Abstract
CRISPR-Cas is a form of adaptive sequence-specific immunity in microbes. This system offers unique opportunities for the study of coevolution between bacteria and their viral pathogens, bacteriophages. A full understanding of the coevolutionary dynamics of CRISPR-Cas requires knowing the magnitude of the cost of resisting infection. Here, using the gram-positive bacterium Streptococcus thermophilus and its associated virulent phage 2972, a well-established model system harbouring at least two type II functional CRISPR-Cas systems, we obtained different fitness measures based on growth assays in isolation or in pairwise competition. We measured the fitness cost associated with different components of this adaptive immune system: the cost of Cas protein expression, the constitutive cost of increasing immune memory through additional spacers, and the conditional costs of immunity during phage exposure. We found that Cas protein expression is particularly costly, as Cas-deficient mutants achieved higher competitive abilities than the wild-type strain with functional Cas proteins. Increasing immune memory by acquiring up to four phage-derived spacers was not associated with fitness costs. In addition, the activation of the CRISPR-Cas system during phage exposure induces significant but small fitness costs. Together these results suggest that the costs of the CRISPR-Cas system arise mainly due to the maintenance of the defence system. We discuss the implications of these results for the evolution of CRISPR-Cas-mediated immunity.
Keywords: CRISPR, immunity, fitness costs
1. Introduction
Parasites and pathogens are ubiquitous in nature and impose strong selection on their hosts [1]. Hosts persist amid this threat with the aid of several resistance mechanisms that have evolved to target invading pathogens [1–3]. Given the fitness benefit to the host of resisting infection, the observation of variable levels of resistance in host populations may appear paradoxical [4,5]. One common explanation for this apparent contradiction is that the fitness advantage conferred by high resistance is often counter-balanced by reductions in other traits important for fitness. In other words, resistance is costly, and these costs are strong determinants of the evolution of both host and parasite life-history traits [6–9]. Understanding the extent and magnitude of resistance costs is therefore central to our overall understanding of host–parasite coevolutionary outcomes [10,11].
While many studies of costs of resistance have focused on eukaryotic hosts, costly resistance in prokaryotic hosts has also been documented [12,13]. However, it is important to note that previous studies often focused on a narrow sense definition of ‘resistance’ involving the ability to limit pathogen entry. Prokaryotes have evolved a diverse arsenal of defence strategies that may act at very different steps of the infection, including blocking pathogen adsorption and entry, or using mechanisms such as restriction–modification (R-M) or CRISPR-Cas to cut pathogen-derived nucleic acids [14,15]. The fitness costs associated with these resistance systems have not been studied extensively. In R-M systems, some studies report the existence of fitness costs associated with the production of enzymes involved in the restriction of foreign DNA [16].
CRISPR-Cas is a recently described microbial defence system [17–20]. All functional CRISPR-Cas systems have two main components: a CRISPR array of short palindromic repeats interspaced by variable, unique spacer sequences of mainly viral and plasmid origin, and CRISPR-associated (Cas) proteins. These systems also have a diverse architecture of cas genes and unique spacer content that are species- and strain-dependent, respectively [20]. Cas proteins interact with invading viral or plasmid genetic material, leading to the addition of a new repeat-spacer unit into the CRISPR array, usually at the 5′-end of the array [17,21]. These spacers are then transcribed, matured and, when associated with Cas proteins, serve as an interference system by recognizing and cleaving the invading dsDNA with identical sequences [22–24]. CRISPR-Cas systems therefore not only confer microbial hosts with acquired and specific immunity against infection by viruses and plasmids, but the spacers present in the host genome confer immune memory and detail the temporally ordered history of genetic encounters experienced by a given bacterial lineage (reviewed in [20]).
These features offer many exciting possibilities to study coevolution between microbial hosts and viral pathogens [25]. Work focusing on the evolutionary dynamics of CRISPR-Cas systems has mostly relied on environmental metagenomic samples and sequenced microbial genomes, revealing high levels of CRISPR diversity as well as patterns of local adaptation consistent with coevolutionary dynamics between hosts and viruses [26–29]. Theoretical evolutionary models have made predictions about the conditions for the maintenance and diversity of CRISPR-Cas systems in microbial populations [30–33]. As with most models of resistance evolution, these predictions depend heavily on assumptions regarding the magnitude of the costs associated with CRISPR-Cas defence [10,11]. However, the fitness costs associated with CRISPR-Cas systems have received little attention, although we might expect such costs to be common [34]. One exception was a recent study of the gram-negative Pseudomonas aeruginosa and a pilus-dependent transducing phage that explored the roles of constitutive receptor-based resistance and inducible type I-F CRISPR-mediated immunity [35]. While inducible CRISPR defence was favoured under low risk of infection, when the infection risk was high, there was stronger selection for receptor-mediated defence. Notably, this advantage was not due to a greater efficacy of this type of resistance relative to CRISPR-Cas, but instead because the inducible nature of CRISPR resistance made the mechanism too costly when infection risk was high [35].
Here we used the gram-positive Streptococcus thermophilus and its highly virulent phage 2972 [36], a well-established microbial laboratory system harbouring functional CRISPR-Cas systems [17,21,24,37], to study the costs of maintaining and deploying a CRISPR-mediated anti-phage defence. Cas protein expression is essential for antiviral defence, with pivotal roles in the acquisition of foreign DNA spacers into the CRISPR array and the targeting of homologous invading DNA. CRISPR 1 (CR1), the most active CRISPR-Cas system in strain S. thermophilus DGCC7710 [23], is classified as a type II-A CRISPR-Cas system due to its (four) Cas proteins content [38]. In addition to the signature proteins Cas1 and Cas2, type II-A systems have two additional Cas proteins: Csn2 has been shown to be involved in the integration of novel spacers following phage exposure [17], while protein Cas9 is necessary during spacer mediated cleavage of homologous foreign DNA [22], as well as spacer acquisition [39,40]. Cas9 is at the centre of recent developments in CRISPR-mediated targeted genome editing [41,42]. We developed different measures of bacterial fitness (in isolation or in competition) and compared different strains of S. thermophilus to estimate the cost of several components of the CRISPR-Cas machinery: the cost of Cas protein expression, the cost of increasing immune memory by incorporating either single or multiple spacers, and finally, how these costs are expressed during phage infection.
