Abstract
Colour discrimination in vertebrates requires cone photoreceptor cells in the retina, and high-acuity colour vision is endowed by a set of four cone subtypes expressing UV-, blue-, green- and red-sensitive opsins. Previous studies identified transcription factors governing cone photoreceptor development in mice, although loss of blue and green opsin genes in the evolution of mammals make it difficult to understand how high-acuity colour vision was organized during evolution and development. Zebrafish (Danio rerio) represents a valuable vertebrate model for studying colour vision as it retains all the four ancestral vertebrate cone subtypes. Here, by RT-qPCR and in situ hybridization analysis, we found that sine oculis homeobox homolog 7 (six7), a transcription factor widely conserved in ray-finned fish, is expressed predominantly in the cone photoreceptors in zebrafish at both the larval and the adult stages. TAL effector nuclease-based six7 knock-out revealed its roles in expression of green, red and blue cone opsin genes. Most prominently, the six7 deficiency caused a loss of expression of all the green opsins at both the larval and adult stages. six7 is indispensable for the development and/or maintenance of the green cones.
Keywords: photoreceptor development, cone opsin, green opsin, six7, zebrafish (Danio rerio)
1. Introduction
In vertebrates, vision is triggered by absorption of photons by visual opsins localized in outer segments of retinal photoreceptor cells. The photoreceptor cells are classified into rod and cone cells, which are distinct from each other in morphology and photoresponse [1–3]. Rods are responsible for twilight vision, while cones function under the daylight condition. Combination of multiple cone subtypes (i.e. UV-, blue-, green- and red-sensitive cones) confers many vertebrates with high-acuity colour vision [4,5], which is based on mutually exclusive expression of one subtype of the opsin in single cells. One of the key vertebrate species that helps us understand visual system evolution is the Southern Hemisphere lamprey, Geotria australis: it is a jawless vertebrate whose ancestor diverged from the lineage leading to jawed vertebrates probably before 500 Ma [6], and it has four subtypes of the cone opsins [7], namely UV (short wavelength sensitive 1: SWS1), blue (short wavelength sensitive 2: SWS2), green (medium wavelength sensitive: RH2) and red (long wavelength sensitive: M/LWS). The ancestral species of the jawed vertebrate is thus thought to have acquired four spectrally distinct cone subtypes by two rounds of chromosome duplications and appears to have had the tetrachromatic colour vision system [8].
The developmental process of visual photoreceptors has been well studied in mice, and important transcription factors governing the development of the photoreceptors have been identified [9,10]. In particular, cone-rod homeobox (Crx) is a master transcriptional regulator for differentiation of retinal progenitor cells into photoreceptor cells [11–13]. Crx determines the cell fate of the photoreceptor progenitor cells in combination with other transcription factors such as thyroid hormone receptor beta (Thrb), which regulates development of the cone photoreceptors expressing M/LWS opsin gene [14]. On the other hand, transcriptional regulatory mechanisms of SWS2 and RH2 opsin genes remain elusive because mammals lost SWS2 and RH2 opsin genes during evolution. Hence, the vertebrate species having all four cone subtypes provide an excellent platform to study detailed mechanisms of the development of the four cone photoreceptors.
Zebrafish (Danio rerio) is a diurnal teleost having four cone subtypes, and it is a valuable animal model to study genetic mechanisms of vertebrate retinal development and diseases [15]. Recent studies isolated the promoter and/or enhancer regions sufficient for exclusive expression of the zebrafish cone opsin genes in a specific subtype of cones, such as UV-(sws1) [16], blue-(sws2) [17], green-(rh2-1, rh2-2, rh2-3 and rh2-4) [18] or red-sensitive (lws1 and lws2) cones [19]. Two transcription factors, T-box 2b (tbx2b) and thrb, were identified to be crucial for expression of a particular subtype of cone opsins in zebrafish: tbx2b is required for gene expression of sws1 [20], while thrb is involved in the expression of lws1 and lws2 [21]. The role of thrb in M/LWS opsin gene expression is conserved between mouse and zebrafish, suggesting that these two vertebrate species share a mechanism underlying exclusive expression of the opsin genes in cone cells. Despite these advances, studies so far have been unable to identify any transcription factors responsible for expression of SWS2 and RH2 in vertebrates.
To understand regulation of sws2 and rh2 opsin genes, we searched for cone-specific transcription factors in zebrafish and found that sine oculis homeobox homolog 7 (six7) is predominantly expressed in cone photoreceptors. The Six family is a group of evolutionarily conserved transcription factors found in diverse organisms ranging from worms to humans [22]. Previous studies reported that six7 is expressed during gastrulation and early segmentation [23], and that combined inactivation of two Six3/6/7 group members, six7 and six3b, results in loss of the eyes [24]. Despite the involvement of six7 in early eye development, it still remains unclear whether and how six7 is engaged in cone photoreceptor development. We investigated roles of six7 in cone development and in cone opsin expression by generating six7 knock-out (KO) zebrafish with TAL effector nucleases (TALENs). The six7 KO zebrafish lost expression of the green opsins at both larval and adult stages, suggesting that six7 is indispensable for the development of the green cones.
2. Material and methods
Full methods and any associated references are available in the electronic supplementary material, methods.
