Abstract
Primary ciliary dyskinesia (PCD) is an autosomal-recessive disorder resulting from loss of normal ciliary function. Symptoms include neonatal respiratory distress, chronic sinusitis, bronchiectasis, situs inversus, and infertility. Clinical features may be subtle and highly variable, making the diagnosis of PCD challenging. The diagnosis can be confirmed with ciliary ultrastructure analysis and/or molecular genetic testing of 32 PCD-associated genes. However, because of this genetic heterogeneity, comprehensive molecular genetic testing is not considered the standard of care, and the most efficient molecular approach has yet to be elucidated. Here, we propose a cost-effective and time-efficient molecular genetic algorithm to solve cases of PCD. We conducted targeted copy number variation (CNV) analysis and/or whole-exome sequencing on 20 families (22 patients) from a subset of 45 families (52 patients) with a clinical diagnosis of PCD who did not have a molecular genetic diagnosis after Sanger sequencing of 12 PCD-associated genes. This combined molecular genetic approach led to the identification of 4 of 20 (20%) families with clinically significant CNVs and 7 of 20 (35%) families with biallelic pathogenic mutations in recently identified PCD genes, resulting in an increased molecular genetic diagnostic rate of 55% (11/20). In patients with a clinical diagnosis of PCD, whole-exome sequencing followed by targeted CNV analysis results in an overall molecular genetic yield of 76% (34/45).
Keywords: primary ciliary dyskinesia, whole-exome sequencing, copy number variation, diagnostic testing
Primary ciliary dyskinesia (PCD, MIM#244400) is a genetically heterogeneous, autosomal-recessive motile ciliopathy. Clinical manifestations include situs inversus, neonatal respiratory distress, progressive bronchiectasis, and respiratory failure in young adulthood. To date, there are 32 genes associated with PCD that account for a genetic diagnosis in approximately 65% of all patients with PCD (Kurkowiak et al. 2015).
The clinical diagnosis of PCD is challenging and relies on patient features and diagnostic investigations. These include diagnostic imaging, nasal nitric oxide levels, cilia beat frequency, and ciliary ultrastructure analysis by electron microscopy (EM) (Leigh et al. 2011). Historically, a diagnosis of PCD has relied on the findings of a classic ciliary ultrastructural defect on EM (Barbato et al. 2009). EM of normal motile cilia consist of nine peripheral microtubule doublets encircling a central microtubular complex supported by microtubular-associated proteins such as outer and inner dynein arms (ODA, IDA, respectively), radial spokes, and nexin links (Knowles et al. 2013a).
A molecular genetic diagnosis of PCD can be established through the identification of biallelic loss-of-function mutations in a PCD-associated gene. Mutations in certain PCD genes result in ciliary dysfunction and can give a specific ultrastructural defect on EM (Knowles et al. 2013a). ODA defects are the ultrastructural defect observed most frequently in PCD and are associated with mutations in DNAH5, DNAI1, DNAI2, DNAL1, TXNDC3, CCDC114, and ARMC4 (Lobo et al. 2015). Defects in both ODA and IDA are seen in patients carrying mutations in LRRC50/DNAAF1, KTU/DNAAF2, DNAAF3, CCDC103, HEATR2, LRRC6, ZMYND10, DYX1C1, C21orf59, and SPAG1. IDA defects with microtubular disorganization are seen with mutations in CCDC39 and CCDC40. Central microtubular abnormalities are observed with mutations in RSPH1, RSPH4A, and RSPH9. Patients with mutations in CCNO and MCIDAS have ciliary aplasia/oligoplasia and a marked reduction of cilia (oligocilia) due to a deficiency in ciliogenesis rather than motility. In addition, patients with mutations in DNAH11, HYDIN, CCDC164/DRC1, and CCDC65/DRC2 do not have obvious ciliary ultrastructural defects and would be not be diagnosed with PCD on the basis of biopsy EM alone. We have shown previously that molecular genetic analysis can complement ciliary ultrastructure analysis and increase the overall diagnostic yield from 57% (28/49) to 69% (36/52) (Kim et al. 2014). However, the molecular genetics of PCD are complex and not considered routine in the diagnostic evaluation of patients (Barbato et al. 2009; Bush and Hogg 2012). Modern advances in molecular genetic technologies may overcome some of these perceived limitations in a cost-effective manner.
In genetically heterogeneous diseases such as PCD, selecting the appropriate gene and technique for molecular analysis is often difficult. Sanger sequencing is available for many, but not all, of the 32 PCD-associated genes. A step-wise approach by prioritizing genes based on ciliary ultrastructure and prevalence of mutations could be conducted using Sanger sequencing (Bush and Hogg 2012; Kim et al. 2014). However, as the number of genes tested increase, Sanger sequencing can become costly and time-consuming. In PCD, many new loci are being discovered on an ongoing basis, quickly out-dating existing available panels. Moreover, newly discovered loci account for a minority of patients with PCD (Knowles et al. 2013a), decreasing the diagnostic value for each gene added to a conventional panel.
