Abstract
A duplication variant within middle-ear-specific gene A2ML1 co-segregates with otitis media in an indigenous Filipino pedigree (LOD score=7.5 at reduced penetrance) and lies within a founder haplotype that is also shared by three otitis-prone European- and Hispanic-American children, but is absent in non-otitis-prone children and >62,000 next-generation sequences. Seven additional A2ML1 variants were identified in six otitis-prone children. Collectively our studies support a role for A2ML1 in the pathophysiology of otitis media.
Keywords: A2ML1, alpha-2-macroglobulin-like 1, otitis media, founder haplotype, linkage analysis, middle ear
Otitis media causes significant morbidity worldwide and hearing loss at any age.1-2 Despite efforts to reduce incidence, otitis media remains an important public health problem within the United States, with otitis media being the most frequent cause of pediatric consults and antibiotic prescription incurring an annual cost of >$5 billion.3 In developing countries including the Philippines the prevalence of chronic suppurative otitis media is 2-6%.2 Strong evidence exists for genetic susceptibility to otitis media,4-6 but only a few loci have been mapped, including genome-wide association at rs10497394 on chromosome 2q31.1.6-7 Here we report rare variants in alpha-2-macroglobulin-like 1 (A2ML1) predisposing to nonsyndromic otitis media within different study populations. A2ML1 encodes a middle-ear-specific protease inhibitor that is 41% identical and 59% similar to alpha-2-macroglobulin (A2M), a known marker for vascular permeability of middle ear mucosa during infection.8
An intermarried, indigenous Filipino community has close to 50% prevalence of otitis media. A pedigree that connects 134 indigenous individuals was constructed (Fig. 1a). DNA samples were obtained from 51 indigenous individuals, of whom 38 have current or previous otitis media. Exome sequencing was performed using samples from two second-cousins with chronic suppurative otitis media. Six exome variants were heterozygous in both indigenous Filipino individuals, passed GATK filters, occur at conserved residues, were predicted to be damaging, and were not in dbSNP or the Exome Aggregation Consortium (ExAC) database (Supplementary Table 1). All six variants were Sanger-sequenced using DNA from 51 indigenous pedigree members. Only the A2ML1 duplication c.2478_2485dupGGCTAAAT (p.(Ser829Trpfs*9)) possibly co-segregates with otitis media (Fig. 1a). Using 95% penetrance and a 5% phenocopy rate, a statistically significant maximum two-point LOD score of 7.5 at Θ=0 was obtained for the A2ML1 variant (Supplementary Table 1).
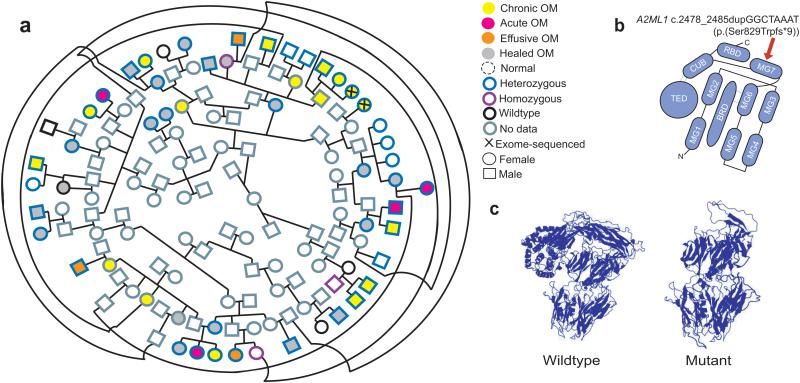
Figure 1. Segregation within the indigenous pedigree, cartoon of A2ML1 domains and molecular modeling for the A2ML1 variant.
