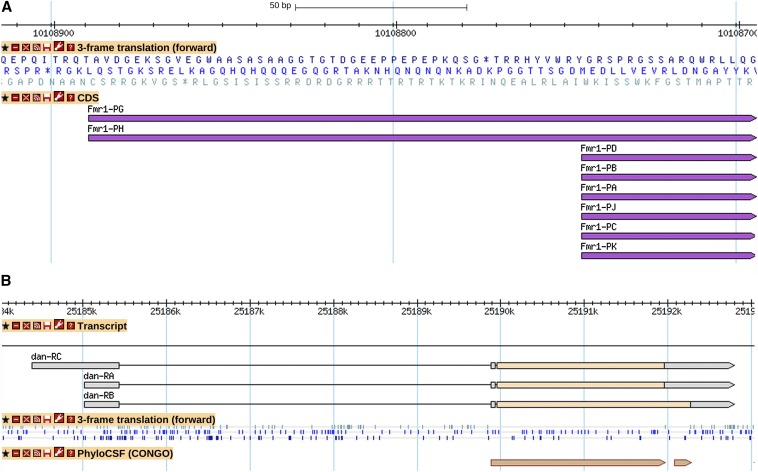
Thus far, no systematic or definitive study of non-AUG translation initiation in Drosophila has been performed. Individual cases have been discovered more or less by chance; some of the more thoroughly characterized include ChAT (Sugihara et al. 1990), ewg (de Simone and White 1993), Eip74EF (Boyd and Thummel 1993), Akt1 (Andjelkovic et al. 1995), and Fmr1 (Figure 3A) (Beerman and Jongens 2011). The majority of the currently annotated cases, including all based on FlyBase analysis (Table S3), are postulated due to the lack of an appropriately placed AUG codon; most are supported by assessment of conservation among Drosophila species or a favorable sequence context for translation initiation, but no additional experimental data.
Figure 3.
Noncanonical terminal extensions of the CDS. (A) CUG start codon in Fmr1 results in a 48-aa N-terminal extension; a GBrowse view showing amino acid sequence and amino ends of annotated polypeptides. Use of this alternative start codon has been confirmed by Western blot, mutagenesis of reported constructs, and rescue constructs (Beerman and Jongens 2011). (B) For the dan gene model, a stop-codon readthrough annotated for dan-RB is supported by PhyloCSF analysis (conservation of protein signatures). A GBrowse view showing (top to bottom): the gene model; stop codons on the plus strand in each of the three open reading frames; and regions of protein conservation among the Drosophila species (tan extents at the bottom). More information on data presented in GBrowse may be found at http://flybase.org/wiki/FlyBase:GBrowse_Tracks#General.
Recently, several systematic studies have allowed a more comprehensive characterization of noncanonical translation initiation in human and mouse. Searching for evolutionary signatures of protein-coding sequences within predicted 5′ UTRs, Ivanov et al. (2011) found 42 new and confirmed 17 previously reported non-AUG starts in humans. All are near-cognates (differing by one base) of AUG; CUG is by far the most common (42%). It may be a requirement that the second base is a pyrimidine, because AAG and AGG starts were not found. Ribosome profiling in mouse embryonic stem cells (Ingolia et al. 2011) indicates that for many transcripts multiple translation starts may be used, primarily additional AUG sites, but also near-cognate codons. Again, CUG was found to be the most common noncanonical start codon. It appears that during translation a non-AUG start codon is still paired with the usual initiating tRNA, Met-tRNAi, and that the protein initiates with a methionine (Peabody 1989; Menschaert et al. 2013). Thus, in FlyBase, the predicted protein sequence from a non-AUG translation start initiates with a methionine, not the amino acid that is usually associated with that codon.
In FlyBase, there are currently 27 genes with transcripts annotated with a non-AUG translation start codon; 11 of these use a CUG start codon (Table S3). However, it seems likely that there are, in fact, many-fold more, especially in the category of alternative translation starts. There are a significant number of gene models for which an alternative splice or an alternative 5′ exon removes all in-frame AUG translation starts within the amino end of the putative protein. Most of these are currently annotated as short protein isoforms (using a downstream AUG), but may in fact be instances in which a non-AUG start is used. It is interesting that the amorphic y1 mutation is caused by a replacement of the AUG start codon with a CUG codon (Matthews et al. 2015). Despite the fact that CUG is the mostly widely used noncanonical translation start, it is unable to function as a start codon in this context.
