Abstract
Multiple mechanisms have been proposed for gene silencing in Saccharomyces cerevisiae, ranging from steric occlusion of DNA binding proteins from their recognition sequences in silenced chromatin to a specific block in the formation of the preinitiation complex to a block in transcriptional elongation. This study provided strong support for the steric occlusion mechanism by the discovery that RNA polymerase of bacteriophage T7 could be substantially blocked from transcribing from its cognate promoter when embedded in silenced chromatin. Moreover, unlike previous suggestions, we found no evidence for stalled RNA polymerase II within silenced chromatin. The effectiveness of the Sir protein–based silencing mechanism to block transcription activated by Gal4 at promoters in the domain of silenced chromatin was marginal, yet it improved when tested against mutant forms of the Gal4 protein, highlighting a role for specific activators in their sensitivity to gene silencing.
Keywords: heterochromatin, transcription, repression, eukaryotic gene regulation, euchromatin
Transcriptional silencing is a form of regional, promoter-independent repression mediated in Saccharomyces cerevisiae by the Silent Information Regulator (SIR) protein complex. Sir proteins are the structural components of a specialized structure of chromatin that is analogous to heterochromatin in other species. Sir protein–based silencing represses transcription of the HML and HMR loci, which contain auxiliary copies of the genes that specify mating-type identity. Transcriptional silencing results in a 1000-fold reduction in transcript levels of the genes at HML and HMR compared to expression of those same genes when at the active MAT locus. Sir-based silencing also operates on some genes close to yeast telomeres (Aparacio et al. 1991; reviewed in Wellinger and Zakian 2012).
The DNA elements and proteins required to establish and maintain silencing have been identified (reviewed in Rusche et al. 2003; Kueng et al 2013). Silencing of transcription requires the recruitment of a complex consisting of Sir2, Sir3, and Sir4 to the silencers and HML and HMR. In addition, silencing also requires the catalytic activity of Sir2, a histone deacetylase acting on acetylated H4K16 (Imai et al. 2000; Landry et al. 2000), and the assembly of a repressive chromatin domain by interactions among multiple copies of the Sir protein complex throughout the domain (Rusche et al. 2002; Thurtle and Rine 2013). The molecular mechanisms by which the binding of the Sir proteins and the establishment of a silenced local chromatin domain repress transcription are the subject of this study.
Three classes of models have been proposed for how Sir proteins silence transcription. A steric occlusion model is inspired by silenced chromatin’s ability to block binding/action of sequence-specific DNA binding proteins, such as HO endonuclease, DNA methylase, and restriction enzymes to their binding sites in silenced chromatin (Singh and Klar 1992; Strathern et al. 1982; Gottschling 1992; Loo and Rine 1994). Thus, Sir proteins create a specialized local chromatin structure that exhibits reduced accessibility to site-specific DNA-binding proteins and, by extension, presumably to the transcription machinery. However, the reduced accessibility in these studies was measured qualitatively. Hence, how much of the 1000-fold repression of HML and HMR expression could be accounted for by this mechanism has been unresolved.
A preinitiation-complex interference model is a refinement of steric occlusion models in response to a possible mismatch between the magnitude of steric occlusion in previous studies and the 1000-fold reduction in transcription in silenced chromatin. The preinitiation-complex interference model is based on chromatin immunoprecipitation (ChIP) analyses indicating the absence of key components of general transcription machinery, TFIIB (Sua7), TFIIE (Tfa2), and RNA Pol II (Rpb1) from silenced chromatin. In contrast, occupancy of Ppr1, the gene-specific activator for a silenced URA3 transgene, is only slightly reduced in silenced chromatin (Chen and Widom 2005). The preinitiation-complex interference model posits that the reduction in transcription achieved in silenced chromatin results from the sensitivity of specific factors within the preinitation-complex to being blocked from accessing their target sites, preventing the formation of a preinitation-complex.
In contrast to the previous two models, the downstream-inhibition model is inspired by ChIP data interpreted as showing that, in addition to RNA Pol II, components of the preinitation-complex, TBP (Spt15) and TFIIH (Tfb1, Kin28) are comparably enriched in silenced and active chromatin, but mRNA capping proteins (Cet1, Abd1) and downstream elongation factors (Spt5, TFIIS, Paf1) are specifically absent (Gao and Gross 2008). In this model, RNA polymerase II is blocked by Sir proteins at the transition between initiation and elongation. In support of this observation, gene-specific activators and RNA Pol II are reported to be localized to HSP82 placed adjacent to HMR-E (Sekinger and Gross 2001).
To provide greater resolution toward the mechanism of silencing, we performed specific tests of predictions made by these models, asking whether the inferred stalled transcription complex at HML exists, and whether the function of a completely heterologous, prokaryotic RNA polymerase can be quantitatively blocked by Sir-based silencing in vivo. Finally, we determined whether transcription activators differed with respect to their sensitivity to Sir-based silencing.
Materials and Methods
Yeast strain construction and media
All strains used in this study were derived from W303-1a and are listed in Table 1. All plasmids used in this study are listed in Table 2 and oligonucleotides used in this study are listed in Table 3. Standard mating and sporulation techniques were used to perform yeast crosses. The T7pro::a1 allele, consisting of a 20-base pair (bp) optimal T7 promoter, as described by Ujvári and Martin (1997), fused to the wild-type a1 ORF including 5 bp directly upstream of the ATG to allow for T7 polymerase initiation, was synthesized with 50 bp of homology matching the native HMRa1 locus upstream and downstream of the T7pro::a1 allele (pJR 3208). The T7pro::a1 allele and homology regions were amplified by PCR using oDS 82 and oDS 83 and transformed into JRY8676. Transformants were counter-selected with 5-FOA. 5-FOA-resistant colonies were screened by PCR, and the structure of the new HMR allele was verified by sequencing. All further strains bearing HMR T7pro::a1 were generated by standard mating and tetrad dissection, and segregates were verified by PCR and sequencing to confirm the presence of T7pro::a1. Strains bearing GAL1pro::a1 alleles were constructed in an identical fashion, with 450 bp of the GAL1 promoter directly upstream of HMRa1 (pJR 3209) (Mr. Gene, GmbH, Germany).
Table 1. Yeast strains used in this study.
| Strain | Parent | Genotype | Source | Plasmid |
|---|---|---|---|---|
| JRY4012 | W303 | MATa his3-11 leu2-3, 112 lys2 trp1-1 ura3-1 can1-100 | R. Rothstein | |
| JRY4579 | W303 | MATa sir4∆::TRP1 his3-11 leu2-3, 112 lys2 trp1-1 ura3-1 can1-100 | ||
| JRY8676 | W303 | MATα HMRa1ORF::K.lactis URA3ORF sir4∆::HIS3 ade2 his3 leu2 trp1 ura3 | ||
| JRY9514 | W303 | MATα HMR:T7pro::a1 ade2 his3 leu2 ura3 [pJR3207] | This study | pJR3207 |
| JRY9515 | W303 | MATα HMR:T7pro::a1 sir4∆::HIS3 ade2 his3 leu2 ura3 [pJR3207] | This study | pJR3207 |
| JRY9516 | W303 | MATα ade2 his3 leu2 ura3 [pJR3207] | This study | pJR3207 |
| JRY9517 | W303 | MATα sir4∆::HIS3 ade2 his3 leu2 ura3 [pJR3207] | This study | pJR3207 |
| JRY9518 | W303 | MATα HMR:T7pro::a1 sir4∆::HIS3 ade2 his3 leu2 ura3 [pJR1237] | This study | pJR1237 |
| JRY9519 | W303 | MATα HMR:a2/a1promoter∆ sir4∆::HIS3 ade2 his3 leu2 trp1 ura3 | This study | |
| JRY9520 | W303 | MATα HMR:T7pro::a1 sir4∆::HIS3 URA3:GAL1promoterNLS-T7polymerase ade2 his3 leu2 ura3 | This study | |
| JRY9521 | W303 | MATα HMR:T7pro::a1 sir4∆::HIS3 URA3:GAL1promoterNLS-T7polymerase ade2 his3 leu2 ura3 / MATa his3-11 leu2-3, 112 lys2 trp1-1 ura3-1 can1-100 | This study | |
| JRY9522 | W303 | MATα HMR:T7pro::a1 sir4∆::HIS3 URA3:GAL1promoterNLS-T7polymerase ade2 his3 leu2 ura3 / mat∆::KANMX hmr∆::HYGMX hml∆::NATMX ade2 his3 leu2 trp1 ura3 | This study | |
| JRY9523 | W303 | MATα HMR:T7pro::a1 13xMYC-SIR3:KANMX ade2 his3 leu2 trp1 ura3 [pJR3207] | This study | pJR3207 |
| JRY9524 | W303 | MATα HMR:T7pro::a1 13xMYC-SIR3:KANMX sir4∆::HIS3 ade2 his3 leu2 trp1 ura3 [pJR3207] | This study | pJR3207 |
| JRY9525 | W303 | mat∆::HYGMX HMR:T7pro::a1 13xMYC-SIR3::KANMX ade2 his3 leu2 trp1 ura3 | This study | |
| JRY9526 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX ade2 his3 leu2 trp1 ura3 | This study | |
| JRY9527 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 ade2 his3 leu2 trp1 ura3 | This study | |
| JRY4013 | W303 | MAT α his3-11 leu2-3, 112 lys2 trp1-1 ura3-1 can1-100 | R. Rothstein | |
| JRY2726 | MATa his4 | |||
| JRY2728 | MAT α his4 | |||
| JRY9528 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3210] | This study | pJR3210 |
| JRY9529 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3211] | This study | pJR3211 |
| JRY9530 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3210] | This study | pJR3210 |
| JRY9531 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3211] | This study | pJR3211 |
| JRY9743 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3376] | This study | pJR3376 |
| JRY9744 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3376] | This study | pJR3376 |
| JRY9745 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3377] | This study | pJR3377 |
| JRY9746 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3377] | This study | pJR3377 |
| JRY9747 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3378] | This study | pJR3378 |
| JRY9748 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3378] | This study | pJR3378 |
| JRY9749 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3379] | This study | pJR3379 |
| JRY9750 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3379] | This study | pJR3379 |
| JRY9751 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3380] | This study | pJR3380 |
| JRY9752 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3380] | This study | pJR3380 |
| JRY9753 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3381] | This study | pJR3381 |
| JRY9754 | W303 | MAT α HMR:GAL1pro::a1 13xMYC-SIR3:KANMX sir4Δ::HIS3 gal4Δ::NATMX ade2 his3 leu2 trp1 ura3 [pJR3381] | This study | pJR3381 |
Unless otherwise noted, strains were from the lab strain collection.