(a). Zebrafish
Zebrafish were treated in accordance with the guidelines of the University of Tokyo. The wild-type (WT) zebrafish strain EkkWill were raised in a 14 L : 10 D cycle, and fed twice per day with living baby brine shrimps. Embryos were raised at 28.5°C in egg water (artificial seawater diluted 1.5 : 1000 in water).
(b). Construction of TAL effector nuclease targeting-vector
We designed TAL effector repeats recognizing exon 1 of zebrafish six7 gene with the Golden Gate method [25]. These repeats were cloned into our original vectors (pTAL7DD and pTAL7RR), which were modified from pTAL3 vector (Addgene plasmid 31034), based on pCS2TAL3DD and pCS2TAL3RR [26]. Information about construction of pTAL7DD and pTAL7RR vectors is in the electronic supplementary material, methods.
(c). Immunohistochemistry and in situ hybridization
Immunohistochemistry and in situ hybridization were performed as described previously [27–30] with some modifications. Detailed information about the methods is in the electronic supplementary material, methods.
(d). Isolation of rods and cones
EGFP-positive rods and cones were isolated by fluorescence activating cell sorter (FACS) from the adult retinas of the two lines of transgenic zebrafish, Tg(rho:egfp)ja2 [31] and Tg(gnat2:egfp)ja23, respectively. These lines express green fluorescent proteins in rods and all the cone subtypes, respectively. Tg(gnat2:egfp)ja23 was generated according to the method described in the previous study [32]. Detailed procedures for isolation of photoreceptors are described in the electronic supplementary material, methods.
(e). RNA extraction and RT-qPCR
Total RNA was extracted and purified using RNeasy Micro Kit (74004, Qiagen) from the eyes of adult or larval zebrafish. The ocular RNA (450 ng for the adult eye or 90 ng for the larval eye) was reverse-transcribed into cDNA using the oligo (dT)15 primer with GoScript Reverse Transcriptase (A5003, Promega). The cDNA was subjected to quantitative PCR using GoTaq qPCR Master Mix (A6001, Promega) with the StepOnePlus Real-time PCR system (Applied Biosystems), following the manufacturer's protocol. The gene-specific primers used for RT-qPCR are listed in the electronic supplementary material, table S3. In all the figures, the mRNA levels were normalized to beta-actin 2 (actb2) mRNA levels.
3. Results
(a). Cone-specific expression of six7
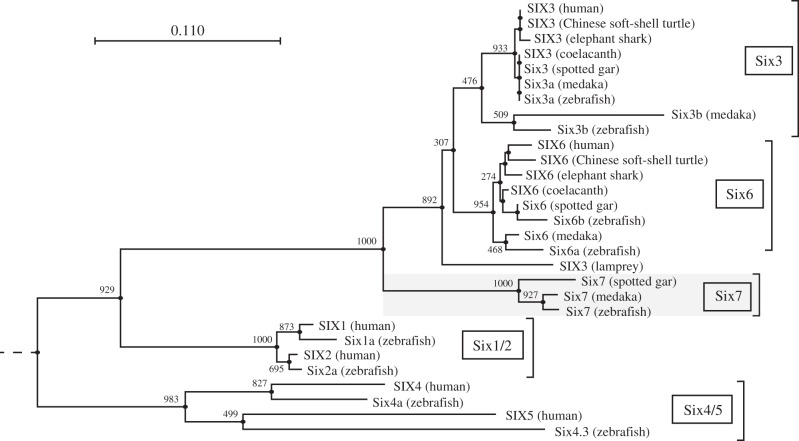
In microarray analysis, we compared gene expression profiles between rod and cone cells purified from the adult zebrafish retina (yet to be published). Among approximately 500 genes that were found to be abundantly expressed in cones (more than 10 times higher than in rods), we focused on sine oculis homeobox homolog 7 (six7), whose signal intensity in cones was the highest among the cone-enriched transcription factors in our microarray analysis. Six7 belongs to Six family, and Six7 is found in ray-finned fish but not in mammals (figure 1). The phylogenetic tree showed that, among Six3/6/7 group, Six7 subfamily was separated before the divergence between Six3 and Six6 subfamilies, both of which are widely conserved in vertebrate species (figure 1).
Figure 1.
Phylogenetic tree of the members in Six family. A neighbour-joining (NJ) tree was constructed from amino acid sequences of SIX domain and homeodomain with 1000 bootstrapping replications. Numerical values indicating bootstrap support are shown at the base of each node. Nematode CEH-34 was employed as an outgroup. Scale bar, 0.11 substitutions per site. A phylogenetic tree including additional members of Six1/2 and Six4/5 groups is presented in the electronic supplementary material, figure S6a.