2. Material and methods
(a). Bacterial strains and plasmids
The wild-type (WT) Streptococcus thermophilus DGCC7710 (STWT) is used as starter culture in the industrial production of yogurt [43,44]. We used 11 derivatives of STWT that differ in the number of spacers in the CRISPR1 locus (electronic supplementary material, table S1). We also used two strains that had disrupted cas9 or csn2 genes, STΔcas9 and STΔcsn2, obtained previously [17] by the integration of a pORI vector construct via homologous recombination of the internal fragment of the respective cas genes (electronic supplementary material, table S1).
Plasmids pBV-YFP and pBV-CFP are derived from pBV5030::P32-ster1357 [45] in which the ster1357 gene was replaced by the yfp and cfp genes, respectively. In both replicative plasmids, the yfp and cfp genes are under the control of P32 constitutive promoter. The construction of the plasmids is detailed in the electronic supplementary material.
(b). Measurement of growth parameters in isolation
We assayed bacterial growth kinetics in 96-well microtitre plates. Prior to measuring growth kinetics, all S. thermophilus strains were taken from −80°C freezer stocks and grown statically overnight at 37°C in 10 ml LM17 medium. Twenty microlitres of each overnight culture was added to wells containing 180 µl of LM17 (which led to an initial concentration of around 107 cfu ml−1) in ffllat-bottom microtitre plates and covered with adhesive film to avoid evaporation, and incubated at 42°C. Optical density (OD) was followed at a wavelength of 630 nm every 15 min (unless otherwise stated) for at least 12 h using a TECAN microplate reader. Plates were shaken linearly for 5 s (amplitude 3.5 mm) before every measurement to homogenize the cultures. Growth assays were split among at least three experimental blocks unless otherwise stated. For each block, we used the same preculture for each strain split between three different replicate wells on the same 96-well microtitre plate.
The OD growth curves obtained from the TECAN microplate reader were analysed using an algorithm developed in Mathematica (electronic supplementary material). Briefly, we used the Mathematica function (BSplineFunction) to smooth the dynamics of each OD growth curve. We retrieved three different parameters from each function (electronic supplementary material, figure S1). First, we obtained the yield, Yi, the maximal OD level reached by each strain i. Second, we maximized the log of the OD growth curve to identify the point where the maximal value, ri, of the growth rate of each strain i. In addition, we retrieved the time at which this maximal growth rate was reached. This value is a measure of the time lag, Li, taken by each strain i to reach its maximal growth rate.
To estimate fitness costs, we calculated how each parameter (Xi = Yi, ri or Li) of the growth kinetics varied among strains, after being normalized to the mean parameter value of STWT ( ) in each experimental block:
) in each experimental block:
| 2.1 |
This normalization yielded three parameter values (
 and
and  ), allowing us to remove some of the variation due to experimental blocks. This normalization provides a more biologically relevant measure of each strain as it is a measure relative STWT. Note, however, that each of these three measures has its own units (
), allowing us to remove some of the variation due to experimental blocks. This normalization provides a more biologically relevant measure of each strain as it is a measure relative STWT. Note, however, that each of these three measures has its own units ( is in log[OD],
is in log[OD],  is in log[OD]/h and
is in log[OD]/h and  is in h).
is in h).
(c). Measurement of competitive ability
We measured the competitive ability of each strain i in competition with the same reference strain STWT carrying the plasmid pBV-YFP. All the strains were grown overnight in 10 ml liquid cultures of LM17 medium at 37°C. The overnight cultures were used to mix each strain with the reference strain (500 µl of strain i and 500 µl of the reference strain) in 9 ml of LM17 medium (10% dilution after the overnight growth). These mixed cultures were allowed to grow for 3 h at 37°C with shaking. One millilitre of each culture was sampled at time t = 0 h (the start of the competition) and t = 3 h (the end of the competition). These samples were centrifuged (10 min, 5000 r.p.m.), and the supernatant was removed and replaced by 0.05 M phosphate-buffered saline (pH 7.4) and left at +4°C to improve the maturation of the fluorescent proteins. Because S. thermophilus forms chains when it replicates, which can alter our ability to use flow cytometry, the cells from each sample were detached using a homogenizer T10 basic ULTRA-TURRAX at intermediate speed (25 000 r.p.m.) for 1 min prior to flow cytometry. CFP was detected at 405 nm excitation and 510/50BP emission, and YFP was detected at 488 nm excitation and 530/30BP emission on a BD FacsCanto II flow cytometer. We measured 50 000 cells in each sample and we used BD FACSDiva software to apply automatic compensation and gating.
Flow cytometry allowed us to obtain the frequency of strain i before and after 3 h of competition with the reference strain: p0 and p3, respectively. Assuming that the competition between the two strains occurs during the exponential phase of the growth we can infer the selection coefficient of strain i relative to the tester strain using
| 2.2 |
As for measurement of the growth parameters in isolation (see equation (2.1)), we normalized this quantity relative to the selection coefficient of STWT that does not carry pBV-YFP and obtained a normalized measure of competitive fitness  which has units of h−1.
which has units of h−1.
In the above procedure, we always used the STWT strain carrying pBV-YFP as the tester strain. In addition to this experimental procedure, we also measured the competition between strains where the tester strain was not the STWT strain, but also when both competing strains (ST+2 and ST+4) carried a pBV plasmid expressing a different fluorescent protein (YFP or CFP). In these experiments, we distinguished between the effects of the strains and the effects of the plasmids carrying different fluorescent markers (see §3d).