FlyBase and GenBank flags:
Genes with transcripts annotated with noncanonical translation starts are flagged with SO terms (Table 1) in the gene model comment section. Comments at the transcript level are included in FlyBase transcript reports (Table S1); a “non-AUG translation initiation” comment and a translation exception flag appear the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “translation exception” type (Table 2).
Exceptional translation (2): few selenoproteins are found in flies
Selenocysteine stop-codon readthrough is another example of rule-bending on a translational level: a UGA codon is read not as a stop signal, but rather as a 21st amino acid, selenocysteine (Sec; “U” in the single-letter code) (Castellano et al. 2001). Selenoprotein synthesis requires a specialized tRNA, Sec-tRNAsec, as well as the proteins involved in the synthesis of Sec-tRNAsec. The selenocysteine UGA codon is distinguished from a UGA stop codon by the presence of downstream stem-loop structure, the Sec insertion sequence (SECIS) in the 3′ UTR of the transcript, which is recognized by SECIS-binding protein (Martin-Romero et al. 2001). Selenocysteine is not only a means to allow a stop-codon readthrough; it also has a catalytic advantage over cysteine in the active site of oxidoreductases (Driscoll and Chavatte 2004).
D. melanogaster has three identified selenoprotein genes: BthD, SelG, and Sps2, which were identified by both in silico analysis of the D. melanogaster genome (Castellano et al. 2001) and by metabolic labeling with 75Se (Martin-Romero et al. 2001). BthD and Sps2 have the Sec insertion quite early in the final protein; SelG, however, extends only two amino acids past the substituted stop codon. Selenoproteins are less well conserved than some of the more rare phenomena: flies have three, humans have 25, worms have only one, and yeast and higher plants lack them altogether. Many of the mammalian selenoproteins themselves are conserved, but the invertebrate nonselenoprotein orthologs have a cysteine in place of Sec (Driscoll and Chavatte 2004). In fact, at least two of the D. melanogaster selenoprotein genes have nonselenoprotein paralogs genomically nearby (Castellano et al. 2001). In contrast, prokaryotes and archaebacteria have a completely different complement of selenoproteins (Driscoll and Chavatte 2004).
FlyBase and GenBank flags:
Selenoprotein genes are identified by a specific SO term (Table 1). At the transcript level, comments are included in FlyBase transcript reports (Table S1) and a translation exception flag appears in the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “translation exception” type (Table 2).
Exceptional translation (3): stop-codon readthrough appears to be common in flies
Stop-codon readthrough is a well-documented regulatory mechanism in viruses (reviewed in Bertram et al. 2001) and has been investigated in S. cerevisiae (reviewed in von der Haar and Tuite 2007). Prior to 2007, a small number of specific cases had been identified in flies, including kel (Xue and Cooley 1993; Robinson and Cooley 1997), oaf (Bergstrom et al. 1995), Syn (Klagges et al. 1996) and hdc (Steneberg et al. 1998). In 2007, using a genome-wide comparative analysis designed to detect regions exhibiting evolutionary signatures specific to protein-coding regions (PhyloCSF) (Lin et al. 2007, 2011), a surprising number of such regions was found immediately beyond annotated stop codons. Because there are other possible explanations for this observation (alternative splicing, A-to-I RNA editing, or polycistronic transcripts, for example), a more thorough analysis was performed by Jungreis et al. (2011). Based primarily on these comparative evolutionary analyses, 328 genes are currently annotated in FlyBase with one or more transcripts subject to stop-codon readthrough (example in Figure 3B); a complete listing is available in File S5. In 22 cases a double readthrough is supported; there even appear to be two cases of triple readthrough (vvl, Ets65A). Two genes exhibit readthrough at two independent sites within alternative exons (Oamb and CG34377). The Oamb gene model is a particularly enthusiastic example of stop-codon readthrough: it has two independent sites, one of which is a double readthrough.