Intron-exon level copy number variations (CNVs) are seen in up to 10.8% of autosomal-recessive Mendelian disorders and are not detectable by Sanger sequencing (Aradhya et al. 2012). Deletions in SPAG1, ARMC4, DYX1C1, LRRC50, and ZYMD10 have been observed in isolated PCD cohorts (Loges et al. 2009; Blanchon et al. 2012; Hjeij et al. 2013; Knowles et al. 2013b; Tarkar et al. 2013; Zariwala et al. 2013), suggesting CNV analysis should be used in the molecular evaluation of patients with PCD. However, the prevalence and clinical significance of CNVs in other PCD-associated genes have not been examined.
Whole-exome sequencing (WES) has been used extensively for gene discovery and is beginning to be used in clinical practice in the diagnosis of genetically heterogeneous diseases (Neveling et al. 2013). The advantages of WES include providing a genomic approach in molecular genetic diagnoses through the analysis of all potential causative genes, including recently discovered genes not available on clinical gene panels (Boycott et al. 2015). Furthermore, this strategy allows for more complete interpretation of variants of uncertain significance (VUS), and can guide further molecular genetic analyses such as targeted CNV analysis. Because of ongoing gene discovery in genetically heterogeneous diseases such as PCD, WES may be a more cost-effective technique (Lucas et al. 2014; Kurkowiak et al. 2015). Here, we describe the complementary role of WES and targeted CNV analysis in solving cases of patients with PCD in a cost and time-effective manner.
Material and Methods
Study subjects
Between the period of August 2011 and December 2012, Sanger sequencing of 12 PCD genes was clinically available and conducted on 45 families with a clinical diagnosis of PCD (Kim et al. 2014). A total of 19 families (42%) were found to have biallelic pathogenic mutations in these 12 PCD genes and thus confirmed a molecular diagnosis. Pathogenic mutations were defined as mutations previously documented in patients with PCD; or nonsense, frameshift and splice-site mutations resulting in loss-of-function. Four families were solved in other research studies (Supporting Information, Table S1) leaving 22 families unsolved. Twenty families consented to have CNV analysis and/or WES. This study was approved by the Research Ethics Boards of the Hospital for Sick Children and St Michael’s Hospital. Study subjects provided written consent and/or assent where appropriate.
Molecular genetic analysis algorithm
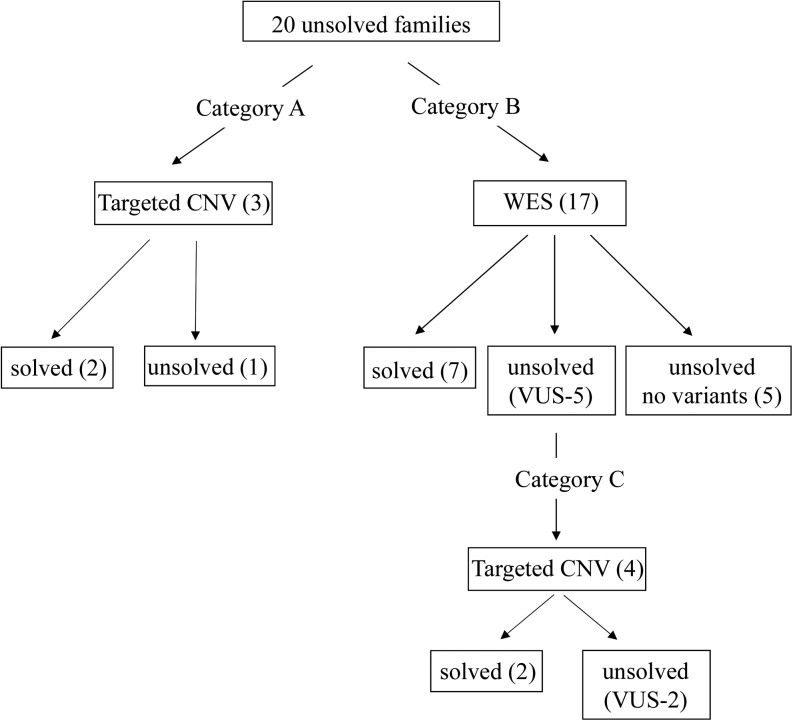
A molecular genetic analysis algorithm was developed to maximize previous molecular genetic data and minimize time and cost. Individuals were divided into three categories depending on the information from previous Sanger sequencing and WES performed in this study (Figure 1). For the individuals who harbored single pathogenic mutations after Sanger sequencing, targeted CNV analysis of the putative causative gene was conducted to ascertain the second allele (category A). Individuals with no pathogenic mutations after Sanger sequencing underwent WES to analyze the remaining 20 genes not covered by Sanger sequencing (category B). WES-sequenced individuals who were not found to have any pathogenic mutations in a PCD gene, further analysis of missense variants occurring at a minor allele frequency of <1% was conducted. Rare missense variants were considered to be contributory if predicted to be pathogenic, disease causing or damaging by in silico prediction programs (MutationTaster, PolyPhen-2, SIFT). These individuals underwent further targeted CNV analysis on the suspected PCD gene (category C).
