Abstract
Research in eco-evolutionary dynamics and community genetics has demonstrated that variation within a species can have strong impacts on associated communities and ecosystem processes. Yet, these studies have centred around individual focal species and at single trophic levels, ignoring the role of phenotypic variation in multiple taxa within an ecosystem. Given the ubiquitous nature of local adaptation, and thus intraspecific variation, we sought to understand how combinations of intraspecific variation in multiple species within an ecosystem impacts its ecology. Using two species that co-occur and demonstrate adaptation to their natal environments, black cottonwood (Populus trichocarpa) and three-spined stickleback (Gasterosteus aculeatus), we investigated the effects of intraspecific phenotypic variation on both top-down and bottom-up forces using a large-scale aquatic mesocosm experiment. Black cottonwood genotypes exhibit genetic variation in their productivity and consequently their leaf litter subsidies to the aquatic system, which mediates the strength of top-down effects from stickleback on prey abundances. Abundances of four common invertebrate prey species and available phosphorous, the most critically limiting nutrient in freshwater systems, are dictated by the interaction between genetic variation in cottonwood productivity and stickleback morphology. These interactive effects fit with ecological theory on the relationship between productivity and top-down control and are comparable in strength to the effects of predator addition. Our results illustrate that intraspecific variation, which can evolve rapidly, is an under-appreciated driver of community structure and ecosystem function, demonstrating that a multi-trophic perspective is essential to understanding the role of evolution in structuring ecological patterns.
Keywords: eco-evolutionary dynamics, community genetics, local adaptation, Gasterosteus aculeatus, Populus trichocarpa
1. Introduction
Emerging interest in eco-evolutionary dynamics has resulted in a growing number of studies documenting the ways by which evolution can influence population dynamics [1–3], community assembly [4–7] and ecosystem function [8]. A critical component of the eco-evolutionary framework is to determine how phenotypic variation from local adaptation affects the way individuals and populations interact with their biotic and abiotic environment [5]. Local adaptation, or the evolution of traits that provide an advantage under local environmental conditions, is a key mode by which evolution shapes contemporary ecological dynamics [5,8–10]. To date, the best case studies detailing the ecological consequences of evolution stem from local adaptation in predators and subsequent shifts in trophic cascades [5,9,11]. For example, species pairs of a predatory fish, three-spined stickleback (Gasterosteus aculeatus; hereafter stickleback), which have repeatedly evolved into sympatric benthivorous and zooplanktivorous ecotypes, cause shifts in the structure of the zooplankton community and alter light availability in a mesocosm environment [8]. Similarly, alewives, guppies and salamanders have been shown to exhibit phenotypic variation stemming from evolution that has impacts on the abundances of associated species and ecosystem processes [5,9,11]. These findings reinforce that top-down control is a fundamental driver of community assembly and ecosystem function [4,12,13] and shed light on the role of intraspecific variation and evolutionary processes in the effects of predation [14,15].
In parallel with the study of the ecological impacts of evolution, there has been significant effort over the past decade to understand how community structure and ecosystem function are altered by genetic and phenotypic variation within dominant or foundation plant species (e.g. community genetics, [16]). This effort has yielded substantial evidence that genetic variation in primary producers can have strong bottom-up effects on community structure, such as the invertebrates and microbes associated with host plants, as well as ecosystem functions, such as primary productivity, decomposition and nutrient cycling [17–22]. For example, variation in the quantity and quality of leaf litter inputs among different genotypes of Populus trichocarpa (hereafter referred to as black cottonwood) modifies phytoplankton abundance, light availability and nutrients in aquatic mesocosms into which leaf litter was deposited [10]. Collectively, previous research demonstrates that genetic and phenotypic variation at the bottom of the food web can fundamentally alter community structure and ecosystem function.
Taken together, eco-evolutionary dynamics and community genetics have explored the role of local adaptation in driving phenotypic variation in the strength of both bottom-up and top-down processes, albeit in independent experiments. Yet, local adaptation is common and is likely present in a multitude of co-occurring species across different trophic levels [23]. As such, extending the eco-evolutionary framework to include intraspecific variation in multiple species within an ecosystem is critical to understanding the role of evolution in ecosystems. Moreover, ecologists have long recognized that bottom-up and top-down forces interact to shape communities [24,25]. For example, the level of productivity of a system can mediate the strength of top-down effects [26,27]. Therefore, our understanding of the impacts of evolutionary change on ecology could benefit greatly from the adoption of a multi-trophic perspective. To fully grasp the link between evolutionary change and ecology, we require a quantitative understanding of the strength of the direct and interactive effects of genetic variation at the top and bottom of food webs on associated communities and ecosystem function.
In this study, we examined the strength of the direct and interactive effects of phenotypic variation stemming from local adaptation within the top and bottom of an aquatic food web. In a large field array, we integrated a common garden containing five genotypes of black cottonwood with aquatic mesocosms (cattle tanks) housing one of two ecotypes of stickleback or a no-fish control (see figure 1 for a picture of the experimental set-up). Stickleback act as important predators in their natal lakes and in aquatic mesocosms [28,29], whereas seasonal pulses of leaf litter by terrestrial riparian plants can dictate trophic interactions and nutrient cycling as a cross ecosystem subsidy in a range of aquatic systems [30,31]. Genotypes of black cottonwood and ecotypes of stickleback both exhibit phenotypic variation from local adaptation to their natal environments [32,33]. To understand the importance of the direct and interactive effects of evolution, we ask two questions: (i) does intraspecific genetic variation in species at different trophic levels (i.e. cottonwood and stickleback) interact to shape community structure and ecosystem function? (ii) How do the strengths of direct and interactive evolutionary effects (black cottonwood genotype and/or stickleback ecotype) compare with the ecological impacts of predator presence?
