Abstract
The Lake Malawi haplochromine cichlid flock is one of the largest vertebrate adaptive radiations. The geographical source of the radiation has been assumed to be rivers to the south and east of Lake Malawi, where extant representatives of the flock are now present. Here, we provide mitochondrial DNA evidence suggesting the sister taxon to the Lake Malawi radiation is within the Great Ruaha river in Tanzania, north of Lake Malawi. Estimates of the time of divergence between the Lake Malawi flock and this riverine sister taxon range from 2.13 to 6.76 Ma, prior to origins of the current radiation 1.20–4.06 Ma. These results are congruent with evaluations of 2–3.75 Ma fossil material that suggest past faunal connections between Lake Malawi and the Ruaha. We propose that ancestors of the Malawi radiation became isolated within the catchment during Pliocene rifting that formed both Lake Malawi and the Kipengere/Livingstone mountain range, before colonizing rivers to the south and east of the lake region and radiating within the lake basin. Identification of this sister taxon allows tests of whether standing genetic diversity has predisposed Lake Malawi cichlids to rapid speciation and adaptive radiation.
Keywords: adaptive radiation, phylogeny, African fishes
1. Introduction
Adaptive radiations make up a high proportion of biodiversity. In many cases, ancestors or sister species of these flocks have been identified, as with Galapagos finches [1], Hawaiian silverswords [2] and Canadian three-spined sticklebacks [3]. Identification of their origins has enabled discussion of events that initiated adaptive radiation, and allowed tests of whether diversification has been promoted by novel mutations that have arisen since colonization, or instead whether adaptation is based primarily on pre-existing genetic variation [4]. This is an important issue to resolve, because it can explain why only some colonizing lineages radiate when provided with ecological opportunity, and how parallel adaptive radiation can take place rapidly in geographically separated habitats.
The evolutionary origins of cichlid fishes radiations in East African lakes are largely elusive or speculative [5–8]. This is partly because of incomplete geographical and genomic sampling of riverine species within and surrounding lake basins. However, it is also due to intrinsic complexity of cichlid evolutionary relationships, as radiations may have been seeded by multiple riverine ancestors [8,9], and rivers can be recolonized by species with lacustrine ancestry [10]. A greater understanding of geographical and phylogenetic ancestry of cichlids is required to test whether functional genetic variation under divergent selection within lake radiations is present within riverine ancestors, and whether this variation has been shared among riverine cichlids through intraspecific gene flow and interspecific hybridization [9].
Lake Malawi contains a radiating flock of at least 450 haplochromine species [11]. Early phylogenetic reconstructions suggested that the lake radiation was monophyletic [12,13]. More recent phylogenies show two species outside the Lake Malawi catchment also fall within the flock, namely Astatotilapia calliptera and Astatotilapia swynnertoni [5,8]. There have been indications that these are sister lineages to the radiating flock [5,8], but the hypothesis has not been well supported by either nuclear or mitochondrial DNA [5,8,14,15]. There is evidence that riverine representatives of the flock outside the catchment have seeded some lacustrine diversity in the radiation [8,16], but preceding this they may have escaped from Lake Malawi into neighbouring drainages. Given such uncertainty, and evidence of recent gene flow across catchment boundaries in A. calliptera [16], there is a need to further resolve relationships of Malawi endemics to cichlids in neighbouring drainages.
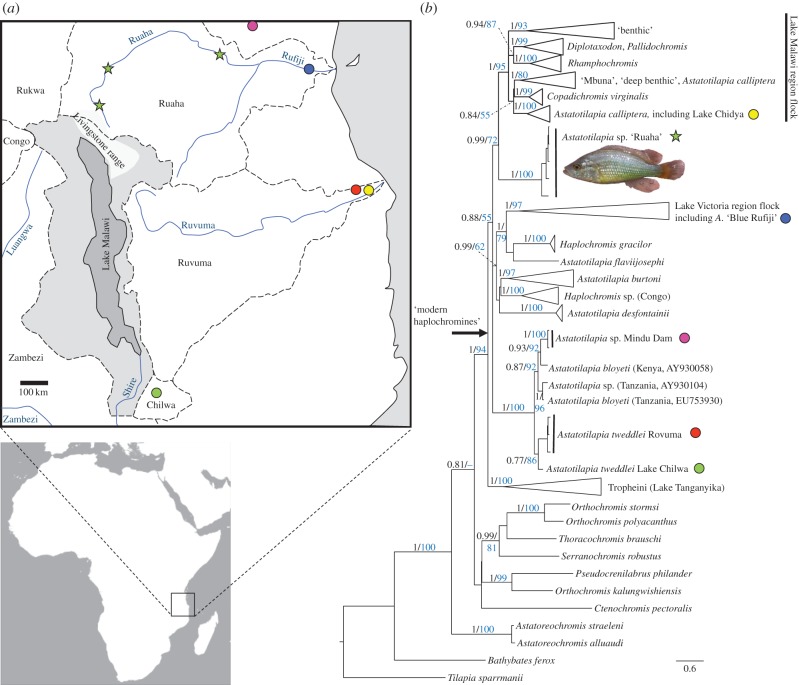
To date, phylogenetic reconstructions have included haplochromines from many of the surrounding catchments (figure 1), including the Zambezi, Lake Chilwa, Ruvuma, Congo and Lake Rukwa catchments [5,8,13,17]. However, no published phylogenies have included haplochromines from the Great Ruaha river [18]. Here, we show phylogenetic reconstructions including an undescribed taxon Astatotilapia sp. ‘Ruaha’ from this region which place it as a sister taxon to the Lake Malawi flock.