A number of predicted cases of stop-codon readthrough have been confirmed by mass spectrometry of wild-type proteins (seven cases) or using reporter constructs in which the readthrough extensions are epitope-tagged (Jungreis et al. 2011). Additionally, an independent ribosome-profiling assay using early embryos and S2 cells has confirmed 43 of the previously predicted readthrough cases (Dunn et al. 2013); it is unclear if unconfirmed cases occur at low levels or only at other developmental stages. Interestingly, this ribosome profiling study identified an additional 307 cases of translation readthrough not identified by Jungreis et al. (2011). These novel cases of translation readthrough exhibit lower conservation scores and lower readthrough rates than those predicted by conservation; the nucleotide character of these novel readthrough regions is intermediate between coding regions and 3′ UTRs. As such, it is unclear if these extensions represent truly functional isoforms of the proteins, or simply provide fodder for the evolution of novel C-terminal variants. For this reason, poorly conserved cases of readthrough are not currently annotated by FlyBase.
With very few exceptions, the annotated readthroughs result in a C-terminal extension of an annotated polypeptide sequence (as opposed to the addition of upstream coding sequences to an annotated polypeptide); note that the analysis of Jungreis et al. (2011) was biased to detect this type of event. This stop-codon readthough phenomenon appears to be distinct from the selenocysteine system and does not require a downstream SECIS motif (Jungreis et al. 2011). The annotated stop-codon readthroughs include cases for all three stop codons, with UGA being the most common. The length of the amino acid extension ranges from four amino acids to more than 1000 amino acids; most are within the range of 8–300 amino acids. Comparisons across 12 Drosophila species for individual genes subject to stop codon readthrough revealed that 97% of readthrough codons were perfectly conserved; substitution of an alternative stop codon was rare and involved only UAA and UAG (Jungreis et al. 2011). This suggests that the three codons are not functionally identical in the context of readthrough events, but that UAA and UAG may be similar.
Many of the longer carboxy readthrough extensions have in common a distinct pattern of conservation: regions of low complexity, variable conservation, and variable length interspersed with regions of protein sequence conservation. They show characteristics of intrinsically disordered protein regions, which confer structural flexibility and may be characteristic of many members of protein complexes (reviewed in Mészáros et al. 2011). For these cases, the carboxy extension due to stop-codon translational readthrough may result in an increased repertoire of protein-protein interactions. An interesting example is caps, which encodes a leucine-rich transmembrane protein and is similar to trn. The 240-aa extension of the Caps protein expands the similarity of the two proteins into a region with this pattern of conservation that is present in the unextended region of the Trn protein (Figure S2).
Jungreis et al. (2011) also used the PhyloCSF algorithm to identify stop-codon readthrough candidates in mammals, nematodes, fungi, and other insects. Although examples of predicted nonselenocysteine readthroughs were found in other phylogenetic groups, only in insects were they found to be common. Use of the three stop codons was not uniform: in many species, only UGA readthroughs were observed and UAA readthroughs were found only in Dipterans. Using a Z-curve reading-frame bias analysis that provided an estimation of the total frequency of readthroughs, Jungreis et al. (2011) assessed 25 species in a broad phylogenetic range. They concluded that within the animal kingdom, abundant readthough may be confined to insects and crustacea.
FlyBase and GenBank flags:
Genes with transcripts subject to stop-codon readthrough are identified by a specific SO term (Table 1). At the transcript level, comments are included in FlyBase transcript reports and double readthroughs are identified (Table S1). A translation exception flag appears in the GenBank RefSeq transcript and protein entries (Table S2). In the sequences of the predicted proteins, an “X” appears at the position(s) of the suppressed stop codon(s). The proposed bulk data flag is of the “translation exception” type (Table 2).