[A] Pedigree connecting 37 variant carriers who have the full spectrum of otitis media (age 3 months to 58 years, median 13 years). An individual with healed otitis media and intellectual disability (ID) is wildtype for the duplication, but her 13-year old son who has chronic otitis media but no ID and her unaffected 4-month old daughter are heterozygous. Nine variant carriers had no evidence of otitis media (median 18 years), while four individuals are wildtype and unaffected. [B] Predicted A2ML1 domains based on alpha 2-macroglobulin structure.9 MG, macroglobulin-like domains 1-7; BRD, bait-region domain; CUB, consists of two four-stranded antiparallel β-sheets; TED, thiol-ester binding domain; RBD, receptor-binding domain. The A2ML1 frameshift variant is expected to occur within the MG7 domain (red arrow). [C] Modeling predicts loss of the receptor-binding and thiol-ester domains due to the A2ML1 duplication.
The A2ML1 duplication is predicted to truncate the protein to <60%, initiate nonsense-mediated decay, and result in loss of thiol-ester and receptor-binding domains (Fig. 1b-c), which are expected to be essential for protease trapping and lysosomal clearance, respectively.9 The duplication was not found in: 61,109 multi-ethnic samples in the ExAC database; 1,385 exome sequences from the University of Washington Center for Mendelian Genomics (UWCMG; Supplementary Fig. 1); and 100 genomes from the Singapore Sequencing Malay Project (SSMP), which includes Southeast Asians of Chinese, Indian and Malayan descent.10
DNA samples were obtained from 123 otitis-prone and 118 non-otitis-prone children that were followed up from birth at the University of Texas Medical Branch (UTMB).5 Among the UTMB children 84 (68.3%) otitis-prone and 79 (66.9%) non-otitis-prone children self-identified as European-American (EA) or Hispanic-American (HA). Sanger sequencing of all A2ML1 coding exons revealed that the same A2ML1 duplication is present in 3 out of 123 otitis-prone children. Two otitis-prone children, one EA and the other HA, are homozygous for the duplication, while a third otitis-prone EA child is heterozygous (Table 1, Supplementary Table 2). Ethnicity for these three otitis-prone carriers was verified by principal components analysis (Supplementary Fig. 2). All three children with the duplication had early-onset severe otitis media requiring tympanostomy tube insertion by age 6 months. Additionally the duplication is absent in 118 non-otitis-prone children (Supplementary Table 2), in 2,756 UWCMG chromosomes of EA/HA descent (Supplementary Fig. 1), and in 67,630 European non-Finnish and 11,606 Latino alleles from the ExAC database (Table 1). Comparing the frequency of this duplication only in individuals of EA/HA descent, this duplication has genome-wide significant association with otitis media (two-sided Fisher’s exact p=3.34×10−14). Moreover the two exome-sequenced indigenous individuals and three otitis-prone children share a haplotype that includes the duplication and three common variants (Supplementary Table 2). The A-dup-A-T haplotype includes 5.2 kb and is estimated to be ~1,800 years old [95%CI: 145, 3462]. This short founder haplotype was most likely introduced to the Americas and the Philippines by colonial Spaniards based on population history.
Table 1.
