Abstract
During the evolution of multicellular organisms, the unit of selection and adaptation, the individual, changes from the single cell to the multicellular group. To become individuals, groups must evolve a group life cycle in which groups reproduce other groups. Investigations into the origin of group reproduction have faced a chicken-and-egg problem: traits related to reproduction at the group level often appear both to be a result of and a prerequisite for natural selection at the group level. With a focus on volvocine algae, we model the basic elements of the cell cycle and show how group reproduction can emerge through the coevolution of a life-history trait with a trait underpinning cell cycle change. Our model explains how events in the cell cycle become reordered to create a group life cycle through continuous change in the cell cycle trait, but only if the cell cycle trait can coevolve with the life-history trait. Explaining the origin of group reproduction helps us understand one of life's most familiar, yet fundamental, aspects—its hierarchical structure.
Keywords: individuality, coevolution, cell cycle, group reproduction, multicellularity, volvocine algae
1. Introduction and model
During the evolution of multicellular organisms, the unit of selection and adaptation, the individual, changes from the single cell to the multicellular group [1–3]. To be an individual, cell groups must reproduce offspring groups; however, it is not understood how group reproduction evolves from cell reproduction [4–6]. Investigations into the origin of group reproduction have run into a chicken-and-egg problem: traits related to reproduction at the group level often appear both to be a result of and a prerequisite for natural selection at the group level [7–9].
Beginning with the basic elements of the cell cycle, we show how group reproduction can emerge through the coevolution of a life-history trait with a trait underpinning life cycle change. We base our model on the life cycles observed in the volvocine green algae, where a wide diversity of colonial forms are observed in the asexual stage [10,11]. In the unicellular Chlamydomonas reinhardtii, a cell grows until it reaches about 2n times its initial size. A series of n rapid rounds of mitotic division occur, leading to 2n offspring cells, which separate to begin the cycle anew (figure 1a, n = 2). This kind of cycle, known as multiple fission, has the basic stages present in the vast majority of unicellular organisms: cell growth, division (mitosis) and separation (of the products of mitosis). The only difference between multiple fission and binary fission is that n = 1 in binary fission and n ≥ 1 in multiple fission. Groups composed of the products of clonal cell division, such as we study here, may be produced by either process. The group life cycle we model is based on the species in the volvocine family Tetrabaenaceae. Thought to be among the simplest multicellular organisms [11], colonies of these species have two or four C. reinhardtii-like cells that stay attached to each other after division, growing as a colony. After growth, cells separate before mitotic cell divisions (figure 1b, n = 2).
Figure 1.
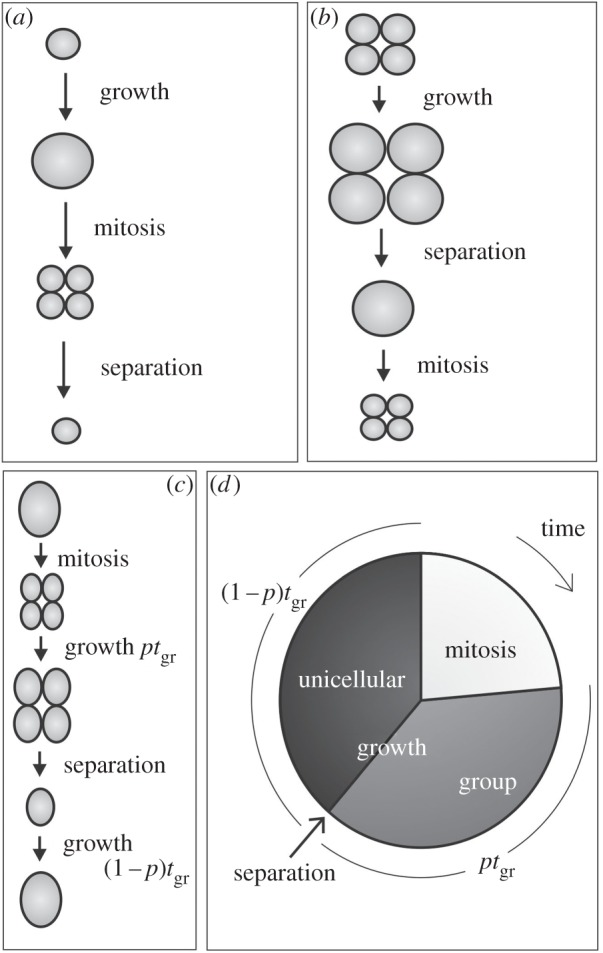
Life cycles. (a) Unicellular life cycle, as seen in species like Chlamydomonas reinhardtii. (b) Group life cycle, as seen in species like Tetrabaena socialis [11]. (c) A cell first grows as a part of a group for a given amount of time ptgr before leaving the group and continuing its life as a unicell. (d) As the life cycle variable p changes from 0 to 1, the order of cell cycle events changes and the separation stage occurs after cell growth in the group life cycle instead of before growth in the unicellular cycle.