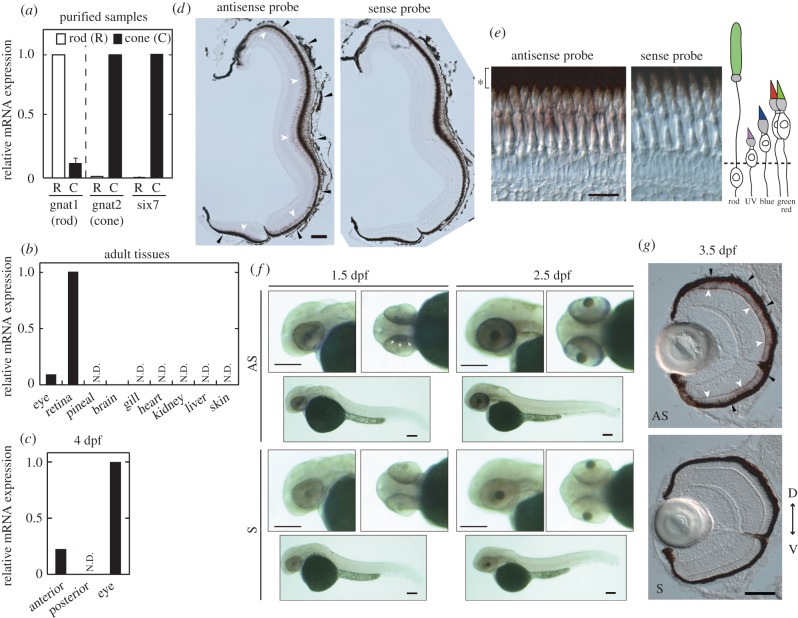
We investigated the expression pattern of six7 in zebrafish. RT-qPCR assay confirmed that six7 was far more abundantly expressed in cones than in rods (more than 10 times; figure 2a). Among the adult tissues, six7 was predominantly expressed in the retina (figure 2b). In situ hybridization analysis of the retina indicated that six7 was expressed specifically in the photoreceptor layer (figure 2d). In particular, the six7-positive cells were mainly located in the layer of the red and green cones (figure 2e), suggesting that six7 regulates transcription in the green and red cones.
Figure 2.
Expression patterns of six7 at the larval and adult stages. (a–c) Relative expression levels of six7 were quantified by RT-qPCR using the following samples: (a) rods and cones isolated from Tg(rho:egfp) and Tg(gnat2:egfp) zebrafish, respectively, at the adult stage (n = 2), (b) adult tissues and (c) larval anterior segments, posterior segments and eyes at 4 days post fertilization (4 dpf). N.D., not detected. (d) In situ hybridization using cryosections of the light-adapted adult zebrafish retina at 7 months post-fertilization (mpf). The six7 expression was detected in the photoreceptor layer (white arrowheads), which is adjacent to the retinal pigmented epithelium (RPE, black arrowheads). Scale bar, 100 µm. (e) Magnified images of (d). The five subtypes of photoreceptor cells are located in distinct layers of the light-adapted retina (as illustrated in the schematic drawing). RPE is indicated by the asterisk. Scale bar, 20 µm. (f) Whole-mount in situ hybridization using larval zebrafish. The six7 expression was observed at both 1.5 dpf (arrowheads) and 2.5 dpf. Scale bars, 100 µm. (g) In situ hybridization using cryosections of the 3.5-dpf larval zebrafish. D, dorsal side; V, ventral side. Scale bar, 50 µm. AS, antisense probe; S, sense probe. (Online version in colour.)
We then analysed expression profiles of six7 during the retinal development. At 4 days post fertilization (4 dpf), six7 expression was detected in the anterior regions of the larvae (including the eyes), but not in the posterior region (figure 2c). Eye-specific expression of six7 was confirmed by whole-mount in situ hybridization (figure 2f). The six7 expression in the eye began at 1.5 dpf, when the retinal development just starts, suggesting the six7 expression in the retinal progenitor cells. In situ hybridization using 3.5 dpf zebrafish ocular sections revealed that six7-positive cells were localized in the photoreceptor cell layer, in which the positive cells were distributed uniformly across the whole retina (figure 2g). Considering that rods are tightly clustered together in the ventral retina at this developmental stage [33] (see also figure 3c), the expression pattern of six7 was similar to that of the cone-specific phototransduction component genes. Although we cannot rule out the possibility that six7 is also expressed in rods at the larval stages, these results imply that six7 is predominantly expressed in cones at 3.5 dpf and suggest that six7 plays a role in the cone development.
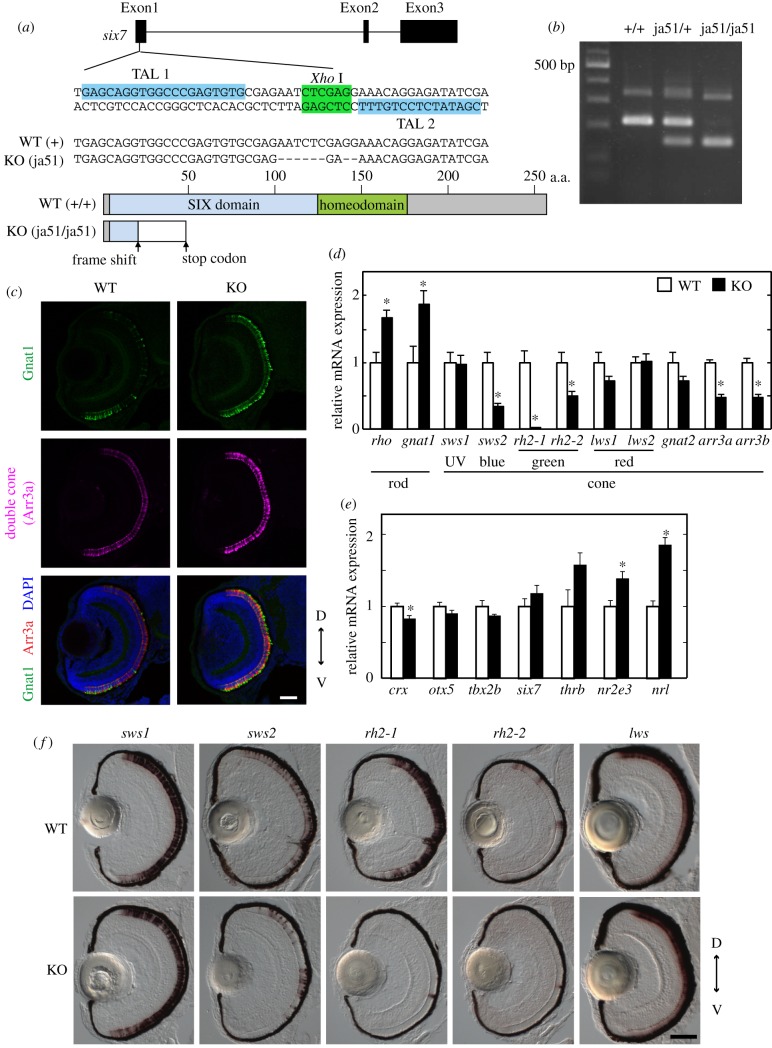
Figure 3.