Rare A2ML1 Variants Identified in Otitis-Prone Children from UTMB
| Chr12 hg19 Coordinate |
PhyloP 100 wayall |
GERP | Nucleotide Change |
Amino Acid Change |
Variant Type |
C- score |
Deleterious Prediction |
N
EA/ HA NOP |
N
EA/ HA OP |
N OP with Variant |
Ethnic Group |
ExAC MAF European non-Finn |
ExAC MAF Latino |
Fisher’s exact p-value |
ExAC MAF Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8,990,070 | 0.40 | 2.60 | c.763C>T | p.(Gln255*) | stop-gained | 29.2 | MT | 118 | 84 | 1 het | HA | 0/67,708 | 0/11,606 | 0.001 | 0/43,010 |
| 8,991,805 | 1.90 | 3.33 | c.1067C>G | p.(Pro356Arg) | missense | 15.1 | MA, PP2, S | 110 | 100 | 1 het | EA | 0/67,250 | 0/11,464 | 0.001 | 0/42,430 |
| 9,004,573 | 7.19 | 2.72 | c.955G>A | p.(Ala810Thr) | missense | 21.3 | LRT, MA, MT, PP2, S |
138 | 154 | 1 het | HA | 0/67,668 | 0/11,606 | 0.002 | 0/43,010 |
| 9,004,827 | 1.38 | 1.94 | c.2478_2485 dupGGCTAAAT |
p.(Ser829Trpfs*9) | frameshift | NA | MT | 158 | 168 | 2 hom, 1 het |
2 EA, 1 HA |
0/67,630 | 0/11,606 | 3.34×10−14 | 0/42,982 |
| 9,006,810 | −1.01 | −4.26 | c.2677C>T | p.(Arg893*) | stop-gained | 36.0 | MT | 98 | 106 | 1 het | HA | 14/67,236 = 0.0002 |
1/11,590 =0.00009 |
0.021 | 4/41,890 =0.0001 |
| 9,009,825 | 9.44 | 3.73 | c.2914G>T | p.(Glu972*) | stop-gained | 44.0 | MT | 140 | 156 | 1 het | HA | 0/67,694 | 0/11,602 | 0.002 | 2/43,008 =0.00005 |
| 9,009,912 | 2.78 | 1.72 | c.3001C>T | p.(Arg1001Trp) | missense | 16.8 | MA, PP2, S | 140 | 156 | 1 het | EA | 0/67,634 | 0/11,606 | 0.002 | 0/42,974 |
| 9,027,091 | 3.41 | 2.39 | c.4292C>T | p.(Ala1431Val) | missense | 19.6 | MT, PP2, S | 114 | 164 | 1 het | EA | 0/67,654 | 0/11,586 | 0.002 | 0/42,934 |
UTMB, University of Texas Medical Branch Galveston; ExAC, Exome Aggregation Consortium; MAF, minor allele frequency; MT, MutationTaster; MA, MutationAssessor; PP2, PolyPhen2 HDIV; S, SIFT; LRT, likelihood ratio test; ; EA, European-American; HA, Hispanic-American; NOP, non-otitis-prone alleles screened for variant; OP, otitis-prone alleles screened for variant; het, heterozygous; hom, homozygous.
UTMB 1031 is heterozygous for two variants, c.955G>A (p.(Ala810Thr)) and c.2914G>T (p.(Glu972*)).
All listed variants were not identified in NOP UTMB children, in 1,385 UWCMG exome sequences and in 100 genomes from the Singapore Sequencing Malay Project.
Two-sided Fisher exact tests were performed by comparing variant frequency in UTMB OP children of EA/HA descent with the combined frequencies in: UTMB NOP children of EA/HA descent; 1,378 UWCMG exomes of EA/HA descent; and European non-Finnish and Latino alleles in ExAC.
For variants that are not identified in ExAC, number of alleles was based on the total alleles reported for the ExAC variant nearest to the nucleotide at which the A2ML1 variant in the UTMB child occurred. For each variant site listed, coverage in ExAC was 50-95× on average and at least 30× in 90-99% of 60,706 samples.
Seven additional variants (three stop-gained and four missense) were each identified as heterozygous in an otitis-prone child but not in non-otitis-prone children (Table 1). With the exception of the A2ML1 duplication, all additional variants identified in UTMB occur in a single proband. All seven single nucleotide variants identified in otitis-prone children from UTMB occur at highly conserved nucleotides, are predicted to be damaging, have C-scores>15, and are absent in UWCMG exomes or SSMP. Five of the seven variants were not in ≥120,716 alleles in the ExAC database (Table 1). Due to the extremely low frequency of these variants, when tested for association comparing their frequency to those found in EA/HA individuals in non-otitis-prone UTMB children, UWCMG and ExAC, although none of these variants are associated with otitis media at a genome-wide significance level, all are nominally significant (two-sided Fisher’s exact p<0.05; Table 1). One HA otitis-prone child is heterozygous for both a stop-gained c.2914G>T (p.(Glu972*)) and a missense c.955G>A (p.(Ala810Thr)) variant: Molecular modeling for these two variants predict domain loss due to the stop-gained variant but no obvious changes due to the missense variant (Supplementary Fig. 3), thus it is possible that only the p.(Glu972*) variant contributes to otitis media susceptibility. For five other variants in UTMB otitis-prone children, stop-gained variants p.(Gln255*) and p.(Arg893*) are likewise predicted to result in domain loss, but of the missense variants only p.(Pro356Arg) and and p.(Arg1001Trp) are revealed by molecular modeling to cause torsional changes and loss of H-bonds (Supplementary Fig. 3).