(d). Phage lysates and exposure
For experiments including phage infection, overnight bacterial cultures and the preparation of the microtitre plates were identical to those described above (§2c), but LM17 medium was supplemented with 10 mM CaCl2 to facilitate phage amplification. Each well received 10 µl of the appropriate phage lysate dilution. For each strain (STWT, ST+2, ST+4), we measured the growth rates of three replicate bacterial cultures for each of four phage titres (0, 103, 105, 107 plaque forming units (pfu)/ml), which allowed us to vary the multiplicity of infection (MOI) between 0 and 1. Growth rate parameters were extracted and analysed as described in §2c. In addition, we performed competition experiments between ST+2 and ST+4 carrying pBV-YFP or pBV-CFP under increasing phage titres as detailed above. The competitive ability of ST+4 against ST+2 was monitored by flow cytometry as explained in §2d.
(e). Statistical analysis
We analysed how fitness costs varied among strains by fitting a linear model to each variable (pairwise competitive ability  growth rate
growth rate  lag
lag 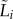 ) with strain as a fixed effect, and experimental block as a random effect. This analysis therefore calculates the variance in each parameter attributable to differences between strains, while accounting for the variance that arises from the different experimental blocks [46]. Variance components were estimated using restricted maximum likelihood (REML) method. Where appropriate, pairwise comparisons were carried out using two-sample t-tests. We also calculated the Pearson correlation coefficient between pairwise competitive ability
) with strain as a fixed effect, and experimental block as a random effect. This analysis therefore calculates the variance in each parameter attributable to differences between strains, while accounting for the variance that arises from the different experimental blocks [46]. Variance components were estimated using restricted maximum likelihood (REML) method. Where appropriate, pairwise comparisons were carried out using two-sample t-tests. We also calculated the Pearson correlation coefficient between pairwise competitive ability  and traits measured during bacterial growth in isolation for each strain. All analyses were carried out using JMP v. 11 (SAS) and R (CRAN).
and traits measured during bacterial growth in isolation for each strain. All analyses were carried out using JMP v. 11 (SAS) and R (CRAN).
3. Results
(a). The correlation between measures of fitness in isolation and in competition
The different parameters used to describe growth in isolation correlated well (between yield  and growth rate
and growth rate  0.90 with R2 = 0.81; between growth rate
0.90 with R2 = 0.81; between growth rate  and lag-time
and lag-time 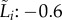 with R2 = 0.73; between lag-time
with R2 = 0.73; between lag-time  and yield
and yield  with R2 = 0.59). Fitness measured as pairwise competitive ability reflects the combined effect of many individual life-history traits. Our experiments allowed us to compare these different experimental measures of host fitness, and we observed that measures of fitness in isolation and in competition correlated well (between yield
with R2 = 0.59). Fitness measured as pairwise competitive ability reflects the combined effect of many individual life-history traits. Our experiments allowed us to compare these different experimental measures of host fitness, and we observed that measures of fitness in isolation and in competition correlated well (between yield 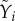 and pairwise competitive ability
and pairwise competitive ability  with R2 = 0.49; between growth rate
with R2 = 0.49; between growth rate 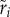 and pairwise competitive ability
and pairwise competitive ability  with R2 = 0.75; between lag-time
with R2 = 0.75; between lag-time  and pairwise competitive ability
and pairwise competitive ability  with R2 = 0.83). This confirms that growth rate parameters and competition assays yield complementary and consistent measures of fitness with the strains used in this study. In the remainder of the study, we have focused on the measures of fitness in competition (
with R2 = 0.83). This confirms that growth rate parameters and competition assays yield complementary and consistent measures of fitness with the strains used in this study. In the remainder of the study, we have focused on the measures of fitness in competition ( ) and on two measures of growth in isolation (
) and on two measures of growth in isolation (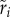 and
and  ) as measures of the fitness of S. thermophilus during the early stage of the competition (figure 1). Note that we focus on the opposite of the lag,
) as measures of the fitness of S. thermophilus during the early stage of the competition (figure 1). Note that we focus on the opposite of the lag, 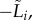 to work with a measure that increases with fitness (a shorter lag-time results in faster increase in population density).
to work with a measure that increases with fitness (a shorter lag-time results in faster increase in population density).
Figure 1.

Relationship between different measures of fitness for 15 strains of S. thermophilus (see the electronic supplementary material, table S1 for strain description). In (a), we plot competitive fitness 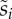 against the maximal growth rate
against the maximal growth rate  In (b), we plot competitive fitness
In (b), we plot competitive fitness  against the lag in growth rate
against the lag in growth rate  (i.e. the shorter the lag, the faster the strain starts to grow). Note that the WT S. thermophilus (STWT) strain is always at the origin (filled circle) because all measures of fitness are normalized to its fitness (see explanations in the main text).
(i.e. the shorter the lag, the faster the strain starts to grow). Note that the WT S. thermophilus (STWT) strain is always at the origin (filled circle) because all measures of fitness are normalized to its fitness (see explanations in the main text).
(b). Costs of Cas protein expression
We studied the costs associated with Cas protein expression using mutant stains of S. thermophilus where cas9 or csn2 had been disrupted in the CR1-Cas system (called STΔcas9 or STΔcsn2, respectively; see the electronic supplementary material, table S1 for details). The cas9 gene in particular is actively involved in both spacer acquisition and cleavage of invading nucleic acids [22,40], and has been shown to be constitutively expressed in S. thermophilus DGCC7710 [47]. In competition, both STΔcas9 and STΔcsn2 strains outcompeted STWT (overall ‘strain’ effect: F2,13 = 15.29, p = 0.0004; mean ± s.e.:  pairwise comparison with STWT: t = −4.60, d.f. = 13, p = 0.0005;
pairwise comparison with STWT: t = −4.60, d.f. = 13, p = 0.0005;  pairwise comparison with STWT: t = −4.96, d.f. = 13, p = 0.0003; figure 2a). Growth in isolation partially confirmed this result since both strains appeared to have a shorter lag phase than the STWT (see the electronic supplementary material, figure S2 for individual growth curves; overall ‘strain’ effect: F2,30 = 16.12, p < 0.0001;
pairwise comparison with STWT: t = −4.96, d.f. = 13, p = 0.0003; figure 2a). Growth in isolation partially confirmed this result since both strains appeared to have a shorter lag phase than the STWT (see the electronic supplementary material, figure S2 for individual growth curves; overall ‘strain’ effect: F2,30 = 16.12, p < 0.0001; 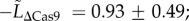 pairwise comparison with STWT: t = −3.31, d.f. = 30, p = 0.0024;
pairwise comparison with STWT: t = −3.31, d.f. = 30, p = 0.0024;  pairwise comparison with STWT: t = 5.65, d.f. = 30, p < 0.0001; figure 2b). However, we did not detect significant differences in maximal growth rates between strains (overall ‘strain’ effect: F2,30 = 0.10, p = 0.903;
pairwise comparison with STWT: t = 5.65, d.f. = 30, p < 0.0001; figure 2b). However, we did not detect significant differences in maximal growth rates between strains (overall ‘strain’ effect: F2,30 = 0.10, p = 0.903; 
 figure 2c).