Figure 1.
Molecular genetic analysis algorithm: study design and diagnostic yield per technology in patients with a clinical diagnosis of primary ciliary dyskinesia (PCD), defined as a ciliary ultrastructure defect and/or clinical features with low nasal nitric oxide (Kim et al. 2014). Sanger sequencing of 12 PCD genes provided a molecular diagnosis in 19 of 45 families. Of the 26 families who remained unsolved, 20 underwent targeted copy number variation (CNV) analysis and/or whole-exome sequencing (WES). Category A families had single pathogenic mutations in PCD genes after Sanger sequencing and underwent targeted CNV analysis alone to ascertain the second allele. Category B families had no pathogenic mutations after Sanger sequencing and underwent WES. Category C families had rare variants of uncertain significance after WES and underwent further targeted CNV analysis to ascertain the second allele. Four families had clinically significant CNVs and seven families were solved with WES alone giving an overall solved rate of 55% (11/20). These 11 families when taken together with 19 families solved through Sanger sequencing, and 4 families solved in other research studies, resulted in a solved rate of 76% (34/45).
Targeted CNV analysis
The selected technique for targeted CNV analysis was based on previously reported intron−exon CNVs in the literature. Exon 7 deletions in DYX1C1 have been documented in other PCD cohorts (Tarkar et al. 2013), and a custom TaqMan copy number assay was designed to detect this specific CNV (File S1). Reported deletions of exon 62 of DNAH5 (Berg et al. 2011) along with the remaining 78 exons were assayed using a commercially available, high-density gene-centric array comparative genomic hybridization (CGH; Prevention Genetics, Marshfield, WI). Similarly for DNAH11, where intron−exon level CNVs have not been reported, CGH was used to assay all 82 exons.
WES and validation
WES was completed with the Illumina Hisequation 2500 platform at The Centre for Applied Genomics (TCAG) at the Hospital for Sick Children following whole-exome capture with the Agilent SureSelectXT Human All Exon V4 capture kit. Sequence reads were aligned to the reference human genome (hg19) with Burrows-Wheelchair Aligner 0.5.9. MarkDuplicates (Picard tools, version 1.79; http://broadinstitute.github.io/picard/) was used to remove any duplicate paired-end reads. Duplicate-free alignments were refined using base space local realignment and quality score recalibration using GATK 1.128. Mean depth of coverage was 138X (range 100−168X) with all cases having >95% of targeted bases sequenced to a depth of greater than 20X.
Single-nucleotide variants (SNVs) and insertion/deletions (indels) were called with default parameter in GATK 1.1.28. SNVs and indels were annotated using SNPEff (http://snpeff.sourceforge.net/) and ANNOVAR and filtered to differentiate novel variants from known polymorphisms by screening against public single nucleotide polymorphism databases (dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; 1000 genomes, www.1000genomes.org; NHLBI Exome Sequencing Project (ESP) Exome Variant Server http://evs.gs.washington.edu/EVS/) and our own internal database consisting of 283 exomes analyzed in the same manner at TCAG. Novel SNVs also were annotated with SIFT, PolyPhen-2, and MutationTaster to assess the putative variant effect on the proteins. SNVs were prioritized based on loss of function mutations (nonsense, frameshift, splice sites) and damaging missense mutations that fit an autosomal recessive disease model.
An in silico gene panel was created by filtered analysis of WES data. WES data were reanalyzed with the publication of new PCD genes and the in silico panel expanded in real-time. At the time of manuscript submission, 32 genes were analyzed for variants (DNAH5, DNAI1, CCDC39, CCDC40, DNAI2, DNAH11, RSPH9, RSPH4A, KTU/DNAAF2, LRRC50/DNAAF1, TXNDC3/NME8, DNAL1, DNAAF3, CCDC103, HEATR2, HYDIN, LRRC6, CCDC114, CCDC164/DRC1, ARMC4, DYX1C1, ZMYND10, CCDC65/DRC2, RSPH1, OFD1, RPGR, C21orf59, SPAG1, DNAH8, CCDC151, MCIDAS, and CCNO). WES coverage of these genes was adequate with >98% of targeted bases sequenced and 92.08% (616/669) of exons having a mean coverage of >20X. Confirmation of variants was conducted on probands and available family members using standard Sanger sequencing methods in a CLIA-CAP diagnostic laboratory and genetic counseling was provided based on these results (Table S2).