Furthermore 622 DNA samples from 143 families ascertained at the University of Minnesota (UMN) were available for study.6 In EA family 123, a missense variant c.887T>C (p.(Val296Ala)) is heterozygous in two siblings and their mother, all three of whom had documented otorrhea, eardrum perforation or abnormalities and/or previous or current otitis media. The unaffected father and a third sibling who had a single prolonged episode of otitis media but no eardrum abnormalities were wildtype for the variant. The missense variant has C-score=15.2, is at a conserved nucleotide and predicted damaging by 2/6 algorithms, and occurs in a domain that is rich in disulfide bonds, which may be needed to link A2ML1 monomers into the active dimer and tetramer formations.9 However this variant is heterozygous in 96 alleles from the ExAC database (European non-Finnish MAF=0.0013), and probably does not confer otitis media susceptibility.
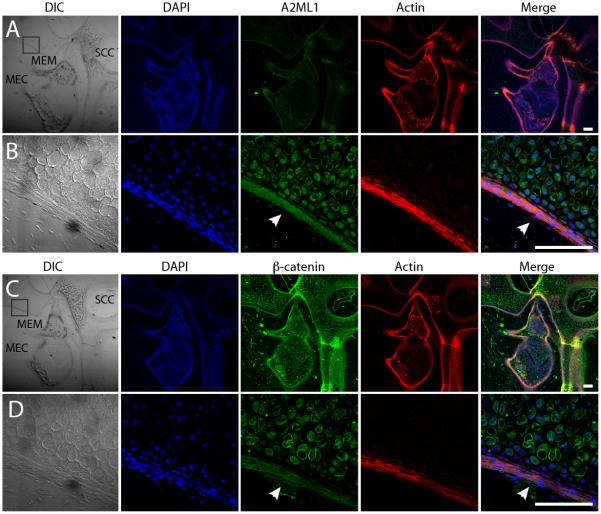
The middle ear localization of A2ML1 was assessed using high-resolution confocal imaging. At P6, immunoreactivity for β-catenin (positive control) and A2ML1 was observed in mucosal epithelium (Fig. 2). In contrast, A2ML1 staining is almost absent in mouse inner ear (Supplementary Fig. 4), which is consistent with no detectable A2ml1 expression within the Shared Harvard Inner-Ear Laboratory Database.
Figure 2. A2ML1 is localized to middle ear epithelium.
[A, C] Sagittal cryosections from P6 wildtype mice showing the middle ear cavity (MEC), the middle ear mucosa (MEM) and semicircular canal of the inner ear (SCC). [B, D] Higher magnification depicting boxed sections in A and C. [A-D] Confocal images of the cryosections immunostained with antibodies for DAPI (blue), A2ML1 (green in A & B), β-catenin (green in C & D) and rhodamine phalloidin (Actin, red). White arrowheads point to MEM with A2ML1 [B] and β-catenin expression [D]. DIC, differential interference contrast. Scale bar: 100 μm.