figure 2c).
Figure 2.

Different fitness measures of different S. thermophilus strains with disrupted cas9 and csn2 genes. In (a), we plot competitive fitness  (mean ± s.e.); in (b), we plot the negative of the lag in growth rate
(mean ± s.e.); in (b), we plot the negative of the lag in growth rate  (mean ± s.e.); and in (c), we plot maximal growth rates
(mean ± s.e.); and in (c), we plot maximal growth rates  (mean ± s.e.).
(mean ± s.e.).
(c). Is increasing CRISPR-Cas immune memory costly?
A unique aspect of the CRISPR-Cas system results from the accumulation of new spacers. With the increasing number of spacers into the CRISPR array, hosts are also left with a memory of past encounters. When older spacers are expressed along with new ones [21], this results in acquired, sequence-specific immune memory. However, it is unknown whether this increasing immune repertoire is costly in S. thermophilus. Costs of increasing CRISPR arrays may arise because as hosts incorporate more spacers that need to be replicated and, perhaps more importantly, expressed as many crRNA molecules [21].
(i). Single spacer
We tested the costs of increasing the CRISPR array. First, we inquired about the fitness on host cells of acquiring a single extra virus-derived spacer (see electronic supplementary material, table S1 for strain details). Comparing our different fitness measures for each of these phage-resistant strains (
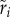 and
and  ) allowed estimating the fitness effects that arise due the acquisition of a single phage-derived spacer. Figure 3 shows that, regardless of the method used to estimate fitness, adding a single spacer does not induce a systematic fitness cost. In fact, while we detected a significant strain effect overall (F11,52 = 6.01, p < 0.0001), the average fitness effect of adding a single spacer is not significantly different from the fitness STWT when measured in competition (
) allowed estimating the fitness effects that arise due the acquisition of a single phage-derived spacer. Figure 3 shows that, regardless of the method used to estimate fitness, adding a single spacer does not induce a systematic fitness cost. In fact, while we detected a significant strain effect overall (F11,52 = 6.01, p < 0.0001), the average fitness effect of adding a single spacer is not significantly different from the fitness STWT when measured in competition ( F1,54 = 1.56, p = 0.282); nor do they differ from STWT in the maximal growth rate achieved (
F1,54 = 1.56, p = 0.282); nor do they differ from STWT in the maximal growth rate achieved ( F1,54 = 0.71, p = 0.491) or in lag time (
F1,54 = 0.71, p = 0.491) or in lag time ( F1,54 = 1.21, p = 0.298).
F1,54 = 1.21, p = 0.298).
Figure 3.
Different fitness measures of 12 phage-resistant S. thermophilus strains. In (a), we plot competitive fitness  (mean ± s.e.); in (b), we plot the opposite of the lag in growth rate
(mean ± s.e.); in (b), we plot the opposite of the lag in growth rate  (mean ± s.e.); and in (c), we plot maximal growth rates
(mean ± s.e.); and in (c), we plot maximal growth rates  (mean ± s.e.). The WT S. thermophilus DGCC7710 is indicated in red, single spacer strains are indicated in blue, and multiple spacers strains are indicated in orange (+2) and light blue (+4).
(mean ± s.e.). The WT S. thermophilus DGCC7710 is indicated in red, single spacer strains are indicated in blue, and multiple spacers strains are indicated in orange (+2) and light blue (+4).
(ii). Multiple spacers
We also measured the fitness of strains acquiring a higher number of additional spacers (+2 and +4 spacers; see the electronic supplementary material, table S1). In contrast to single spacers, we found that acquiring two or four spacers seemed to result in a small but positive selective advantage in competition (F1,12 = 8.55, p = 0.026; figure 3a). This difference in competitive ability did not appear to be due to the maximal growth rate, which did not differ between the multiple spacer-resistant strains and the WT strain (F1,33 = 1.30, p = 0.287). The difference in competitive ability is more likely to be due to differences in lag time, which was shorter in the strains carrying two or four spacers relative to the WT strain (F1,33 = 5.718, p = 0.0081; figure 3).
(d). Are costs contingent on the presence of phage?
We also inquired whether fitness costs could be revealed under the stressful conditions of surviving a phage infection. While harbouring and expressing additional phage-derived spacers in the genome might not be very costly (as suggested by our results in §3c), additional costs may occur when challenged with virulent phages. A recent study reported that in P. aeruginosa, type I-F CRISPR-Cas system was particularly costly under high phage exposure [35]. Specifically, if the acquisition of additional spacers is costly under phage exposure, we hypothesized that such inducible costs of immune deployment should be higher for strains carrying a higher number of spacers. To test this hypothesis, we used two types of comparisons.