Figure 1.
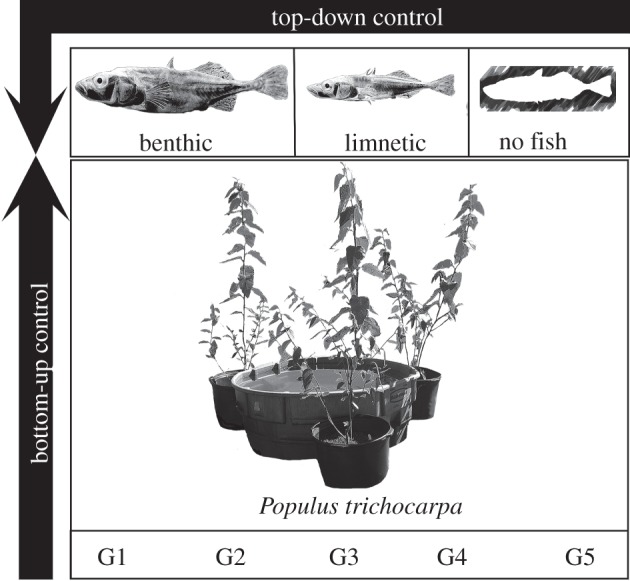
A schematic of the experimental design, demonstrating the use of genotypic variation in black cottonwood and ecotypic variation in three-spined stickleback. Each cottonwood genotype was planted in monoculture around 12 aquatic mesocosms (three trees per mesocosm). Leaf litter from these trees was collected as it senesced in the autumn, weighed and added to the aquatic environment. Benthic, limnetic or a no stickleback treatment was assigned to four mesocosms of each tree genotype and adult fish were added in the spring. The amount of the leaf litter contribution from each cottonwood genotype is illustrated in electronic supplementary material, figure S1.
The top-down effects that result from the addition of fish predators are well documented and led us to the prediction that presence of a fish predator alone will have cascading effects on both aquatic communities and ecosystem function [29,34]. Ecologists have recognized that the level of productivity of a system can mediate the strength of these top-down effects [26,27]. As such, we predict that terrestrial subsidies from the most productive black cottonwood genotypes with the greatest litter inputs to the aquatic habitat will lead to stronger impacts of fish presence compared with less productive clones. Finally, we predict that local adaptation in black cottonwood and stickleback ecotypes will interact to effect aquatic ecosystem function. This interaction will stem from benthic ecotypes, which feed primarily in the littoral area on benthic invertebrates that are strongly influenced by leaf litter [28,30], having the largest effect when paired with the most productive black cottonwood genotypes. Ultimately, our goal is to gain a deeper understanding of the community and ecosystem consequences of phenotypic evolution by exploring the interplay of ecology and evolution at multiple trophic levels.
2. Methods
(a). Aquatic mesocosms and cottonwood trees
To manipulate intraspecific variation from the bottom-up, we selected five genotypes of black cottonwood (P. trichocarpa), a dominant riparian tree species native to western North America that exhibits local adaptation to site conditions in leaf chemistry, growing season and primary productivity [32,35,36]. These five genotypes originated from southern localities of British Columbia (BC), Canada, and were equally related [10]. In June 2012, we placed five black cottonwood genotypes, raised in a common garden, in monoculture around 1136-l aquatic mesocosms with three replicate trees per mesocosm and 12 replicate mesocosms per genotype (180 trees, 60 mesocosms total; see figure 1 for graphic of experimental design). Although trees were placed in pots directly next to mesocosms, there was little leaf area overhanging the tanks, so the effects of shade were likely minimal on the aquatic environment. During autumn senescence (end of September 2012), we bagged each black cottonwood individual with vineyard netting (15 mm diameter, Smart Net Systems, Comox, BC, Canada) staked with a 3 m bamboo pole to contain leaf litter. From October to December, fallen leaf litter was collected weekly from each tree, air-dried for 48 h, weighed and 75% was deposited into mesocosms. The remaining 25% was used to estimate litter nutrient content and decomposition rates. At the end of December, any remaining leaves on trees were collected by hand, weighed and deposited into mesocosms. Cumulative leaf litter production varied by over twofold among genotypes, or approximately 70–150 g per tank, whereas litter decomposition varied by approximately 20% among genotypes [10]. To estimate the rates of decomposition for each genotype, 2 g of leaf litter was placed in mesh bags and submerged in the mesocosm at the time of peak litter fall. Leaf litter bags were removed one month later to determine total mass loss [10].