Similarity of A2ML1 to A2M suggests that these protease inhibitors might have similar and overlapping protective functions within the middle ear. A2M is detected at low levels in serous middle ear effusions, but is increased in acute purulent otitis media with a greater proportion found in complex with protease, which indicates passage of high-molecular-weight A2M due to increased mucosal permeability and greater inhibitory A2M activity during infection.8,11 Additionally low serum A2M levels were detected in children with recurrent otitis media.12 The tetrameric A2M structure is an effective trap for proteases,9 which if left unchecked can damage middle ear mucosa. Using A2M as template, A2ML1 variants are predicted to affect thiol-protease trapping, tetramer formation, and/or receptor-mediated clearance of A2ML1-protease complexes. Interestingly bacitracin, which is an antibiotic component of ear drops used in Europe for acute otitis media with otorrhea,13 competitively inhibits binding of A2M–protease complexes to macrophages or fibroblasts for clearance.14 If bacitracin dampens A2M activity which might compensate for dysfunctional A2ML1, it can be hypothesized that for A2ML1 variant carriers, bacitracin might not be the antibiotic of choice for otitis media.
A2ML1 variants were previously found in Noonan-like syndrome cases, however otitis media was not described in these cases and it was not mentioned whether the hearing loss in two A2ML1 variant carriers with Noonan-like syndrome was conductive or sensorineural.15 On the other hand, the A2ML1 carriers reported here do not have cardinal features of Noonan syndrome. We therefore present a rare genetic cause of susceptibility to nonsyndromic otitis media which affects a total of 37 indigenous Filipino, 1 HA and 2 EA individuals. A2ML1 localizes specifically to middle ear epithelium, thus supporting a role in the pathophysiology of otitis media.
ONLINE METHODS
STUDY APPROVAL
Prior to study onset, approval was obtained from the following institutional review boards: Baylor College of Medicine and Affiliated Hospitals; the National Commission on Indigenous Peoples, Philippines; University of Minnesota (UMN); University of the Philippines (UP) Manila; University of Texas Medical Branch (UTMB) Galveston; University of Virginia; and University of Washington (UW). Informed consent was obtained from all adult subjects and from parents of children enrolled in the study. In addition, approval was obtained from the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital Medical Center.
SUBJECT ASCERTAINMENT
The indigenous population comprises the original inhabitants of an island within the central Philippines and is of Negrito stock, which is historically known to consist of the first wave of migrants to the Philippines 20,000-30,000 years ago. The indigenous peoples in the same region had early contacts with colonial Spaniards in the late 16th century, and some of the earliest Spanish settlements were built in the main island. Due to characteristic Negrito features such as short stature, curly hair, flat nose and dark skin, the indigenous population has been subjected to racial and socio-economic discrimination over the centuries, which fostered intermarriage within the same community. Genetic comparison with other populations showed that this indigenous population has multiple elements of admixture, but remain primarily of mixed Negrito and Indo-Malay ethnicity.16
Out of a current population of ~250 indigenous individuals, we performed otoscopy on 175 members of the indigenous community, of whom 82 (46.9%) were diagnosed with current or previous otitis media. A pedigree that connects 134 indigenous individuals, of whom 51 provided DNA samples, was constructed (Fig. 1a). For the indigenous pedigree, diagnosis was based on these otoscopic findings: chronic otitis media, eardrum perforation with well-defined borders, usually with thickened middle ear mucosa and mucopurulent discharge; acute otitis media, hyperemic eardrum with or without perforation or discharge; otitis media with effusion, dull or retracted, intact, non-hyperemic eardrum with middle ear fluid; intact non-hyperemic eardrum with healed perforation or scarring and/or previously documented otitis media. None of the ascertained individuals had craniofacial deformities, cardiopulmonary defects, or immunodeficiency that would cause recurrent infections. No dermatologic diseases or intellectual disabilities co-segregate with otitis media. From 38 affected and 13 unaffected pedigree members, saliva was collected and DNA extracted using Oragene kits (DNA Genotek).