Results
Four individuals from three families harbored one pathogenic mutation after Sanger sequencing (category A). Two families (111, 113) each harbored two different nonsense mutations in DNAH5 and CNV analysis of DNAH5 was conducted using array CGH to ascertain the second allele. Two clinically significant CNVs in DNAH5 were found and likely causative second allele, corresponding to the ODA defect observed on ciliary EM (Table 1). The third family harbored a previously reported nonsense mutation in DNAH11, c.8698C > T (p.R2900*) (Lucas et al. 2012). However, DNAH11 CNV analysis with array CGH did not detect a CNV and this case remained unsolved.
Table 1. Biallelic pathogenic variants in known PCD genes through CNV and WES analysis.
| Family | Patient | Ethnicity | Sex | Dx Age | Situs Status | Ciliary EM | nNO nL/min | Gene | hg 19 Genomic Coordinates | Transcript | Exon/ Intron | Base Changes | Predicted Effect | Segregation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound Heterozygous by CNV | ||||||||||||||
| 111 | 4 | White | M | 4 yr | I | ODA | 8.1 | DNAH5 | Chr5: 13701398 | NM_001369.2 | Ex 77 | c.13486C > T | p.R4496* (1) | Unknown |
| DNAH5 | Chr5: 13816665- | NM_001369.2 | Int 42 | Ex 41-42 4.4kB del | Unknown | |||||||||
| 13821065 | Int 40 | |||||||||||||
| 113 | 12 | Portuguese | F | 6 yr | A | ODA | 22 | DNAH5 | Chr5: 13865895 | NM_001369.2 | Ex 27 | c.4237C > T | p.Q1413* | Maternal |
| DNAH5 | Chr5: 13931161- | NM_001369.2 | Int 2 | Ex 2 1.6kB del | Paternal | |||||||||
| 13932789 | Int 1 | |||||||||||||
| 113 | 36 | Portuguese | M | 14 yr | S | ODA | 8.9 | DNAH5 | Chr5: 13865895 | NM_001369.2 | Ex 27 | c.4237C > T | p.Q1413* | Maternal |
| DNAH5 | Chr5: 13931161- | NM_001369.2 | Int 2 | Ex 2 1.6kB del | Paternal | |||||||||
| 13932789 | Int 1 | |||||||||||||
| Homozygous by WES | ||||||||||||||
| 115 | 14 | East Indian | M | 7 yr | S | Inadequate | 37.5 | LRRC6 | Chr8: 133645009 | NM_012472.4 | Ex 5 | c.630delG | p.W210Cfs*12 (2) | Paternal |
| 115 | 26 | East Indian | M | 9 yr | S | ODA+IDA | 5.8 | LRRC6 | Chr8: 133645009 | NM_012472.4 | Ex 5 | c.630delG | p.W210Cfs*12 (2) | Maternal |
| 141 | 50 | Pakistani | F | 5 yr | I | Inconclusive | 17.1 | SPAG1 | Chr8: 101226130 | NM_003114.4 | Ex 12 | c.1519dupA | p.I507Nfs*5 | Unknown |
| 140 | 49 | Portuguese | F | 18 yr | I | Not done | 6.9 | ARMC4 | Chr10: 28276450 | NM_018076.2 | Ex 3 | c.247G > T | p.E83* | Unknown |
| Chr10: 28151418 | NM_018076.2 | Ex 18 | c.2744del | p.L915* | Unknown | |||||||||
| 135 | 39 | Pakistani | F | 12 yr | S | Oligocilia | 22.1 | CCNO | Chr5: 54527295 | NM_021147.3 | Ex 3 | c.961C > T | p.Q321* (3) | Trans |
| 138 | 46 | White | F | 35 yr | S | Normal | 53.2 | HYDIN | Chr16: 71008480 | NM_001270974.1 | Ex 32 | c.4866del | p.P1623Qfs*20 | Unknown |
| Chr16: 70866971 | NM_001270974.1 | Int 79 | c.13680-1G > T | splice site | Unknown | |||||||||
| Compound Heterozygous by WES | ||||||||||||||
| 120 | 24 | White | M | 2 mo | I | ODA+IDA | 11.2 | ZMYND10 | Chr3: 50382964 | NM_015896.2 | Ex 1 | c.47T > G | p.V16G (4) | Maternal |
| Chr3: 50381183 | NM_015896.2 | Ex 3 | c.300del | p.F101Sfs*38 (2) | Unknown | |||||||||
| 137 | 45 | Hispanic | F | 30 yr | S | Inadequate | 17.1 | RSPH1 | Chr21: 43913159 | NM_080860.3 | Ex 2 | c.85G > T | p.E29* | Unknown |
| Chr21: 43906558 | NM_080860.3 | Ex 4 | c.287dup | p.N96Kfs*2 Knowles (5) | Child carrier | |||||||||
Genomic coordinates are approximate for copy number variations. Further clinical characteristics previously described (Kim et al. 2014). Previously described mutations are in bold (1) (Hornef et al. 2006); (2) (Zariwala et al. 2013); (3) (Wallmeier et al. 2014); (4) (Moore et al. 2013); (5) (Knowles et al. 2014). PCD, primary ciliary dyskinesia; CNV, copy number variation; WES, whole-exome sequencing; Dx, diagnosis; EM, electron microscopy; nNO, nasal nitric oxide; M, male; I, situs inversus; ODA, outer dynein arms; F, female; A, situs ambiguous; S, situs solitus; inadequate = inadequate ciliated cells to analyze, IDA, inner dynein arms inconclusive EM; inconclusive = adequate sample.
