Abstract
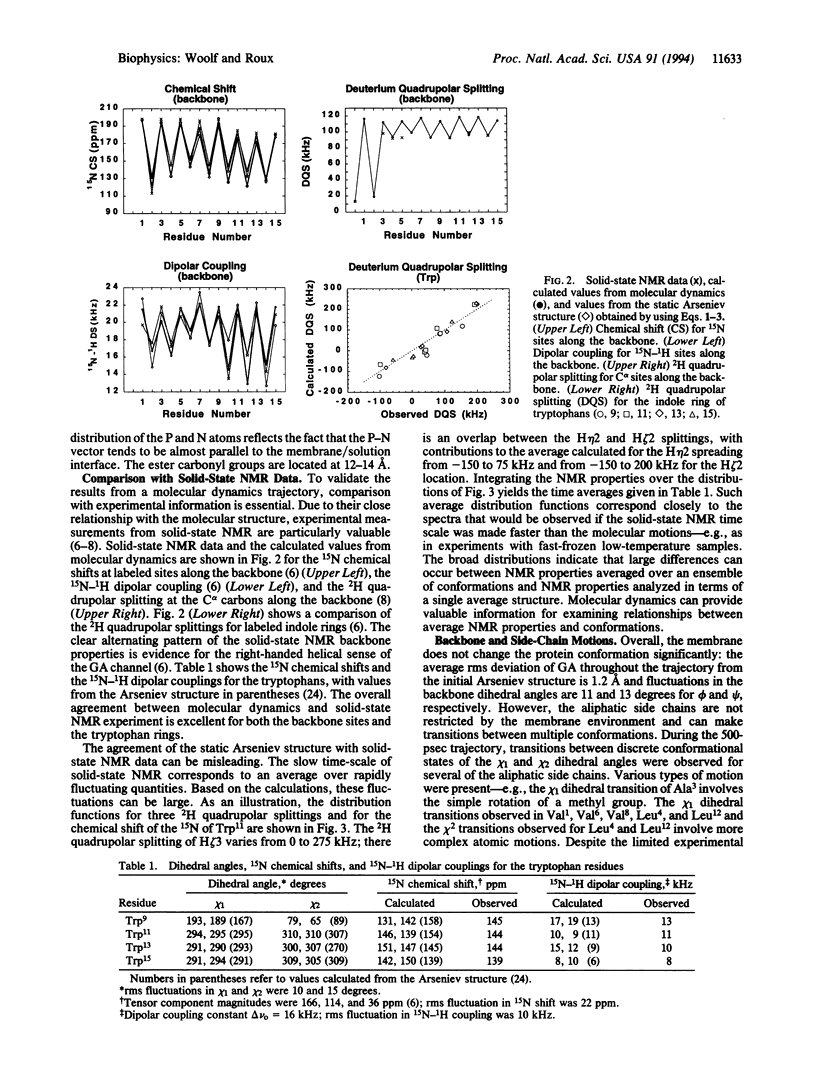
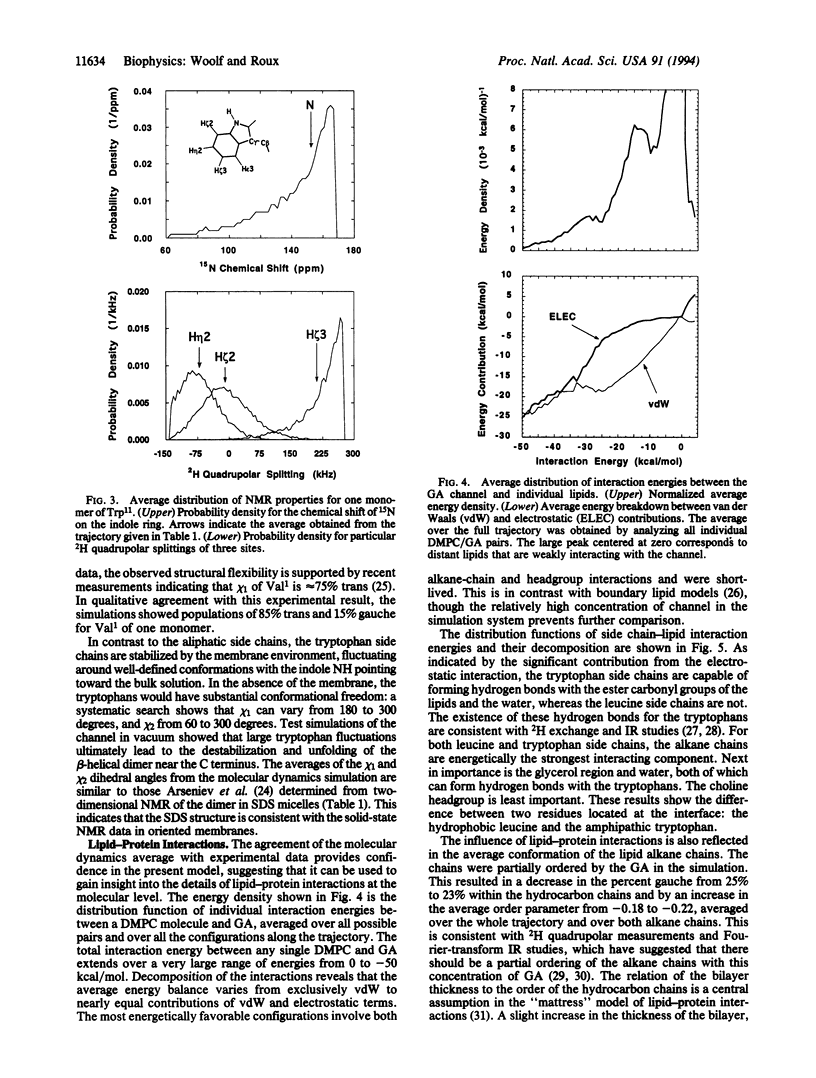
A molecular dynamics simulation of the gramicidin A channel in an explicit dimyristoyl phosphatidylcholine bilayer was generated to study the details of lipid-protein interactions at the microscopic level. Solid-state NMR properties of the channel averaged over the 500-psec trajectory are in excellent agreement with available experimental data. In contrast with the assumptions of macroscopic models, the membrane/solution interface region is found to be at least 12 A thick. The tryptophan side chains, located within the interface, are found to form hydrogen bonds with the ester carbonyl groups of the lipids and with water, suggesting their important contribution to the stability of membrane proteins. Individual lipid-protein interactions are seen to vary from near 0 to -50 kcal/mol. The most strongly interacting conformations are short-lived and have a nearly equal contribution from both van der Waals and electrostatic energies. This approach for performing molecular dynamics simulations of membrane proteins in explicit phospholipid bilayers should help in studying the structure, dynamics, and energetics of lipid-protein interactions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Bechinger B., Seelig J. Conformational changes of the phosphatidylcholine headgroup due to membrane dehydration. A 2H-NMR study. Chem Phys Lipids. 1991 May-Jun;58(1-2):1–5. doi: 10.1016/0009-3084(91)90105-k. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Greathouse D. V., Koeppe R. E., 2nd, Andersen O. S. Amino acid sequence modulation of gramicidin channel function: effects of tryptophan-to-phenylalanine substitutions on the single-channel conductance and duration. Biochemistry. 1991 Sep 10;30(36):8830–8839. doi: 10.1021/bi00100a015. [DOI] [PubMed] [Google Scholar]
- Bouchard M., Auger M. Solvent history dependence of gramicidin-lipid interactions: a Raman and infrared spectroscopic study. Biophys J. 1993 Dec;65(6):2484–2492. doi: 10.1016/S0006-3495(93)81300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Damodaran K. V., Merz K. M., Jr, Gaber B. P. Structure and dynamics of the dilauroylphosphatidylethanolamine lipid bilayer. Biochemistry. 1992 Aug 25;31(33):7656–7664. doi: 10.1021/bi00148a029. [DOI] [PubMed] [Google Scholar]