Table 2. Plasmids used in this study.
| Plasmid | Backbone | Bacteria Selection | Yeast Selection | Insert | Source |
|---|---|---|---|---|---|
| pJR3207 | pUC18 | Amp | LEU2 | GAL1pro:NLS-T7 Polymerase | Benton et al. (1990) |
| pJR1237 | pRS425 | Amp | LEU2 | Empty vector | Brachmann et al. (1998) |
| pJR3208 | pMA-T | Amp | None | T7pro::a1 | This study, Mr. Gene |
| pJR3209 | pMK-RQ | Kan | None | GAL1pro::a1 | This study, Mr. Gene |
| pJR2781 | pRS41H | Amp | HYG | Empty vector | Taxis and Knop (2006) |
| pJR3210 | pRS41H | Amp | HYG | gal4L331P | This study |
| pJR3211 | pRS41H | Amp | HYG | GAL4 | This study |
| pJR3376 | pRS41H | Amp | HYG | gal4-147∆768 | This study |
| pJR3377 | pRS41H | Amp | HYG | gal4-238∆768 | This study |
| pJR3378 | pRS41H | Amp | HYG | gal4-763∆851 | This study |
| pJR3379 | pRS41H | Amp | HYG | gal4-238∆851 | This study |
| pJR3380 | pRS41H | Amp | HYG | gal4-848∆ | This study |
| pJR3381 | pRS41H | Amp | HYG | gal4-844∆ | This study |
Table 3. Oligonucleotides used in this study.
| Purpose | Name | Sequence |
|---|---|---|
| LM-PCR | LMPCR linker A (oDS 35) | GCGGTGATTTAAAAGATCTGAATTC |
| LMPCR Linker B (oDS 36) | GAATTCAGATC | |
| HMLα1-LM-PCR-1 (oDS 37) | TGCTCAGCTAGACGTTTTTC | |
| HMLα1-LM-PCR-2 (oDS 38) | CGTTTTTCTTTCAGCTTTTTTGA | |
| HMLα1-LM-PCR-3 (oDS 39) | CAGCTTTTTTGAAACCGCTGTG | |
| Strain construction | T7pro::a1/GAL1pro::a1 knock in primer at HMR F (oDS 82) | TTTTTCTGTGTAAGTTGATAATTACTTCTATCGTTTTCTATGCTGCGCAT |
| T7pro::a1/GAL1pro::a1 knock in primer at HMR R (oDS 83) | GAAACTAAAAGAAAAACCCGACTATGCTATTTTAATCATTGAAAACGAAT | |
| GAL4 KO F | ATCATTTTAAGAGAGGACAGAGAAGCAAGCCTCCTGAAAGCGGATCCCCGGGTTAATTAA | |
| GAL4 KO R | GAAGTGAACTTGCGGGGTTTTTCAGTATCTACGATTCATTCGATGAATTCGAGCTCGTTT | |
| qPCR | a1 qPCRr F | TGGATGATATTTGTAGTATGGCGGA |
| a1 qPCR R | TCCCTTTGGGCTCTTCTCTT | |
| ACT1 qPCR F | TGTCCTTGTACTCTTCCGGT | |
| ACT1 qPCR R | CCGGCCAAATCGATTCTCAA | |
| ARS504 qPCR F | GTCAGACCTGTTCCTTTAAGAGG | |
| ARS504 qPCR R | CATACCCTCGGGTCAAACAC | |
| TEL VIR 1.2 kb qPCR F | GTGCTAAAGGAATCCCCAGAGA | |
| TEL VIR 1.2 kb qPCR R | TCTGTCCATTTTCCCTCTGCTC | |
| HMR E qPCR F | CGAACGATCCCCGTCCAAGTTATGAGC | |
| HMR E qPCR R | CAGGAGTACCTGCGCTTATTCTCAAAC | |
| HMR I qPCR F | AGTTTCAGCTTTCCGCAACAGT | |
| HMRa1 3′ qPCR F | CCAACATTTTCGTATATGGCG | |
| HMRa1 3′ qPCR R | CTTGTGCAAATTCCAACTAAAGG | |
| HMR a2 C qPCR F | CTTCTATCGTTTTCTATGCTGCG | |
| GAL1 promoter qPCR F | GAGCCCCATTATCTTAGCCTAAAAAAAC | |
| GAL1 promoter qPCR R | TACTGCCAATTTTTCCTCTTCATAACC | |
| GAL1 3′ ORF qPCR F | GAACGAGTCTCAAGCTTCTTGC | |
| GAL1 3′ ORF qPCR R | GCTGGTTTAGAGACGATGATAGC |
The gal4L331P allele (gift from M. Johnston) was cloned into pJR 2781 using BamHI and HindIII digestion and ligation, producing plasmid pJR3210. Wild-type GAL4 was also cloned into pJR2781 using an identical protocol, resulting in pJR3211. Further gal4 alleles were cloned by QuikChange site-directed mutagenesis on pJR3211, resulting in pJR3376-pJR3381. The resulting gal4 alleles were verified by sequencing the entire ORF in both directions to ensure only the desired changes were created.
For galactose induction experiments, cells were grown to OD600 ≈ 0.7 in CSM raffinose or CSM-Leu raffinose (2%); then, prewarmed 20% galactose was added to a final concentration of 2%. All solution percentages are wt/vol. Fresh CSM galactose or CSM-Leu galactose media were added to samples for kinetic experiments to maintain culture volume and OD over time. All media in nonkinetic experiments were 2% of the indicated sugar.
Potassium permanganate transcription bubble assay
Ten OD units of exponentially growing cells (OD600 ≈ 0.9–1) grown in CSM or CSM-5 mM nicotinamide were harvested and resuspended in 1 ml cold PBS. These whole cells or 12 μg genomic DNA were reacted with 20 mM KMnO4 on ice for intervals as depicted in Figure 1. The reaction was stopped with an excess of a 20 mM ED (Tris-Cl, 20 mM EDTA, 1% SDS, 0.4 M β-ME stop solution). DNA was extracted from whole cells using glass bead lysis, phenol-chloroform extraction, and sodium-acetate and ethanol precipitation. Both genomic DNA reacted with KMnO4 and DNA extracted from KMnO4-treated cells was resuspended in TE and stored at 4°. A G/A genomic DNA ladder was generated by 3-min reaction of 5 μg yeast genomic DNA with 95% formic acid. The resulting modified bases in the naked DNA and in vivo–treated DNA samples were converted to double-strand breaks using piperidine (Gilmour and Fan 2009). LM-PCR analysis was performed as described by Gilmour and Fan (2009) using HPLC-PAGE–purified linker primers oDS 35 and oDS 36, as well as oDS 37, oDS 38, and oDS 39 to amplify and label HMLα1 LM-PCR products. Before resolving and separating LM-PCR products by gel electrophoresis, ODS 39 was labeled using radioactive 10 mCi/ml gamma-32P-ATP (Perkin Elmer). Radioactive LM-PCR products were resolved on a 19:1 Acrylamide:Bis-Acrylamide 6% urea sequencing gel. Radioactivity patterns in the gel were visualized by exposure in a phosphor-imager cassette and scanned on a Typhoon scanner (GE Healthcare).
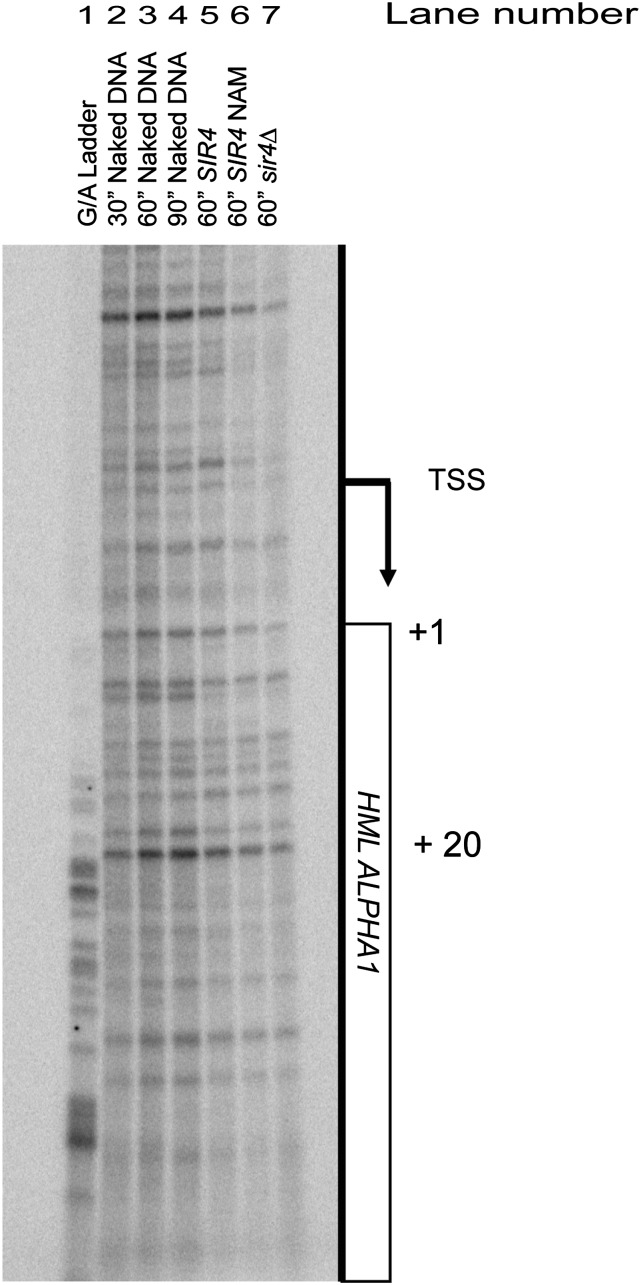
Figure 1.