A total of 18 individuals from 17 families who did not have any pathogenic mutations detected on initial Sanger sequencing (category B and C) underwent WES. Seven category B families were solved by WES alone where two pathogenic variants were found in one of the additional 20 PCD genes not analyzed by Sanger sequencing (Table 1). Molecular genetic results were congruent with ultrastructural analysis when available. For those cases with available family members for segregation analysis, phase was determined to be trans.
WES on individual 46 revealed a splice site (c.13680-1G > T) and frameshift mutation, c.4866del (p.P1623Qfs*20) in HYDIN. Because of the presence of a pseudogene on chromosome 1 (HYDIN2), further characterization of both variants was conducted using Sanger sequencing (Olbrich et al. 2012) (File S1 and Figure S1). Primers mapping exclusively to HYDIN on chromosome 16 were designed (Table S2) which further resolved the next generation data from WES. The frameshift was found to be homozygous, and likely the causative allele. The splice site variant was heterozygous, but its pathogenicity is uncertain. Determination of segregation would help further resolve these variants, however, family members were not available. Ciliary biopsy of this individual was normal, as seen in other individuals with HYDIN mutations, further supporting the pathogenicity of the frameshift variant (Olbrich et al. 2012).
Individuals who did not have biallelic pathogenic mutations after WES were reanalyzed for rare missense variants of uncertain significance in the 32 known PCD genes. Five individuals from five families were found to harbor such VUS.
Individual 44 was found to have a homozygous missense c.1555G > C (p.A519P) variant in DNAAF3 (Table 2) on WES, which was not reported in other PCD patients nor present in our control databases. This transversion resulted in an amino acid change which is predicted to be tolerated by SIFT and MutationTaster, and only probably damaging by Polyphen-2. DNAAF3 mutations are associated with ODA + IDA defects, not seen in this patient. We concluded this variant was not likely contributing to this patient’s PCD phenotype and this case remained unsolved.
Table 2. Variants of uncertain significance in known PCD genes through CNV and WES analysis.
| Family | Patient | Ethnicity | Sex | Age at Dx | Situs Status | Ciliary EM | nNO nL/min | Gene | hg 19 Genomic coordinates | Transcript | Exon/ Intron | Base Changes | Predicted Effect | Segregation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound heterozygous, likely pathogenic (solved) | ||||||||||||||
| 114 | 9 | White | M | 4 yr | S | ODA+IDAa | 13.4 | DYX1C1 | Chr15: 55727162 | NM_130810.3 | Ex 8 | c.988C > T | p.R330W | Maternal |
| DYX1C1 | Chr15: 55729606- | NM_130810.3 | Int 7 | Ex 7 2.1Kb del (6) | Paternal | |||||||||
| 55731727 | Ex 7 | |||||||||||||
| 132 | 21 | White | M | 6 yr | I | Normal | 4.9 | DNAH11 | Chr7: 21847620 | NM_001277115.1 | Ex 63 | c.10285C > A | p.R3429S | Unknown |
| DNAH11 | Chr7: 21604615- | NM_001277115.1 | Int 6 | Ex7-14 32.29Kb dup | Unknown | |||||||||
| 21636907 | Int 14 | |||||||||||||
| Homozygous VUS | ||||||||||||||
| 142 | 51 | Sri Lankan Tamil | F | 18 yr | S | Inconclusive | 14.5 | DNAH11 | Chr7: 21639487 | NM_001277115.1 | Ex 15 | c.2750A > T | p.E917V | Unknown |
| DNAH11 | Chr7: 21847621 | NM_001277115.1 | Ex 63 | c.10286G > T | p.R3429L | Unknown | ||||||||
| 136 | 44 | Portuguese | M | 8 yr | S | Normal | 20.1 | DNAAF3 | Chr19: 55670702 | NM_001256714.1 | Ex 12 | c.1555G > C | p.A519P | Unknown |
| Single VUS | ||||||||||||||
| 134 | 38 | Hispanic | M | 15 yr | S | Normal | 17 | DNAH11 | Chr7: 21940666 | NM_001277115.1 | Ex 82 | c.13345C > T | p.R4449C | Unknown |
Genomic coordinates are approximate for copy number variations. Further clinical characteristics previously described (Kim et al. 2014). Previously described mutations are in bold (6) (Tarkar et al. 2013). Cases with two VUS, likely pathogenic were considered solved cases. PCD, primary ciliary dyskinesia; CNV, copy number variation; WES, whole-exome sequencing; Dx, diagnosis; EM, electron microscopy; M, male; S, situs solitus; I, situs inversus; ODA, outer dynein arms, IDA inner dynein arms; F, female; inconclusive = adequate sample inconclusive EM.
