Abstract
Variation among parasite strains can affect the progression of disease or the effectiveness of treatment. What maintains parasite diversity? Here I argue that competition among parasites within the host is a major cause of variation among parasites. The competitive environment within the host can vary depending on the parasite genotypes present. For example, parasite strategies that target specific competitors, such as bacteriocins, are dependent on the presence and susceptibility of those competitors for success. Accordingly, which parasite traits are favoured by within-host selection can vary from host to host. Given the fluctuating fitness landscape across hosts, genotype by genotype (G×G) interactions among parasites should be prevalent. Moreover, selection should vary in a frequency-dependent manner, as attacking genotypes select for resistance and genotypes producing public goods select for cheaters. I review competitive coexistence theory with regard to parasites and highlight a few key examples where within-host competition promotes diversity. Finally, I discuss how within-host competition affects host health and our ability to successfully treat infectious diseases.
Keywords: within-host competition, parasite diversity, bacteriocins, coexistence, public goods, G×G interactions
1. Introduction
The maintenance of diversity, be it at the species or genotypic level, is a fundamental problem in biology. The root of the issue is that, in a given environment, the fittest type should competitively exclude all others. How diversity is maintained, therefore, has been the focus of a rich body of empirical and theoretical work in ecology and evolutionary biology. Here, I examine how these works inform our understanding of parasite diversity. I start by describing the complex selective environment faced by parasites, focusing on the varied forms of competitive interaction that can occur within a single host individual. Then, I review theoretical treatments of competition and diversity to gain insights into how diversity can be maintained in the face of competition. I then highlight a few key systems where within-host competitive interactions are thought to be crucial in maintaining parasite diversity. Finally, I conclude by discussing the implications of parasite diversity and within-host competitive interactions for host health and the treatment of infectious diseases.
Parasite fitness is dependent on both within- and among-host selection [1]. Within the host, competition with the host's native microbiota or other co-infecting parasites is a key determinant of parasite fitness and successful transmission to new hosts. These competitive interactions can be categorized using classic terms from the fields of ecology and social behaviour [2–4]. Most fundamentally, genotypes or species consuming the same resources compete via exploitative competition (figure 1a). Within the host, pathogen traits that enable faster use of host resources can allow a pathogen to outgrow others within the host and to be numerically dominant upon transmission (e.g. [5]). These traits can be exclusive to the pathogen, such as a greater number of nutrient receptors or increased replication machinery (e.g. [6,7]). Alternatively, faster growth rate can be achieved by the release of compounds that enable degradation of host tissues or sequestration of nutrients [8,9]. If these released products can benefit other members of the community, they can be viewed as public goods, and the interactions between the producers can be viewed as cooperation [4,10]. As these public goods increase access to host resources, producing genotypes will be able to exploit hosts better than genotypes with lower levels of production (figure 1b). However, when within the same host, non-cooperating cheaters that avoid cost of production may have a competitive advantage.
Figure 1.
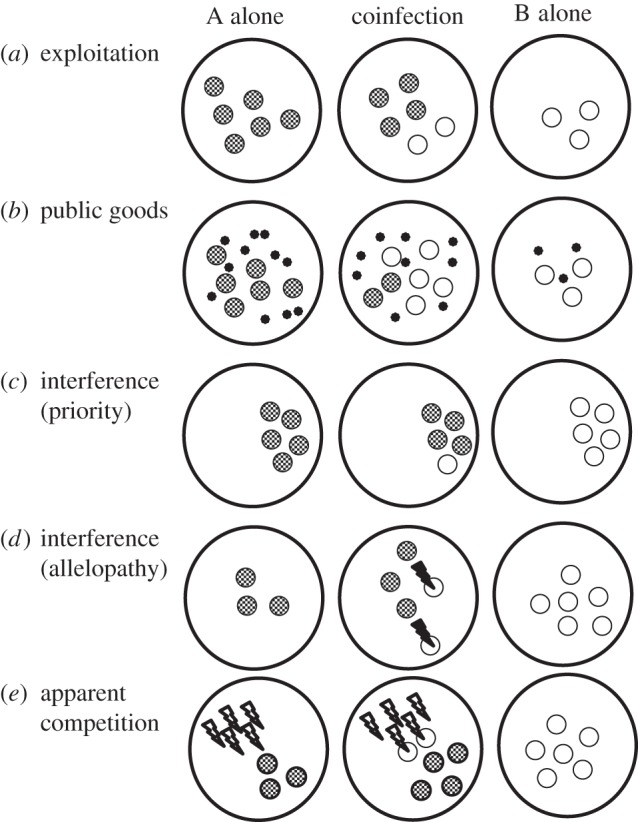
Mechanisms of within-host competition. Each large circle represents a host individual either infected with parasite A (smaller grey circles) alone, parasite B (smaller white circles) alone or simultaneously infected with both parasites. The relative growth rate of each parasite is indicated by the number of parasites present within each host individual. (a) When parasites compete through faster or more efficient exploitation of host resources, within-host selection favours parasites with superior exploitation (here, parasite A). (b) If parasites achieve greater host exploitation through the release of public goods (smallest black circles), then within-host selection favours parasites that produce less of the public good (here, parasite B). (c) When parasites compete over access to host infection sites, the parasite that occupies a site first (here, parasite A) can interfere with the attachment of later arriving parasites. (d) When parasites compete through the production of alleopathic agents (black lightning bolts), within-host selection favours parasites that produce costly competitive toxins (here, parasite A). (e) When parasites compete via the host immune systems, parasites who benefit when infecting alone by being resistant to the host immune response they elicit (white lightning bolts) may also benefit via within-host selection by eliminating immune-sensitive competitors.