The KMnO4 reactivity of HMLα1 in vivo. The pattern of KMnO4 reactivity is shown for the promoter and 5′ region of HMLα1 coding region. Genomic sequences of A+G are shown as a G/A ladder in lane 1. Naked genomic DNA reacted with 20 mM KMnO4 for various times is shown in lanes 2–4. The pattern of reactivity of this region in cells reacted with KMnO4 is shown in lanes 5–7. The reactivity pattern for cells with HMLα repressed lane 5, cells in which HML was derepressed with 5 mM nicotinamide (lane 6) or genetically by sir4∆ (lane 7) all lack enhanced cleavage sites characteristic of paused/stalled RNA polymerase. The arrow denotes the transcription start site (TSS) (Zhang and Dietrich 2005). The numbers on the right side of the panel identify bases in HMLα1 beginning at the initiation ATG codon as +1.
mRNA extraction, cDNA preparation, and analysis
RNA was purified from cultures at OD600 ≈ 0.75 using the Qiagen RNAeasy kit (Qiagen) and on-column DNAse digestion (Qiagen). cDNA was prepared from 2 μg total RNA using random hexamer primers, and in some experiments oligo dT primers, and the Superscript III cDNA synthesis kit (Invitrogen). cDNA was then quantified by qPCR using the Dynamo SYBR green qPCR kit (NEB) and detected on a Stratagene MX3000 quantitative PCR system. All primer sets were normalized to ACT1 amplification levels. Samples were analyzed in technical triplicate for each of at least three independent RNA preparations.
Whole-cell extract preparation and immunoblotting
Protein was extracted from cells grown to an OD600 ≈ 0.75 using 20% trichloroacetic acid and solubilized in SDS sample buffer. Extracts were fractionated on standard 10% SDS-PAGE gels and transferred to nitrocellulose membranes by wet-transfer at 150 V for 1 hr. Immunoblotting followed standard protocols and blots were imaged on a Li-COR odyssey imager using secondary antibodies conjugated to IR dyes. Antibodies used in immunoblots were anti-T7 RNA polymerase (Millipore 70566) and anti-Pgk1 (Invitrogen 459250).
Chromatin immunoprecipitation assay
Cells were cross-linked with 1% formaldehyde at OD600 ≈ 0.6 to 0.9 for 20 min at room temperature and then quenched with glycine to a final concentration of 300 mM for 5 min at room temperature. Cells were washed twice with cold Tris-buffered saline and lysed with 0.5 mm zirconia beads in FA-lysis buffer (Aparicio et al. 2005) with protease inhibitors (Roche) in a MP Fastprep-24. Chromatin was isolated as described (Aparicio et al. 2005). For 13XMyc-Sir3 immunoprecipitations, 50 μl of anti-Myc agarose beads (Sigma E6654) were incubated overnight at 4° with sonicated chromatin from 42.5 ml of culture. For Gal4 immunoprecipitations, 4 μl of anti-Gal4 antibody (Abcam 1396) and 25 μl protein A sepharose (GE Healthcare) were incubated with sonicated chromatin under identical conditions. Resin washes, elution, and DNA purification were performed as described (Aparicio et al. 2005). Precipitated DNA fragments were analyzed by qPCR, as described previously. The negative-control primer set for 13XMyc-Sir3 ChIP, ARS504, was chosen because it gave a consistently low IP/Input signal indistinguishable from a no-tag control and has been shown not to be bound by Sir3. Gal4 ChIP was normalized to a region downstream of the GAL1 gene not bound by Gal4. ChIP values are presented as enrichment relative to a control locus [(IP(primer)/IN(primer)]/[IP(control)/IN(control)].
Results
No evidence of a stalled polymerase at HML
In larger eukaryotes, expression of some genes is regulated by transcriptional pausing in which RNA polymerase engages a promoter, separates the strands, and produces a short transcript before stopping, awaiting a signal that restarts transcription (reviewed in Adelman and Lis 2012). Others have reported, and we have confirmed, that a ChIP experiment with an antibody against RNA polymerase II indicates a detectable level of recovery of HML and HMR from Sir+ cells above background levels (Gao and Gross 2008). However, we have previously reported various sources of artifacts of ChIP signals. In this case, the relative resistance of silenced chromatin to shearing (Teytelman et al. 2009) results in the precipitated chromatin being contaminated with larger flanking DNA carrying adjacent active genes. Hence, ChIP is prone to overestimate the occupancy of silenced genes with factors typically associated with transcription.
The most definitive hallmark of a gene with a paused polymerase is a transcription bubble at a specific position detected by the reactivity of the unpaired bases in the bubble to potassium permanganate (Zeitlinger et al. 2007). To date, there has been no example of a canonical transcriptionally paused polymerase in Saccharomyces detected by a permanganate assay. However, the ChIP data in support of the downstream-inhibition model suggested that the first transcription factors in the progression of gene activation, which were missing from silent chromatin but present in euchromatin, are Cet1 and Abd1, members of the mRNA capping checkpoint (Gao and Gross 2008). These data would imply that RNA polymerase has assembled the preinitiation complex in silenced chromatin and is stalled on melted DNA strands, awaiting the action of Cet1 and Abd1 to move into productive elongation. If so, then evidence of that stall should be detected by the potassium permanganate assay for paused polymerases.
Potassium permanganate preferentially reacts with and modifies T residues in single-stranded DNA (Rubin and Schmid 1980) and can be used to detect stalled RNA polymerases that have melted the DNA template and opened a characteristic transcription bubble (Giardina et al. 1992). Positions of greater or lesser permanganate reactivity are revealed by piperidine cleavage of the reacted DNA and electrophoretic separation of the resulting fragments. The T positions in melted DNA show up as positions of enhanced cleavage relative to naked double-stranded DNA. No positions of increased cleavage, characteristic of a stalled RNA polymerase, were detected anywhere in the upstream or early region of HMLα1 (Figure 1, lanes 2, 3, and 4 vs. lane 5). Similarly, there was no significant increase in reactivity, relative to the signal in naked DNA, in samples from cells treated with nicotinamide (NAM), a chemical inhibitor of Sir2, or in samples from a sir4∆ mutant (Figure 1, lanes 2, 3, and 4 vs. lane 7). The 5 mM NAM was sufficient for complete derepression of HMRa1. Cells grown in 5 mM NAM showed comparable levels of HMRa1 expression to sir4∆ cells (data not shown). The absence of evidence of separated DNA strands, which would be required of an engaged RNA polymerase, was inconsistent with the mechanism of silencing operating after transcription initiation.
The recent genome-wide datasets on ChIP-exo-seq of preinitiation-complexes and nascent transcript sequencing, performed in MATa cells, allowed independent evaluation of nascent transcripts from HMLα (Churchman and Weissman 2012; Rhee and Pugh 2013). We found no evidence of RNA polymerase binding or early elongation at HML in those datasets.
T7 RNA polymerase was repressed by Sir proteins at HMR
The difference between the steric occlusion model and the preinitiation-complex interference model hinges on whether the restriction of access of a protein to its recognition sequence in silenced chromatin is sufficient to account for the 1000-fold repression of silenced chromatin. Moreover, to date, evidence of the steric occlusion model rests on the ability of simple DNA-binding proteins, with no source of energy from nucleotide hydrolysis, to access and modify their target site within silenced chromatin vs. euchromatin. One could argue that it is easier to inhibit a simple binding reaction than it is to inhibit a process powered by nucleotide hydrolysis and, hence, the steric occlusion model rests on inadequate tests. Conversely, the preinitiation-complex inhibition invites speculation that key components of the core transcription apparatus are unexpectedly sensitive to occlusion, relative to the transcription factor Ppr1, perhaps having evolved sensitivity to the effect of the Sir proteins.
To challenge the steric occlusion model more rigorously, and to determine whether the yeast core transcription machinery is preternaturally sensitive to blockage by silenced chromatin, we tested whether the RNA polymerase from bacteriophage T7, a single subunit RNA polymerase that evolved independently of native eukaryotic chromatin, was able to transcribe a silenced template. For this experiment, the native promoter of HMRa1 was replaced with a minimum optimal T7 promoter (HMR T7pro::a1), as defined by Ujvári and Martin (1997), in cells with a plasmid-encoded, galactose-inducible T7 RNA polymerase endowed with a nuclear localization sequence (Figure 2A). Such a T7 RNA polymerase is able to transcribe a euchromatic locus in yeast (Benton et al. 1990).