Exceptional translation (4): a single translational frameshift is highly conserved
D. melanogaster has a single example of translational frameshifting, involving the gene Oda (Ornithine decarboxylase antizyme) (Ivanov et al. 1998). This event is part of a mechanism of polyamine autoregulation that is conserved from yeast through mammals (Ivanov et al. 2000; Olsen and Zetter 2011). The ornithine decarboxylase antizyme regulates the activity of the enzyme ornithine decarboxylase (ODC), a key enzyme in the synthesis of polyamines. The Oda transcript does not code for a functional antizyme without an additional regulatory step: the reading frame changes in the second exon. High polyamine concentrations drive a +1 ribosomal frameshift, thus the production of functional Oda protein is sensitive to polyamine levels.
FlyBase and GenBank flags:
The Oda gene report includes an appropriate SO term (Table 1), and reports for the transcripts include an explanatory comment (Table S1). A “ribosomal_slippage” flag appears in the GenBank RefSeq transcript and protein entries (Table S2). The proposed bulk data flag is of the “translation exception” type (Table 2).
Exceptional translation (5): mitochondria play by their own rules
There are 13 protein-coding genes mapped to the D. melanogaster mitochondrial genome (mitochondrial genes are flagged by the SO term “mt_gene,” SO:0000088). Because mitochondria use an alternative genetic code, all mitochondrial protein-coding transcripts are flagged as translation exceptions (see http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG5 for nonstandard codon usage specific to invertebrate mitochondria). In addition, some mitochondrial genes have an incomplete stop codon. This is a stop codon that is not entirely encoded in the genome: it is completed upon post-transcriptional 3′ polyadenylation. The transcripts of three mitochondrial genes (mt:CoII, mt:ND5, mt:ND4) have this type of structure and have an additional translation exception comment referring to the stop codon exception. For one mitochondrial gene (mt:CoI) the start codon has not been identified. Mitochondrial translation is known to use noncanonical start codons, but the sequence preceding mt:CoI does not contain any of the identified alternative codons in the correct ORF. The transcript for this gene is flagged with the comment “start codon not determined."
Conclusion
The annotation of the D. melanogaster genome has been greatly facilitated by access to both extensive sets of high throughput data and a large body of gene-specific research. Combined with expert manual assessment of each gene model, this has allowed FlyBase to compile a uniquely detailed and nuanced gene model annotation set, which continues to improve. Unfortunately, some of the most interesting aspects of our knowledge of the D. melanogaster genome are difficult to leverage to inform the annotations of other species.
The subjects of this article, the rule-benders or exceptional cases, largely identify biological phenomena that create problems for automated gene prediction algorithms. Although some cases among the rule-benders would present more difficulties than others, it may be feasible to incorporate automated second-pass steps to identify many exceptional cases. A number of highly conserved phenomena are well defined; they affect relatively few genes, but these may be straightforward to identify.
A parallel approach would be a gene model pipeline that is largely automated, but that allows incorporation of manual corrections and additions. Ideally, this would be an ongoing process as more is learned about the variability and flexibility of the genome. If an efficient system for expert review of submissions were developed, then input from a variety of sources, such as researchers interested in a group of genes across many species, undergraduate annotation projects, and even specialist crowd-sourcing, could be encouraged.
Supplementary Material
Acknowledgments
We thank Stephanie Mohr and Yanhui Hu of the DRSC and our colleagues at FlyBase for their helpful comments on the manuscript. FlyBase is supported by National Human Genome Research Institute at the National Institutes of Health (U41 HG00739 to W.G., PI] and Medical Research Council (UK) (G1000968 to N.B., PI).
Notes added in proof: It has been brought to our attention that a second case of translational frameshifting, for the gene Apc, has been described by Baranov et al., 2011 RNA Biol. 8: 637–647 (PMID:21593603).
Additionally, see Matthews et al. 2015 (pp. 1721–1736) in this issue for a related work.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.018937/-/DC1
The FlyBase Consortium members at the time of writing include the following: William Gelbart,* Nicholas H. Brown,† Thomas Kaufman,‡ Maggie Werner-Washburne,§ Richard Cripps,§ Kris Broll,* Madeline Crosby,* Gilberto dos Santos,* David Emmert,* L. Sian Gramates,* Kathleen Falls,* Beverley B. Matthews,* Susan Russo,* Andrew Schroeder,* Pinglei Zhou,* Mark Zytkovicz,* Boris Adryan,† Giulia Antonazzo,† Helen Attrill,† Marta Costa,† Steven Marygold,† Peter McQuilton,† Gillian Millburn,† Laura Ponting,† Alix Rey,† Nicole Staudt,† Raymund Stefancsik,† Josh Goodman,‡ Gary Grumbling,‡ Victor Strelets,‡ Jim Thurmond,‡ and Phillip Baker§.