Competition over limited resources can also select for strategies that block or attack potential competitors (i.e. interference competition) [11]. Multiple types of interference competition have been recognized among parasites. For example, in a study on acanthocephalan parasites of fish [12], the number of parasites that can establish within an individual host is limited by the amount of space left unoccupied by pre-established parasites (figure 1c). Similarly, microbes in a biofilm can pre-empt the attachment of other cells, or alternatively, overgrow established layers, depriving them access to nutrients [13]. More aggressive forms of interference include contact-dependent killing [14] and allelopathy (i.e. the release of growth-inhibiting compounds). Examples of allelopathy abound in the microbial world, as numerous antibiotics and other secondary metabolites have been found to have inhibitory effects [15]. A key class of allelopathic compounds, bacteriocins, target even closely related strains of the same species and are likely a key mechanism maintaining within-species diversity [16,17]. Critically, allelopathy is only beneficial in the presence of a susceptible competitor (figure 1d). In the absence of a suitable target, the costs of production (or even the carriage of production machinery) may lower the fitness of aggressive mechanisms of interference.
Finally, competition between two parasites within a host can be mediated by the host immune system [18,19]. In ecology, this is termed apparent competition, because the negative effect that two species have on each other (i.e. competition) is not due to shared resource use, but due to a shared predator [20]. Under classic apparent competition, increasing density of one prey species promotes population growth of a shared predator, thereby increasing predation on a second prey species. In the case of parasites, the presence of one parasite can stimulate the host immune system. If the immune response is non-specific, it may cross-react with other co-infecting parasite strains or species [2]. This immune-system mediated competition thereby favours pathogens that can escape the immune response by hiding from or being immune to its effects. In fact, the ability to withstand a severe inflammatory host response can favour the emergence of a parasite that provokes the immune response and subsequently benefits from the clearing of less resistant competitors (figure 1e) [21].
How do the complex competitive interactions within the host balance with among-host selection? Empirical work has demonstrated that each of the mechanisms described above can alter parasite within-host reproduction and transmission success. However, how different mechanisms of within-host competition affect each other and the full extent to which host epidemiology influences parasite evolution is still an open question [22]. Nevertheless, it is clear that the complex competitive environment within the host can favour different strategies in different host individuals: the competitive strategy that is successful in one host may depend on the presence of a specific competitor, and may be costly in the next host due to absence of that competitor. One result of this complex selective landscape is that parasites can evolve adaptive plastic responses to deal with the varied competitive environments encountered within hosts [23,24]. Host populations can, however, support many parasite species, and considerable genetic diversity within a given parasite species. So how is this diversity maintained given that competition often leads to exclusion of one competitor by another? I next review our understanding of competitive coexistence in relation to parasites.
2. Effects of competition on parasite diversity
Competitive coexistence requires that each species limits its own growth more than it limits the growth of another species [25]. Ecologically, this can be achieved by three main mechanisms: niche partitioning, competition–colonization trade-offs and heterogeneity in the competitive environment. Niche partitioning is the best understood mechanism of coexistence. Niches can be described in numerous dimensions involving essential resources, abiotic tolerances and susceptibility to natural enemies. If specialization in a given niche dimension is costly, then one species will become competitively dominant in each dimension, and species diversity will be limited by the number of niche dimensions. Thus over evolutionary time, competition is expected to lead to character displacement among species, lessening competition between species and thereby stabilizing the community [26]. Accordingly, one predicted outcome of within-host competition among parasites is specialization of parasites to different host tissues or different host species [27]. Within a host species, niche partitioning can occur due to specific immunity [28]. A successful strain elicits an immune response thereby limiting its own success, while antigenically different strains can still invade. Not surprisingly, fewer strains can be maintained if there are large intrinsic fitness differences among strains or if the immune response is cross-reactive (i.e. resulting in apparent competition). However, relatively simple models of strain competitive ability and immune response can explain the incredible antigenic variation seen within some human pathogens [28,29].
In the absence of niche partitioning, coexistence can occur across homogeneous patches via a competition–colonization trade-off [30,31]. Thus, even if one species is dominant within a patch, the other can persist by reaching more unoccupied patches. For parasites, host individuals form discrete patches, and a strain that is inferior in within-host competition may be better able to disperse to competitor-free hosts. One remarkable example of this trade-off is the mitigation of a competitive asymmetry between two species of pigeon lice by the unique ability of the competitively inferior species to disperse to new hosts by attaching to a fly [32]. Similarly, although the mechanism is not clear, HIV strains favoured by within-host selection are at a disadvantage in transmission to new hosts [33]. Additionally, the competition–colonization trade-off can arise from a trade-off between within-host competition and persistence outside the host [34]. For example, a parasite that is better at free-living survival may be slower to recover or grow within the host, and thus competitively inferior. In support of this idea, a comparative study of the phages of Escherichia coli found that slower growing phages were more persistent in the environment, suggesting a fundamental trade-off between these properties [35]. Further supporting a trade-off, experimental evolution of an arbovirus found that higher extracellular survival was associated with reduced fecundity [36], although such trade-offs are not necessarily universal [37] and should be evaluated over both ecological and evolutionary time scales.
A final category for coexistence requires heterogeneity in the competitive environment [25,38]. A corollary to the maxim that species coexistence requires each species to limit its own growth more than it limits the growth of another species, is that species can coexist if each species can increase when rare [39]. Even in the absence of local niche partitioning, negative-frequency dependence can occur if species experience competition on different spatial scales [40] or if species differ in how they respond competitively to spatial and temporal variation in the environment (e.g. through a storage effect [41]). Empirical demonstrations of these mechanisms are less well established than niche partitioning or competition–colonization trade-offs; nevertheless, they suggest that variation in how species experience competition is an important mechanism of coexistence [42,43]. For parasites, transient changes in the environment or among hosts can result in different pathogen genotypes being successful at different times or in different places. If this variation dampens otherwise successful strains and favours strains that were struggling, then it may promote coexistence. Such equalizing effects are inherent in among-host selection and can be more forceful when epidemiological feedbacks occur. Additionally, within-host selection is expected to vary depending on the composition of the within-host community. For example, parasite traits that increase the speed of host exploitation will be favoured within the host, but parasites with these traits may be susceptible to invasion by parasites that can attack them.