First, we measured the potential reduction in fitness measured among hosts that were not exposed to phage 2972, and with hosts that were exposed but survived due to successful defence. To test this, we used our measures of fitness when phage-resistant strains (i.e. ST+4 containing four spacers derived from phage 2972 and ST+2 containing +2 spacers derived from phage 2972) were grown in isolation (ri and −Li) and challenged with a range of phage titres. As expected, phage titres had a dramatic impact on the growth rate of the STWT strain that is sensitive to the highly virulent phage 2972, while both resistant ST+4 and ST+2 reached high density, confirming that these strains are phage-resistant (figure 4a). To test whether phage titres affected the cost of resistance, we calculated normalized quantities for growth rate  lag time
lag time and yield
and yield  for ST+4 and ST+2 relative to the WT strain in the absence of phage exposure. Regardless of phage titre, the maximal growth rate of strain ST+2 was always higher than the growth rate of strain ST+4 (tRatio = −4.91, d.f. = 54, p ≤ 0.0001). This analysis showed that the growth rate of both strains decreased with increasing phage titre (‘titre effect’; F1.54 = 24.02, p = 0.0001), but overall this reduction was roughly equivalent for both strains (strain × titre interaction, F1,54 = 0.09, p = 0.763). Neither the lag time (F1,54 = 0.04, p = 0.846) nor the yield (F1,54 = 3.48, p = 0.559) were affected in either strain when infected with increasing phage concentrations.
for ST+4 and ST+2 relative to the WT strain in the absence of phage exposure. Regardless of phage titre, the maximal growth rate of strain ST+2 was always higher than the growth rate of strain ST+4 (tRatio = −4.91, d.f. = 54, p ≤ 0.0001). This analysis showed that the growth rate of both strains decreased with increasing phage titre (‘titre effect’; F1.54 = 24.02, p = 0.0001), but overall this reduction was roughly equivalent for both strains (strain × titre interaction, F1,54 = 0.09, p = 0.763). Neither the lag time (F1,54 = 0.04, p = 0.846) nor the yield (F1,54 = 3.48, p = 0.559) were affected in either strain when infected with increasing phage concentrations.
Figure 4.
Different fitness measures of the WT S. thermophilus and multiple spacer strains in the presence of phages. In (a), we plot the maximal growth rates of the WT S. thermophilus (STWT) (black line), a two spacer-resistant strain (dotted line) and a four spacer-resistant strain (grey line) after being exposed to various titres of phage. In (b), we plot the competitive fitness of strain ST+4 (+4 additional spacers derived from phage 2972) versus strain ST+2 (+2 additional spacers derived from phage 2972) after being exposed to various titres of phage.
As a second approach, we measured the competition between ST+4 and ST+2 phage-resistant strains challenged with a range of phage titres. Pairwise competition assays were made possible by adding fluorescent pBV-YFP (or pBV-CFP) plasmids in each of the two strains (see §2d), and monitoring their frequency when challenged with variable amounts of phages using flow cytometry. The inversion of the fluorescent markers in independent experiments also allowed us to distinguish the effect of the marker from the effect of carrying different numbers of spacers. In spite of a large effect of the marker (carrying pBV-CFP is more costly than carrying pBV-YFP: s = −0.32 ± 0.08), our analysis showed that strain ST+4, carrying the highest number of spacers, was a poorer competitor relative to ST+2 (competitive fitness of ST+4 is s = −0.26 ± 0.02; one-sample t-test, t = 15.09, d.f. = 23, p = 0.0001; figure 4b), confirming the results obtained for individual growth rate under phage challenge (figure 4a). We did not find a significant effect of phage titre on the competition between the two strains (F1,21 = 0.77, p = 0.39). Because of the deleterious effect of carrying pBV-CFP we repeated this experiment where we used only the pBV-YFP plasmid in one of the competing strains. We detected a weaker plasmid effect (s = −0.08 ± 0.003) and obtained very similar results regarding the strain effect (ST+4 has a lower fitness than ST+2). Again, we did not detect an effect of phage exposure on the competition between ST+2 and ST+4 (F1,21 = 2.08, p = 0.16).
4. Discussion
One of the fascinating features of CRISPR-Cas-mediated phage resistance is its similarity to the immune system of metazoans. The development of such homologous functions probably resulted from convergent evolution: the immune system is an adaptation to the temporally variable selection imposed by a diverse community of pathogens. Immunity may take different forms among species, but it rarely comes for free. Ecological immunology explores the fitness trade-offs between investment into immune function and other costly life-history traits [48]. Costs may result from pleiotropy with other key host life-history traits [49,50], or reflect a loss in performance that derives from the energetic requirements of immune deployment or from their immunopathological effects [51,52]. Evidence for costs of resistance is common for both invertebrate [52–54] and vertebrate hosts [8,55,56], although they are not necessarily ubiquitous [7]. Some work has also measured the relative costs of constitutive and inducible forms of defence in microbes. For example, in P. aeruginosa, while inducible type I-F CRISPR-Cas immunity is useful under low infection risk, it is simply too costly when virus numbers are high, where constitutive receptor-based defence is more effective [35]. Westra et al.'s study [35] suggests a cost for CRISPR, but it is unclear from where these costs arise. In the current work, we explored these different costs of CRISPR-Cas-mediated phage resistance in S. thermophilus, a classical model microbe for the study of CRISPR-Cas, particularly type II-A.
(a). Measuring fitness in Streptococcus thermophilus
We developed two different approaches to evaluate potential fitness costs associated with CRISPR-Cas. The growth assay is relatively easy to carry out because it does not require marking individual strains. By contrast, the use of a competitive growth assay was only feasible after developing a specific experimental procedure involving the use of fluorescence markers. The use of fluorescent markers in gram-positive bacteria can be challenging [57] and our study yields a new biological molecular tool that can be used in S. thermophilus and other gram-positive bacteria. These competitive assays allow fitness estimates when strains are competing for a common resource. Alternatively, measures on isolated strains may be more convenient if the aim is an absolute measure of the performance of a specific strain [58]. The measures of fitness based on the maximal growth rate are only expected to be equal in isolation and in competition during the early stages of the growth assay when density- and frequency-dependent selection are negligible [59]. Hence, in our opinion, the best option is to adopt and compare the results obtained from both approaches.