In order to replicate study findings from the indigenous population, DNA samples were obtained from two previously established otitis media cohorts: (1) a case-control cohort from UTMB that includes 123 otitis-prone and 118 non-otitis-prone children;5 and (2) a family cohort from UMN that consists of 622 DNA samples from 143 families.6 For the UTMB cohort, children were considered otitis-prone based on any of these criteria: first episode of acute otitis media at <6 months; ≥3 episodes of acute otitis media within a six-month period; ≥4 episodes of acute otitis media within a 12-month period; ≥6 episodes by age six years; or tympanostomy tube placement for recurrent or persistent otitis media.5 Non-otitis-prone children included those with 0-2 episodes of acute otitis media by age two years. Children were excluded from study if they had ear or nasopharyngeal defects, immunologic abnormalities, major medical conditions or treatment for chronic diseases.
At UMN, each family (median size four with two affected individuals) was recruited when the proband underwent tympanostomy tube insertion for chronic or recurrent otitis media.6 All family members were examined by an otolaryngologist and tested by tympanometry. Family members were considered affected if ≥2 data sources, whether otoscopy, tympanometry, medical record or personal history, were positive for otitis media. Family members and probands who have Down syndrome, craniofacial anomalies, genetic syndromes with otitis media, or tympanostomy tubes not due to otitis media were excluded from study.
DNA SEQUENCING AND GENOTYPING
Exome sequencing was performed at the UW Center for Mendelian Genomics (CMG) on an Illumina HiSeq, sequencing to an average read depth ~60×. Sequence capture was performed using the Roche NimbleGen Big Exome 2011 Library. Sequence alignment and variant detection, calling and annotation were performed as described.17
Selected variants that were observed in both exomes (Supplementary Table 1) were Sanger-sequenced using 51 DNA samples from the indigenous Filipino pedigree. All coding exons of A2ML1 were sequenced using DNA from 123 otitis-prone children from UTMB and 143 probands from UMN families. Exons containing variants were also sequenced using samples from four members of UMN family 123 (exon 9) and 118 non-otitis-prone children from UTMB (exons 8, 9, 10, 19, 20, 21, 24 and 34).
The occurrence of nine A2ML1 variants was also checked in exome sequence data from 1,385 unrelated individuals whose DNA samples were submitted to UWCMG for gene identification for various phenotypes (Supplementary Fig. 1). None of these individuals were reported to have otitis media. Ethnic background was inferred for these individuals using principal components analysis (PCA).
To verify ethnic background of UTMB children, DNA samples from three otitis-prone children who carried the A2ML1 duplication variant as well as 38 non-otitis-prone children with sufficient DNA content were submitted to UWCMG for genotyping using the Illumina HumanCoreExome-24v1 BeadChip which includes ~540,000 markers. The generated genotypes were used to perform PCA while comparing to HapMap3 genotypes (Supplementary Fig. 2).
BIOINFORMATIC, LINKAGE AND HAPLOTYPE ANALYSES
For variant identification from exome data, sequences were analyzed using Variant Mendelian Tools, a modification of Variant Association Tools,18 which facilitates variant selection based on sharing among relatives and annotation from RefSeq, dbSNP, SeattleSeq, ExAC, Combined Annotation Dependent Depletion (CADD) and dbNSFP, among others. The dbNSFP database includes nucleotide conservation scores (GERP, phastCons and phyloP) and prediction for nonsynonymous variants from six different algorithms (fathmm, likelihood ratio test, MutationAssessor, MutationTaster, PolyPhen-2 and SIFT). Shared nonsynonymous variants in the two indigenous Filipino exomes were selected for further study if they: passed GATK filters (ABFilter, HRunFilter, QDFilter, QUALFilter, SBFilter, SnpCluster); are absent in dbSNP and ExAC; occur at conserved nucleotides; have scaled C-scores≥15; and are predicted to be damaging by ≥3 algorithms. Due to database limitations for indel annotations, shared indel variants within coding region that passed GATK filters were kept under consideration.