The interplay of strategies favouring faster growth and strategies depending on attack has been modelled extensively with regard to bacteriocins. Strains that produce bacteriocins are able to attack sensitive strains, but they pay a cost of production. Thus, producers cannot displace sensitive strains unless they are sufficiently numerous and toxic, or unless the competitive interactions are local [44,45]. In order for both producer and sensitive strains to coexist stably, there must be underlying spatial heterogeneity in resource availability [46] or stochastic environmental disturbances that hit some patches but not others [47]. This spatial heterogeneity allows sensitive strains to gain an advantage in either low resource or empty patches. Notably, when more than two strains are examined, coexistence can occur in the absence of spatial heterogeneity. Resistant strains can evolve either from sensitive strains [48] or represent non-producing cheaters [45]. If resistant strains have higher growth rates than producers (because they are not paying the cost of production) and lower growth rates than sensitives (because they are paying a cost of resistance), then resistant strains can out-compete producers, but are out-competed by sensitives in a rock–paper–scissors-like dynamic (RPS). As each strain can be invaded by another, these non-transitive interactions can maintain diversity through negative-frequency dependence. Theoretical, as well as experimental, evaluations of this dynamic show that stability is contingent upon spatial structure facilitating local interactions [49]. Furthermore, a study using E. coli strains in mice found that strains could be transmitted between hosts and replaced each other as predicted by the RPS model, although coexistence was not achieved in the relatively small communities of the experimental set-up [50].
How do non-transitive ecological dynamics play out on an evolutionary timescale? Intriguingly, theory has suggested that spatial structure coupled with non-transitive pairwise interactions favours the evolution of competitive restraint [51,52]. This result can be understood intuitively as more rapacious competitors more effectively eliminate their subordinate partner (be it Resistant > Producer, P > S or S > R), placing them more frequently in contact with a partner that can out-compete them. For example, if a more rapacious form of the resistant strain arises, say by mutations that lower the cost of resistance, it will more effectively out-compete the producer strain. However, the resistant strain will then find itself more likely to encounter sensitive strains which limit the spread of the rapacious resistant strain. Thus, restrained behaviour towards ‘the enemy of my enemy’ is beneficial to a strain subject to community-level feedbacks. Such feedbacks favour community coexistence [53]. The importance of spatial structure and non-transitive interactions on the evolution of competitive restraint has been experimentally demonstrated in vitro with E. coli [54]. Furthermore, ecological feedbacks on community stability extend beyond the three-player game [53]. Allowing mutation and recombination leads to the prediction that a diverse number of strains can be maintained, with each strain producing one or more allelopathic agents [55,56]. This result is consistent with the diversity of bacteriocins carried among hosts in a natural population of E. coli [57].
In summary, although competition often leads to competitive exclusion, there are numerous mechanisms whereby coexistence can occur. Importantly, competition is not a transient effect that quickly becomes minimized due to character displacement. The patchy structure inherent in the parasite lifestyle sets the stage for heterogeneity across hosts to favour coexistence and for competition to act as a continual selective force on parasite traits. The interplay between ecological and evolutionary forces in maintaining parasite diversity is already well recognized with respect to host genetic diversity [58–60]. Analogously, competitive interactions within the host can shape the evolution of parasite phenotypes and can even facilitate the coexistence of multiple parasite types.
3. Key examples linking within-host competitive interactions and parasite diversity
Several parasite systems suggest that competitive interactions within the host are important for the maintenance of parasitic diversity. Here I will focus on three systems where we have a strong understanding of the diversifying effect of within-host competition. Diplostomum trematodes have a complex life cycle involving two intermediate hosts (snails and fish) and a definitive bird host. When a snail host is co-infected with multiple genotypes of D. pseudospathaceum, transmission success of each genotype is lower relative to a single genotype infection. However, when a fish host is exposed to multiple parasite genotypes simultaneously, the probability of parasite establishment is higher relative to single genotype exposures [61]. Thus, exploitative competition in the snail host reduces diversity, while interactions with the immune system of the fish host can maintain diversity. Exactly how parasite diversity interacts with the host immunity in this system is still under investigation, but there is evidence for cross-reactivity in adaptive immunity as well as specificity in innate immunity, a pattern contrary to the standard paradigm [62]. Furthermore in cross-species infections, some strains of D. pseudospathaceum perform better in mixed exposures than when infecting a fish alone, while others do worse [63,64]. Therefore, predicting the outcome of competition among species is dependent on knowing the specific parasite genotypes present within the host: a strain that is disfavoured in one host individual may have a fitness advantage in another due to differences in the parasite community.
Similar genotypic effects are seen in competition among species of entomopathogenic nematodes. These insect-killing nematodes (genus Steinernema) are involved in a mutualistic symbiosis with bacteria in the genus Xenorhabdus. Xenorhabdus bacteria are known to produce a suite of compounds that attack the insect immune system and prevent the insect from being colonized by fungal and other bacterial competitors. Additionally, Xenorhabdus produce bacteriocins that are effective at inhibiting the growth of Xenorhabdus genotypes distinct from the producing genotype. A diversity of bacteriocin phenotypes have been isolated from a small geographical area [65–67] and distributed in a pattern which suggests that within-insect interactions are important for maintaining this diversity. Specifically, one bacteriocin phenotype was found to dominate within an insect host, yet on a larger scale, consistent with nematode movement, multiple bacteriocin phenotypes could be detected. Additionally, laboratory-based co-infections have demonstrated a fitness benefit to the inhibiting phenotype that is conditional on the presence of a sensitive competitive partner within the insect host [68]. In the absence of a sensitive competitor, exploitative competition seems paramount: parasites that establish an infection faster are competitively dominant [69]. Thus, the competitive context determines which traits selection favours. Furthermore, transmission in this system requires a free-living stage. Species differ in longevity in this stage, which can influence their relative competitive success (F. Bashey 2011, unpublished data). Conditions within the host can also influence the size of the free-living stage, which influences the probability of successful establishment in a new host [70]. Hence, diverse within-host competitive environments can select for diverse parasite genotypes due to both within- and among-host fitness components.