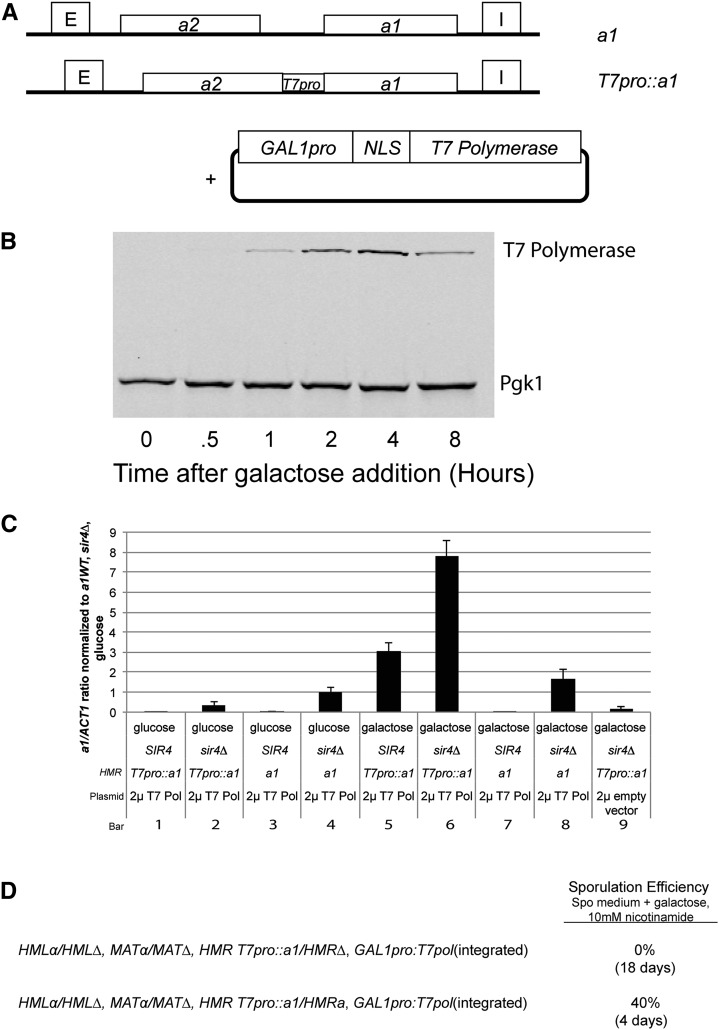
Figure 2.
Quantifying transcription and translation of a1 transcripts from HMR T7pro::a1. (A) A schematic of T7pro::a1 at HMR in comparison to wild-type HMRa1. Twenty bp of the T7 minimal optimum promoter (Ujvári and Martin 1997) replaced the region between a2 and 5 bp upstream of the initiation codon for a1 at HMR. The schematic of the 2 μ plasmid (pJR3207) carrying the nuclear localization signal–enhanced T7 RNA polymerase gene is also shown. (B) Protein immunoblot of T7 RNA polymerase protein levels before and upon galactose induction in CSM-leu medium. Pgk1 levels served as a loading control. (C) Quantitation of a1 transcripts as determined by qRT-PCR of wild-type a1 and T7pro::a1 at HMR in SIR4 and sir4∆, with or without T7 RNA polymerase. All a1 expression values were normalized to ACT1 mRNA from the same sample, and that ratio was further normalized to the a1/ACT1 mRNA ratio in HMRa1 sir4∆ strains grown in glucose. All cultures were seeded from saturated overnight growth in the indicated media. cDNA synthesis was primed using random hexamers. Each bar represents the average and SE of three biological replicates. (D) Diagram of strain genotypes used to test possible translation of a1 transcripts made by T7 RNA polymerase. The top line shows a diploid whose only source of a1 mRNA would be transcribed from T7pro::a1 by T7 RNA polymerase. The bottom line shows an isogenic control diploid with a source of wild-type a1 mRNA at HMR. Ten mM nicotinamide was used to derepress HML and HMR and allow for expression of mating-type information normally silenced by Sir proteins. Sporulation requires both a1 and α2 proteins and the formation of a heterodimer to proceed. Sporulation efficiency was measured as a percentage of cells that formed tetrads.
To quantify the amount of T7 RNA polymerase produced per cell over time, we performed immunoblotting experiments with an antibody against T7 polymerase in a time course following induction in galactose-containing medium (Figure 2B) and compared the signal to that from a dilution series of purified T7 polymerase. T7 RNA polymerase protein levels increased over time, reaching a peak by 4 hr, followed by a gradual decline to steady-state levels. We estimate that the number of T7 polymerase molecules per cell at steady state was approximately 10,000 (data not shown). This value is approximately equal to the number of active RNA Pol II complexes per cell, although there was only one promoter in the genome for T7 RNA polymerase compared to the thousands of promoters for RNA polymerase II (Borggrefe et al. 2001).
Upon induction, T7 RNA polymerase was able to transcribe T7pro::a1 at more than five-fold the level of wild-type HMRa1 in sir4∆ cells (Figure 2C, bar 6 vs. 8). Transcription induced from the T7pro::a1 required T7 RNA polymerase (Figure 2B, bars 5, 6 vs. bars 1, 2, 9), although a background level of a1 transcripts, approximately 10% the level from a wild-type HMRa locus in sir4∆ strains, was detected in cells lacking T7 RNA polymerase (Figure 2C, bars 2, 9). A low background level of transcripts was also detected in sir4∆ strains deleted for the entire promoter region between a1 and a2 of HMR (data not shown). Transcription by the T7 RNA polymerase was specific to the T7 promoter: there was no significant increase in a1 transcription from the a1 promoter at HMR (Figure 2C, bar 4 vs. bar 8). At steady state, T7 RNA polymerase was able to transcribe HMR T7pro::a1 in SIR4 cells at approximately 40% of the level in sir4∆ cells (Figure 2C, bar 5 vs. bar 6).
Some fraction of the T7pro::a1 transcripts were apparently 3′ end-processed and polyadenylated based on their ability to be primed for cDNA synthesis by oligo dT primers, although random hexamers primers detected more transcripts. Previous work has also detected polyadenylated transcripts produced by T7 RNA polymerase in yeast (Sathyanarayana et al. 1999). To test whether the transcripts produced by T7 RNA polymerase were translated, we created diploid strains in which the only source of a1 messages was at HMR T7pro::a1. Sporulation requires a1 protein and can proceed with even small amounts present (reviewed in Piekarska et al. 2010). Such strains were not able to sporulate (0 tetrads among thousands surveyed) in sporulation medium containing galactose and 10 mM nicotinamide to derepress HML and HMR. In contrast, 40% of cells from an isogenic diploid strain bearing wild-type HMRa1 incubated in identical medium sporulated (Figure 2D). Therefore, the T7pro::a1 transcripts were not translated into functional a1 protein. Moreover, these data indicated that the background level of T7 polymerase-independent transcripts in T7pro:a1 in sir4∆ strains also did not produce functional a1 protein. Previous work indicates that 5′-transcript capping is defective on T7 transcripts in yeast, which may have provided the block to a1 translation in our assays (Pinkham et al. 1994; Dower and Rosbash 2002).
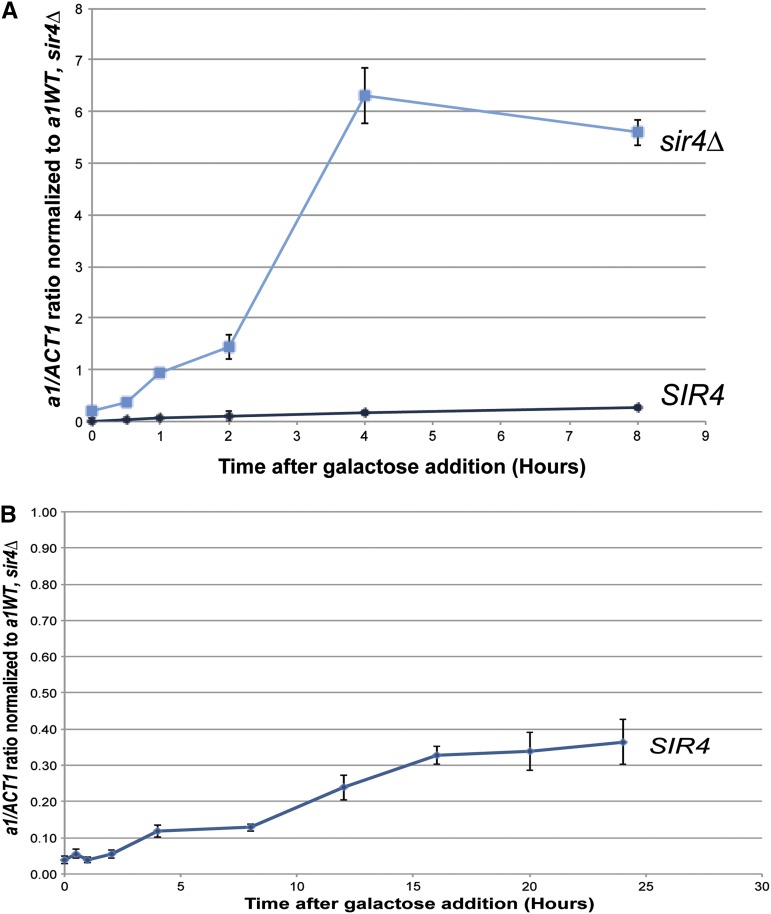
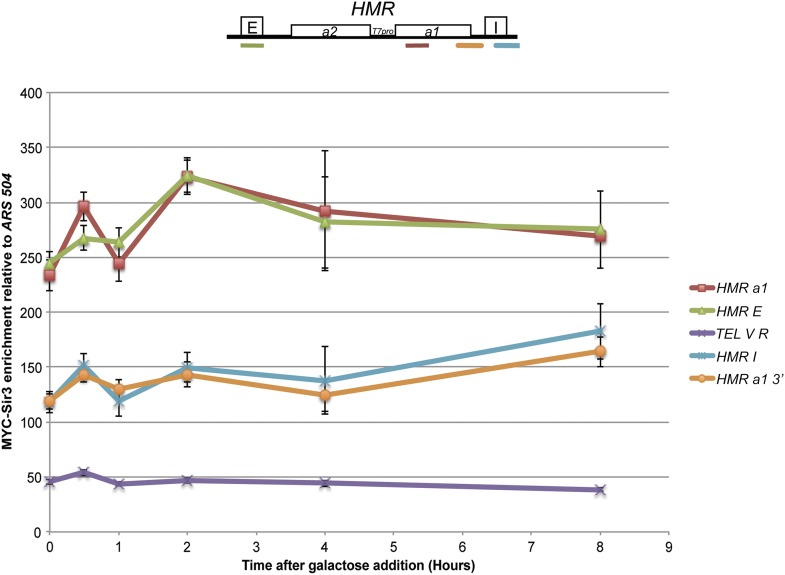
The ability of T7 RNA polymerase to transcribe from its single promoter at HMR in cells that had been grown for many generations with a vast excess of that polymerase was interesting, but not particularly comparable to the challenge faced by RNA polymerase II in trying to transcribe HML and HMR. Therefore, we measured the HMR T7pro::a1 transcript levels in a time course of T7 RNA polymerase induction. As expected, in sir4∆ cells, T7 RNA polymerase robustly transcribed T7pro::a1 transcripts beginning as early as 30 min after the beginning of induction, peaking at approximately 4 hr (Figure 3A, sir4∆). In contrast, transcription by T7 RNA polymerase was barely detectable in SIR4 strains at 1 hr, and then increased only slightly over the next 7 hr (Figure 3B). At 2 hr of induction, expression in SIR4 cells was 200-fold less than in the sir4∆ mutant. The magnitude of the difference in expression between SIR4 cells and sir4∆ cells decreased slightly at later time points, as discussed further below. Nevertheless, these data established that silenced chromatin had a profound ability to inhibit transcription by T7 RNA polymerase within five-fold of the 1000-fold repression by Sir proteins of the native promoters at HML and HMR. ChIP analysis revealed that Sir3 protein remained bound throughout the HMRa1 gene and the entire HMR locus in SIR4 cells throughout the 8-hr induction, in further support of the conclusion that silenced chromatin is largely refractory to transcription by T7 RNA polymerase (Figure 4).