(b). Fish addition and experimental timing
The following spring (4 April 2013; four months after the final black cottonwood litter additions), we added stickleback ecotypes to mesocosms. Stickleback were added in the spring as adults to mimic the life cycle in natural systems, where both ecotypes grow quickly and reach adulthood in late spring before reproducing throughout the summer [28]. Three benthic or four limnetic stickleback individuals were added to separate tanks of each tree genotype (n = 4 tanks per ecotype; 40 tanks total containing fish; electronic supplementary material, figure S2). Some tanks were too turbid to adequately allow for a thorough assessment of fish mortality. Therefore, we added one additional benthic and three limnetic stickleback to each respective mesocosm one week after our initial additions (11 April 2013) based on visual surveys of tanks that were clear enough to observe dead individuals. We added three deceased benthic individuals to each control (fishless) mesocosm to account for nutrients introduced as a result of fish mortality and decomposition. The additional limnetics were added, so that fish biomass was consistent, as limnetics at the time of fish addition were approximately 0.5 g, and benthics were approximately 1.0 g. Benthics and limnetics were obtained from Priest Lake, British Columbia in June 2012 and reared for one generation in separate 750 000 gallon experimental ponds to eliminate the possibility of hybridization. As such, the fish used for this experiment were F1 progeny from wild-caught parents. At the end of the experiment (5 June 2013), we drained each mesocosm and searched for all living stickleback and captured 110 individuals (mean of 2.65 for benthic tanks and 2.85 for limnetic tanks). This search was difficult owing to the turbidity of the water and the abundance of leaf litter, so some fish may not have been recovered. There were no significant differences between benthic and limnetic stickleback treatments in average fish biomass in the mesocosms at the end of the study (electronic supplementary material, table S1). Given that fish were added in the spring and leaf litter from cottonwood genotypes was added as it senesced in the fall, we have included some figures that illustrate the effect of genotype over time and the effect of fish addition (electronic supplementary material, figure S2).
(c). Measuring the community and ecosystem responses
To determine community-level responses, we measured the amount of chlorophyl a (CHLA) in vivo using a fluorometer (Trilogy Designs) in each mesocosm on a weekly basis to estimate the abundance of phytoplankton. For consumers, we measured zooplankton abundance and biomass from columns of water at the beginning and end of the study. Macro-invertebrates were collected from the water column using a sweep net at the end of study. Finally, benthic invertebrates were collected by live-sorting individuals found in benthic substrate at the end of study (see electronic supplementary material, S1 for sampling details). We analysed the biomass of the phytoplankton community and the richness of the zooplankton, macro-invertebrate and benthic invertebrate communities. In addition, we examined the abundance of the most common zooplankton taxa that occurred in at least 40% of mesocosms (Bosmina sp. 34% of total zooplankton abundance, calanoid copepods 11%), macro-invertebrates (notonectids 48% of total macro-invertebrate abundance, Menetus sp. 32%) and benthic invertebrates (chironomids 44%, of total benthic invertebrate abundance, oligochaetes 31%, mayflies 17%).
To estimate ecosystem-level responses, we measured most parameters three times at regular intervals (two to three weeks) throughout the experiment. We measured gross primary productivity (GPP) using diurnal fluctuations in oxygen concentration. We measured the light extinction in mesocosms as the amount of photosynthetically active radiation (PAR) extinguished per centimetre of water depth ((PAR5 cm below surface – PARmaximum depth)/difference in depths). Finally, we measured soluble reactive phosphorus (SRP) and total ammonium to determine nutrient availability in the water (see electronic supplementary material, S1 for sampling details).
(d). Analysis of community and ecosystem data
To test for effects of both cottonwood and stickleback treatments on community and ecosystem data, we first checked each variable for normality and equal variance between treatments. If data were not normal or had unequal variances between groups they were log transformed before using full factorial ANOVA. If log-transformation did not yield a normally distributed dataset with roughly equal variances, we analysed data with a generalized linear model (GLM) and specified a Poisson error distribution. All GLMs with a Poisson distribution were tested for overdispersion. If data were overdispersed, we used a GLM with a negative binomial error distribution and compared AIC values of GLMs with Poisson and negative binomial error distributions to ensure the best model was used for the analysis.
(e). Direct and interactive evolutionary effects versus the ecological impact of predators
To compare the relative effects of different treatment levels on community and ecosystem responses, we calculated Hedge's g for the direct effects of fish presence, black cottonwood genotype and stickleback ecotype by dividing the difference of the most divergent treatment means by the pooled standard deviation. We also calculated Hedge's g for the interaction between black cottonwood genotype and fish presence and for the interaction between black cottonwood and stickleback ecotype by dividing the most divergent interactive treatment means by the pooled standard deviation [5].
3. Results
(a). Does intraspecific genetic variation in species at different trophic levels interact to shape community structure and ecosystem function?
We found pervasive genotype by ecotype interactions at the community level, as four of the eight most common taxa showed significant interactions (Bosmina, notonectids, mayflies and oligochaetes; electronic supplementary material, table S2). For example, mesocosms containing the benthic ecotypes had 66% greater oligochaete abundances than those containing the limnetic ecotype. The effect of ecotype was present only in tanks containing the most productive cottonwood genotypes (G2 and G3), with tanks containing limnetics having an average of approximately 0.13 oligochaetes compared with approximately 14.13 in samples from tanks containing benthic stickleback (electronic supplementary material, table S2 and figure 2). In addition, the amount of available phosphorous in the mesocosms at the end of the experiment depended on an interaction between black cottonwood genotypes and stickleback ecotypes. On average, tanks containing benthic ecotypes had twofold more SRP, a measure of the biologically available phosphorous [37], compared with limnetic ecotypes (electronic supplementary material, table S2 and figure 2). Yet, the effect of benthic sticklebacks on SRP was disproportionately high in tanks containing G2 and G3 genotypes (figure 2), which were the genotypes contributing the most litter resources (electronic supplementary material, figure S1). For example, mesocosms containing the most productive genotype (G3) had approximately eightfold more SRP when benthic fish were present than when limnetic fish were present. We also observed large effect sizes for the interactive effects between cottonwood genotype and stickleback ecotype, both at the community and ecosystem level (figure 4).
Figure 2.