Revised after pathology review.
The four other individuals were found to have rare missense VUS predicted to be damaging, disease-causing, or deleterious by at least one of the in silico prediction programs, MutationTaster, PolyPhen-2 and SIFT. These individuals underwent targeted CNV analysis to ascertain a second hit (category C, Figure 1).
Individual 9 was initially found to have a single variant in DNAH5 (c.3890A > G; p.D1297G) that was predicted to be disease-causing by Poly-Phen, SIFT, and MutationTaster and ciliary ultrastructure was interpreted as ODA (Kim et al. 2014). However, array CGH analysis of all exons of DNAH5 did not yield a second allele, prompting further review of WES data. In addition to the rare variant in DNAH5, a single rare missense variant in DYX1C1 (c.988C > T; p.R330W) was found in WES data. This variant was not observed in control populations and was also predicted to be disease causing by SIFT and MutationTaster. Targeted CNV analysis by TaqMan copy number assay of a previously reported exon 7 deletion in DYX1C1 was conducted and was found in this individual (Tarkar et al. 2013). Subsequent segregation analysis confirmed trans orientation of the rare missense variant and deletion. This prompted further pathologic review and revision of the initial ciliary EM to ODA + IDA consistent with other DYX1C1 families (Table 2) (Tarkar et al. 2013), suggesting this is the causative gene in this family.
Individual 21 was found to have a rare missense VUS (c.10285C > A) in DNAH11 on initial Sanger sequencing and WES confirmed this VUS. MutationTaster, PolyPhen-2 and SIFT algorithms predict the p.R3429S change to be deleterious, probably damaging and damaging respectively. Additionally, this is a highly conserved amino-acid residue residing in the conserved Helix 2 domain (Schwabe et al. 2008). Array CGH analysis on DNAH11 was conducted and demonstrated a 32.29-kb duplication spanning exons 7−14 and is likely the causative second allele in this individual (Table 2).
Individual 51 was found to have two previously unreported homozygous missense variants in DNAH11, which also were detected through WES. One variant, c.10286G > T (p.R3429L) involves the same highly conserved amino-acid residue in the Helix 2 domain observed in individual 21. The other variant, c.2750A > T (p.E917V) also was predicted to be possibly damaging by Polyphen-2, deleterious by SIFT, and deleterious by MutationTaster. Subsequent array CGH of DNAH11 analysis did not reveal a deletion or duplication and this case remained unsolved (Table 2).
Patient 38 was heterozygous in exon 82 of the DNAH11 gene for an undocumented missense variant defined as c.13366C > T and predicted to result in p.R4456C substitution. The SIFT and PolyPhen-2 protein function algorithms predicts this change to be tolerated and benign, whereas the MutationTaster program indicates that this c.13366C > T variant is deleterious. However, DNAH11 array CGH did not yield a second allele de-prioritizing this variant leaving this case unsolved (Table 2).
Through Sanger sequencing of the first 12 PCD genes, we found that five individuals who had a clinical diagnosis of PCD did not harbor a disease-causing mutation in a known PCD gene, nor did we find, using WES, a pathogenic mutation or rare missense VUS in any of the known PCD genes (Table S3). We suspect these patients either have a mutation not detectable by either technique or a novel PCD locus.
Overall, the combination of targeted CNV and WES analysis allowed for a molecular diagnosis in 55% of our unsolved families (11/20). Clinically significant CNVs were detected in 8.8% (4/45) of our total patient population, which is consistent with other autosomal-recessive disorders (Aradhya et al. 2012). Of the 45 families, Sanger sequencing solved 19 (42%) families whereas subsequent targeted CNV analysis with WES solved an additional 11, yielding an overall diagnostic rate of 30/45 (67%). Four further families were enrolled in other studies, such that in our cohort, 34/45 (76%) had a molecular diagnosis of PCD. As all variants detected by Sanger sequencing were covered in our WES data, if our molecular genetic approach was modified to WES followed by targeted CNV analysis, the overall diagnostic rate would approach 76%.