Figure 3.
Quantitative transcript analysis of HMR T7pro::a1 upon induction of T7 RNA polymerase. (A) a1 transcript levels from T7pro::a1 as determined by qRT-PCR in SIR4 (JRY9523) and sir4∆ (JRY9524) strains upon induction of T7 RNA polymerase and media switch from noninducing (raffinose) to inducing (galactose) carbon sources. All a1 expression values were normalized as in Figure 2 except for galactose cultures being the reference. All cultures were seeded from saturated overnight growth in the indicated media. cDNA synthesis was primed using random hexamers. Each bar represents the average and SE of three biological replicates. (B) a1 transcript levels from T7pro::a1 as determined by qRT-PCR in SIR4 cells over an extended induction of T7 RNA polymerase. Note that even at the late time points the level of a1 expression in SIR4 cells has still not achieved the same level as in cells chronically grown in galactose medium as in Figure 2. The levels of a1 expression in the sir4∆ cells were consistent with the values in Figure 2.
Figure 4.
13xMYC-Sir3 enrichment at HMR and telomere V R upon induction of T7 RNA polymerase in SIR4 strains. 13xMYC-Sir3 enrichment as assayed by ChIP followed by qRT-PCR at HMR and telomere V R from the same cultures used in Figure 3. Values are displayed as 13xMYC-Sir3 enrichment at the color-coded positions relative to an ARS504 negative control. The cartoon above the plot shows the location of the primer sets at HMR. Each point represents the average and SE of three biological replicates, except the HMRa1 8-hr time point, which is an average of two biological replicates.
To investigate further transcription by T7 polymerase from T7pro::a1 in SIR4 strains, we performed an additional identical experiment in SIR4 cells and tracked a1 expression over a longer time period. We detected a consistent increase in T7pro::a1 transcripts over time (Figure 3B). As previously shown, when cells were assayed after entering stationary phase, a1 expression levels increased beyond what was seen at the longest time point in our induction experiment (Figure 2C, bar 5).
The Gal4 activator was largely insensitive to Sir protein silencing
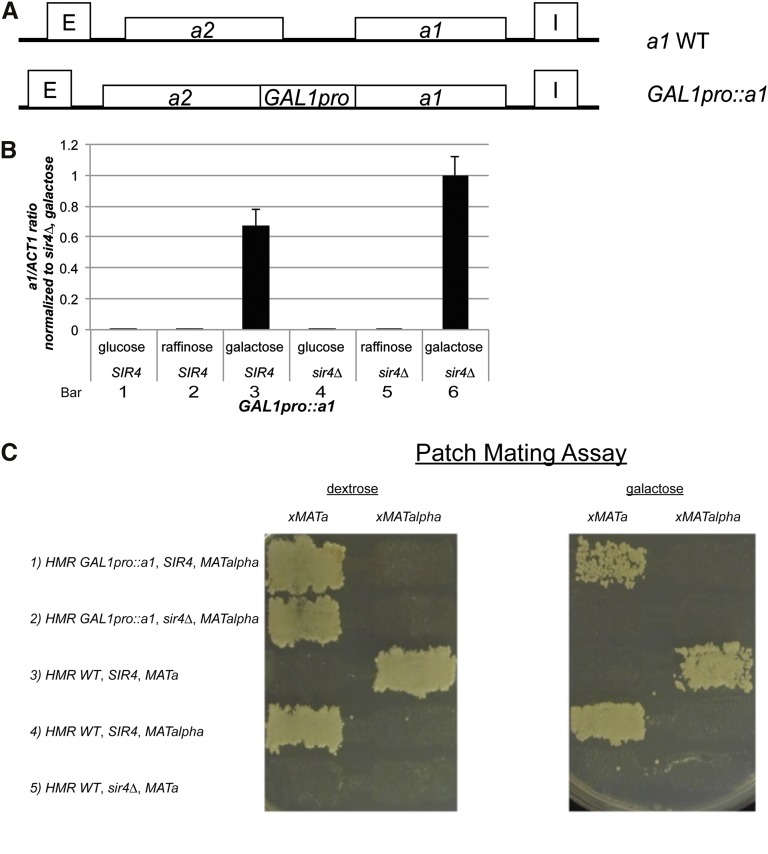
When placed at HML or HMR, yeast genes vary with respect to their sensitivity to silencing (Brand et al. 1985; Chen and Widom 2005; Ren et al. 2010). To explore the parameters influencing the sensitivity of a gene to Sir-based silencing, we replaced the promoter of HMRa1 with the full 450-bp GAL1 promoter (HMR GAL1pro::a1), maintaining the approximate size of the HMR locus and the distance from the promoter to silencers (Figure 5A). As expected, in SIR4 and sir4∆ strains HMR GAL1pro::a1 was not expressed in repressing (glucose medium) or noninducing conditions (raffinose medium) (Figure 5B, bars 1, 2, 4, 5). Again, as expected, GAL1pro::a1 was expressed in sir4∆ strains grown in galactose medium but not glucose medium (Figure 5B, bar 4 vs. 6). In SIR4 strains grown in inducing medium, GAL1pro::a1 expression was 60% of the level of the sir4∆ strain (Figure 5B, bar 3 vs. bar 6). These results verified that GAL1pro::a1 was expressed at steady state and was largely insensitive to Sir-based silencing.
Figure 5.
Analysis of expression of HMR GAL1pro::a1. (A) A schematic of GAL1pro::a1 at HMR in comparison to wild-type a1. Four hundred fifty bp of the GAL1 promoter containing the four binding sites of Gal4 (4 UASg sites) replaced the region between the ORFs of a2 and a1 at HMR. (B) mRNA expression as determined by qRT-PCR of GAL1pro::a1 at HMR in SIR4 and sir4∆ on various carbon sources. All a1 expression values were normalized to ACT1 mRNA from the same sample and further to the a1/ACT1 mRNA ratio in GAL1pro::a1, sir4∆ strains in galactose medium. Each bar represents the average and SEM of three biological replicates. (C) Patch mating assay of various strains with either HMR GAL1pro::a1 or native HMRa1 on dextrose or galactose media. Query strains were mated to tester strains prototrophic for all markers expect for his4 and any resulting diploids were replica plated onto minimal medium lacking histidine and containing dextrose or galactose. Growth of diploids demonstrated the ability to mate.
As expected, the transcripts produced by GAL1pro::a1 were translated based on the a/α diploid mating phenotype of MATα, HMR GAL1pro::a1, sir4∆ strains when grown on galactose-containing medium (Figure 5C, row 2, right-most panel). Likewise, MATα, HMR GAL1pro::a1, SIR4 strains exhibited reduced mating efficiency in cells grown on galactose-containing medium but not in cells grown on glucose-containing medium (Figure 5C, row 1 vs. 2).