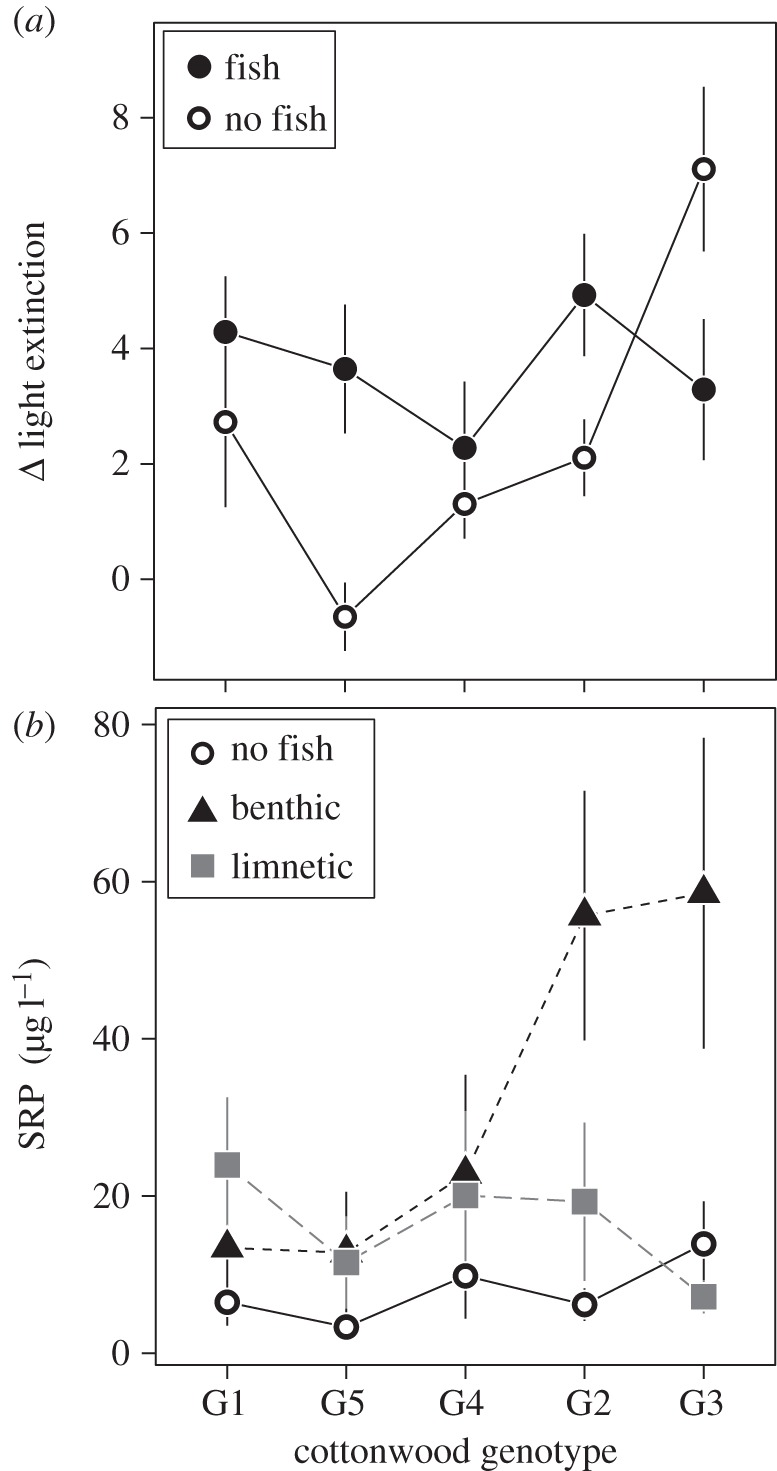
The effects of black cottonwood genotype (shown in ascending order of productivity (electronic supplementary material, figure S1) and stickleback presence and ecotype on ecosystem responses: (a) light extinction, measured as the number of photons extinguished per centimetre in the water column and (b) soluble reactive phosphorous, a measure of the biologically available phosphorous. Data are shown as means with standard errors. Full ANOVA results for all ecosystem responses are in electronic supplementary material, table S2. Electronic supplementary material, figure S2 illustrates the both SRP and light extinction both before and after the addition of fish.
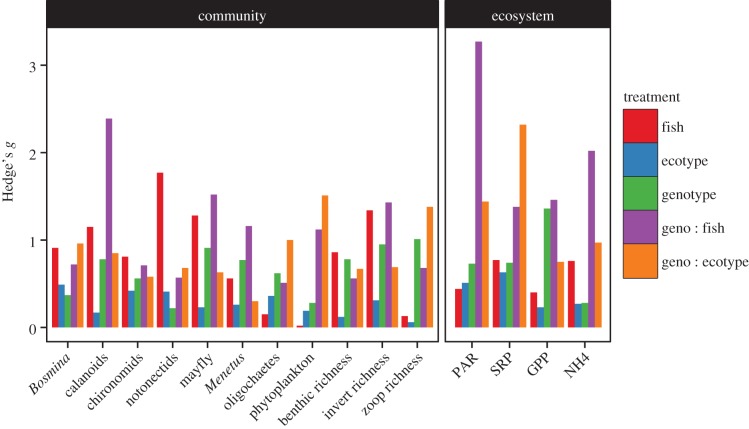
Figure 4.
Hedge's g effect sizes for each treatment for response variables at community and ecosystem levels. Community-level calculations are based on effects on abundance of the most common species in the zooplankton, macro-invertebrate and benthic invertebrate communities as well as richness abundance and richness calculations for these communities. Ecosystem function effect sizes are calculated from nutrient, productivity and light environment data.