Figure 1.
(a) Lake Malawi and surrounding major river systems; (b) Bayesian phylogeny based on 544 mtDNA NADH2 sequences. Numbers above branches indicate posterior probabilities (black, values greater than 0.7 shown) and maximum-likelihood bootstrap support (blue, values greater than 70% shown). (Online version in colour.)
2. Material and methods
Genetic samples (fin clips) were collected from riverine haplochromines (electronic supplementary material, table S1; figure 1) and preserved in 95% ethanol. DNA was isolated using the Promega Wizard kit. Sequences of the mitochondrial gene NADH2 [7] were generated and aligned with sequences of other haplochromines and outgroup taxa, using ClustalW in DAMBE [19]. This resulted in an alignment of 1047 bp with 544 sequences (electronic supplementary material, table S2). Bayesian phylogenies were generated in MrBayes v. 3.2.4 [20], using Partitionfinder [21] models, and two runs of 10 million generations. Resultant trees were combined after removal of 50% as burn-in. Maximum-likelihood phylogenetic analysis were conducted in RaxML [22], Partitionfinder models and 100 bootstrap replicates. We dated divergence times using a subset of 40 NADH2 sequences (electronic supplementary material, table S2). Time-calibrated trees were generated with BEAST v. 1.8.0 [23] using Partitionfinder models and two sets of calibrations [24,25] employed independently (electronic supplementary material, table S3). Random local clocks were used in runs of 50 million generations, with 20% of trees removed as burn-in. Consensus trees from TreeAnnotator [23] were viewed in Figtree (http://tree.bio.ed.ac.uk/software/figtree).
3. Results and discussion
An as yet undescribed representative of the ‘modern haplochromine’ group, Astatotilapia sp. ‘Ruaha’, was present at three Great Ruaha sites (figure 1). On the basis of mitochondrial NADH2 DNA sequences, the species was resolved as an immediate sister taxon to the radiating flock (figure 1; electronic supplementary material, figure S1). The results of the analyses suggest they diverged between 2.13 Ma (95% highest posterior density (HPD) 1.52–2.84 Ma; using non-cichlid fossil derived calibrations from Friedman et al. [24]) and 6.76 Ma (95% HPD 3.76–10.12 Ma; using non-cichlid fossil derived calibrations from Schwarzer et al. [25]). This divergence took place before initial divergence of extant representatives of the Lake Malawi flock estimated at 1.2 Ma (95% HPD 1.52–2.84 Ma) or 4.06 Ma (95% HPD 2.02–6.59 Ma), from Friedman et al. [24] and Schwarzer et al. [25] calibrations, respectively. The Astatotilapia sp. ‘Ruaha’ lineage is geographically separated from the Malawi catchment by the Livingstone/Kipengere mountain range. This comprises steep mountainous areas and high altitude plateau, and it is plausible that both geography and low temperatures impose barriers to habitat occupancy and dispersal across the boundary [16]. The range was formed during Pliocene rifting that initiated formation of Lake Malawi [26], perhaps driving simultaneous population division and ecological opportunity for species flock formation.
Close evolutionary relationships between Malawi and upper Ruaha haplochromines are mirrored by recent observations from fish fossils of fluviatile deposits of the Chiwondo beds dated to between 2 and 3.75 Ma [18]. The Chiwondo fauna includes claroteid catfishes and tigerfish (Hydrocynus) [18], but geographically the nearest system containing extant representatives of these non-cichlid families is the Ruaha. It has been proposed on the basis of these fossils that rivers currently in Lake Malawi catchment were once extensions of the Great Ruaha system in pre-rift times [18]. Our results are compatible with this concept and imply further molecular studies may identify this region as a source of genetic diversity of other elements of the Malawi fauna. Notably, although the Chiwondo fauna includes representatives of Cichlidae, it has not been possible to identify remains to a lower taxonomic level [18].