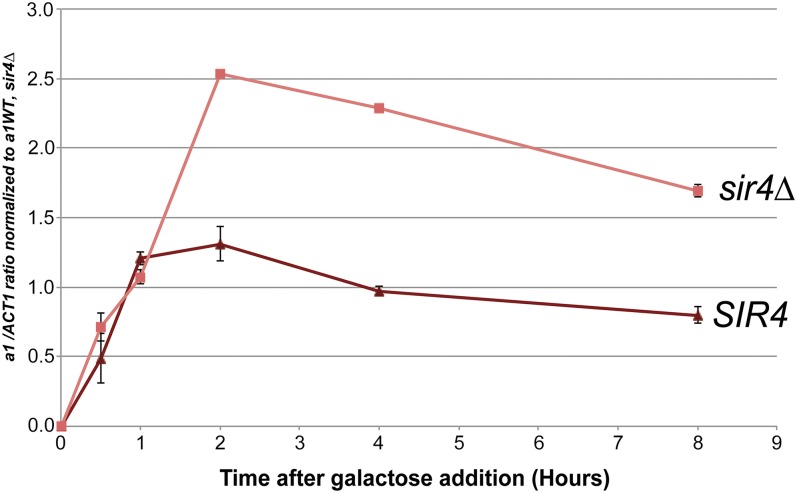
Steady-state analyses of the impact of silencing on T7 RNA polymerase obscured the sensitivity to Sir-based silencing that was revealed in kinetic experiments. Hence, we performed analogous kinetic experiments measuring HMR GAL1pro::a1 transcripts in Sir+ and Sir− cells at time points after induction. Remarkably, in the first hour after induction there was no detectable difference in transcription from HMR GAL1pro::a1 between sir4∆ cells and SIR4 cells (Figure 6). After 2 hr, sir4∆ strains exhibited approximately two-fold higher levels of a1 mRNA levels than SIR4 strains, and this difference in level was relatively unchanged at later time points (Figure 6). The level of a1 transcripts produced in the SIR4, GAL1pro::a1 strain was approximately equal to the level in sir4∆ strains with a wild-type HMRa1 locus. Thus, the magnitude of silencing escape of the GAL1pro::a1 allele was significant.
Figure 6.
Quantitation of a1 transcripts from HMR GAL1pro::a1 upon induction with galactose in SIR4 and sir4∆ strains. a1 mRNA expression of GAL1pro::a1 as determined by qRT-PCR in SIR4 and sir4∆ strains upon induction with galactose. Strains were initially grown in CSM noninducing (raffinose) medium and galactose was added to induce transcription from the GAL1 promoter. All a1 expression values were normalized as in Figure 5. All cultures were seeded from saturated overnight growth in the indicated media. cDNA synthesis was primed using random hexamers. Each bar represents the average and SE of three biological replicates.
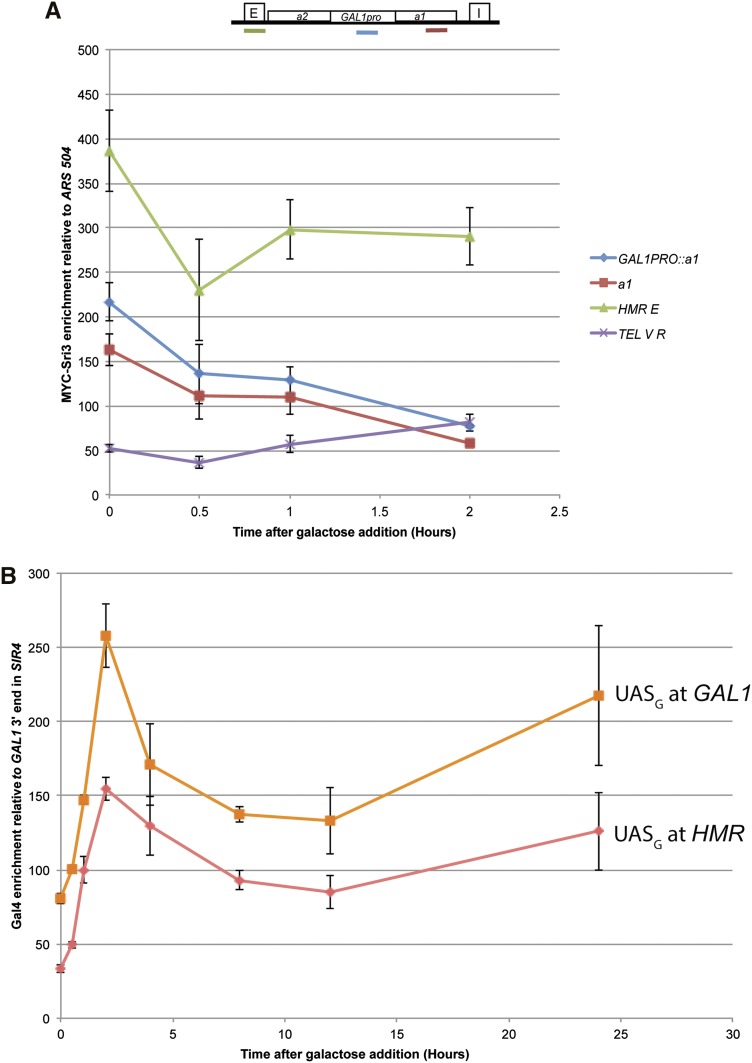
Sir protein enrichment at the GAL1 promoter at HMR (GAL1pro::a1) at an internal region of a1 and at HMR-E significantly decreased between t = 0 and t = 2 hr (paired Student’s t-test P = 0.012, P = 0.009, P = 0.021, respectively) (Figure 7A). Although it is possible that transcription caused some loss of Sir proteins from the nucleosomes within the a1 gene, the reduction at HMR-E could not be due to transcription. Despite the considerable transcription of a1 from the GAL1 promoter in Sir+ cells (Figure 7A, SIR4 t = 2), Sir protein enrichment at the a1 gene remained considerably above background (Figure 7A). We emphasize that at present it is unknown whether Sir proteins occupied the a1 gene in the same cells that produced a1 transcripts, or whether these assays reflected two different populations in the same culture.
Figure 7.
13xMYC-Sir3 and Gal4 enrichment at HMR GAL1pro::a1 upon kinetic galactose induction in SIR4 strains. (A) 13xMYC-Sir3 enrichment as assayed by Sir3 ChIP followed by qRT-PCR at HMR from the identical SIR4 kinetic cultures described in Figure 6. Values are displayed as 13xMYC-Sir3 enrichment relative to an ARS504 negative control primer set. All HMR primer sets showed a statistically significant reduction in Sir3 occupancy between 0 and 2 hr (see text for p-values). The cartoon above the plot shows the location of the primers sets at HMR. Each point represents the average and SEM for three biological replicates. (B) Gal4 enrichment at HMR GAL1pro::a1 and at GAL1 as assayed by ChIP followed by qRT-PCR. Values are displayed as Gal4 enrichment relative to a negative control primer set mapping to the intergenic region 3′ to the GAL1 locus. Each point represents three biological replicates and SEM.
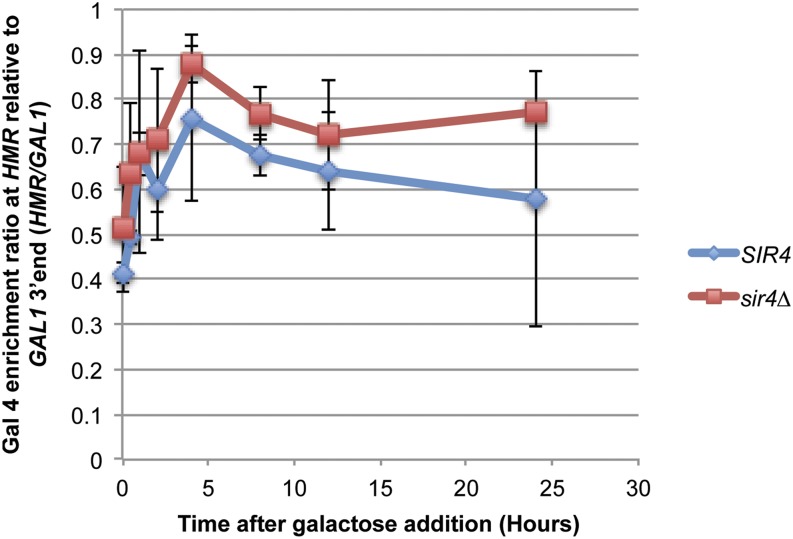
The rapid and identical induction kinetics for transcription in Sir+ and Sir− cells raised the possibility that Gal4 protein was bound to the GAL1 UAS in (GAL1pro::a1) prior to induction, just as it is known to bind these sequences at the native GAL gene cluster (Lohr and Hopper 1985; Selleck and Majors 1987). Chromatin immunoprecipitation analysis of Gal4 in SIR4 and sir4∆ strains revealed that at t = 0, there was considerable enrichment of Gal4 protein at both sites, albeit with two-fold greater enrichment at the native GAL1 locus (Figure 7B). Upon induction, Gal4 enrichment levels increased at both HMR GAL1pro::a1 and at the native GAL1 locus, presumably reflecting the well-documented induction of Gal4 protein in galactose medium. At least part of the difference between the enrichment of Gal4 protein at the GAL1 UAS at the native GAL gene cluster vs. HMR was due to the presence of Sir proteins at HMR, as evidenced by comparing the relative enrichment of Sir3 at HMR in SIR4 vs. sir4∆ cells (Figure 8).
Figure 8.
Gal4 enrichment at HMR GAL1pro::a1 and GAL1 upon galactose induction in SIR4 and sir4∆ strains. Gal4 enrichment as assayed by ChIP followed by qRT-PCR at HMR from the same SIR4 and sir4∆ cultures described in Figure 6 and Figure 7. Values are displayed as the ratio of Gal4 enrichment at HMR GAL1pro::a1 relative to enrichment at GAL1 and a negative control from the intergenic region 3′ to the GAL1 locus to account for the reduced ability of HMR GAL1pro::a1 to recruit Gal4 compared to native GAL1. Each point represents three biological replicates with SEM.