Perhaps the best understood example of within-host competition from a mechanistic basis comes from work on the opportunistic pathogen Pseudomonas aeruginosa. In response to low levels of iron in the within-host environment, this species releases pyoverdin, its primary siderophore or iron scavenging molecule [71]. Pyoverdin constitutes a public good: non-producing cells can bind to this molecule and thereby gain iron. In the absence of competition, increased pyoverdin production increases population growth; however, in the presence of a competitor that produces less pyoverdin, fitness is reduced due to the higher cost of pyoverdin production [72]. Thus, selection on this trait depends upon the social context. Strains also compete through interference competition. Pseudomonas produces several types of bacteriocins [73] as well as possessing the ability to attack neighbouring cells in a contact-dependent manner [74]. The fitness benefit of bacteriocin production has also been shown to be contingent upon the nature of the competitive environment, depending not just on the presence of susceptible competitors, but also on the local density of kin [75]. Moreover, selection on bacteriocin production shifts depending on the level of pyoverdin production of the susceptible strain [76], demonstrating further the context dependence of within-host fitness. Similarly, contact-dependent killing has been found to be responsive to the behaviour of competitors in a tit-for-tat fashion [77]. Although most of this work has been elucidated in an in vitro setting, multiple competitive phenotypes can be isolated from a host population [78] or a host individual [79]. Moreover, receptors for these molecules demonstrate high levels of diversifying selection [80].
4. Implications of within-host competition to host health
Parasite growth within the host is a major cause of the negative effects that parasites have on host health [81,82]. As outlined above (figure 1), different mechanisms of within-host competition select for different parasite traits, some of which may increase or decrease within-host growth. Accordingly, a high prevalence of multiple infections is predicted to have varied effects on host health [22,83]. The earliest theoretical treatment of the effect of multiple infections on the evolution of virulence assumed that pathogens compete within the host via exploitative competition. Thus, they predicted that due to within-host selection for increased growth, pathogen virulence should evolve to be higher in mixed infections relative to single genotype infections [84,85]. While this prediction has been upheld in some systems [5], other systems are dominated by different forms of within-host interactions. When public goods allow greater host exploitation [8,86] or virulent immuno-provoking forms of apparent competition [87], mixed infections are predicted to result in lower virulence due to within-host selection favouring cheats. However, incorporating epidemiological feedbacks can change these predictions [88,89]. Similarly, spiteful forms of interference competition have been shown to lower virulence [90], but they can raise virulence depending on the scale of competition and the kin structure within the host [91]. As models more explicitly incorporate multiple forms of interactions within the host into an epidemiological framework [92], we will further improve our ability to connect knowledge of the competitive environment to host health outcomes.
Already, understanding within-host competition has helped to explain strains with puzzling phenotypes. For example, highly virulent forms of Streptococcus pneumonia are not transmitted from the host once they invade the body, yet they persist because they can withstand apparent competition that occurs in the nasal cavity [93]. Additionally, coupling selection in the within-host environment with selection occurring outside the host can provide a more accurate understanding of how variation in virulence is maintained in nature [94,95]. Within-host competition has also explained why some forms of methicillin-resistant Staphylococcus aureus (MRSA) do not respond to treatment with vancomycin [96]. Unlike other cases where antibiotic resistance occurs after antibiotic treatment or due to high levels of resistance circulating in the community, some forms of vancomycin resistance in MRSA appear to occur due to within-host competition. Specifically, resistant variants were found to arise spontaneously within hosts due to competition favouring a bacteriocin-producing form that could attack the wild-type, followed by the evolution of a bacteriocin-resistant strain. Traits conferring bacteriocin resistance also provided partial resistance to vancomycin. These examples illustrate that within-host selection pressures are profound and of significant clinical relevance.
While clinicians have mainly noted when the presence of one pathogen species complicates treatment of another [97], the presence of a competitor can improve the outcome for the host when interference or apparent competition slows disease progression [90,98,99]. In fact, the health benefits of many probiotics are thought to be due in part to their effectiveness in interference competition [100]. Interest in using bacteriocins as an alternative to traditional, broad-spectrum antibiotics is growing [101–103]. Historically, the narrow killing range of bacteriocins limited their utility; however, faster diagnostic tests, the ability to engineer bacteriocins to target novel sites [104], and our awareness of the consequences of using broad-spectrum antibiotics has made the therapeutic use of bacteriocins more viable.
As within-host competition alters selection on parasite traits and increases parasite diversity, it is further important to realize that variation in disease outcomes could be due as much to the within-host parasite community as due to host genotypic effects. Indeed, some cases that we have viewed as parasite interactions with the host genotype (i.e. Gparasite × Ghost) may in fact be due to differences in the within-host competitive environment [105]. Critically, variation in disease that is due to Gparasite × Gparasite may be more amenable to intervention than variation caused by host genotypic differences. Capitalizing on within-host selection is a major tenet of Hamiltonian medicine [106] and understanding how parasite traits influence parasite fitness across spatial scales is crucial to predicting when novel therapies, such as targeting virulence factors, will be successful [107,108].