Analysis of the WT zebrafish and the six7 KO zebrafish at the larval stage. (a) Top: exon–intron organization and partial nucleotide sequences of zebrafish six7. The binding sites of the left and right TALENs are highlighted in blue. The recognition site of the endonuclease XhoI is highlighted in green. Middle: the nucleotide sequence of the KO (ja51 mutant) fish is compared with the WT sequence. Deletions are indicated by dashes. Bottom: schematic of Six7 and its mutant protein. (b) Genotyping of the six7 KO zebrafish. See details in the electronic supplementary material, methods. (c) Immunofluorescence of ocular sections of the WT and KO larvae at 3.5 dpf. The ocular sections were immunostained with the anti-Gnat1 antibody for rods (green) and with the zpr1 antibody for double cones (i.e. red and green cones; magenta). DAPI staining identified all the cell nuclei (blue). Scale bar, 40 µm. (d,e) Expression profiles of the phototransduction component genes (d) and the transcription factors contributing to the photoreceptor development in mouse and/or zebrafish (e) in the WT and six7 KO larval eyes at 3.5 dpf. The mRNA levels were quantified by RT-qPCR. The data were represented by mean + s.e.m. (n = 5). Statistical significance between WT and six7 KO data is shown by the asterisks (*p < 0.05 by Student's t-test). (f) Expression profiles of the cone opsin genes by in situ hybridization using the larval eyes of the WT and KO at 3.5 dpf. The lws probe was designed to recognize both of the lws1 and lws2 opsin genes. D, dorsal side; V, ventral side. Scale bar, 50 µm. (Online version in colour.)
(b). Decrease of green and blue cones in the larva of six7 knock-out zebrafish
To examine the role of six7 in photoreceptor development, we generated KO zebrafish with TALENs. The KO zebrafish had a mutation in the six7 coding region: the ja51 mutation caused a frame shift of the amino acid sequence of Six7 by 8 bp loss, thereby introducing an immature stop codon (figure 3a,b). The KO zebrafish apparently had the normal body shape throughout the developmental stages (data not shown).
We examined expression levels of phototransduction component genes in the eyes of the WT zebrafish and six7ja51/ja51 KO zebrafish larvae by RT-qPCR. Notably, mRNA levels of the middle wavelength-sensitive opsins, green opsins (rh2-1 and rh2-2) and blue opsin (sws2) were all reduced in 3.5 dpf six7ja51/ja51 larvae (figure 3d). Like other teleosts, zebrafish has multiple copies of green opsin genes, rh2-1, rh2-2, rh2-3 and rh2-4, among which rh2-3 and rh2-4 are expressed at extremely low levels during larval stage in WT [34] and also in six7ja51/ja51 (data not shown).
In situ hybridization analysis (figure 3f) revealed that green opsin expression was hardly detected in the eye of six7ja51/ja51 larvae, while blue opsin expression was reduced approximately by half in six7ja51/ja51 larvae, suggesting decreases in the numbers of middle wavelength-sensitive cones (i.e. green and blue cones). Consistently, six7ja51/ja51 larvae exhibited reduced mRNA levels of arrestin 3a (arr3a) and arrestin 3b (arr3b) (figure 3d), which are expressed in the double cones (green and red cones) and the single cones (UV and blue cones), respectively [35]. This observation further supports the reduction of the number of blue and green cones in six7ja51/ja51 larvae at 3.5 dpf.
The reduction in larval mRNA levels of blue and green opsins was also observed in another six7 mutant line, six7ja52/ja52(electronic supplementary material, figure S1b). Using this line, we generated six7ja52/ja52;six3bja53/ja53 double KO larvae and found that the double mutant had extremely small eyes (electronic supplementary material, figure S1e): the loss of the eyes recapitulated the morpholino-induced phenotypes of six7/six3b-deficient larvae reported in the previous study [24], supporting that the six7 deficiency is caused by the ja52 mutation. The expression profiles of opsin genes in six7ja52/ja52 larvae were quite similar to those in six7ja51/ja51 larvae, suggesting that the phenotype of six7 mutants did not arise from any off-target effect of TALENs but from the six7 deficiency. In the following experiments, we used six7ja51/ja51 zebrafish, which we refer to as six7 KO.