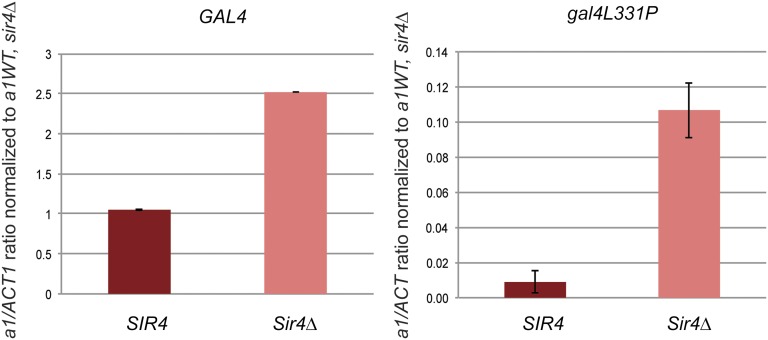
Abf1 is the transcription factor responsible for activating transcription from MATa (McBroom and Sadowski 1995) and therefore is susceptible to silencing at HMR, whose regulatory region sequence is identical to MATa. To explore why Gal4 was largely immune from silencing at HMR whereas Abf1 is sensitive, we tested whether mutant forms of Gal4 that compromise some aspect of its function would become sensitive to silencing. A point mutant in the central domain of Gal4 (gal4L331P) compromises its ability to activate transcription but not its ability to bind DNA (Johnston and Dover 1988; Mylin et al. 1990). As expected, relative to wild-type Gal4, the gal4L331P mutant protein showed a 25-fold reduced ability to activate transcription of HMR GAL1pro::a1 in sir4∆ strains (Figure 9, note the y-axis scale). In SIR4 cells a1 transcript levels were even further reduced (Figure 9, right panel), with 12-fold instead of the 2.5-fold reduction of wild-type Gal4. Thus, in this example, a weaker activator exhibited a greater degree of sensitivity to silencing than a stronger version of that same activator. This relationship between sensitivity to silencing and activator strength of Gal4 was observed for some other alleles of GAL4, but exceptions such as gal4-763∆851 indicate that silencing sensitivity is influenced by more than simply activator strength (Table 4).
Figure 9.
Analysis of HMR Gal1pro:a1 transcription by wild-type Gal4 vs. gal4L331P. a1 mRNA expression from GAL1pro::a1 as determined by qRT-PCR of mRNA from strains bearing Gal4 or the gal4L331P mutant in SIR4 and sir4∆ in galactose medium. All a1 expression values were normalized as in Figure 5B. All cultures were seeded from saturated overnight growth. Each bar represents the average and SE of three biological replicates.
Table 4. Analysis of expression of HMR GAL1pro::a1 in gal4 mutants.
| Gal4 Allele | DNA Binding Affinitya (% of WT) | GAL1pro::a1 mRNA in sir4∆ as a Percentage of Gal4, sir4∆, GAL1pro::a1 mRNA Expression (%) | Ratio of Gal1pro::a1, sir4∆ mRNA /GAL1pro::a1, SIR mRNA (sir4∆/SIR) | Gal1pro::a1, sir4∆ mRNA |
|---|---|---|---|---|
| WT (GAL4) | 100 | 100 | 1.7 | 2.51 ± 0.12 |
| gal4-147∆768 | 51.7 | 118 | 2.5 | 2.96 ± 0.08 |
| gal4-238∆768 | 75.6 | 63.8 | 1.8 | 1.60 ± 0.06 |
| gal4-763∆851 | 19.1 | 47 | 11.5 | 1.79 ± 0.21 |
| gal4-238∆851 | 28.7 | 8.9 | 10.9 | 0.223 ± 0.01 |
| gal4-848∆ | 85.1 | 5 | 22 | 0.125 ± 0.2 |
| gal4-844∆ | 46.6 | 4.8 | 22.8 | 0.121 ± 0.21 |
| gal4∆ | 0 | 0.4 | ND | 0 ± 0.01 |
a1 transcript levels from GAL1pro::a1 as determined by qRT-PCR in SIR4 and sir4∆ strains upon GAL induction and media switch from noninducing (raffinose) to inducing (galactose) carbon sources. All a1 expression values were normalized to ACT1 mRNA and further to the a1/ACT1 mRNA ratio in HMRa1 sir4∆ strains. Each mRNA expression number represents the average and SEM for three biological replicates.
DNA binding affinity of gal4 mutants calculated based on filter binding assay described in Ma and Ptashne (1987).
Discussion
Steric occlusion was a major contributor to the mechanism of gene silencing
To date, the T7 RNA polymerase assay represents the most stringent test of the steric occlusion model for several reasons. First, the assay used a transcription protein rather than a simple DNA binding protein as a quantitative test of the Sir protein transcriptional repression mechanism. Second, the assay provided a strong challenge to Sir protein repression because within 2 hr of induction, the level of T7 RNA polymerase was comparable to the level of RNA polymerase II. Yet these cells had only a single T7 promoter, in contrast to thousands of targets for RNA Pol II. Therefore, the T7 polymerase transcription assay stringently measured how effective steric occlusion was at blocking transcription by an RNA polymerase present at a concentration more than 1000-times that of its target sequence. Third, because of its prokaryotic origin, T7 polymerase offered immunity from potential complications resulting from interactions among eukaryotic chromatin proteins.
Sir-based silencing was a significant impediment to T7 polymerase transcription (Figure 2 and Figure 3). The difference between the T7-mediated transcription of HMR T7pro::a1 in SIR4 vs. sir4∆ cells was 200-fold at 2 hr after induction, although somewhat lower at longer time points. A 200-fold repression of T7 RNA polymerase due to Sir proteins was in reasonably close quantitative agreement with the 1000-fold magnitude of repression of RNA Pol II by Sir proteins at HMRa1 and might be accountable by differences in mRNA half-lives.
By 8 hr of induction, a gradual increase in the low-level expression of a1 transcripts from the T7pro::a1 in SIR4 cells was detected (Figure 3B). The cause of this low-level escape from silencing was unclear, but the vast excess of T7 RNA polymerase offered a simple possibility. During each cell cycle, silenced chromatin has to be replicated at HMR. At least one and probably both chromatids have to assemble new Sir protein complexes. Therefore, replication could be a target of opportunity when the chromatin at the T7 promoter would not yet be completely decorated by Sir proteins and, hence, available to T7 RNA polymerase. The higher level of expression from the T7 promoter in SIR4 cells at steady state might reflect the additive effect of many successful competitions between T7 RNA polymerase for its promoter over the binding of Sir protein complexes to the chromatin on that promoter.
The issue of what happens to Sir proteins on HMR at the later time points after induction of T7 RNA polymerase, when a1 RNA transcripts were detected, is not fully resolved. ChIP data were consistent with Sir proteins remaining associated with HMR T7pro::a1 in SIR4 strains even when low-level transcription occurred (Figure 4). However, we could not exclude the simpler possibility that the cells that produced the a1 transcripts were a different subset of the culture from the cells that contributed this ChIP signal of Sir proteins at HMR. It is possible that in batch culture most cells had fully silenced HMR T7pro::a1 and those cells drove the ChIP signal, whereas a small population of cells in the culture, if derepressed, could account for the small a1 transcript signal.
The mechanism of Sir-based silencing was largely independent of any special feature of the native transcription machinery. Occlusion of a specific eukaryotic transcription factor or coactivator would have no effect on blocking transcription by T7 RNA polymerase. It remains unclear whether T7 RNA polymerase was physically prevented from binding its promoter in silenced chromatin, or whether the 200-fold difference in repression between SIR4 and sir4∆ strains reflects a mixture of reduced polymerase binding and reduced postbinding events. We have not been able to produce an epitope-tagged T7 RNA polymerase that still functions as a polymerase. Existing antibodies, although serviceable for immunoblots, have not provided sufficient sensitivity in ChIP assays to test whether Sir-based inhibition of promoter binding could account for the full magnitude of the effect.
Histone deactylation as a mechanism of gene repression is shared among many taxa. However, the use of Sir proteins as structural components of heterochromatin is specific to yeast. Surprisingly, in analogous experiments using Gal4 and T7 polymerase to investigate repression of transcription by Drosophila polycomb complex, Polycomb blocks Gal4 activation but not T7 polymerase from productive transcription in flies (McCall and Bender 1996; Fitzgerald and Bender 2001).
No evidence of RNA polymerase II in silenced chromatin
Our results were incompatible with the downstream-inhibition model for silencing in yeast and call into question the data supporting that model. Specifically, the critical prediction of this model is that there would be a position near the promoter of α1, where RNA Pol II is found within Sir-silenced heterochromatin bound on the template and with the two strands of the template held apart in what is equivalent to a paused transcription bubble. The nearly identical cleavage pattern of the permanganate-labeled DNA from this region in SIR4 and sir4 cells indicated a lack of any position at HMLα1 in silenced cells at which RNA polymerase II was stalled (Figure 1). Likewise, the pattern of permanganate reactivity was indistinguishable between HMLα1 silenced by Sir proteins and HMLα1 repressed by Tup1/Ssn6 corepressor acting through the a1/α2 repressor (Strathern et al. 1981; Komachi et al. 1994) . The mechanism of repression by a1/α2 through Ssn6/Tup1 does not involve a stalled polymerase, but rather acts at the level of transcription initiation (Parnell and Stillman 2011). It should be noted that, in contrast to other organisms, there has not been a paused polymerase in yeast detected by the permanganate assay. Hence, formally there was no direct positive control for this result. Nevertheless, together with the other results presented here, the most parsimonious interpretation is that the mechanism of silencing is one that operates prior to any engagement by RNA polymerase.
ChIP-exo-seq (Rhee and Pugh 2013) and nascent transcript-seq datasets (Churchman and Weissman 2012) also contained no evidence of RNA PolI II at silenced HML. The lack of a stalled RNA Pol II in silenced chromatin led us to conclude that the primary repressive mechanism of Sir protein–mediated silencing acts before DNA melting in the transcription cascade. Previous reports of ChIP (Gao and Gross 2008) and ChIP genome-wide array data (Steinmetz et al. 2006) that inspired the downstream-inhibition model were likely misled by the relative shearing resistance of silenced chromatin (Teytelman et al. 2009) resulting in artifactual precipitation of HML and HMR with RNA polymerase engaged on flanking genes in inadequately sheared chromatin.