In summary, the lifestyle of pathogens where selection occurs both within the host and in transmission between hosts creates opportunities for selection to maintain diverse pathogen phenotypes. This occurs in two main ways. First, the rich diversity of competitive strategies that parasites employ within the host can engender reciprocal selection between parasites. Parasites more successful at exploiting the host than competitors can be invaded by attacking or freeloading strategies, which then select for resistance and a restarting of this cycle. Second, heterogeneity among hosts in the composition of the within-host competitive environment itself can favour different parasite traits across hosts. As the relative costs and benefits of different forms of competition depend on the identity of the competitors, selection due to competition can be continual and within-host competition should be viewed as a potentially major driver of parasite diversity. Better understanding this driving force on parasite traits will enable us to more effectively treat infectious diseases.
Acknowledgements
I thank C. Lively and the various iterations of his laboratory group for thought-provoking discussion on the maintenance of diversity. Additionally, I thank A. Gibson, A. Bhattacharya, O. Restif and two anonymous reviewers for their insightful comments on previous versions of this manuscript.
Competing interests
I declare I have no competing interests.
Funding
This study was supported by NSF DEB 0919015.
References
- 1.Ewald PW. 1983. Host-parasite relations, vectors, and the evolution of disease severity. Ann. Rev. Ecol. Syst. 14, 465–485. ( 10.1146/annurev.es.14.110183.002341) [DOI] [Google Scholar]
- 2.Mideo N. 2009. Parasite adaptations to within-host competition. Trends Parasitol. 25, 261–268. ( 10.1016/j.pt.2009.03.001) [DOI] [PubMed] [Google Scholar]
- 3.Brown SP, Inglis RF, Taddei F. 2009. Evolutionary ecology of microbial wars: within-host competition and (incidental) virulence. Evol. Appl. 2, 32–39. ( 10.1111/j.1752-4571.2008.00059.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607. ( 10.1038/nrmicro1461) [DOI] [PubMed] [Google Scholar]
- 5.de Roode JC, et al. 2005. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA 102, 7624–7628. ( 10.1073/pnas.0500078102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton JP, Skelly P, Halton DW. 2004. Role of the tegument and gut in nutrient uptake by parasitic platyhelminths. Can. J. Zool. 82, 211–232. ( 10.1139/z03-213) [DOI] [Google Scholar]
- 7.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol. 84, 1597–1606. ( 10.1128/JVI.01783-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West SA, Buckling A. 2003. Cooperation, virulence and siderophore production in bacterial parasites. Proc. R. Soc. Lond. B 270, 37–44. ( 10.1098/rspb.2002.2209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown SP, Hochberg ME, Grenfell BT. 2002. Does multiple infection select for raised virulence? Trends Microbiol. 10, 401–405. ( 10.1016/S0966-842X(02)02413-7) [DOI] [PubMed] [Google Scholar]
- 10.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. 2004. The evolution of cooperation. Q. Rev. Biol. 79, 135–160. ( 10.1086/383541) [DOI] [PubMed] [Google Scholar]
- 11.Schoener TW. 1983. Field experiments on interspecific competition. Am. Nat. 122, 240–285. ( 10.1086/284133) [DOI] [Google Scholar]
- 12.Brown AF. 1986. Evidence for density-dependent establishment and survival of Pomphorhynchus laevis (Müller, 1776) (Acanthocephala) in laboratory-infected Salmo gairdneri Richardson and its bearing on wild populations in Leuciscus cephalus (L.). J. Fish Biol. 28, 659–669. ( 10.1111/j.1095-8649.1986.tb05201.x) [DOI] [Google Scholar]
- 13.An DD, Danhorn T, Fuqua C, Parsek MR. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl Acad. Sci. USA 103, 3828–3833. ( 10.1073/pnas.0511323103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol. 21, 230–237. ( 10.1016/j.tim.2013.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez JL. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. ( 10.1126/science.1159483) [DOI] [PubMed] [Google Scholar]
- 16.Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7, 129–133. ( 10.1016/S0966-842X(99)01459-6) [DOI] [PubMed] [Google Scholar]
- 17.Kerr B. 2007. The ecological and evolutionary dynamics of model bacteriocin communities. In Bacteriocins: ecology and evolution (eds Riley MA, Chavan MA.), pp. 112–134. Berlin: Springer. [Google Scholar]
- 18.Read AF, Taylor LH. 2001. The ecology of genetically diverse infections. Science 292, 1099–1102. ( 10.1126/science.1059410) [DOI] [PubMed] [Google Scholar]
- 19.Roode LR, Jacobus CD, Bell AS, Stamou P, Gray D, Read AF. 2006. The role of immune-mediated apparent competition in genetically diverse malaria infections. Am. Nat. 168, 41–53. ( 10.1086/505160) [DOI] [PubMed] [Google Scholar]
- 20.Holt R. 1977. Predation, apparent competition and the structure of prey communities. Theor. Pop. Ecol. 12, 197–229. ( 10.1016/0040-5809(77)90042-9) [DOI] [PubMed] [Google Scholar]
- 21.Brown SP, Le Chat L, Taddei F. 2008. Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol. Lett. 11, 44–51. ( 10.1111/j.1461-0248.2007.01125.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alizon S, de Roode JC, Michalakis Y. 2013. Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567. ( 10.1111/ele.12076) [DOI] [PubMed] [Google Scholar]
- 23.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat. Rev Microbiol. 11, 285–293. ( 10.1038/nrmicro2977) [DOI] [PubMed] [Google Scholar]