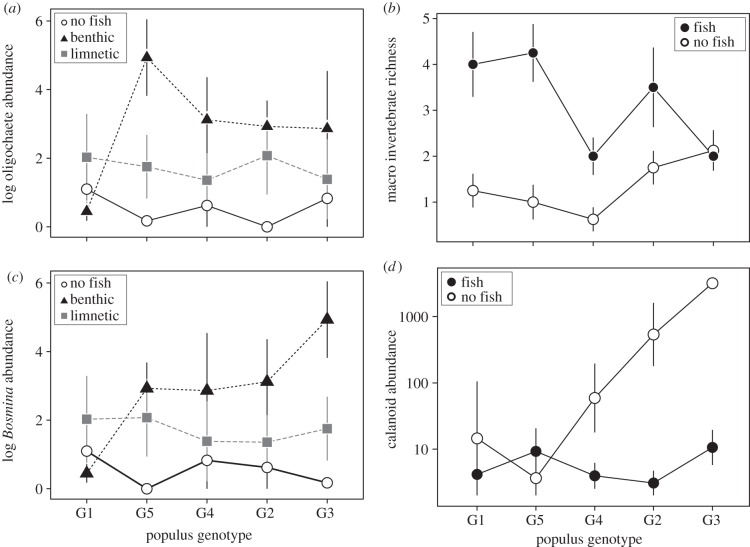
We also tested for an interaction between black cottonwood genotypes and stickleback presence/absence to determine how intraspecific variation in a bottom-up resource shapes top-down control by predators. We found that intraspecific genotypic variation in black cottonwood often dictated the strength of top-down control by the presence of fish (electronic supplementary material, table S1; figures 2 and 3). Specifically, top-down control by fish was often strongest in mesocosms containing litter inputs from the most productive black cottonwood genotypes (G2 and G3). For example, fish predation reduced the abundance of calanoid copepods by 71% in tanks containing litter from cottonwood genotypes G2 and G3, but fish had no effect in tanks containing litter from less productive tree genotypes. In contrast, macro-invertebrate species richness was two- to 4.5-fold lower in tanks containing fish for four of the five black cottonwood genotypes, whereas there was no effect of fish presence on richness in tanks containing the G3 genotype (electronic supplementary material, table S1 and figure 3).
Figure 3.
The effects of black cottonwood genotype (shown in ascending order of productivity (electronic supplementary material, figure S1) and stickleback ecotype on community metrics: (a) oligochaete abundance, (b) macro invertebrate richness, (c) Bosmina abundance, and (d) calanoid abundance. Data are shown as means with standard errors. Full ANOVA results for community responses are in electronic supplementary material, table S1.
We observed similar interactive effects between fish and black cottonwood genotypes at the ecosystem level, also driven primarily by stronger top-down control when coupled with the most productive black cottonwood genotypes. Specifically, whether fish presence had a positive or negative effect on light extinction in mesocosms depended on the genotypic identity of the litter inputs (e.g. –53% for G3, +661% for G5). For nutrient availability in mesocosms, the presence of fish increased SRP for some genotypes (+506% for G2) more so than others (+118% for G4; figure 2).
(b). How does the ecological impact of intraspecific variation compared with the impact of a traditional ecological driver?
The effects of top-down control by fish predators are well documented in aquatic ecosystems [12,34]. As such, we calculated standard effect sizes to examine the impact of fish presence/absence (pooling benthic and limnetic treatments) compared with the direct and interactive effects of top-down and bottom-up intraspecific variation (tree genotype and fish ecotype). We found that the addition of fish had an average Hedge's g effect size of 0.76 across both community and ecosystem responses (figure 4). Fish had strong impacts on the macro-invertebrate, benthic invertebrate and zooplankton communities, fitting with predictions of the trophic effects of predators (electronic supplementary material, table S1). The addition of fish also had effects on ecosystem function, particularly in nutrient availability (SRP and ammonia).
The direct effect of intraspecific variation within black cottonwood alone was similar in magnitude to that of fish addition, with an average Hedge's g of 0.69 for genotype effects averaged across community and ecosystem responses. The genotype of black cottonwood litter had a significant effect on the number and community composition of substrate dwelling benthic invertebrates (electronic supplementary material, table S1). At the ecosystem level, we found that cottonwood genetic variation influenced tank-level productivity (GPP) over the course of the experiment (range: 1.29–0.74). In contrast, the direct effect of ecotypic variation in stickleback ecotypes was comparatively small (average Hedge's g of 0.31). At the community level, stickleback ecotype had significant effects on notonectid and oligochaete abundances. At the ecosystem level, stickleback ecotype had a significant effect on SRP, with tanks containing the benthic ecotype having twofold higher SRP on average than tanks containing the limnetic ecotype.
We found that the effect sizes for the interaction of black cottonwood genotypes and stickleback ecotypes was an average Hedge's g of 0.98, higher than both that of predator addition and the impacts of intraspecific variation in either taxa alone. Similarly, the interaction between black cottonwood genotype and fish presence was exceptionally strong relative to both fish addition and the direct impacts of intraspecific variation in either black cottonwood or stickleback, with an average Hedge's g of 1.30 across community and ecosystem metrics. In sum, these results indicate that the interactive effects of genetic variation between trophic levels are strong relatively to the effects of predator addition.