- Davies M. A., Brauner J. W., Schuster H. F., Mendelsohn R. A quantitative infrared determination of acyl chain conformation in gramicidin/dipalmitoylphosphatidylcholine mixtures. Biochem Biophys Res Commun. 1990 Apr 16;168(1):85–90. doi: 10.1016/0006-291x(90)91678-l. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. The Photosynthetic Reaction Center from the Purple Bacterium Rhodopseudomonas viridis. Science. 1989 Sep 29;245(4925):1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Jacobs R. E., White S. H. The nature of the hydrophobic binding of small peptides at the bilayer interface: implications for the insertion of transbilayer helices. Biochemistry. 1989 Apr 18;28(8):3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Lee K. C., Cross T. A. Side-chain structure and dynamics at the lipid-protein interface: Val1 of the gramicidin A channel. Biophys J. 1994 May;66(5):1380–1387. doi: 10.1016/S0006-3495(94)80928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. A., Engelman D. M. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983 May 15;166(2):211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- Moll F., 3rd, Cross T. A. Optimizing and characterizing alignment of oriented lipid bilayers containing gramicidin D. Biophys J. 1990 Feb;57(2):351–362. doi: 10.1016/S0006-3495(90)82536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouritsen O. G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys J. 1984 Aug;46(2):141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owicki J. C., Springgate M. W., McConnell H. M. Theoretical study of protein--lipid interactions in bilayer membranes. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1616–1619. doi: 10.1073/pnas.75.4.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascher I., Lundmark M., Nyholm P. G., Sundell S. Crystal structures of membrane lipids. Biochim Biophys Acta. 1992 Dec 11;1113(3-4):339–373. doi: 10.1016/0304-4157(92)90006-v. [DOI] [PubMed] [Google Scholar]
- Pastor R. W., Venable R. M., Karplus M. Model for the structure of the lipid bilayer. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):892–896. doi: 10.1073/pnas.88.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser R. S., Davis J. H., Dahlquist F. W., Lindorfer M. A. 2H nuclear magnetic resonance of the gramicidin A backbone in a phospholipid bilayer. Biochemistry. 1991 May 14;30(19):4687–4696. doi: 10.1021/bi00233a008. [DOI] [PubMed] [Google Scholar]