Here we found a positive correlation between these different measures of fitness (figure 1). We retained three different parameters to characterize the growth trajectories of our bacterial strains (yield, maximum growth rate and lag time). Interestingly, the correlation between selection coefficients (in competition) and growth parameters (in isolation) was lowest for the yield and higher for the other two parameters. A similar result has been observed in yeast [60]. This is likely to be due to the fact that both maximal growth rate and lag time (when measured in the context of a growth assay) are closer to the selection coefficient measured during the exponential phase of the competition assay. Taken together these data suggest that one may use measures of population growth in isolation to infer selection coefficients [61].
As in all classical fluctuating assays [62], we obtained our strains with additional spacers after exposing the WT S. thermophilus strain to a new environment (exposure to virulent/lytic phages here). In principle, this yields mutants with the same genetic background, but this procedure may result in additional mutations elsewhere in the genome [63]. In addition, for the strains used in the current study involving the inactivation of specific Cas genes (Δcas9 and Δcsn2), this issue may have been even more acute because it involved the addition of an antibiotic resistance gene in the focal gene. Therefore, the potential presence of additional mutations must be kept in mind for the interpretation of our results below.
(b). Fitness costs and their implications for the evolution of CRISPR-Cas
The evolutionary dynamics of host resistance depend crucially on the fitness costs associated with host defence systems [10,11,30,31,33]. Recent work has been particularly fruitful in elucidating the potential causes and consequences of host–pathogen coevolution at both phenotypic and molecular levels. Westra et al. [35] found that knocking out a single cas gene (csy3) in P. aeruginosa had no appreciable effect on the bacteria's competitive ability against an WT strain, while knocking out the entire type I-F CRISPR-Cas system (six cas genes and two CRISPR arrays) resulted in a fitness cost. This would imply not only that CRISPR is not costly, but that it may in fact have an additional beneficial functional in the absence of phage exposure [35]. This contrasts with our findings in S. thermophiles and its type II-A CRISPR1-Cas system (four cas genes and one CRISPR array), as we found evidence of a sizable fitness cost of expressing either cas9 or csn2. Of note, both genes are not found in the CRISPR-Cas system found in P. aeruginosa type I-F system. The endonuclease Cas9 has been found to be constitutively expressed following phage infection [47]. Given the fast time scales within which viral infection occurs, particularly in S. thermophilus, constitutive expression of immunity is more likely to guarantee a quick response to invading viral pathogens, but these results show that there may also be substantial fitness costs associated with Cas protein expression. Another potential source of constitutive cost may involve the regulation of the host genome by CRISPR-Cas systems [64] or arise due to the acquisition of self-targeting spacers (spacers targeting the bacterial genome), which may impose a fitness load on CRISPR-Cas systems [40,65].
We did not find any evidence for the existence of fitness costs associated with the acquisition of up to four new phage-targeting spacers (from 32 spacers in the CRISPR1 of the WT strain to 36 spacers in the phage-resistant strain ST+4) in the absence of phage infection. In fact, we found that the two strains carrying more spacers had an even higher fitness than the WT. One explanation for this effect may be that the experimental accumulation of multiple spacers, as discussed above, introduced beneficial mutations elsewhere in the genome of the bacteria. Information on the genetic background of these phage-resistant strains is required to evaluate the potential contributions of these additional mutations to their fitness [66].
We also measured the fitness costs of these additional spacers in the presence of increasing amounts of phage, to explore the potential effect of launching the immune system. We detected a significant effect of phage exposure on the bacterial growth rate, especially at the higher dose of phage (figure 4a). The fact that the fitness of both the ST+2 and ST+4 strains is equally reduced when challenged with 107 pfu phage compared with lower phage titres suggests an inducible cost (albeit small) of immune deployment. As this inducible cost is similar for both strains, this also suggests that recognition of different phage genomic sites does not trigger a more costly immune response. Although the inducible cost we detect is relatively weak, our results are in line with work in P. aeruginosa, where costs of CRISPR were mostly apparent in the presence of high phage titres. It is possible that we could not detect a stronger cost in the presence of phage because we used lower MOIs in our experiment in order to avoid the potentially confounding toxic effects of phage-derived endolysin found in phage preparations. Note that the inducible cost of CRISPR immunity measured in P. aeruginosa is relative to other constitutive forms of host defence, such as receptor-mediated resistance (loss of pili). It would be particularly interesting to compete the S. thermophilus mutants we used in this study with other S. thermophilus resistance mutants based on cell-surface receptors. Tracking the change between CRISPR and cell-surface mutants would thus provide relevant measures of fitness of these alternative resistance mechanisms, and constitutes an exciting avenue for future work.
Our results have important implications for the evolution of CRISPR-Cas immunity, and why the occurrence of functional CRISPR-Cas systems varies widely among archaea and bacteria [67]. Some bacterial species also appear to lose this system relatively easily [27,34]. Losing a CRISPR-Cas system may be beneficial when it allows the acquisition of new adaptations by foreign genetic elements [32]. But it may also be beneficial if it allows elimination of the constitutive cost associated with this immunity in situations where the risk of phage infection has been reduced. This may be the explanation for the rapid loss of CRISPR observed in Mycoplasma gallisepticum after a recent host shift from chicken to house finch [29]. The release from its ancestral phage community and the existence of constitutive costs of immunity could lead to selection against CRISPR-Cas immunity [29,32].
There are also more practical implications of our results. Streptococcus thermophilus is a commercially important bacterial species, heavily used to produce fermented dairy products, and the efficiency of this process is at risk by phage contamination. One of the current industrial practices to prevent phage-induced fermentation failures in the dairy industry consists in using several defined starter cultures [68]. An attractive solution would be to select S. thermophilus strains with strong CRISPR-mediated defences, but this strategy is likely to fail if CRISPR is too costly. Our study indicates that in the absence of phage, strains with additional spacers may not be easily outcompeted by susceptible strains.