- 24.Leggett HC, Brown SP, Reece SE. 2014. War and peace: social interactions in infections. Phil. Trans. R. Soc. B 369, 20130365 ( 10.1098/rstb.2013.0365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chesson P. 2000. Mechanisms of maintenance of species diversity. Ann. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 26.Lawlor LR, Smith JM. 1976. The coevolution and stability of competing species. Am. Nat. 110, 79–99. ( 10.2307/2459878) [DOI] [Google Scholar]
- 27.Karvonen A, Terho P, Seppälä O, Jokela J, Valtonen ET. 2006. Ecological divergence of closely related Diplostomum (Trematoda) parasites. Parasitology 133, 229–235. ( 10.1017/S0031182006000242) [DOI] [PubMed] [Google Scholar]
- 28.Cobey S, Lipsitch M. 2012. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science 335, 1376–1380. ( 10.1126/science.1215947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins ER, Grad YH, Gupta S, Buckee CO. 2014. Contrasting within- and between-host immune selection shapes Neisseria Opa repertoires. Sci. Rep. 4, 6554 ( 10.1038/srep06554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kneitel JM, Chase JM. 2004. Trade-offs in community ecology: linking spatial scales and species coexistence. Ecol. Lett. 7, 69–80. ( 10.1046/j.1461-0248.2003.00551.x) [DOI] [Google Scholar]
- 31.Amarasekare P. 2000. Coexistence of competing parasitoids on a patchily distributed host: local versus spatial mechanisms. Ecology 81, 1286–1296. ( 10.1890/0012-9658(2000)0811286:COCPOA]2.0.CO;2) [DOI] [Google Scholar]
- 32.Harbison CW, Bush SE, Malenke JR, Clayton DH. 2008. Comparative transmission dynamics of competing parasite species. Ecology 89, 3186–3194. ( 10.1890/07-1745.1) [DOI] [PubMed] [Google Scholar]
- 33.Alizon S, Fraser C. 2013. Within and between host evolutionary rates across the HIV genome. Retrovirology 10, 49–59. ( 10.1186/1742-4690-10-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caraco T, Wang IN. 2008. Free-living pathogens: life-history constraints and strain competition. J. Theor. Biol. 250, 569–579. ( 10.1016/j.jtbi.2007.10.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Paepe M, Taddei F. 2006. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4, e193 ( 10.1371/journal.pbio.0040193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasik BR, Bhushan A, Ogbunugafor CB, Turner PE. 2015. Delayed transmission selects for increased survival of vesicular stomatitis virus. Evolution 69, 117–125. ( 10.1111/evo.12544) [DOI] [PubMed] [Google Scholar]
- 37.Gallet R, Lenormand T, Wang IN. 2012. Phenotypic stochasticity protects lytic bacteriophage populations from extinction during the bacterial stationary phase. Evolution 66, 3485–3494. ( 10.1111/j.1558-5646.2012.01690.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amarasekare P. 2003. Competitive coexistence in spatially structured environments: a synthesis. Ecol. Lett. 6, 1109–1122. ( 10.1046/j.1461-0248.2003.00530.x) [DOI] [Google Scholar]
- 39.Snyder R, Chesson P. 2004. How the spatial scales of dispersal, competition, and environmental heterogeneity interact to affect coexistence. Am. Nat. 164, 645–650. ( 10.1086/424969) [DOI] [PubMed] [Google Scholar]
- 40.Murrell DJ, Law R. 2003. Heteromyopia and the spatial coexistence of similar competitors. Ecol. Lett. 6, 48–59. ( 10.1046/j.1461-0248.2003.00397.x) [DOI] [Google Scholar]
- 41.Chesson P. 2000. General theory of competitive coexistence in spatially-varying environments. Theor. Popul. Biol. 58, 211–237. ( 10.1006/tpbi.2000.1486) [DOI] [PubMed] [Google Scholar]
- 42.Sears ALW, Chesson P. 2007. New methods for quantifying the spatial storage effect: an illustration with desert annuals. Ecology 88, 2240–2247. ( 10.1890/06-0645.1) [DOI] [PubMed] [Google Scholar]
- 43.Angert AL, Huxman TE, Chesson P, Venable DL. 2009. Functional tradeoffs determine species coexistence via the storage effect. Proc. Natl Acad. Sci. USA 106, 11 641–11 645. ( 10.1073/pnas.0904512106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao L, Levin BR. 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl Acad. Sci. USA 78, 6324–6328. ( 10.1073/pnas.78.10.6324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durrett R, Levin S. 1997. Allelopathy in spatially distributed populations. J. Theor. Biol. 185, 165–171. ( 10.1006/jtbi.1996.0292) [DOI] [PubMed] [Google Scholar]
- 46.Frank SA. 1994. Spatial polymorphism of bacteriocins and other allelopathic traits. Evol. Ecol. 8, 369–386. ( 10.1007/BF01238189) [DOI] [Google Scholar]
- 47.Czaran TL, Hoekstra RF. 2003. Killer-sensitive coexistence in metapopulations of micro-organisms. Proc. R. Soc. Lond. B 270, 1373–1378. ( 10.1098/rspb.2003.2338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feldgarden M, Riley MA. 1999. The phenotypic and fitness effects of colicin resistance in Escherichia coli K-12. Evolution 53, 1019–1027. ( 10.2307/2640807) [DOI] [PubMed] [Google Scholar]
- 49.Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174. ( 10.1038/nature00823) [DOI] [PubMed] [Google Scholar]
- 50.Kirkup BC, Riley MA. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock–paper–scissors in vivo. Nature 428, 412–414. ( 10.1038/nature02429) [DOI] [PubMed] [Google Scholar]