- Rice D., Oldfield E. Deuterium nuclear magnetic resonance studies of the interaction between dimyristoylphosphatidylcholine and gramicidin A'. Biochemistry. 1979 Jul 24;18(15):3272–3279. doi: 10.1021/bi00582a012. [DOI] [PubMed] [Google Scholar]
- Roux B., Karplus M. Molecular dynamics simulations of the gramicidin channel. Annu Rev Biophys Biomol Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- Seelig J. 31P nuclear magnetic resonance and the head group structure of phospholipids in membranes. Biochim Biophys Acta. 1978 Jul 31;515(2):105–140. doi: 10.1016/0304-4157(78)90001-1. [DOI] [PubMed] [Google Scholar]
- Smith R., Thomas D. E., Separovic F., Atkins A. R., Cornell B. A. Determination of the structure of a membrane-incorporated ion channel. Solid-state nuclear magnetic resonance studies of gramicidin A. Biophys J. 1989 Aug;56(2):307–314. doi: 10.1016/S0006-3495(89)82677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swairjo M. A., Seaton B. A. Annexin structure and membrane interactions: a molecular perspective. Annu Rev Biophys Biomol Struct. 1994;23:193–213. doi: 10.1146/annurev.bb.23.060194.001205. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Nemoto Y., Harada I. Environments and conformations of tryptophan side chains of gramicidin A in phospholipid bilayers studied by Raman spectroscopy. Biochemistry. 1990 Feb 13;29(6):1572–1579. doi: 10.1021/bi00458a031. [DOI] [PubMed] [Google Scholar]
- Venable R. M., Zhang Y., Hardy B. J., Pastor R. W. Molecular dynamics simulations of a lipid bilayer and of hexadecane: an investigation of membrane fluidity. Science. 1993 Oct 8;262(5131):223–226. doi: 10.1126/science.8211140. [DOI] [PubMed] [Google Scholar]
- Weiss M. S., Schulz G. E. Structure of porin refined at 1.8 A resolution. J Mol Biol. 1992 Sep 20;227(2):493–509. doi: 10.1016/0022-2836(92)90903-w. [DOI] [PubMed] [Google Scholar]
- Zaccai G., Büldt G., Seelig A., Seelig J. Neutron diffraction studies on phosphatidylcholine model membranes. II. Chain conformation and segmental disorder. J Mol Biol. 1979 Nov 15;134(4):693–706. doi: 10.1016/0022-2836(79)90480-7. [DOI] [PubMed] [Google Scholar]