Discussion
In genetically heterogeneous diseases such as PCD, a step-wise molecular genetic approach has been proposed based on ciliary ultrastructure and the prevalence of certain mutations. If genes are assayed in a cost-efficient, step-wise tiered fashion, only one gene may be assayed at one time, taking up to 4−6 months to complete, costing up to $12,000 USD. In addition, expert EM ciliary ultrastructure analysis may not be available to guide such tiered analysis. Newly developed next-generation sequencing (NGS) targeted panels are less costly, at $1900−$4200 USD, but do not include all 32 PCD genes (Prevention Genetics; Ambry Genetics, Aliso Viejo, CA; Center for Genomics and Transcriptomics, Tübingen, Germany). However, unlike tiered Sanger sequencing, NGS concurrently assays all genes. If a patient had previous genetic testing, these genes would be unnecessarily retested. Furthermore CNV data from NGS is unreliable and not possible with Sanger sequencing.
Here, we describe the utility of filtered WES data and targeted CNV analysis to circumvent the limitations of targeted Sanger and NGS panel sequencing in PCD. Because of the cost-efficiency of WES ($2−$5000 USD) (Neveling et al. 2013), we propose that WES could be conducted as a first-line molecular diagnostic test through the analysis of an in silico gene panel, followed by targeted CNV analysis. In addition to providing a definitive molecular diagnosis when biallelic pathogenic mutations are found, WES provides additional information on all PCD genes and aids in the interpretation of rare missense VUS. In the absence of convincing pathogenic mutations in all known PCD genes, rare missense VUS could result in loss of function and potentially be disease-causing prompting further targeted CNV analysis. CNV analysis should be targeted to the candidate PCD-associated gene on the basis of sequencing results. CNV analysis initially could begin with CNVs reported other PCD cohorts, followed by more extensive full exon analysis using array CGH or multiplex ligation-dependent probe amplification (Stuppia et al. 2012). Further studies are required to determine if the novel CNVs reported in our cohort are private mutations or common to other PCD patients. In addition, as CNV algorithms from WES data improve, they may replace other CNV analyses and provide a genome-wide CNV approach (Samarakoon et al. 2014). If a PCD case remains unsolved after WES and targeted CNV analysis, WES data can be further analyzed as novel PCD loci become published expanding the in silico panel in real-time, instead of expending further sequencing consumables. In our population, if WES was followed by targeted CNV analysis, 76% of patients with PCD would have had a molecular genetic diagnosis and WES should be considered the most cost-efficient molecular genetic technique in such genetically heterogeneous disorders.
Supplementary Material
Acknowledgments
The authors thank Natasha Jivraj, Eriskay Liston, Shaidah Deghan Manshadi, Melody Miki, Donna Wilkes, Jennifer Welsh, Dr Peter Bikangaga, and Dr David Chitayat for assisting in patient recruitment. We are grateful to Dr Ted Young and Dr Dimitri J. Stavropoulos for designing and conducting TaqMan assays. Thank you to the PCD patients and their family members for their participation and support, the investigators, and co-ordinators of the “Genetic Disorders of Mucociliary Clearance Consortium” (GMDCC), which is part of the Rare Disease Clinical Research Network, Dr Margaret Leigh, Susan Minnix, and Whitney Wolf (University of North Carolina at Chapel Hill) for their continued support. This work was selected for study by the FORGE Canada Steering Committee, which consists of: Kym Boycott (leader; University of Ottawa), Jan Friedman (co-lead; University of British Columbia), Jacques Michaud (co-lead; Université de Montréal), Francois Bernier (University of Calgary), Michael Brudno (University of Toronto), Bridget Fernandez (Memorial University), Bartha Knoppers (McGill University), Mark Samuels (Université de Montréal), and Stephen W. Scherer (University of Toronto). Funding was provided by the McLaughlin Centre, University of Toronto. Funding for support for research was provided to M.R.K., M.A.Z, S.D.D., and D.A.H. by US National Institutes of Health (NIH) grant U54HL096458, which is a part of the NCATS Rare Diseases Clinical Research Network (RDCRN). RDCRN is an initiative of the Office of Rare Diseases Research (ORDR), NCATS, funded through a collaboration between NCATS and NHLBI; to M.R.K. and M.A.Z. by NIH/NHLBI grant 5R01HL071798NIH-NHLBI. This work also was supported by NIH/NCATS grant UL1 TR000083 to the University of North Carolina at Chapel Hill. The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.019851/-/DC1
All work was conducted at Hospital for Sick Children and St Michael’s Hospital, Toronto, Ontario, Canada.