For six variants identified from exome sequence data, two-point linkage analysis was performed for the indigenous Filipino pedigree using Superlink19 (Supplementary Table 1). All individuals with chronic otitis media, acute otitis media, otitis media with effusion or healed otitis media were considered affected. For otitis media susceptibility variants, the penetrance is unknown. In addition adults with normal otoscopic findings may have undocumented otitis media in childhood or some individuals might have recurrent otitis media but are unaffected at time of examination. Therefore affecteds-only linkage analysis, in which all unaffected pedigree members were assumed to have unknown affection status, was performed using an autosomal dominant mode of inheritance and a disease allele frequency of 0.0001. Affecteds-only analysis was performed under two conditions: (1) full penetrance with no phenocopies, and (2) 95% penetrance with 5% phenocopy rate (Supplementary Table 1).
For A2ML1 variants, molecular modeling was performed using Phyre220 with human A2M structure as template (Protein Data Bank ID 4ACQ). The age of the haplotype which includes the A2ML1 duplication was estimated using DMLE+.21 Genotypes from 241 unrelated UTMB children for three SNPs surrounding the A2ML1 variant (rs1860927, rs1860926 and rs78452682) were included in the analysis (Supplementary Table 2). A population growth rate of 1.2% based on annual US estimates was used. Galveston Island, Texas has a population of ~48,000 while the proportion of otitis-prone children among all children followed from birth is 4.4%. Thus the proportion of the population sampled was specified to be 0.001 in the analysis. Each generation was assumed to occur at 25-year intervals.
IMMUNOLOCALIZATION STUDIES
Six wild type C57BL/6J mice (3 male, 3 female) were used. For the middle ear, hemi-dissected heads of P6 mice were fixed overnight at 4°C with 4% paraformaldehyde (PFA), followed by incubation first in a solution of 1× phosphate buffer saline (PBS), and 0.25M EDTA for 24 hours and then in 30% sucrose/1×PBS solution for 4 hours. Samples were embedded in OCT and 14μm-sections were cut using a cryostat. For the organ of Corti, P7 mice were euthanized and decapitated. Temporal bones were isolated and fixed overnight at 4°C with 4% PFA followed by fine dissection of cochleae to isolate organ of Corti.
Sections were permeabilized with PBS containing 0.25% Triton X-100 for 1 hour, followed by blocking with 10% normal goat serum (Vector Labs) diluted in PBS for 1 hour. Sections were incubated overnight at 4°C with A2ML1 antibody (abcam ab72872, 1:200 dilution in PBS + 3% NGS) or for middle ear sections β-catenin antibody (1:200), then washed and incubated with a secondary antibody in PBS + 3% NGS for 1 hour at room temperature. Rhodamine phalloidin was used at a 1:300 dilution for F-actin labeling (Invitrogen). Nuclei were stained with DAPI (Molecular Probes). Samples were mounted using Prolong Gold antifade mounting medium (Molecular Probes) and imaged using a 100× objective and a confocal microscope (LSM 700, Carl Zeiss).
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to the indigenous community, families and individuals who participated in this study. We also thank Melquiadesa Pedro, Sheryl Mae Lagrana, Vic Ostan, surgical residents and audiology students who assisted data collection, and Drs. John Belmont and Seema Lalani for overall support. This study was supported by: the Hearing Health Foundation, Action On Hearing Loss and the National Organization for Hearing Research Foundation (to R.L.P.S.C.); the University of the Philippines Manila – National Institutes of Health (to G.T.A. and R.L.P.S.C.); and National Institutes of Health grants U54 HG006493 (to D.A.N.), R01 DK084350 (to M.M.S.), R01 DC003166 (to K.D.), R01 DC005841 (to T.C.), R01 DC011803 and R01 DC012564 (to S.R. and Z.M.A.), and R01 DC011651 and R01 DC003594 (to S.M.L.).