- 51.Prado F, Kerr B. 2008. The evolution of restraint in bacterial biofilms under nontransitive competition. Evolution 62, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frean M, Abraham ER. 2001. Rock–scissors–paper and the survival of the weakest. Proc. R. Soc. Lond. B 268, 1323–1327. ( 10.1098/rspb.2001.1670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson CR, Seinen I. 2002. Selection for restraint in competitive ability in spatial competition systems. Proc. R. Soc. Lond. B 269, 655–663. ( 10.1098/rspb.2001.1948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nahum JR, Harding BN, Kerr B. 2011. Evolution of restraint in a structured rock–paper–scissors community. Proc. Natl Acad. Sci. USA 108, 10 831–10 838. ( 10.1073/pnas.1100296108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Czaran TL, Hoekstra RF, Pagie L. 2002. Chemical warfare between microbes promotes biodiversity. Proc. Natl Acad. Sci. USA 99, 786–790. ( 10.1073/pnas.012399899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pagie L, Hogeweg P. 1999. Colicin diversity: a result of eco-evolutionary dynamics. J. Theor. Biol. 196, 251 ( 10.1006/jtbi.1998.0838) [DOI] [PubMed] [Google Scholar]
- 57.Gordon DM, O'Brien CL. 2006. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152, 3239–3244. ( 10.1099/mic.0.28690-0) [DOI] [PubMed] [Google Scholar]
- 58.Kirchner JW, Roy BD. 2002. Evolutionary implications of host–pathogen specificity: fitness consequences of pathogen virulence traits. Evol. Ecol. Res. 4, 27–48. [Google Scholar]
- 59.Marston MF, Pierciey FJ, Shepard A, Gearin G, Qi J, Yandava C, Schuster SC, Henn MR, Martiny JBH. 2012. Rapid diversification of coevolving marine Synechococcus and a virus. Proc. Natl Acad. Sci. USA 109, 4544–4549. ( 10.1073/pnas.1120310109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paterson S, et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278. ( 10.1038/nature08798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karvonen A, Rellstab C, Louhi KR, Jokela J. 2012. Synchronous attack is advantageous: mixed genotype infections lead to higher infection success in trematode parasites. Proc. R. Soc. B 279, 171–176. ( 10.1098/rspb.2011.0879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rellstab C, Karvonen A, Louhi KR, Jokela J. 2013. Genotype-specific versus cross-reactive host immunity against a macroparasite. PLoS ONE 8, e78427 ( 10.1371/journal.pone.0078427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seppala O, Karvonen A, Valtonen ET, Jokela J. 2009. Interactions among co-infecting parasite species: a mechanism maintaining genetic variation in parasites? Proc. R. Soc. B 276, 691–697. ( 10.1098/rspb.2008.1229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seppälä O, Karvonen A, Rellstab C, Louhi K-R, Jokela J. 2012. Reciprocal interaction matrix reveals complex genetic and dose-dependent specificity among coinfecting parasites. Am. Nat. 180, 306–315. ( 10.1086/666985) [DOI] [PubMed] [Google Scholar]
- 65.Hawlena H, Bashey F, Lively CM. 2010. The evolution of spite: population structure and bacteriocin-mediated antagonism in two natural populations of Xenorhabdus bacteria. Evolution 64, 3198–3204. ( 10.1111/j.1558-5646.2010.01070.x) [DOI] [PubMed] [Google Scholar]
- 66.Hawlena H, Bashey F, Mendes Soares H, Lively CM. 2010. Spiteful interactions in a natural population of the bacterium Xenorhabdus bovienii. Am. Nat. 175, 374–381. ( 10.1086/650375) [DOI] [PubMed] [Google Scholar]
- 67.Hawlena H, Bashey F, Lively CM. 2012. Bacteriocin-mediated interactions within and between coexisting species. Ecol. Evol. 2, 2521–2526. ( 10.1002/ece3.354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bashey F, Young SK, Hawlena H, Lively CM. 2012. Spiteful interactions between sympatric natural isolates of Xenorhabdus bovienii benefit kin and reduce virulence. J. Evol. Biol. 25, 431–437. ( 10.1111/j.1420-9101.2011.02441.x) [DOI] [PubMed] [Google Scholar]
- 69.Bashey F, Hawlena H, Lively CM. 2013. Alternative paths to success in a parasite community: within-host competition can favor higher virulence or direct interference. Evolution 67, 900–907. ( 10.1111/j.1558-5646.2012.01825.x) [DOI] [PubMed] [Google Scholar]
- 70.Therese MO, Bashey F. 2012. Natal-host environmental effects on juvenile size, transmission success, and operational sex ratio in the entomopathogenic nematode Steinernema carpocapsae. J. Parasitol. 98, 1095–1100. ( 10.1645/GE-3069.1) [DOI] [PubMed] [Google Scholar]
- 71.Kummerli R, Jiricny N, Clarke LS, West SA, Griffin AS. 2009. Phenotypic plasticity of a cooperative behaviour in bacteria. J. Evol. Biol. 22, 589–598. ( 10.1111/j.1420-9101.2008.01666.x) [DOI] [PubMed] [Google Scholar]
- 72.Ghoul M, West SA, Diggle SP, Griffin AS. 2014. An experimental test of whether cheating is context dependent. J. Evol. Biol. 27, 551–556. ( 10.1111/jeb.12319) [DOI] [PubMed] [Google Scholar]
- 73.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84, 499–510. ( 10.1016/S0300-9084(02)01422-0) [DOI] [PubMed] [Google Scholar]
- 74.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. ( 10.1038/nature10244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Inglis RF, Roberts PG, Gardner A, Buckling A. 2011. Spite and the scale of competition in Pseudomonas aeruginosa. Am. Nat. 178, 276–285. ( 10.1086/660827) [DOI] [PubMed] [Google Scholar]