Communicating editor: J. M. Cherry
Literature Cited
- Alioto T. S., 2007. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 35(Database issue): D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjelkovic M., Jones P. F., Grossniklaus U., Cron P., Schier A. F., et al. , 1995. Developmental regulation of expression and activity of multiple forms of the Drosophila RAC protein kinase. J. Biol. Chem. 270: 4066–4075. [DOI] [PubMed] [Google Scholar]
- Andrews J., Smith M., Merakovsky J., Coulson M., Hannan F., et al. , 1996. The stoned locus of Drosophila melanogaster produces a dicistronic transcript and encodes two distinct polypeptides. Genetics 143: 1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton R. J., Tsai L. T. Y., Schwabe T., DeSalvo M., Gaul U., et al. , 2005. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell 123: 145–156. [DOI] [PubMed] [Google Scholar]
- Batut P., Dobin A., Plessy C., Carninci P., Gingeras T. R., 2013. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman R. W., Jongens T. A., 2011. A non-canonical start codon in the Drosophila fragile X gene yields two functional isoforms. Neuroscience 181: 48–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar Y., Nannapaneni K., Casavant T. L., Scheetz T. E., Welsh M. J., 2007. Eukaryotic operon-like transcription of functionally related genes in Drosophila. Proc. Natl. Acad. Sci. USA 104: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom D. E., Merli C. A., Cygan J. A., Shelby R., Blackman R. K., 1995. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics 139: 1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Innes S., Minella O., Richardson J., Stansfield I., 2001. Endless possibilities: translation termination and stop codon recognition. Microbiology 147: 255–269. [DOI] [PubMed] [Google Scholar]
- Boyd L., Thummel C. S., 1993. Selection of CUG and AUG initiator codons for translation depends on downstream sequences. Proc. Natl. Acad. Sci. USA 90: 9164–9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S., Ashburner M., 1997. The Adh-related gene of Drosophila melanogaster is expressed as a functional dicistronic messenger RNA: Multigenic transcription in higher organisms. EMBO J. 16: 2023–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H., et al. , 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S., Morozova N., Morey M., Berry M. J., Serras F., et al. , 2001. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2: 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Wheeler D. A., Kronmiller B., Carlson J. W., Halpern A., et al. , 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Nash H. A., 2007. Drosophila TRP channels require a protein with a distinctive motif encoded by the inaF locus. Proc. Natl. Acad. Sci. USA 104: 17730–17734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines B., Wang H., Wang L., Li Y., Han Y., et al. , 2011. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 21: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Simone S. M., White K., 1993. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol. Cell. Biol. 13: 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Reuter G., Loewendorf A., 2001. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. USA 98: 9724–9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos G., Schroeder A. J., Goodman J. L., Strelets V. B., Crosby M. A., et al. , 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43(Database issue): D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D. L., Chavatte L., 2004. Finding needles in a haystack. In silico identification of eukaryotic selenoprotein genes. EMBO Rep. 5: 140–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. G., Foo C. K., Belletier N. G., Gavis E. R., Weissman J. S., 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K., Lewis S. E., Mungall C. J., Yandell M., Stein L., et al. , 2005. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 6: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabler M., Volkmar M., Weinlich S., Herbst A., Dobberthien P., et al. , 2005. Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species Drosophila melanogaster and D. virilis. Genetics 169: 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. L., Fan Y. J., Wang X. Y., Zhang Y., Pu J., et al. , 2015. A conserved intronic U1 snRNP-binding sequence promotes trans-splicing in Drosophila. Genes Dev. 29: 760–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron D., Gevaert K., Van Damme P., 2014. The proteome under translational control. Proteomics 14: 2647–2662. [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. A., Nicholls R. D., 2000. Diverse splicing mechanisms fuse the evolutionarily conserved bicistronic MOCS1A and MOCS1B open reading frames. RNA 6: 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden C. A., Bosco G., 2008. Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C. U., Sarnow P., 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hooks K. B., Griffiths-Jones S., 2011. Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol. 8: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Giniger E., Aigaki T., 2003. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 17: 2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Aigaki T., 2006. Alternative trans-splicing: A novel mode of pre- mRNA processing. Biol. Cell 98: 135–140. [DOI] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M., et al. , 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Landolin J. M., Brown J. B., Sandler J. E., Takahashi H., et al. , 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Roesel C., Flockhart I., Perkins L., Perrimon N., et al. , 2013. UP-TORR: Online Tool for Accurate and Up-to-Date Annotation of RNAi Reagents. Genetics 195: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibnsouda S., Ferrer P., Vincent A., 1998. Conservation of read-through transcription of the Drosophila serendipity genes during evolution is gratuitous. Mol. Gen. Genet. 259: 484–490. [DOI] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S., 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. P., Simin K., Letsou A., Atkins J. F., Gesteland R. F., 1998. The Drosophila gene for antizyme requires ribosomal frameshifting for expression and contains an intronic gene for snRNP Sm D3 on the opposite strand. Mol. Cell. Biol. 18: 1533–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. P., Gesteland R. F., Atkins J. F., 2000. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 28: 3185–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. P., Firth A. E., Michel A. M., Atkins J. F., Baranov P. V., 2011. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39: 4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Lin M. F., Spokony R., Chan C. S., Negre N., et al. , 2011. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 21: 2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagges B. R. E., Heimbeck G., Godenschwege T. A., Hofbauer A., Pflugfelder G. O., et al. , 1996. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J. Neurosci. 16: 3154–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V., Dorn R., 2004. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene 331: 165–176. [DOI] [PubMed] [Google Scholar]
- Krauss V., Fassl A., Fiebig P., Patties I., Sass H., 2006. The evolution of the histone methyltransferase gene Su(var)3–9 in metazoans includes a fusion with and a re-fission from a functionally unrelated gene. BMC Evol. Biol. 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M., Mongelard F., Plata-Rengifo P., Baxter E. M., Corces V. G., et al. , 2001. Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Lasda E. L., Blumenthal T., 2011. Trans-splicing. Wiley Interdiscip. Rev. RNA 2: 417–434. [DOI] [PubMed] [Google Scholar]
- Lin C. F., Mount S. M., Jarmołowski A., Makałowski W., 2010. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. F., Carlson J. W., Crosby M. A., Matthews B. B., Yu C., et al. , 2007. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17: 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. F., Jungreis I., Kellis M., 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Graham J., Nycz K., McKim K. S., 2000. Two genes required for meiotic recombination in Drosophila are expressed from a dicistronic message. Genetics 154: 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan S. J., Edeen P., Esnayra J., McKeown M., 1996. att, a target for regulation by tra2 in the testes of Drosophila melanogaster, encodes alternative RNAs and alternative proteins. Mol. Cell. Biol. 16: 4222–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallela A., Nishikura K., 2012. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 47: 493–501. [DOI] [PubMed] [Google Scholar]
- Martin-Romero F. J., Kryukov G. V., Lobanov A. V., Carlson B. A., Lee B. J., et al. , 2001. Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J. Biol. Chem. 276: 29798–29804. [DOI] [PubMed] [Google Scholar]
- Matthews B. B., dos Santos G., Crosby M. A., Emmert D. B., St. Pierre S. E., et al. , 2015. FlyBase Gene Model Annotations for Drosophila melanogaster. Impact of High-throughput Data G3 (Bethesda) 1721–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Duff M. O., Eipper-Mains J., Graveley B. R., 2010. Global analysis of trans-splicing in Drosophila. Proc. Natl. Acad. Sci. USA 107: 12975–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy K., Emrich S. J., Lawson D., Campbell D., Dialynas E., et al. , 2012. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 40(Database issue): D729–D734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., et al. , 2013. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12: 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros B., Simon I., Z. Dosztányi, 2011. The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys. Biol. 8: 035003. [DOI] [PubMed] [Google Scholar]