(c). Increase of rod cells in the larva of six7 knock-out zebrafish
In contrast to the remarkable reduction in the number of blue and green cones, the mRNA levels of rod-specific phototransduction component genes, rhodopsin (rho) and rod transducin α-subunit (gnat1), were upregulated in six7 KO larvae as compared with WT (figure 3d). The number of Gnat1-positive cells was increased in KO larvae when assessed by immunohistochemistry with anti-Gnat1 antibody (figure 3c). It is also evident that six7 KO exhibited increased mRNA levels of rod-specific transcription factors, neural retina leucine zipper (nrl) and nuclear receptor subfamily 2, group E, member 3 (nr2e3) (figure 3e). These two transcription factors are both essential for rod development in mice [36,37]. Together, these data support the notion that KO larvae had many more rods than WT larvae at 3.5 dpf.
(d). Decrease of green cones in adult six7 knock-out zebrafish
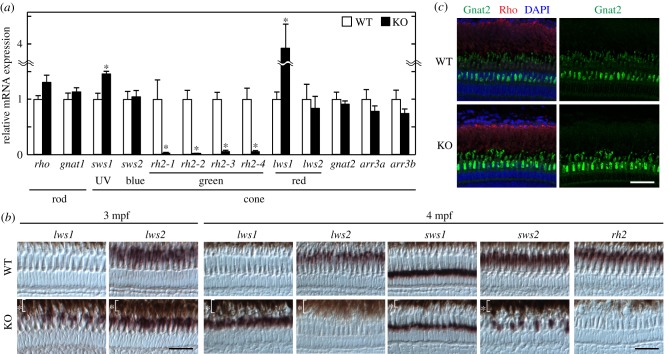
To examine whether the number of photoreceptors expressing green opsins are reduced throughout developmental stages, we analysed expression profiles of opsin genes in adult zebrafish. The number of green cones expressing rh2-1, rh2-2, rh2-3 or rh2-4 was severely reduced in six7 KO at the adult stage (figure 4a,b), indicating that transcription of all the green opsin genes was suppressed throughout developmental stages in KO. The reduction of the number of green cone cells in KO at both adult and larval stages suggests that six7 is indispensable for development of green cones.
Figure 4.
Analysis of the WT zebrafish and the six7 KO zebrafish at the adult stage. (a) mRNA expression levels of phototransduction component genes in the WT and six7 KO eyes at 3 mpf. The RT-qPCR data were represented by mean + s.e.m. (n = 4). Statistical significance between WT and six7 KO data is shown by the asterisks (*p < 0.05 by Student's t-test). (b) Expression profiles of cone opsin genes in the central retina of the adult WT and KO by in situ hybridization. All subsets of green cones were stained with a mixture of four different RNA probes, each of which specifically recognizes the corresponding subtype of the green opsin (rh2) gene. RPE is indicated by the asterisks. Demagnified images are shown in the electronic supplementary material, figure S2. Scale bars, 30 µm. (c) Fluorescent images in the central retina of the adult WT and KO at 3 mpf. The cryosections of the adult zebrafish retina were immunostained with the anti-Gnat2 antibody for cones (green) and with the anti-rhodopsin antibody for rods (red). DAPI staining identified all the cell nuclei (blue). Scale bar, 40 µm. (Online version in colour.)
(e). Developmental stage-dependent phenotypes of blue cones and rods in six7 knock-out zebrafish
The mRNA levels of rod-specific genes, rho and gnat1, were upregulated in six7 KO larvae (figure 3d), but at the adult stage, they were not significantly different between WT and KO (figure 4a). Similarly, the suppressed expression of blue opsin gene (sws2) in six7 KO was observed only at the larval stage (figure 3d) but not at the adult stage (figure 4a). The number of blue cones was reduced in the adult KO central retina, where early differentiated photoreceptors were located (figure 4b; electronic supplementary material, figure S2), suggesting that six7 is necessary for the development of blue cones only during the larval stage.
(f) Ectopic expression of red opsin gene lws1 in the central retina of adult six7 knock-out zebrafish
Zebrafish have two duplicated red opsin genes, lws1 and lws2, and their expression is spatially regulated in the retina [19,34]. In the eyes of adult six7 KO, the lws1 mRNA level was markedly upregulated (figure 4a), in contrast to its normal expression in six7 KO larvae (figure 3d). Spatial expression pattern of lws1 in the adult KO retina was also distinct from that in the adult WT retina. In WT, the lws1 expression was confined to the peripheral retina, while the lws2 expression was more localized in the central retina (figure 4b; electronic supplementary material, figures S2 and S4b). In six7 KO, the lws1-expressing cells were observed across the whole retina (figure 4b; electronic supplementary material, figures S2 and S4b). At this adult stage (3 mpf), the number of lws2-expressing cells in the central retina was mostly unaffected by the loss of six7 (figure 4b), as is consistent with the RT-qPCR analysis (figure 4a), but surprisingly, at the later stage (4 mpf), lws2 showed a severely reduced expression in the central retina of KO (figure 4b; electronic supplementary material, figure S2). Considering that the loss of six7 does not affect the development of red cone, at the larval stage (figure 3) and that six7 KO maintains the laminar structure of the retina (figure 4), we suppose that six7 controls only the expression pattern of the duplicated red opsin genes in the red cones. We further validated the number of the red cones in six7 KO by whole-mount immunohistochemistry with 1D4 antibody as a red cone marker [38] and with anti-Gnat2 antibody to stain all the cone subtypes. The loss of six7 caused reduction in the number of the Gnat2-positive cells in the double cone layer, while apparent increase or decrease in the red cones was not observed in six7 KO retina at 6 mpf (electronic supplementary material, figure S4c). Collectively, the deficiency of six7 is likely to cause gradual switching of the expression of red opsins from lws2 to lws1 in the central retina without affecting the total number of the red cones. These results suggest that six7 also plays an important role in spatio-temporally regulated expression of red opsin subtypes.