It has been proposed that the ancestor of the Lake Malawi haplochromine flock is a riverine haplochromine similar to Astatotilapia bloyeti or A. calliptera [27]. Our study places specimens assigned to A. bloyeti in a sister clade to Astatotilapia tweddlei, consistent with previous analyses of both nuclear and mitochondrial markers [8], and our results suggest both taxa are more distantly related to Malawi cichlids than Astatotilapia sp. ‘Ruaha’. Our results also show that A. calliptera outside the Lake Malawi catchment are part of a geographically broader ‘Lake Malawi region’ flock. It remains equivocal whether the species secondarily colonized external rivers from Lake Malawi, or instead whether there have been multiple colonizations of A. calliptera from outside the catchment along with maintenance of the ancestral riverine phenotype [5,8,14]. In either case, given mitochondrial DNA evidence suggesting that Astatotilapia sp. ‘Ruaha’ is a sister species to the flock, and fossil evidence of historic connectivity of the Ruaha and Lake Malawi, it seems plausible that extant representatives of the Malawi flock are biogeographically derived from a species with a former distribution that encompassed both the Ruaha and Lake Malawi catchments. Further phylogenetic analyses based on nuclear genome data will help to provide further resolution of the relationship between Astatotilapia sp. ‘Ruaha’ and Malawi cichlids. Genome-wide data will also help to resolve whether A. calliptera occupy a basal, sister or derived position in the flock, which may force reconsideration of the biogeographic scenario suggested here.
Recent results show a high proportion of genomic diversity present within Lake Malawi cichlids is also present in riverine cichlids [9]. It has been proposed that riverine species may be active transporters of genomic material enabling rapid adaptation within lacustrine flocks. However, such situations require introgression among riverine taxa at contact zones, and gene flow across catchment boundaries. There is support for the concept of intraspecific gene flow across watersheds within Africa [16], but currently only indirect evidence of interspecific hybridization among river cichlids [27], and there is no evidence of interspecific hybridization among riverine haplochromines in the region surrounding Lake Malawi. A greater understanding of taxonomic and spatial patterns of genetic diversity within and among potentially ancestral riverine cichlids is required, including Astatotilapia sp. ‘Ruaha’. This would enable tests of the importance of active transport of genes through via riverine species and hybridization events for explaining shared genomic diversity among lacustrine radiations [9].
Supplementary Material
Acknowledgements
We thank TAFIRI staff and J. Swanstrom for field assistance.
Ethics
Tanzania Commission for Research and Technology (COSTECH) issued a permit for this study (no. 2011-205-NA-2011-103).
Data accessibility
DNA sequences can be accessed from Genbank (KR010448–KR010461).
Authors' contributions
M.J.G., B.P.N. and G.F.T. conceived the study and wrote the manuscript. M.J.G., B.P.N., S.M., A.S. and M.J.G. collected field samples and data. M.J.G. generated and analysed sequence data. All authors critically revised and approved the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was financially supported by the Royal Society-Leverhulme Trust Africa Award AA100023.
References
- 1.Sato A, Tichy H, O'hUigin C, Grant PR, Grant BR, Klein J. 2001. On the origin of Darwin's finches. Mol. Biol. Evol. 18, 299–311. ( 10.1093/oxfordjournals.molbev.a003806) [DOI] [PubMed] [Google Scholar]
- 2.Barrier M, Baldwin BG, Robichaux RH, Purugganan MD. 1999. Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol. Biol. Evol. 16, 1105–1113. ( 10.1093/oxfordjournals.molbev.a026200) [DOI] [PubMed] [Google Scholar]
- 3.Taylor EB, McPhail JD. 1999. Evolutionary history of an adaptive radiation in species pairs of threespine sticklebacks (Gasterosteus): insights from mitochondrial DNA. Biol. J. Linn. Soc. 3, 271–291. ( 10.1111/j.1095-8312.1999.tb01891.x) [DOI] [Google Scholar]
- 4.Feulner PG, et al. 2013. Genome-wide patterns of standing genetic variation in a marine population of three-spined sticklebacks. Mol. Ecol. 22, 635–649. ( 10.1111/j.1365-294X.2012.05680.x) [DOI] [PubMed] [Google Scholar]