5. Conclusion
In summary, our study demonstrates that costs of CRISPR defence arise mainly due to the maintenance of the defence system, while increasing immune memory to a level of very high protection (up to four new spacers) does not entail a considerable fitness cost. In addition, we did not find evidence for the existence of large fitness costs associated with the deployment of immunity when resistant bacteria are exposed to phages. Further experimental work is required to evaluate the magnitude of these different types of cost in other prokaryotes.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank M. E. Dupuis, D. Tremblay and M. Villion for advice and technical assistance in the early stages of the project.
Data accessibility
The full dataset is available on Dryad (http://dx.doi.org/10.5061/dryad.11r07).
Authors' contributions
Conceived and designed the experiments: P.F.V., S.M. and S.G. Performed the experiments and developed molecular tools: P.F.V., G.L., R.G. and F.G. Analysed the data: P.F.V. and S.G. Wrote the paper: P.F.V., S.M. and S.G.
Competing interests
The authors declare we have no competing interests.
Funding
P.F.V. was funded by an EMBO short-term fellowship (ASTF 7-2012) and by a strategic award from the Wellcome Trust for the Centre for Immunity, Infection and Evolution (http://ciie.bio.ed.ac.uk grant reference no. 095831). S.M. acknowledges funding from NSERC of Canada (Discovery program). S.M. also holds a Tier 1 Canada Research Chair in Bacteriophages. This work was funded by an ERC Starting grant no. 243054 to S.G.
References
- 1.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32, 569–577. ( 10.1038/ng1202-569) [DOI] [PubMed] [Google Scholar]
- 2.Gandon S, Michalakis Y. 2000. Evolution of parasite virulence against qualitative or quantitative host resistance. Proc. R. Soc. Lond B 267, 985–990. ( 10.1098/rspb.2000.1100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross J, Sanders MF. 2009. The development of genetic resistance to myxomatosis in wild rabbits in Britain. Epidemiol. Infect. 92, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazzaro BP, Sackton TB, Clark AG. 2006. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics 174, 1539–1554. ( 10.1534/genetics.105.054593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auld SKJR, Scholefield JA, Little TJ. 2010. Genetic variation in the cellular response of Daphnia magna (Crustacea: Cladocera) to its bacterial parasite. Proc. R. Soc. B 277, 3291–3297. ( 10.1098/rspb.2010.0772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boots M, Haraguchi Y. 1999. The evolution of costly resistance in host–parasite systems. Amer. Nat. 153, 359–370. ( 10.1086/303181) [DOI] [PubMed] [Google Scholar]
- 7.Labbe P, Vale P, Little T. 2010. Successfully resisting a pathogen is rarely costly in Daphnia magna. BMC Evol. Biol. 10, 355 ( 10.1186/1471-2148-10-355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonneaud C, Balenger SL, Hill GE, Russell AF. 2012. Experimental evidence for distinct costs of pathogenesis and immunity against a natural pathogen in a wild bird. Mol. Ecol. 21, 4787–4796. ( 10.1111/j.1365-294X.2012.05736.x) [DOI] [PubMed] [Google Scholar]
- 9.Susi H, Laine A-L. 2015. The effectiveness and costs of pathogen resistance strategies in a perennial plant. J. Ecol. 103, 303–315. ( 10.1111/1365-2745.12373) [DOI] [Google Scholar]
- 10.Boots M, Best A, Miller MR, White A. 2009. The role of ecological feedbacks in the evolution of host defence: what does theory tell us? Phil. Trans. R. Soc. B 364, 27–36. ( 10.1098/rstb.2008.0160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy MA, Forde SE. 2009. Ecological feedbacks and the evolution of resistance. J. Anim. Ecol. 78, 1106–1112. ( 10.1111/j.1365-2656.2009.01568.x) [DOI] [PubMed] [Google Scholar]
- 12.Lennon JT, Khatana SAM, Marston MF, Martiny JBH. 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 1, 300–312. ( 10.1038/ismej.2007.37) [DOI] [PubMed] [Google Scholar]
- 13.Jessup CM, Bohannan BJ. 2008. The shape of an ecological trade-off varies with environment. Ecol. Lett. 11, 947–959. ( 10.1111/j.1461-0248.2008.01205.x) [DOI] [PubMed] [Google Scholar]
- 14.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. ( 10.1038/nrmicro2315) [DOI] [PubMed] [Google Scholar]
- 15.Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687. ( 10.1038/nrmicro3096) [DOI] [PubMed] [Google Scholar]
- 16.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72. ( 10.1128/MMBR.00044-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. ( 10.1126/science.1138140) [DOI] [PubMed] [Google Scholar]
- 18.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64, 475–493. ( 10.1146/annurev.micro.112408.134123) [DOI] [PubMed] [Google Scholar]
- 19.Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, van der Oost J. 2012. The CRISPRs, they are A-Changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–339. ( 10.1146/annurev-genet-110711-155447) [DOI] [PubMed] [Google Scholar]
- 20.Van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Micro 12, 479–492. ( 10.1038/nrmicro3279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carte J, et al. 2014. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 93, 98–112. ( 10.1111/mmi.12644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garneau JE, et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. ( 10.1038/nature09523) [DOI] [PubMed] [Google Scholar]
- 23.Magadán AH, Dupuis M-È, Villion M, Moineau S. 2012. Cleavage of Phage DNA by the Streptococcus thermophilus CRISPR3-Cas System. PLoS ONE 7, e40913 ( 10.1371/journal.pone.0040913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupuis M-È, Villion M, Magadán AH, Moineau S. 2013. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun. 4, 2087. [DOI] [PubMed] [Google Scholar]
- 25.Vale PF, Little TJ. 2010. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc. R. Soc. B 277, 2097–2103. ( 10.1098/rspb.2010.0055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320, 1047–1050. ( 10.1126/science.1157358) [DOI] [PubMed] [Google Scholar]