4. Discussion
Ecologists have long recognized that productivity can impact the strength of top-down control, as the effects of predators depend largely on the abundance of prey species, which are often influenced by nutrient limitation [26,27,38]. Specifically, nutrient subsidies have been suggested to enhance prey quality and abundance, ultimately leading to stronger top-down effects [39]. In this study, we found that intraspecific variation stemming from local adaptation is sufficient to drive similar patterns. For example, pelagic crustaceans, such as calanoid copepods, have been shown to rely heavily on terrestrial material in oligotrophic lakes [40] and are also a common prey for sticklebacks [28]. We observed that mesocosms containing litter from the most productive cottonwood genotypes fostered a greater abundance of calanoid copepods, which ultimately led to a larger decrease in calanoid abundance when fish were present (figure 3). Furthermore, we found that intraspecific variation in a predator species can alter the relationship between primary productivity and top-down control. For example, the abundance of oligochaetes in our study depended both on the genotype of leaf litter added and the ecotype of stickleback present. In mesocosms containing litter from the most productive tree genotypes, we observed a significant reduction in oligochaetes when the limnetic ecotype was present, but an increase in oligochaetes when the benthic ecotype was present (figure 3). Taken together, these findings demonstrate that intraspecific variation can create sufficiently different ecological conditions or impact rates of prey consumption that drive changes in the strength of biotic control. These results also suggest that combinations of locally adapted populations of co-occurring species present within an ecosystem can dictate the abundance of species found within that ecosystem.
Our results provide general insights into how genetic variation at the bottom and top of a food web interacts to shape the structure of communities and functioning of an aquatic ecosystem. At the ecosystem level, we found that phosphorous availability in aquatic mesocosms depended on both the tree genotype and the fish ecotype present in a given mesocosm. This interaction emerged when the benthic sticklebacks were added to tanks containing litter from the most productive tree genotypes (figure 2). Benthic sticklebacks have been hypothesized to excrete substantially more phosphorous than limnetic ecotypes as a function of local adaptation in their diet preferences [28] and extensive reduction of bony armour (R.El-S. 2014, unpublished data). Previous research has suggested that differences in bony structures may underlie differences in both organismal stoichiometry and excretion, as bone requires comparatively large amounts of phosphorous [41,42]. These ecotypic differences in phosphorous excretion, when coupled with the measured variation between cottonwood genotypes in leaf litter phosphorous content [10], present a potential mechanism underlying SRP values that vary based on pairwise combinations of stickleback ecotype and black cottonwood genotype. This interaction demonstrates that population-level local adaptation in two species can impact ecosystem function in an intuitive way. The rapid rate at which evolution has occurred in these two species [28,32] suggests that these interactions could arise over relatively short, ecologically relevant timescales.
When we compared the standard effect sizes of our experimental treatments, we found that community and ecosystem responses to combinations of phenotypic variation at the top and bottom of the food web were directly comparable in magnitude to the ecological effects of predator addition (fish presence/absence). Moreover, the interactive effects, both from genetic variation at two trophic levels and from genetic variation interacting with fish presence, were at least as strong as many of the direct effects of predator presence alone. These findings are noteworthy, given the well-documented impacts of fish on many aspects of freshwater ecosystems and particularly as a driver of trophic cascades [12,29,34]. Our study posits that the role of genetic and phenotypic variation has been under-appreciated in aquatic food web context and warrants further consideration moving forward.
The fields of eco-evolutionary dynamics and community genetics have made tremendous strides in integrating ecology and evolution by demonstrating that intraspecific variation that emerges from local adaptation can impact communities and ecosystems. However, experiments that have investigated the ecological impacts of evolutionary change have invariably focused on phenotypic shifts within a single focal taxa. In the study species presented here, there is strong evidence that variation among black cottonwood genotypes and stickleback ecotypes is adaptive. For example, reciprocal transplant experiments have demonstrated that ecotypic variation within stickleback is locally adaptive [33]. Similarly, there is considerable evidence that black cottonwood populations exhibit high levels of heritable phenotypic differentiation in tree phenology (growing season) and primary productivity as a result of adaptation to local site conditions [32,35,36]. In this study, we did not explicitly incorporate the evolutionary processes driving phenotypic variation among cottonwood genotypes and stickleback ecotypes. Yet, given the widespread nature of local adaptation [23], it seems highly likely that multiple species within a given ecosystem show patterns of phenotypic adaptation to local conditions. As such, interactions between rapidly evolving species that show ecologically important phenotypic trait variation may be common. Systems where predators show phenotypic evidence of adaptation to local prey species [3] and prey show variation in predator avoidance phenotypes are particularly strong candidates for study. Future work directed at investigating the ecological impacts of the adaptive variation in these interactions, perhaps using multi-trophic reciprocal transplant experiments, could continue to develop our understanding of how local evolutionary processes impact ecology and further our understanding of the interplay between ecology and evolution.
5. Conclusion
Previous work in community genetics has demonstrated that intraspecific variation in productivity of a foundation plant species can have cascading effects. Additionally, experiments on eco-evolutionary dynamics have illustrated the differential effects locally adapted predators have on prey communities. Our study demonstrates that combinations of genetic variation within species can have profound effects on contemporary ecological processes, even shifting the strength of top-down control. Moreover, the strength of these effects can rival those of more classical ecological drivers, such as the presence of predators. Consequently, including the interactive effects of genetic variation within co-occurring species and across trophic levels will ultimately help us to determine how profoundly evolution dictates ecological dynamics and yield a more complete understanding of the factors that shape community structure and ecosystem function.
Supplementary Material
Acknowledgements
We thank A.A. MacDonald and E.M. Hart for their assistance in this research. T. Fukami, the Schluter Laboratory, and one anonymous reviewer provided valuable comments on the manuscript.