- 5.Seehausen O, et al. 2003. Nuclear markers reveal unexpected genetic variation and a Congolese-Nilotic origin of the Lake Victoria cichlid species flock. Proc. R. Soc. Lond. B 270, 129–137. ( 10.1098/rspb.2002.2153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verheyen E, Salzburger W, Snoeks J, Meyer A. 2003. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300, 325–329. ( 10.1126/science.1080699) [DOI] [PubMed] [Google Scholar]
- 7.Salzburger W, Mack T, Verheyen E, Meyer A. 2005. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 5, 17 ( 10.1186/1471-2148-5-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce DA, Lunt DH, Genner MJ, Turner GF, Bills R, Seehausen O. 2011. Repeated colonization and hybridization characterize the Lake Malawi cichlid fish species flock. Curr. Biol. 21, R108–R109. ( 10.1016/j.cub.2010.11.029) [DOI] [PubMed] [Google Scholar]
- 9.Loh YH, et al. 2013. Origins of shared genetic variation in African cichlids. Mol. Biol. Evol. 30, 906–917. ( 10.1093/molbev/mss326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturmbauer C, Salzburger W, Duftner N, Schelly R, Koblmüller S. 2010. Evolutionary history of the Lake Tanganyika cichlid tribe Lamprologini (Teleostei: Perciformes) derived from mitochondrial and nuclear DNA data. Mol. Phylogenet. Evol. 57, 266–284. ( 10.1016/j.ympev.2010.06.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genner MJ, Seehausen O, Cleary DFR, Knight ME, Michel E, Turner GF. 2004. How does the taxonomic status of allopatric populations influence species richness within African cichlid fish assemblages? J. Biogeogr. 31, 93–102. ( 10.1046/j.0305-0270.2003.00986.x) [DOI] [Google Scholar]
- 12.Meyer A, Kocher TD, Basasibwaki P, Wilson AC. 1990. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 347, 550–553. ( 10.1038/347550a0) [DOI] [PubMed] [Google Scholar]
- 13.Nagl S, Tichy H, Mayer WE, Takezaki N, Takahata N, Klein J. 2000. The origin and age of haplochromine fishes in Lake Victoria, east Africa. Proc. R. Soc. Lond. B 267, 1049–1061. ( 10.1098/rspb.2000.1109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genner MJ, Turner GF. 2012. Ancient hybridization and phenotypic novelty within Lake Malawi's cichlid fish radiation. Mol. Biol. Evol. 29, 195–206. ( 10.1093/molbev/msr183) [DOI] [PubMed] [Google Scholar]
- 15.Hulsey CD, Keck BP, Alamillo H, O'Meara BC. 2013. Mitochondrial genome primers for Lake Malawi cichlids. Mol. Ecol. Res. 13, 347–353. ( 10.1111/1755-0998.12066) [DOI] [PubMed] [Google Scholar]
- 16.Nichols P, Genner MJ, Van Oosterhout C, Smith A, Swanstrom J, Parsons P, Sungani H, Joyce DA. 2015. Secondary contact seeds phenotypic novelty in cichlid fishes. Proc. R. Soc. B 282, 20142272 ( 10.1098/rspb.2014.2272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce DA, Lunt DH, Bills R, Turner GF, Katongo C, Duftner N, Sturmbauer C, Seehausen O. 2005. An extant cichlid fish radiation emerged in an extinct Pleistocene lake. Nature 435, 90–95. ( 10.1038/nature03489) [DOI] [PubMed] [Google Scholar]
- 18.Stewart KM, Murray AM. 2013. Earliest fish remains from the Lake Malawi Basin, Malawi, and biogeographical implications. J. Vert. Paleontol. 33, 532–539. ( 10.1080/02724634.2013.741086) [DOI] [Google Scholar]
- 19.Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 30, 1720–1728. ( 10.1093/molbev/mst064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. ( 10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 21.Lanfear R, Calcott B, Ho SYH, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 22.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman M, Keck BP, Dornburg A, Eytan RI, Martin CH, Hulsey CD, Wainwright PC, Near TJ. 2013. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. R. Soc. B 280, 20131733 ( 10.1098/rspb.2013.1733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarzer J, Misof B, Tautz D, Schliewen UK. 2009. The root of the East African cichlid radiations. BMC Evol. Biol. 9, 186 ( 10.1186/1471-2148-9-186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danley PD, Husemann M, Ding B, Dipietro LM, Beverly EJ, Peppe DJ. 2012. The impact of the geologic history and paleoclimate on the diversification of East African cichlids. Int. J. Evol. Biol. 2012, 574851 ( 10.1155/2012/574851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarzer J, Swartz ER, Vreven E, Snoeks J, Cotterill FP, Misof B, Schliewen UK. 2012. Repeated trans-watershed hybridization among haplochromine cichlids (Cichlidae) was triggered by Neogene landscape evolution. Proc. R. Soc. B 279, 4389–4398. ( 10.1098/rspb.2012.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences can be accessed from Genbank (KR010448–KR010461).