Communicating editor: R. M. Cantor
Literature Cited
- Aradhya S., Lewis R., Bonaga T., Nwokekeh N., Stafford A., et al. , 2012. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet. Med. 14: 594–603. [DOI] [PubMed] [Google Scholar]
- Barbato A., Frischer T., Kuehni C. E., Snijders D., Azevedo I., et al. , 2009. Primary ciliary dyskinesia: a consensus statement on diagnostic and treatment approaches in children. Eur. Respir. J. 34: 1264–1276. [DOI] [PubMed] [Google Scholar]
- Berg J. S., Evans J. P., Leigh M. W., Omran H., Bizon C., et al. , 2011. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: implications for application to clinical testing. Genet. Med. 13: 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchon S., Legendre M., Copin B., Duquesnoy P., Montantin G., et al. , 2012. Delineation of CCDC39/CCDC40 mutation spectrum and associated phenotypes in primary ciliary dyskinesia. J. Med. Genet. 49: 410–416. [DOI] [PubMed] [Google Scholar]
- Boycott K., Hartley T., Adam S., Bernier F., Chong K., et al. , 2015. The clinical application of genome-wide sequencing for monogenic diseases in Canada: Position Statement of the Canadian College of Medical Geneticists. J. Med. Genet. 52: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A., Hogg C., 2012. Primary ciliary dyskinesia: recent advances in epidemiology, diagnosis, management and relationship with the expanding spectrum of ciliopathy. Expert Rev. Respir. Med. 6: 663–682. [DOI] [PubMed] [Google Scholar]
- Hjeij R., Lindstrand A., Francis R., Zariwala M. A., Liu X., et al. , 2013. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 93: 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornef N., Olbrich H., Horvath J., Zariwala M. A., Fliegauf M., et al. , 2006. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 174: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. H., A Hall D., Cutz E., Knowles M. R., Nelligan K. A., et al. , 2014. The role of molecular genetic analysis in the diagnosis of primary ciliary dyskinesia. Ann. Am. Thorac. Soc. 11: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M. R., Daniels L. A., Davis S. D., Zariwala M. A., Leigh M. W., 2013a Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 188: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- doi: 10.1164/rccm.201311-2047OC. Knowles, M. R., L. E. Ostrowski, M. W. Leigh, P. R. Sears, S. D. Davis et al., 2014 Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care. Med. 15: 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M. R., Ostrowski L. E., Loges N. T., Hurd T., Leigh M. W., et al. , 2013b Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 93: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkowiak M., Zietkiewicz E., Witt M., 2015. Recent advances in primary ciliary dyskinesia genetics. J. Med. Genet. 52: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh M. W., O’Callaghan C., Knowles M. R., 2011. The challenges of diagnosing primary ciliary dyskinesia. Proc. Am. Thorac. Soc. 8: 434–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo J., Zariwala M. A., Noone P. G., 2015. Primary ciliary dyskinesia. Semin. Respir. Crit. Care Med. 36: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loges N. T., Olbrich H., Becker-Heck A., Haffner K., Heer A., et al. , 2009. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 85: 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J. S., Adam E. C., Goggin P. M., Jackson C. L., Powles-Glover N., et al. , 2012. Static respiratory cilia associated with mutations in Dnahc11/DNAH11: a mouse model of PCD. Hum. Mutat. 33: 495–503. [DOI] [PubMed] [Google Scholar]
- Lucas J. S., Burgess A., Mitchison H. M., Moya E., Williamson M., et al. , 2014. Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 99: 850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. J., Onoufriadis A., Shoemark A., Simpson M. A., Zur Lage P. I., et al. , 2013. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 93: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K., Feenstra I., Gilissen C., Hoefsloot L. H., Kamsteeg E. J., et al. , 2013. A post-hoc comparison of the utility of Sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum. Mutat. 34: 1721–1726. [DOI] [PubMed] [Google Scholar]
- Olbrich H., Schmidts M., Werner C., Onoufriadis A., Loges N. T., et al. , 2012. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 91: 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzi J. R., Becker-Heck A., Castleman V. H., Al-Mutairi D. A., Liu Y., et al. , 2012. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 44: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon P. S., Sorte H. S., Kristiansen B. E., Skodje T., Sheng Y., et al. , 2014. Identification of copy number variants from exome sequence data. BMC Genomics 15: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe G. C., Hoffmann K., Loges N. T., Birker D., Rossier C., et al. , 2008. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 29: 289–298. [DOI] [PubMed] [Google Scholar]
- Stuppia L., Antonucci I., Palka G., Gatta V., 2012. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int. J. Mol. Sci. 13: 3245–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkar A., Loges N. T., Slagle C. E., Francis R., Dougherty G. W., et al. , 2013. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 45: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier J., Al-Mutairi D. A., Chen C. T., Loges N. T., Pennekamp P., et al. , 2014. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Genet. 46: 646–651. [DOI] [PubMed] [Google Scholar]
- Zariwala M. A., Gee H. Y., Kurkowiak M., Al-Mutairi D. A., Leigh M. W., et al. , 2013. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 93: 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.