Ethics
Handling of stickleback in this experiment adhered to an approved University of British Columbia animal care protocol (no. 11-0402). Fish were collected with permission from Fisheries and Oceans Canada (SARA permit no. 197).
Data accessibility
Data for this paper have been uploaded to dryad http://dx.doi.org/10.5061/dryad.0h4f8.
Author contributions
S.M.R., M.A.R.C. and G.M.C. conceived the study. All authors contributed substantially to data collection. S.M.R. and A.S. carried out the analysis. S.M.R. wrote the first draft of the manuscript and all authors contributed to revisions.
Competing interests
We declare we have no competing interests.
Funding
S.M.R. was supported by the University of British Columbia. G.M.C. was supported by an NSERC Discovery Grant, the Canadian Foundation for Innovation and the University of British Columbia. A.S. was supported by the Killam Trust at UBC and the Gordon and Betty Moore Foundation as part of the Ocean Tipping Points project (www.oceantippingpoints.org). T.S. was supported by The Hakubi Project, Kyoto University.
References
- 1.Hairston NG, Ellner SP, Geber MA, Yoshida T, Fox JA. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127. ( 10.1111/j.1461-0248.2005.00812.x) [DOI] [Google Scholar]
- 2.Ellner SP, Geber MA, Hairston NG. 2011. Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol. Lett. 14, 603–614. ( 10.1111/j.1461-0248.2011.01616.x) [DOI] [PubMed] [Google Scholar]
- 3.Hiltunen T, Becks L. 2014. Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat. Commun. 5, 5226 ( 10.1038/ncomms6226) [DOI] [PubMed] [Google Scholar]
- 4.Post DM, Palkovacs EP, Schielke EG, Dodson SI. 2008. Intraspecific phenotypic variation in a predator affects zooplankton community structure and cascading trophic interactions. Ecology 89, 2019–2032. ( 10.1890/07-1216.1) [DOI] [PubMed] [Google Scholar]
- 5.Bassar RD, et al. 2010. Local adaptation in Trinidadian guppies alters ecosystem processes. Proc. Natl Acad. Sci. USA 107, 3616–3621. ( 10.1073/pnas.0908023107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas TE, Mononen T, Comeault AA, Hanksi I, Nosil P. 2013. Evolution of camouflage drives rapid ecological change within an insect community. Curr. Biol. 23, 1835–1843. ( 10.1016/j.cub.2013.07.067) [DOI] [PubMed] [Google Scholar]
- 7.Lundsgaard-Hansen B, Matthews B, Seehausen O. 2014. Ecological speciation and phenotypic plasticity affect ecosystems. Ecology 95, 2723–2735. ( 10.1890/13-2338.1) [DOI] [Google Scholar]
- 8.Harmon LJ, Matthews B, DesRoches S, Chase J, Shurin J, Schluter D. 2009. Evolutionary diversification in stickleback affects ecosystem functioning. Nature 458, 1167–1170. ( 10.1038/nature07974) [DOI] [PubMed] [Google Scholar]
- 9.Palkovacs EP, Post DM. 2009. Experimental evidence that phenotypic divergence in predators drives community divergence in prey. Ecology 90, 300–305. ( 10.1890/08-1673.1) [DOI] [PubMed] [Google Scholar]
- 10.Crutsinger GM, et al. 2014. Testing a ‘genes-to-ecosystems’ approach to understanding aquatic-terrestrial linkages. Mol. Ecol. 23, 5888–5903. ( 10.1111/mec.12931) [DOI] [PubMed] [Google Scholar]
- 11.Urban MC. 2013. Evolution mediates the effects of apex predation on aquatic food webs. Proc. R. Soc. B 280, 20130859 ( 10.1098/rspb.2013.0859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks JL, Dodson SI. 1965. Predation, body size, and composition of the plankton. Science 150, 28–35. ( 10.1126/science.150.3692.28) [DOI] [PubMed] [Google Scholar]
- 13.Sih AP, Crowley P, McPeek M, Petranka J, Strohmeier K. 1985. Predation, competition, and prey communities: a review of field experiments. Annu. Rev. Ecol. Syst. 16, 269–311. ( 10.1146/annurev.es.16.110185.001413) [DOI] [Google Scholar]
- 14.Bolnick DI, Svanbäck R, Fordyce JA, Yang LH, Davis JM, Hulsey CD, Forister ML. 2003. The ecology of individuals: incidence and implications of individual specialization. Am. Nat. 161, 1–28. ( 10.1086/343878) [DOI] [PubMed] [Google Scholar]
- 15.Bolnick DI, et al. 2011. Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 26, 183–192. ( 10.1016/j.tree.2011.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitham TG, et al. 2006. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523. ( 10.1038/nrg1877) [DOI] [PubMed] [Google Scholar]
- 17.Schweitzer JA, Bailey JK, Hart SC, Wimp GM, Chapman S, Whitham TG. 2005. The interaction of plant genotype and herbivory decelerate leaf litter decomposition and alter nutrient dynamics. Oikos 110, 133–145. ( 10.1111/j.0030-1299.2005.13650.x) [DOI] [Google Scholar]
- 18.Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ. 2006. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313, 966–968. ( 10.1126/science.1128326) [DOI] [PubMed] [Google Scholar]