- 76.Inglis RF, Brown SP, Buckling A. 2012. Spite versus cheats: competition among social strategies shapes virulence in Pseudomonas aeruginosa. Evolution 66, 3472–3484. ( 10.1111/j.1558-5646.2012.01706.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. ( 10.1016/j.cell.2013.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bakkal S, Robinson SM, Ordonez CL, Waltz DA, Riley MA. 2010. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 156, 2058–2067. ( 10.1099/mic.0.036848-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Köhler T, Buckling A, van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl Acad. Sci. USA 106, 6339–6344. ( 10.1073/pnas.0811741106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 187, 2138–2147. ( 10.1128/JB.187.6.2138-2147.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Roode JC, Yates AJ, Altizer S. 2008. Virulence-transmission trade-offs and population divergence in virulence in a naturally occurring butterfly parasite. Proc. Natl Acad. Sci. USA 105, 7489–7494. ( 10.1073/pnas.0710909105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ebert D. 1994. Virulence and local adaptation of a horizontally transmitted parasite. Science 265, 1084–1086. ( 10.1126/science.265.5175.1084) [DOI] [PubMed] [Google Scholar]
- 83.Buckling A, Brockhurst M. 2008. Kin selection and the evolution of virulence. Heredity 100, 484–488. ( 10.1038/sj.hdy.6801093) [DOI] [PubMed] [Google Scholar]
- 84.Bremermann HJ, Pickering J. 1983. A game-theoretical model of parasite virulence. J. Theor. Biol. 100, 411–426. ( 10.1016/0022-5193(83)90438-1) [DOI] [PubMed] [Google Scholar]
- 85.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37–78. ( 10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 86.Chao L, Hanley KA, Burch CL, Dahlberg C, Turner PE. 2000. Kin selection and parasite evolution: higher and lower virulence with hard and soft selection. Q. Rev. Biol. 75, 261–275. ( 10.1086/393499) [DOI] [PubMed] [Google Scholar]
- 87.Ewald PW. 1994. Evolution of infectious disease, p. 298 Oxford, UK: Oxford University Press. [Google Scholar]
- 88.Alizon S, van Baalen M. 2008. Multiple infections, immune dynamics, and the evolution of virulence. Am. Nat. 172, E150–E168. ( 10.1086/590958) [DOI] [PubMed] [Google Scholar]
- 89.Alizon S, Lion S. 2011. Within-host parasite cooperation and the evolution of virulence. Proc. R. Soc. B 278, 3738–3747. ( 10.1098/rspb.2011.0471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vigneux F, Bashey F, Sicard M, Lively CM. 2008. Low migration decreases interference competition among parasites and increases virulence. J. Evol. Biol. 21, 1245–1251. ( 10.1111/j.1420-9101.2008.01576.x) [DOI] [PubMed] [Google Scholar]
- 91.Gardner A, West SA, Buckling A. 2004. Bacteriocins, spite and virulence. Proc. R. Soc. Lond. B 271, 1529–1535. ( 10.1098/rspb.2004.2756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sofonea MT, Alizon S, Michalakis Y. 2015. From within-host interactions to epidemiological competition: a general model for multiple infections. Phil. Trans. R. Soc. B 370, 20140303 ( 10.1098/rstb.2014.0303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lysenko ES, Lijek RS, Brown SP, Weiser JN. 2010. Within-host competition drives selection for the capsule virulence determinant of Streptococcus pneumoniae. Curr. Biol. 20, 1222–1226. ( 10.1016/j.cub.2010.05.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barrett LG, Bell T, Dwyer G, Bergelson J. 2011. Cheating, trade-offs and the evolution of aggressiveness in a natural pathogen population. Ecol. Lett. 14, 1149–1157. ( 10.1111/j.1461-0248.2011.01687.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mikonranta L, Friman VP, Laakso J. 2012. Life history trade-offs and relaxed selection can decrease bacterial virulence in environmental reservoirs. PLoS ONE 7, e43801 ( 10.1371/journal.pone.0043801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koch G, Yepes A, Forstner KU, Wermser C, Stengel ST, Modamio J, Ohlsen K, Foster KR, Lopez D. 2014. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158, 1060–1071. ( 10.1016/j.cell.2014.06.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singer M. 2010. Pathogen-pathogen interaction: a syndemic model of complex biosocial processes in disease. Virulence 1, 10–18. ( 10.4161/viru.1.1.9933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Balmer O, Stearns SC, Schötzau A, Brun R. 2009. Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology 90, 3367–3378. ( 10.1890/08-2291.1) [DOI] [PubMed] [Google Scholar]
- 99.Massey R, Buckling A, ffrench-Constant R. 2004. Interference competition and parasite virulence. Proc. R. Soc. Lond. B 271, 785–788. ( 10.1098/rspb.2004.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gillor O, Etzion A, Riley MA. 2008. The dual role of bacteriocins as anti- and probiotics. Appl. Microbiol. Biotechnol. 81, 591–606. ( 10.1007/s00253-008-1726-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. ( 10.1038/nrmicro2937) [DOI] [PubMed] [Google Scholar]
- 102.Riley MA, Robinson SM, Roy CM, Dennis M, Liu V, Dorit RL. 2012. Resistance is futile: the bacteriocin model for addressing the antibiotic resistance challenge. Biochem. Soc. Trans. 40, 1438–1442. ( 10.1042/BST20120179) [DOI] [PubMed] [Google Scholar]
- 103.Scholl D, Martin DW. 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob Agents Chemother 52, 1647–1652. ( 10.1128/AAC.01479-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74, 3868–3876. ( 10.1128/aem.00141-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 15, 1095–1103. ( 10.1111/j.1461-0248.2012.01831.x) [DOI] [PubMed] [Google Scholar]
- 106.Crespi B, Foster K, Ubeda F. 2014. First principles of Hamiltonian medicine. Phil. Trans. R. Soc. B 369, 20130366 ( 10.1098/rstb.2013.0366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Köhler T, Perron GG, Buckling A, van Delden C. 2010. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 6, e1000883 ( 10.1371/journal.ppat.1000883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Allen RC, Popat R, Diggle SP, Brown SP. 2014. Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12, 300–308. ( 10.1038/nrmicro3232) [DOI] [PubMed] [Google Scholar]