(g). Expression levels of transcription factors responsible for photoreceptor development in six7 knock-out zebrafish
Two transcription factors (tbx2b and thrb) are known to contribute to development of UV and red cone cells in zebrafish, respectively [20,21]. The mRNA levels of these transcription factors were mostly unaffected by the loss of six7 at both the larval and adult stages (figure 3e; electronic supplementary material, figure S3). The unaltered expression of these transcription factors indicates that the developmental process of the green cones might be independent of that of the UV and red cones. Importantly, no significant change was observed between WT and KO in the mRNA levels of crx and orthodenticle homolog 5 (otx5) (figure 3e; electronic supplementary material, figure S3), the master transcriptional regulators of photoreceptor cells in zebrafish [39,40]. It should be noted that the loss of six7 did not affect the laminar structure of the larval retina (figure 3c,f), nor the positions and the morphologies of each photoreceptor cells in the adult retina (figure 4c; electronic supplementary material, figure S2). Together, our results indicate that six7 governs the expression of green opsin genes without affecting gene expression of the key transcription factors.
4. Discussion
We investigated the role of six7, which is predominantly expressed in cone photoreceptors throughout the development of zebrafish (figure 2). We generated six7 KO zebrafish and found that six7 was essential for the expression of all subsets of the green opsin genes (figures 3 and 4), which have been duplicated and diverged in the teleost lineage. The loss of six7 caused expression switching between the two red opsin genes, from lws2 to lws1, in the central retina at the adult stage (figure 4). These results demonstrate that six7 is important for the effective detection of the middle- and long-wavelength light by controlling the expression of the green and red opsins in zebrafish. In six3b KO larvae, on the other hand, no significant alteration of mRNA levels of the opsin genes was observed (electronic supplementary material, figure S5), suggesting that six7 plays specialized roles in regulating the expression of these opsin genes, in contrast to the redundant role of six3b and six7 in eye formation [24] (electronic supplementary material, figure S1).
We observed an increase in the number of rods and reduced number of blue cones in six7 KO at the larval stage (figure 3). Recently, Saade et al. [41] also reported an increased number of rods due to the loss of six7. In their report, a forward genetic screen of the genes involved in rod development led to isolation of a mutant called lots-of-rods jr (ljr), which was characterized by an increased number of rods at the larval stage. They mapped the ljr mutation on chromosome 7, in which six7 is located. The morpholino-mediated knockdown of six7 phenocopied the ljr mutation [41]. In this study, by using the six7 KO zebrafish, we analysed the role of six7 in the photoreceptor development at the adult stage. The six7 KO caused the increase in number of rods as well as the suppression of the mRNA level of the blue opsin only at the larval stage (figure 3d), while the number of the green cones was reduced by six7 KO at both the larval and adult stages (figures 3d and 4a). By contrast, the ectopic expression of the red opsin lws1 in the central retina of six7 KO was observed only at the adult stage (figure 4a). These phenotypes observed at either the larval or the adult stage suggest that the differentiation programmes of the retinal progenitor cells change as zebrafish grow, and that six7 contributes to stage-dependent differentiation in photoreceptor development.
So far, six7 has been found widely in the ray-finned fish genomes but not in birds nor mammals. The emergence of the Six7 subfamily at the early evolutionary stage (figure 1; electronic supplementary material, figure S6a) suggests that six7 is lost in some vertebrate species. We noticed a gene termed SIX7 in the Ensembl Genome database of reptilian species, green anole and Chinese soft-shell turtle, and our subsequent tBLASTn searches identified SIX7-like sequences in the NCBI genome database of green sea turtle and Indian python. These reptilian SIX7 sequences are quite diverged from those of ray-finned fish Six7, as is evident from too many amino acid substitutions and gaps in the sequence alignment (electronic supplementary material, figure S6b). Additional genome information will be required to make clear whether these SIX7 genes are true orthologues of ray-finned fish six7.
Although it is likely that six7 directs expression of rh2 (the green opsin) genes in ray-finned fish, SIX7 is lost in birds and elephant shark, both of which retain RH2 genes. In these species, expression of the RH2 gene could be regulated by a six7-independent mechanism. To gain insight into evolutionary aspects of the regulatory mechanisms, we compared the gene synteny around the RH2 gene locus among various vertebrate species (electronic supplementary material, figure S7). Intriguingly, the gene synteny can be classified into two types: (i) the non-teleost type and (ii) the teleost type (electronic supplementary material, figure S7). The non-teleost type was found in the species ranging from elephant shark to tetrapods, suggesting that this synteny represents an ancient type. In the teleost lineage, the genes around RH2 are likely to have been rearranged into the teleost type, in which six7 could have acquired the important role in the expression of rh2 genes.