In silenced chromatin, there remained a five-fold difference between the 200-fold repression of T7 RNA polymerase and the 1000-fold repression of transcription of the native HMRa1 that has not been accounted for. We cannot exclude the possibility that in a small subset of cells RNA polymerase II occasionally does engage the a1 promoter at HMR, perhaps when the chromatin is replicated, yet fails to elongate all the way through silenced chromatin. There is a precedent for a different mechanism of epigenetic silencing that blocks transcriptional elongation as has been shown for methylated genes in Neurospora (Rountree and Selker 1997). If so, then our data indicated that there would be no unique arrest point of such polymerase molecules within the detection sensitivity of the assay. A simpler model to explain the discrepancy between the 200-fold repression of T7 RNA polymerase and 1000-fold repression of transcription of the native promoters at HML and HMR might be differences in the half-lives of the different RNAs, which to date have not been evaluated.
Implications of the Gal4 activator escape from silencing at HMR
The results of the experiments with the HMR GAL1pro::a1 allele highlight that a simple steric occlusion model alone cannot explain all aspects of the mechanism of transcriptional silencing. Gal4-activated transcription was repressed, at most, two-fold due to Sir proteins (Figure 6).
The affinities of Abf1, Gal4, and T7 polymerase for their binding sites appear to be similar (in the low nM range) (Taylor et al. 1991; Ujvári and Martin 1997; Beinoravičiūtė-Kellner et al. 2005). Yet, T7 RNA polymerase and Abf1-dependent activation are dramatically repressed, but Gal4 can both access its binding site and promote transcription in the presence of Sir proteins.
In addition, Gal4 was, by inference, able to recruit coactivators to silenced regions in vivo, in contrast to in vitro data claiming activator-interference is a primary silencing mechanism (Johnson et al. 2013). In the first hour of galactose induction, there was no difference in transcription at HMR GAL1pro::a1 between SIR4 and sir4∆ strains (Figure 7). The identical transcript induction kinetics in SIR4 and sir4∆ strains over the first hour suggests that Gal4 had already occupied its binding site in cells of either genotype before induction began. Moreover, the rate of mRNA production in the two strains was similar for that first hour. Hence, by these criteria, Gal4 was largely immune from Sir-based silencing. At later time points there was a consistent approximately two-fold higher level of transcripts in sir4∆ than in SIR4. The reason for this difference remains unknown. It is striking to us that Gal4 appeared to be largely immune to silencing given that the expression level from HMR Gal1pro::a1 was only 2-fold to 2.5-fold greater than achieved by the native a1 promoter at wild-type HMR in sir4 cells. A recent article concludes that transcription factors that are weak activators are sensitive to Sir-based silencing, whereas those that are stronger activators are not (Wang et al. 2015). It would be striking if such a small difference in activator strength could explain the difference in silencing sensitivity of Abf1 relative to Gal4.
Conceptually, being able to determine whether Rap1 occupies its binding site at HML and whether Abf1 occupies its binding site at HMR (McBroom and Sadowski 1995) would provide useful information regarding the mechanism of silencing. Unfortunately, the quality of the reagents needed for this assessment, combined with the relative shearing resistance of HML and HMR (Teytelman et al. 2009), and the inaccessibility of key positions within HML and HMR to antibodies (Thurtle and Rine 2013) have, to date, prevented a clear resolution.
Acknowledgments
We thank David Gross, Jonathan Widom, and the members of our lab, including Meru Sadhu and Deborah Thurtle, for helpful discussions during the course of these experiments. We thank William Studier for anti-T7 RNA polymerase antibodies. This work was supported by an NSF predoctoral fellowship (DGE 1106400 to D.L.S.), NIH Genetics Training Grant (T32 GM 007127), and a grant from the National Institutes of Health (GM31105 JR).
Footnotes
Communicating editor: S. L. Jaspersen
Literature Cited
- Adelman K., Lis J. T., 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparacio O. M., Billington B. L., Gottschling D. E., 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Aparicio, O., J. V. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi et al., 2005 Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol 21: 21.3. [DOI] [PubMed] [Google Scholar]
- Beinoravičiūtė-Kellner R., Lipps G., Krauss G., 2005. In vitro selection of DNA binding sites for ABF1 protein from Saccharomyces cerevisiae. FEBS Lett. 579: 4535–4540. [DOI] [PubMed] [Google Scholar]
- Benton B. M., Eng W. K., Dunn J. J., Studier F. W., Sternglanz R., et al. , 1990. Signal-mediated import of bacteriophage T7 RNA polymerase into the Saccharomyces cerevisiae nucleus and specific transcription of target genes. Mol. Cell. Biol. 10: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T., Davis R., Bareket-Samish A., Kornberg R. D., 2001. Quantitation of the RNA polymerase II transcription machinery in yeast. J. Biol. Chem. 276: 47150–47153. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K., 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41: 41–48. [DOI] [PubMed] [Google Scholar]
- Chen L., Widom J., 2005. Mechanism of transcriptional silencing in yeast. Cell 120: 37–48. [DOI] [PubMed] [Google Scholar]
- Churchman L. S., Weissman J. S., 2012. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K., Rosbash M., 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald D. P., Bender W., 2001. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 21: 6585–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Gross D. S., 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol. Cell. Biol. 28: 3979–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C., Perez-Riba M., Lis J. T., 1992. Promoter melting and TFIID complexes on Drosophilia genes in vivo. Genes Dev. 6: 2190–2200. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Fan R., 2009. Detecting transcriptionally engaged RNA polymerase in eukaryotic cells with permanganate genomic footprinting. Methods 48: 368–374. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA 89: 4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L., 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Johnson A., Wu R., Peetz M., Gygi S. P., Moazed D., 2013. Heterochromatic gene silencing by activator interference and a transcription elongation barrier. J. Biol. Chem. 288: 28771–28782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J., 1988. Mutational analysis of the GAL4-encoded transcriptional activator protein of Saccharomyces cerevisiae. Genetics 120: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komachi K., Redd M. J., Johnson A. D., 1994. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 8: 2857–2867. [DOI] [PubMed] [Google Scholar]
- Kueng S. M. Oppikofer, and Gasser S. M., 2013. Sir protein and the assembly of silent chromatin in budding yeast. Annu. Rev. Genet. 47: 275–306. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., et al. , 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E., 1985. Organization of the GAL1–GAL10 intergenic control region chromatin. Nucleic Acids Res. 13: 8409–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S., Rine J., 1994. Silencers and domains of generalized repression. Science 264: 1768–1771. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M., 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48: 847–853. [DOI] [PubMed] [Google Scholar]
- McBroom L. D., Sadowski P. D., 1995. Functional analysis of the ABF1-binding sites within the Ya regions of the MATa and HMRa loci of Saccharomyces cerevisiae. Curr. Genet. 28: 1–11. [DOI] [PubMed] [Google Scholar]
- McCall K., Bender W., 1996. Probes of chromatin accessibility in the Drosophila bithorax complex respond differently to Polycomb-mediated repression. EMBO J. 15: 569–580. [PMC free article] [PubMed] [Google Scholar]
- Mylin L. M., Johnston M., Hopper J. E., 1990. Phosphorylated forms of GAL4 are correlated with ability to activate transcription. Mol. Cell. Biol. 10: 4623–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarska I., Rytka J., Rempola B., 2010. Regulation of sporulation in the yeast Saccharomyces cerevisiae. Acta Biochim. Pol. 57: 241–250. [PubMed] [Google Scholar]
- Parnell E. J., Stillman D. J., 2011. Shields up: The Tup1-Cyc8 repressor complex blocks coactivator recruitment. Genes Dev. 25: 2429–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham J. L., Dudley A. M., Mason T. L., 1994. T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol. Cell. Biol. 14: 4643–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wang C. L., Sternglanz R., 2010. Promoter strength influences the S phase requirement for establishment of silencing at the Saccharomyces cerevisiae silent mating type loci. Genetics 186: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. S., Pugh B. F., 2013. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W., 1980. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 8: 4613–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree M. R., Selker E. U., 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11: 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Sathyanarayana U. G., Freeman L. A., Lee M. S., Garrard W. T., 1999. RNA polymerase-specific nucleosome disruption by transcription in vivo. J. Biol. Chem. 274: 16431–16436. [DOI] [PubMed] [Google Scholar]
- Sekinger E. A., Gross D. S., 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. M., 1987. In vivo DNA-binding properties of a yeast transcription activator protein. Mol. Cell. Biol. 7: 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Klar A. J., 1992. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 6: 186–196. [DOI] [PubMed] [Google Scholar]
- Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., et al. , 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24: 735–746. [DOI] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I., 1981. Control of cell type in yeast by the mating type locus: The α1-α2 hypothesis. J. Mol. Biol. 147: 357–372. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., et al. , 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31: 183–192. [DOI] [PubMed] [Google Scholar]
- Taxis C., Knop M., 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 40: 73–78. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E., 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 5: 1285–1298. [DOI] [PubMed] [Google Scholar]
- Teytelman L., Ozaydin B., Zill O., Lefrançois P., Snyder M., et al. , 2009. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One 4: e6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurtle D., Rine J., 2013. The molecular topology of silenced chromatin in Saccharomyces cerevisiae. Genes Dev. 28: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvári A., Martin C. T., 1997. Identification of a minimal binding element within the T7 RNA polymerase promoter. J. Mol. Biol. 273: 775–781. [DOI] [PubMed] [Google Scholar]
- Wang X., Bryant G., Zhao A., Ptashne M., 2015. Nuclesome avidities and transcriptional silencing in yeast. Curr. Biol. 25: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Wellinger R. J., Zakian V. A., 2012. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: Beginning to end. Genetics 191: 1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.-W., Nechaev S., et al. , 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39: 1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S., 2005. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 33: 2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]