- 19.Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. ( 10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 20.Bailey JK, et al. 2009. From genes to ecosystems: a synthesis of the effects of plant genetic factors across levels of organization. Phil. Trans. R. Soc. B 364, 1607–1616. ( 10.1098/rstb.2008.0336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson MT, Vellend M, Stinchcombe JR. 2009. Evolution in plant populations as a driver of ecological changes in arthropod communities. Phil. Trans. R. Soc. B 364, 1593–1605. ( 10.1098/rstb.2008.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackrel SL, Wootton JT. 2013. Local adaptation of stream communities to intraspecific variation in a terrestrial ecosystem subsidy. Ecology 95, 37–43. ( 10.1890/13-0804.1) [DOI] [PubMed] [Google Scholar]
- 23.Hereford J. 2009. A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 173, 579–588. ( 10.1086/597611) [DOI] [PubMed] [Google Scholar]
- 24.Hunter MD, Price P. 1992. Playing chutes and ladders. Heterogeneity and the relative roles of bottom-up and top-down forces in natural communities. Ecology 73, 724–732. [Google Scholar]
- 25.Power ME. 1992. Top-down and bottom-up forces in food webs—do plants have primacy? Ecology 73, 733–746. ( 10.2307/1940153) [DOI] [Google Scholar]
- 26.Rosemond AD, Mulholland PJ, Elwood JW. 1993. Top-down and bottom-up control of stream periphyton: effects of nutrients and herbivores. Ecology 74, 1264–1280. ( 10.2307/1940495) [DOI] [Google Scholar]
- 27.Forkner RE, Hunter MD. 2000. What goes up must come down? Nutrient addition and predation pressure on oak herbivores. Ecology 81, 1588–1600. ( 10.1890/0012-9658(2000)081%5B1588%3AWGUMCD%5D2.0.CO%3B2) [DOI] [Google Scholar]
- 28.Schluter D, McPhail JD. 1992. Ecological character displacement and speciation in sticklebacks. Am. Nat. 140, 85–108. ( 10.1086/285404) [DOI] [PubMed] [Google Scholar]
- 29.Kratina P, Greig HS, Thompson PL, Carvalho-Pereira TSA, Shurin JB. 2012. Warming modifies trophic cascades and eutrophication in experimental freshwater communities. Ecology 93, 1421–1430. ( 10.1890/11-1595.1) [DOI] [PubMed] [Google Scholar]
- 30.Wallace JB, Eggert SL, Meyer JL, Webster JR. 1997. Multiple trophic levels of a forest stream linked to terrestrial litter inputs. Science 277, 102–104. ( 10.1126/science.277.5322.102) [DOI] [Google Scholar]
- 31.Gessner, et al. 2010. Diversity meets decomposition. Trends Ecol. Evol. 25, 372–380. ( 10.1016/j.tree.2010.01.010) [DOI] [PubMed] [Google Scholar]
- 32.McKown AD, et al. 2014. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 201, 1263–1276. ( 10.1111/nph.12601) [DOI] [PubMed] [Google Scholar]
- 33.Schluter D. 1993. Adaptive radiation in sticklebacks—size, shape, and habitat use efficiency. Ecology 74, 699–709. ( 10.2307/1940797) [DOI] [Google Scholar]
- 34.Carpenter SR, Kitchell JF, Hodgson JR. 1985. Cascading trophic interactions and lake productivity: fish predation and herbivory can regulate lake ecosystems. Bioscience 35, 634–638. ( 10.2307/1309989) [DOI] [Google Scholar]
- 35.Weber JC, Stettler RF, Heilman PE. 1985. Genetic variation and productivity of Populus trichocarpa and its hybrids. I. Morphology and phenology of 50 native clones. Can. J. For. Res. 15, 376–383. ( 10.1139/x85-060) [DOI] [Google Scholar]
- 36.Xie C-Y, Carlson MR, Ying CC. 2012. Ecotypic mode of regional differentiation of black cottonwood (Populus trichocarpa) due to restricted gene migration: further evidence from a field test on the northern coast of British Columbia. Can. J. For. Res. 42, 400–405. ( 10.1139/x11-187) [DOI] [Google Scholar]
- 37.Nürnberg GK, Peters RH. 1984. Biological availability of soluble reactive phosphorus in anoxic and toxic freshwaters. Can. J. Fish. Aq. Sci. 41, 757–765. ( 10.1139/f84-088) [DOI] [Google Scholar]
- 38.Throop HL, Lerdau MT. 2004. Effects of nitrogen deposition on insect herbivory: implications for community and ecosystem processes. Ecosystems 7, 109–133. ( 10.1007/s10021-003-0225-x) [DOI] [Google Scholar]
- 39.Polis GA, Anderson WB, Holt RD. 1997. Toward an integration of landscape ecology and food web ecology: the dynamics of spatially subsidized food webs. Annu. Rev. Ecol. Syst. 28, 289–316. ( 10.1146/annurev.ecolsys.28.1.289) [DOI] [Google Scholar]
- 40.Cole JJ, Carpenter SR, Kitchell J, Pace ML, Solomon CT, Weidel B. 2011. Strong evidence for terrestrial support of zooplankton in small lakes based on stable isotopes of carbon, nitrogen, and hydrogen. Proc. Natl Acad. Sci. USA 108, 1975–1980. ( 10.1073/pnas.1012807108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanni MJ. 2002. Nutrient cycling by animals in freshwater ecosystems. Annu. Rev. Ecol. Syst. 33, 341–370. ( 10.1146/annurev.ecolsys.33.010802.150519) [DOI] [Google Scholar]
- 42.Hendrixson HA, Sterner RW, Kay AD. 2007. Elemental stoichiometry of freshwater fishes in relation to phylogeny, allometry and ecology. J. Fish Biol. 70, 121–140. ( 10.1111/j.1095-8649.2006.01280.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this paper have been uploaded to dryad http://dx.doi.org/10.5061/dryad.0h4f8.