Previously, Tsujimura et al. [18] identified a 0.5 kbp enhancer region (termed locus control region; LCR) located 15 kbp upstream of the tandem array of rh2 genes, as a regulator essential for expression of all the duplicated rh2 genes in zebrafish [18]. Interestingly, LCR contains the binding sites of both Six7 (TAATGTC) [42] and Crx (TAATC) [11]. The loss of expression of all subsets of rh2 genes in the six7 KO zebrafish (figure 4a,b) suggests that six7 may activate expression of the rh2 genes through the LCR. Teleosts possess a variable number of duplicated rh2 genes [43] (electronic supplementary material, figure S8), whose spectral sensitivities are diverged from each other [44]. The six7 regulation of the duplicated rh2 expression may enable teleosts to adapt to diverse aquatic environments.
Supplementary Material
Acknowledgements
We thank the members of Fukada lab for valuable discussion and also thank S. Kuraku (RIKEN) for valuable comments. We thank members of FACS core laboratory, the University of Tokyo, for support of cell sorting. We also thank D. F. Voytas (University of Minnesota) and B. N. Kennedy (UCD) for supplying plasmids, and ZIRC for providing zpr1 antibody.
Ethics
All research described here adhered to local guidelines and all appropriate ethical approval and licences were obtained.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
T.S. and Y.F. conceived and designed the research. Y.O., supported by T.S., carried out the molecular laboratory work, participated in data analysis and carried out sequence alignments. T.S. performed flow cytometry. D.K. and Y.F. supervised the project. Y.O., T.S., D.K. and Y.F. wrote the manuscript. All authors approved the manuscript for publication.
Competing interests
The authors declare no conflict of interest.
Funding
This work was supported by Grants-in-Aid for scientific research from MEXT, Japan (to D.K. and Y.F.), by Grant for Basic Science Research Projects from the Sumitomo Foundation (to D.K.) and by Research Foundation for Opto-Science and Technology (to D.K.).
References
- 1.Ebrey T, Koutalos Y. 2001. Vertebrate photoreceptors. Prog. Retin. Eye Res. 20, 49–94. ( 10.1016/S1350-9462(00)00014-8) [DOI] [PubMed] [Google Scholar]
- 2.Kawamura S, Tachibanaki S. 2008. Rod and cone photoreceptors: molecular basis of the difference in their physiology. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 150, 369–377. ( 10.1016/j.cbpa.2008.04.600) [DOI] [PubMed] [Google Scholar]
- 3.Lamb TD. 2013. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119. ( 10.1016/j.preteyeres.2013.06.001) [DOI] [PubMed] [Google Scholar]
- 4.Okano T, Fukada Y, Yoshizawa T. 1995. Molecular basis for tetrachromatic color vision. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 112, 405–414. ( 10.1016/0305-0491(95)00085-2) [DOI] [PubMed] [Google Scholar]
- 5.Goldsmith TH. 2013. Evolutionary tinkering with visual photoreception. Vis. Neurosci. 30, 21–37. ( 10.1017/S095252381200003X) [DOI] [PubMed] [Google Scholar]
- 6.Shu D-G, et al. 2003. Head and backbone of the Early Cambrian vertebrate Haikouichthys. Nature 421, 526–529. ( 10.1038/nature01264) [DOI] [PubMed] [Google Scholar]
- 7.Collin SP, Knight MA, Davies WL, Potter IC, Hunt DM, Trezise AEO. 2003. Ancient colour vision: multiple opsin genes in the ancestral vertebrates. Curr. Biol. 13, R864–R865. ( 10.1016/j.cub.2003.10.044) [DOI] [PubMed] [Google Scholar]
- 8.Lagman D, Ocampo Daza D, Widmark J, Abalo XM, Sundström G, Larhammar D. 2013. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol. Biol. 13, 238 ( 10.1186/1471-2148-13-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaroop A, Kim D, Forrest D. 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576. ( 10.1038/nrn2880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adler R, Raymond PA. 2008. Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain Res. 1192, 134–150. ( 10.1016/j.brainres.2007.03.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furukawa T, Morrow EM, Cepko CL. 1997. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531–541. ( 10.1016/S0092-8674(00)80439-0) [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. 1997. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030. ( 10.1016/S0896-6273(00)80394-3) [DOI] [PubMed] [Google Scholar]
- 13.Freund CL, et al. 1997. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91, 543–553. ( 10.1016/S0092-8674(00)80440-7) [DOI] [PubMed] [Google Scholar]
- 14.Ng L, Hurley JB, Dierks B, Srinivas M, Saltó C, Vennström B, Reh TA, Forrest D. 2001. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98. ( 10.1038/83829) [DOI] [PubMed] [Google Scholar]
- 15.Fadool JM, Dowling JE. 2008. Zebrafish: a model system for the study of eye genetics. Prog. Retin. Eye Res. 27, 89–110. ( 10.1016/j.preteyeres.2007.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo W, Williams J, Smallwood PM, Touchman JW, Roman LM, Nathans J. 2004. Proximal and distal sequences control UV cone pigment gene expression in transgenic zebrafish. J. Biol. Chem. 279, 19 286–19 293. ( 10.1074/jbc.M400161200) [DOI] [PubMed] [Google Scholar]
- 17.Takechi M, Seno S, Kawamura S. 2008. Identification of cis-acting elements repressing blue opsin expression in zebrafish UV cones and pineal cells. J. Biol. Chem. 283, 31 625–31 632. ( 10.1074/jbc.M806226200) [DOI] [PubMed] [Google Scholar]