In the model, a unicell grows for a time, tgr, and then reproduces. Larger parental cells produce more offspring. Mortality is assumed to be constant and growth slows as cells become larger. We assume that cells need time to switch from growing to reproducing. As tgr increases, fecundity increases, but the probability of survival to reproduction decreases. Therefore, we expect growing time to be optimized in unicells to create a balance between fecundity and survival.
We study a life cycle trait, p, which affects how ephemeral the groups of offspring cells are. This trait is based on a major difference between unicellular and the simplest colonial volvocines: the amount of time that offspring cells adhere to each other and grow. When p = 0, the unicellular cycle described above is in effect and groups of offspring cells exist momentarily following divisions. When p is greater than zero, a focal cell spends some of its life in the immediate company of its siblings. Conditions are favourable for kin or group selection as within-group conflict is low. We assume a group cost α and a group benefit β: α is the reduction in cell growth and β is the increase in survival when living in a group. Our focus is on how these group-dependent changes affect the optimality of pre-existing life-history traits (i.e. tgr) and the implications of this for the evolution of the group cycle (via changes in p). Whether the fully group (p = 1) or fully unicellular (p = 0) is optimal depends mainly on α and β. See figure 1 and table 1 for all the variables and parameters in the model and the electronic supplementary material for analysis.
Table 1.
Model parameters and variables.
| symbol | interpretation | constraints |
|---|---|---|
| p | life cycle trait: proportion of time that the products of cell division stay together growing in group before separating to finish growth as single cells | 0 ≤ p ≤ 1 |
| tgr | life-history trait: time for growth | 0 ≤ tgr |
| r(p,tgr) | intrinsic rate of cell population increase is a function of both p and tgr | |
| tsw | time needed to switch between cell growth and cell division and to undergo mitotic cell divisions. High switching times can favour longer growing times whereas certain growth parameters can favour shorter growing times | 0 < tsw |
| c0 | initial size of a cell | |
| b | exponent in the expression of the metabolic rate | b is typically ¾ |
| k | growth parameter for unicells | |
| K | 1/K is the time needed for the cell size to be multiplied by 21/(1−b) | 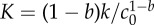 |
| α | effect of group-living on the growth parameter. Smaller values mean that living in a group has a larger cost (decreased cell growth) | 0 < α < 1 |
| m | death rate for unicells, assumed constant for simplicity | |
| β | effect of group-living on the death rate. Smaller values mean that living in a group has a larger benefit (reduced death rate) | 0 < β < 1 |
| r | population growth rate of cells. We maximize the cell-level population growth rate even when the group cycle emerges, p > 0. Because we assume within-group homogeneity, calculations of cell fitness are equivalent to calculations involving cell-group fitness [12]. The choice of using cell fitness in our calculations does not mean that the cell is the unit of selection and bearer of adaptions when there is a group life cycle |
2. Results
The simplest group cycle is characterized by a change in the order of stages in the cell cycle (figure 1). Unicells grow, divide and separate; however, cells in colonies grow, separate and divide. Our model shows how this discontinuous change can come about gradually through continuous evolution of a life cycle trait, p (figure 1d). We take the intrinsic growth rate of the population r = ln(fecundity × survival)/(generation time) as fitness. The number of offspring depends on cell size, (final size)/(initial size), using a growth model described in the electronic supplementary material. Fecundity is given in equation (2.1), and viability in equation (2.2).
| 2.1 |
and
| 2.2 |
The intrinsic population growth rate is given in the following equation:
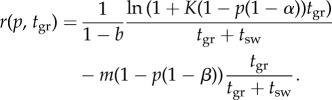 |
2.3 |
We give qualitative results in figure 2. If the life cycle and life-history traits co-evolve, the fitness maximum is reached either for a fully colonial or a fully unicellular life cycle. In the case where unicellularity (p = 0) is not optimal, p will evolve to one if the life-history trait is free to evolve. If we assume that the life-history trait, tgr, is fixed at its unicellular optimum, then p may evolve to an intermediate optimum value (grey box in figure 2). This means that the transition from to a fully group life cycle can become stymied, if the life-history trait is fixed. Such groups would be ephemeral aggregations of well-adapted unicellular organisms. By contrast, if we also let the life-history trait, tgr, evolve, then intermediate values of p are no longer optimal and the life cycle will go to the fully grouped state (p = 1) (figure 2).