- Misra S., Crosby M. A., Mungall C. J., Matthews B. B., Campbell K. S., et al. , 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., et al. , 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Yokoyama H., Goto A., Beck K., Kitagawa Y., 1999. A Drosophila gene encoding multiple splice variants of Kazal-type serine protease inhibitor-like proteins with potential destinations of mitochondria, cytosol and the secretory pathway. Eur. J. Biochem. 266: 282–292. [DOI] [PubMed] [Google Scholar]
- Olsen R. R., Zetter B. R., 2011. Evidence of a role for antizyme and antizyme inhibitor as regulators of human cancer. Mol. Cancer Res. 9: 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T., Zsindely N., Vamos E. E., Komonyi L., Bodai L., et al. , 2013. Functional characterization and gene expression profiling of Drosophila melanogaster short dADA2b isoform-containing dSAGA complexes. BMC Genomics 14: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada G. E., Munita R., Cerda C. A., Gysling K., 2014. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 42: 10564–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli D., Tonka C. H., Ayme-Southgate A., 1988. An unusual split Drosophila heat shock gene expressed during embryogenesis, pupation and in testis. J. Mol. Biol. 200: 47–53. [DOI] [PubMed] [Google Scholar]
- Peabody D. S., 1989. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264: 5031–5035. [PubMed] [Google Scholar]
- Phillips A. M., Smith M., Ramaswami M., Kelly L. E., 2000. The products of the Drosophila stoned locus interact with synaptic vesicles via synaptotagmin. J. Neurosci. 20: 8254–8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plongthongkum N., Kullawong N., Panyim S., Tirasophon W., 2007. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 354: 789–794. [DOI] [PubMed] [Google Scholar]
- Robinson D. N., Cooley L., 1997. Examination of the function of two kelch proteins generated by stop codon suppression. Development 124: 1405–1417. [DOI] [PubMed] [Google Scholar]
- Rodriguez J., Menet J. S., Rosbash M., 2012. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell 47: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H. D., Domingos P. M., Kang M. J., Steller H., 2007. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 26: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Will C. L., Brosius J., Frilander M. J., Luhrmann R., 2004. Identification of an evolutionarily divergent U11 small nuclear ribonucleoprotein particle in Drosophila. Proc. Natl. Acad. Sci. USA 101: 9584–9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R. A., Miksch J. L., Xie X. L., Cornish J. A., Galewsky S., 1990. Expression of the Drosophila gonadal gene: alternative promoters control the germ-line expression of monocistronic and bicistronic gene transcripts. Development 108: 613–622. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P., 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039. [DOI] [PubMed] [Google Scholar]
- Steneberg P., Englund C., Kronhamn J., Weaver T. A., Samakovlis C., 1998. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the Drosophila trachea. Genes Dev. 12: 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G., Tackett M. R., Nechkin S., Shtokalo D., Antonets D., et al. , 2013. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat. Struct. Mol. Biol. 20: 1333–1339. [DOI] [PubMed] [Google Scholar]
- Sugihara H., Andrisani V., Salvaterra P. M., 1990. Drosophila choline acetyltransferase uses a non-AUG initiation codon and full length RNA is inefficiently translated. J. Biol. Chem. 265: 21714–21719. [PubMed] [Google Scholar]
- Szafranski K., Schindler S., Taudien S., Hiller M., Huse K., et al. , 2007. Violating the splicing rules: TG dinucleotides function as alternative 3′ splice sites in U2-dependent introns. Genome Biol. 8: R154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T., Tuite M. F., 2007. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 15: 78–86. [DOI] [PubMed] [Google Scholar]
- Walker D. L., Wang D., Jin Y., Rath U., Wang Y., et al. , 2000. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J. Cell Biol. 151: 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall A. A., Phillips A. M., Kelly L. E., 2005. Effective translation of the second cistron in two Drosophila dicistronic transcripts is determined by the absence of in-frame AUG codons in the first cistron. J. Biol. Chem. 280: 27670–27678. [DOI] [PubMed] [Google Scholar]
- Xue F., Cooley L., 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.