- 18.Tsujimura T, Chinen A, Kawamura S. 2007. Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl Acad. Sci. USA 104, 12 813–12 818. ( 10.1073/pnas.0704061104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsujimura T, Hosoya T, Kawamura S. 2010. A single enhancer regulating the differential expression of duplicated red-sensitive opsin genes in zebrafish. PLoS Genet. 6, e1001245. ( 10.1371/journal.pgen.1001245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Delfin K, et al. 2009. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc. Natl Acad. Sci. USA 106, 2023–2028. ( 10.1073/pnas.0809439106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, Wong ROL. 2013. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl Acad. Sci. USA 110, 15 109–15 114. ( 10.1073/pnas.1303551110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami K, Sato S, Ozaki H, Ikeda K. 2000. Six family genes—structure and function as transcription factors and their roles in development. Bioessays 22, 616–626. () [DOI] [PubMed] [Google Scholar]
- 23.Seo HC, Drivenes O, Ellingsen S, Fjose A. 1998. Transient expression of a novel Six3-related zebrafish gene during gastrulation and eye formation. Gene 216, 39–46. ( 10.1016/S0378-1119(98)00328-X) [DOI] [PubMed] [Google Scholar]
- 24.Inbal A, Kim S-H, Shin J, Solnica-Krezel L. 2007. Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron 55, 407–415. ( 10.1016/j.neuron.2007.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cermak T, et al. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. ( 10.1093/nar/gkr218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861. ( 10.1371/journal.pgen.1002861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima D, Mano H, Fukada Y. 2000. Vertebrate ancient-long opsin: a green-sensitive photoreceptive molecule present in zebrafish deep brain and retinal horizontal cells. J. Neurosci. 20, 2845–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima D, Torii M, Fukada Y, Dowling JE. 2008. Differential expression of duplicated VAL-opsin genes in the developing zebrafish. J. Neurochem. 104, 1364–1371. ( 10.1111/j.1471-4159.2007.05093.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barthel LK, Raymond PA. 2000. In situ hybridization studies of retinal neurons. Methods Enzymol. 316, 579–590. [DOI] [PubMed] [Google Scholar]
- 30.Mano H, Kojima D, Fukada Y. 1999. Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland. Brain Res. Mol. Brain Res. 73, 110–118. ( 10.1016/S0169-328X(99)00242-9) [DOI] [PubMed] [Google Scholar]
- 31.Asaoka Y, Mano H, Kojima D, Fukada Y. 2002. Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl Acad. Sci. USA 99, 15 456–15 461. ( 10.1073/pnas.232444199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy BN, Alvarez Y, Brockerhoff SE, Stearns GW, Sapetto-Rebow B, Taylor MR, Hurley JB. 2007. Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Invest. Ophthalmol. Vis. Sci. 48, 522–529. ( 10.1167/iovs.06-0975) [DOI] [PubMed] [Google Scholar]
- 33.Raymond PA, Barthel LK, Curran GA. 1995. Developmental patterning of rod and cone photoreceptors in embryonic zebrafish. J. Comp. Neurol. 359, 537–550. ( 10.1002/cne.903590403) [DOI] [PubMed] [Google Scholar]
- 34.Takechi M, Kawamura S. 2005. Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J. Exp. Biol. 208, 1337–1345. ( 10.1242/jeb.01532) [DOI] [PubMed] [Google Scholar]
- 35.Renninger SL, Gesemann M, Neuhauss SCF. 2011. Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur. J. Neurosci. 33, 658–667. ( 10.1111/j.1460-9568.2010.07574.x) [DOI] [PubMed] [Google Scholar]
- 36.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. 2001. Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447–452. ( 10.1038/ng774) [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Rattner A, Nathans J. 2005. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 25, 118–129. ( 10.1523/JNEUROSCI.3571-04.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin J, Brocher J, Linder B, Hirmer A, Sundaramurthi H, Fischer U, Winkler C. 2012. The 1D4 antibody labels outer segments of long double cone but not rod photoreceptors in zebrafish. Investig. Ophthalmol. Vis. Sci. 53, 4943–4951. ( 10.1167/iovs.12-9511) [DOI] [PubMed] [Google Scholar]
- 39.Gamse JT, Shen Y-C, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JO. 2002. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat. Genet. 30, 117–121. ( 10.1038/ng793) [DOI] [PubMed] [Google Scholar]
- 40.Shen Y, Raymond PA. 2004. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Dev. Biol. 269, 237–251. ( 10.1016/j.ydbio.2004.01.037) [DOI] [PubMed] [Google Scholar]
- 41.Saade CJ, Alvarez-Delfin K, Fadool JM. 2013. Rod photoreceptors protect from cone degeneration-induced retinal remodeling and restore visual responses in zebrafish. J. Neurosci. 33, 1804–1814. ( 10.1523/JNEUROSCI.2910-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh CS, Ellingsen S, Austbø L, Zhao X-F, Seo H-C, Fjose A. 2010. Autoregulatory binding sites in the zebrafish six3a promoter region define a new recognition sequence for Six3 proteins. FEBS J. 277, 1761–1775. ( 10.1111/j.1742-4658.2010.07599.x) [DOI] [PubMed] [Google Scholar]
- 43.Rennison DJ, Owens GL, Taylor JS. 2012. Opsin gene duplication and divergence in ray-finned fish. Mol. Phylogenet. Evol. 62, 986–1008. ( 10.1016/j.ympev.2011.11.030) [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama S. 2008. Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282. ( 10.1146/annurev.genom.9.081307.164228) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.