Figure 2.
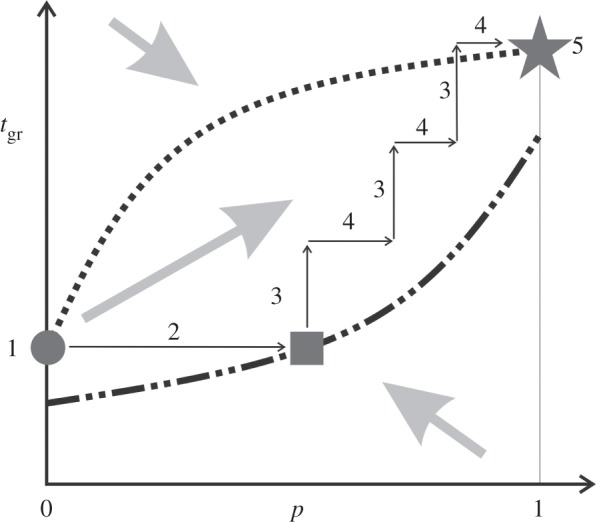
Fitness landscape and steps during an evolutionary transition in individuality. Selection gradient indicated by thick grey arrows. ∂pr = 0 shown by the dash-dotted line indicates the stable equilibria for a given value of tgr and 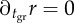 shown by the dotted line indicates the stable equilibria for a given value of p. If both parameters are allowed to change, there are two equilibria (grey circle and star), however, only one is stable. In the example, the equilibrium represented by the star is stable, and the circle is unstable, so a group life cycle evolves. The grey box indicates internal equilibrium, possible if tgr is not allowed to change. Numbered steps in an evolutionary transition are described in the text. Figure indicates qualitative results based on the conditions α > β, m(1 − β) < K(1 − α) and tsw is slightly above
shown by the dotted line indicates the stable equilibria for a given value of p. If both parameters are allowed to change, there are two equilibria (grey circle and star), however, only one is stable. In the example, the equilibrium represented by the star is stable, and the circle is unstable, so a group life cycle evolves. The grey box indicates internal equilibrium, possible if tgr is not allowed to change. Numbered steps in an evolutionary transition are described in the text. Figure indicates qualitative results based on the conditions α > β, m(1 − β) < K(1 − α) and tsw is slightly above 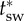 .
.
In figure 2, stages in evolutionary transitions in individuality [12] are indicated by numerals. (1) Features, such as multiple fission, that facilitate the emergence of group structure are present in unicells. (2) Group structure emerges. (3) Group-specific adaptations arise in traits already subject to selection. (4) Selective pressure for increased group cohesiveness. Steps (3) and (4) may repeat themselves as needed to complete the life cycle transition. (5) Further evolution of group-specific adaptations as group selection on cohesive groups is now established.
3. Discussion
The results of the model highlight the fact that the evolution of a more-grouped life cycle (increasing p) affects fitness components that are already being affected by previously optimized life-history traits, such as growing time tgr. Life-history traits are closely related to fitness and subject to a constraining trade-off. A fundamental trade-off faced by all organisms is between reproduction and survival. As p increases, all else equal, fecundity decreases and viability increases (because of α and β). Even if the net effect of this change in p is positive, the contrasting effect of changing p on fitness components that trade off with life-history traits such as tgr can be sub-optimal. When p changes, a cell does better (existing as it does for some time in a group context) if it can also adjust tgr to bring the fitness components back into balance. In addition to its direct effects on fitness, increasing tgr increases fecundity and decreases viability, essentially counteracting the lack of balance (between fitness components that trade off) induced by the change in p. Whether optimal tgr increases or decreases as groups start forming is likely to be sensitive to some of our assumptions (especially the assumption that number of cells per colony does not affect cell growth). However, changing these kind of assumptions would not affect the general lessons drawn from this model.
Because of previously optimized life-history traits, it is not enough for the products of cell division to simply stick together to evolve a group life cycle. Without the coevolution of previously optimized life-history traits (tgr), the evolutionary transition is stymied part way through (figure 2, grey box) and a complete group-level life cycle cannot emerge. The specific choice of growing time as the life-history trait is not critical for the more general results. Other life-history traits, such as amount of a limiting resource allocated to growth, are also likely to be important in unicellular algae, and we would expect such traits to behave similarly to our focal life-history trait tgr in terms of the potential to interact with life cycle evolution.