Footnotes
URL
Combined Annotation Dependent Depletion, http://cadd.gs.washington.edu/
dbNSFP, https://sites.google.com/site/jpopgen/dbNSFP
dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
Exome Aggregation Consortium, http://exac.broadinstitute.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
SeattleSeq Annotation, http://snp.gs.washington.edu/SeattleSeqAnnotation137/
Shared Harvard Inner-Ear Laboratory Database, https://shield.hms.harvard.edu/
UCSC Genome Bioinformatics, http://genome.ucsc.edu/
United States Census Bureau Population Estimates, http://www.census.gov/popest/
Variant Mendelian Tools, varianttools.sf.net/VMT
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Monasta L, et al. Burden of disease caused by otitis media: systematic review and global estimates. PLoS One. 2012;7:e36226. doi: 10.1371/journal.pone.0036226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acuin J. Chronic suppurative otitis media. Burden of Illness and Management Options. World Health Organization; Geneva: 2004. [Google Scholar]
- 3.Casey JR, Pichichero ME. Payment analysis of two diagnosis and management approaches of acute otitis media. Clin. Pediatr. 2014;53:865–873. doi: 10.1177/0009922814533592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvestad E, et al. Otitis media: genetic factors and sex differences. Twin Res. 2004;7:239–244. doi: 10.1375/136905204774200514. [DOI] [PubMed] [Google Scholar]
- 5.Patel JA, et al. Association of proinflammatory cytokine gene polymorphisms with susceptibility to otitis media. Pediatrics. 2006;118:2273–2279. doi: 10.1542/peds.2006-0764. [DOI] [PubMed] [Google Scholar]
- 6.Daly KA, et al. Chronic and recurrent otitis media: a genome scan for susceptibility loci. Am. J. Hum. Genet. 2004;75:988–997. doi: 10.1086/426061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen EK, et al. A genome-wide association study of chronic otitis media with effusion and recurrent otitis media identifies a novel susceptibility locus on chromosome 2. J. Assoc. Res. Otolaryngol. 2013;14:791–800. doi: 10.1007/s10162-013-0411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakagami M, Harada T, Juhn SK, Duvall AJ., 3rd Morphologic and biochemical study of vascular permeability of the middle ear mucosa in experimental otitis media. Ann. Otol. Rhinol. Laryngol. 1990;99:654–659. doi: 10.1177/000348949009900813. [DOI] [PubMed] [Google Scholar]
- 9.Marrero A, et al. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew. Chem. Int. Ed. Engl. 2012;51:3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- 10.Wong LP, et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013;92:52–66. doi: 10.1016/j.ajhg.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson B, Lundberg C, Ohlsson K. Granulocyte protease inhibition in acute and chronic middle ear effusion. Acta Otolaryngol. 1983;95:341–349. doi: 10.3109/00016488309130952. [DOI] [PubMed] [Google Scholar]
- 12.Bondestam M, Foucard T, Gebre-Medhin M. Serum albumin, retinol-binding protein, thyroxin-binding prealbumin and acute phase reactants as indicators of undernutrition in children with undue susceptibility to acute infections. Acta Paediatr. Scand. 1988;77:94–98. doi: 10.1111/j.1651-2227.1988.tb10605.x. [DOI] [PubMed] [Google Scholar]
- 13.van Dongen TM, van der Heijden GJ, Venekamp RP, Rovers MM, Schilder AG. A trial of treatment for acute otorrhea in children with tympanostomy tubes. N. Engl. J. Med. 2014;370:723–733. doi: 10.1056/NEJMoa1301630. [DOI] [PubMed] [Google Scholar]
- 14.Marynen P, Van Leuven F, Cassiman JJ, Van den Berghe H. Solubilization and affinity purification of the alpha 2-macroglobulin receptor for human fibroblasts. J. Biol. Chem. 1984;259:7075–7079. [PubMed] [Google Scholar]
- 15.Vissers LE, et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur. J. Hum. Genet. 2014;23:317–324. doi: 10.1038/ejhg.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The HUGO Pan-Asian SNP Consortium et al. Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 17.Santos-Cortez RLP, et al. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am. J. Hum. Genet. 2013;93:132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang GT, Peng B, Leal SM. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(Suppl1):S189–198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 20.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 21.Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–895. doi: 10.1093/bioinformatics/18.6.894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.