Should a life-history trait value such as tgr be considered a group-specific adaptation—and thus an indication of some degree of group-level individuality—once it has evolved away from the unicellular optimum? We think so. Maynard Smith & Szathmáry [1, p. 6] boiled down the issue of transitions in individuality as follows: ‘entities that were capable of independent replication before the transition can replicate only as part of a larger whole after it’. The capacity for independent replication is critical. Changes in a life-history trait, tgr, can be detrimental to the functionality that cells would have on their own, were they to leave the group. Groups of cells that evolve away from the unicellular optimal value of tgr are no longer merely spatio-temporal collections of entities with the full capacity to function on their own. These cells now require the group context, the larger whole, to reproduce most effectively.
Our results imply it is not necessary for a fully group life cycle to precede the evolution of adaptation at the group level. The two can emerge together and reinforce one another during their coevolution (figure 2). We do not claim that group adaptations can evolve without any group selection (i.e. without any group-level life cycle). Rather, we argue that a small step towards a group life cycle, that is, say, an initial increase in cell adhesiveness so that p moves away from zero (triggered perhaps for non-adaptive reasons [13]), is sufficient to favour a shift away from values of cell life-history traits that are optimal at the cell level towards values that are adaptive at the group level. These changed trait values can, in turn, set the stage for further adaptive change in the group life cycle (i.e. further increase in cell adhesiveness), so as to restrict cells from living alone. This process of coevolution between life cycle and life-history traits was suggested previously [12], and our model shows how and why it works (figure 2).
Supplementary Material
Acknowledgements
We thank P. Ferris, Z. Grochau-Wright, E. R. Hanschen, M. Leslie and A. Potticary.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Competing interests
We declare we have no competing interests.
Authors' contributions
O.M. carried out mathematical analysis, and participated in the design of the study and writing of the manuscript; D.S. participated in the design of the study and writing of the manuscript; R.E.M. conceived of the study, led the design of the study, coordinated the study and led the writing of the manuscript. All authors gave final approval for publication.
Funding
This work was supported by NASA award NNX13AH41G and by NSF award MCB-1412395.
References
- 1.Maynard Smith J, Szathmáry E. 1995. The major transitions in evolution. San Francisco, CA: W.H. Freeman. [Google Scholar]
- 2.Bonner JT. 1998. The origins of multicellularity. Integr. Biol. Issues News Rev. 1, 27–36. () [DOI] [Google Scholar]
- 3.Michod RE. 1999. Darwinian dynamics: evolutionary transitions in fitness and individuality. Princeton, NJ: Princeton University Press. [Google Scholar]
- 4.Griesemer J. 2001. The units of evolutionary transition. Selection 1, 67–80. ( 10.1556/Select.1.2000.1-3.7) [DOI] [Google Scholar]
- 5.Rainey PB, Kerr B. 2010. Cheats as first propagules: a new hypothesis for the evolution of individuality during the transition from single cells to multicellularity. BioEssays 32, 872–880. ( 10.1002/bies.201000039) [DOI] [PubMed] [Google Scholar]
- 6.Libby E, Ratcliff WC. 2014. Ratcheting the evolution of multicellularity. Science 346, 426–427. ( 10.1126/science.1262053) [DOI] [PubMed] [Google Scholar]
- 7.Rainey PB. 2007. Unity from conflict. Nature 446, 616 ( 10.1038/446616a) [DOI] [PubMed] [Google Scholar]
- 8.Clarke E. 2014. Origins of evolutionary transitions. J. Biosci. 39, 1–14. ( 10.1007/s12038-013-9375-y) [DOI] [PubMed] [Google Scholar]
- 9.Godfrey-Smith P. 2009. Darwinian populations and natural selection. Oxford, UK: Oxford University Press. [Google Scholar]
- 10.Kirk DL. 1998. Volvox: molecular-genetic origins of multicellularity and cellular differentiation. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 11.Arakaki Y, Kawai-Toyooka H, Hamamura Y, Higashiyama T, Noga A, Hirono M, Olson BJSC, Nozaki H. 2013. The simplest integrated multicellular organism unveiled. PLoS ONE 8, e81641 ( 10.1371/journal.pone.0081641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shelton DE, Michod RE. 2014. Group selection and group adaptation during a major evolutionary transition: insights from the evolution of multicellularity in the volvocine algae. Biol. Theory 9, 452–469. ( 10.1007/s13752-014-0159-x) [DOI] [Google Scholar]
- 13.Newman SA, Forgacs G, Muller GB. 2006. Before programs: the physical origination of multicellular forms. Int. J. Dev. Biol. 50, 289–299. ( 10.1387/ijdb.052049sn) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.