- 27.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1, e00227 ( 10.1128/mBio.00227-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touchon M, Rocha EPC. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS ONE 5, e11126 ( 10.1371/journal.pone.0011126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8, e1002511 ( 10.1371/journal.pgen.1002511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin BR. 2010. Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 6, pe1001171 ( 10.1371/journal.pgen.1001171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Childs LM, Held NL, Young MJ, Whitaker RJ, Weitz JS. 2012. Multiscale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution 66, 2015–2029. ( 10.1111/j.1558-5646.2012.01595.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandon S, Vale PF. 2013. The evolution of resistance against good and bad infections. J. Evol. Biol. 27, 303–312. ( 10.1111/jeb.12291) [DOI] [PubMed] [Google Scholar]
- 33.Levin BR, Moineau S, Bushman M, Barrangou R. 2013. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 9, e1003312 ( 10.1371/journal.pgen.1003312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 9, e1003844 ( 10.1371/journal.pgen.1003844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westra ER, et al. 2015. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr. Biol. 25, 1043–1049. ( 10.1016/j.cub.2015.01.065) [DOI] [PubMed] [Google Scholar]
- 36.Lévesque C, Duplessis M, Labonté J, Labrie S, Fremaux C, Tremblay D, Moineau S. 2005. Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 71, 4057–4068. ( 10.1128/AEM.71.7.4057-4068.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. ( 10.1128/JB.01412-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makarova KS, et al. 2011. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Micro 9, 467–477. ( 10.1038/nrmicro2577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202. ( 10.1038/nature14245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, Terns RM, Terns MP. 2015. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 29, 356–361. ( 10.1101/gad.257550.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. ( 10.1016/j.cell.2014.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotech. 32, 347–355. ( 10.1038/nbt.2842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hols P, et al. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29, 435–463. [DOI] [PubMed] [Google Scholar]
- 44.Quiberoni A, Moineau S, Rousseau GM, Reinheimer J, Ackermann H-W. 2010. Streptococcus thermophilus bacteriophages. Int. Dairy J. 20, 657–664. ( 10.1016/j.idairyj.2010.03.012) [DOI] [Google Scholar]
- 45.Fleuchot B, et al. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119. ( 10.1111/j.1365-2958.2011.07633.x) [DOI] [PubMed] [Google Scholar]
- 46.Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White J-SS. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. 24, 127–135. ( 10.1016/j.tree.2008.10.008) [DOI] [PubMed] [Google Scholar]
- 47.Young JC, et al. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS ONE 7, e38077 ( 10.1371/journal.pone.0038077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulenburg H, Kurtz J, Moret Y, Siva-Jothy MT. 2009. Introduction: ecological immunology. Phil. Trans. R. Soc. B 364, 3–14. ( 10.1098/rstb.2008.0249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boots M, Begon M. 1993. Trade-offs with resistance to a granulosis virus in the Indian meal moth, examined by a laboratory evolution experiment. Funct. Ecol. 7, 528–534. ( 10.2307/2390128) [DOI] [Google Scholar]
- 50.Kraaijeveld AR, Godfray HC. 1997. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature 389, 278–280. ( 10.1038/38483) [DOI] [PubMed] [Google Scholar]
- 51.Moret Y, Schmid-Hempel P. 2000. Survival for immunity: the price of immune system activation for bumblebee workers. Science 290, 1166–1168. ( 10.1126/science.290.5494.1166) [DOI] [PubMed] [Google Scholar]
- 52.Sadd BM, Siva-Jothy MT. 2006. Self-harm caused by an insect's innate immunity. Proc. R. Soc. B 273, 2571–2574. ( 10.1098/rspb.2006.3574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraaijeveld AR, Ferrari J, Godfray HCJ. 2002. Costs of resistance in insect–parasite and insect–parasitoid interactions. Parasitology 125, S71–S82. ( 10.1017/S0031182002001750) [DOI] [PubMed] [Google Scholar]
- 54.Boots M. 2011. The evolution of resistance to a parasite is determined by resources. Am. Nat. 178, 214–220. ( 10.1086/660833) [DOI] [PubMed] [Google Scholar]
- 55.Bonneaud C, Mazuc J, Gonzalez G, Haussy C, Chastel O, Faivre B, Sorci G. 2003. Assessing the cost of mounting an immune response. Am. Nat. 161, 367–379. ( 10.1086/346134) [DOI] [PubMed] [Google Scholar]
- 56.Eraud C, Jacquet A, Faivre B. 2009. Survival cost of an early immune soliciting in nature. Evolution 63, 1036–1043. ( 10.1111/j.1558-5646.2008.00540.x) [DOI] [PubMed] [Google Scholar]
- 57.Aymanns S, Mauerer S, van Zandbergen G, Wolz C, Spellerberg B. 2011. High-level fluorescence labeling of gram-positive pathogens. PLoS ONE 6, e19822 ( 10.1371/journal.pone.0019822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bataillon T, Bailey SF. 2014. Effects of new mutations on fitness: insights from models and data. Ann. N. Y. Acad. Sci. 1320, 76–92. ( 10.1111/nyas.12460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevin L-M. 2011. On measuring selection in experimental evolution. Biol. Lett. 7, 210–213. ( 10.1098/rsbl.2010.0580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bell G. 2010. Experimental genomics of fitness in yeast. Proc. R. Soc. B 277, 1459–1467. ( 10.1098/rspb.2009.2099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol. Biol. Evol. 31, 232–238. ( 10.1093/molbev/mst187) [DOI] [PubMed] [Google Scholar]
- 62.Luria SE, Delbrück M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrangou R, Coûté-Monvoisin A-C, Stahl B, Chavichvily I, Damange F, Romero DA, Boyaval P, Fremaux C, Horvath P. 2013. Genomic impact of CRISPR immunization against bacteriophages. Biochem. Soc. Trans. 41, 1383–1391. [DOI] [PubMed] [Google Scholar]
- 64.Westra ER, Buckling A, Fineran PC. 2014. CRISPR-Cas systems: beyond adaptive immunity. Nat. Rev. Micro 12, 317–326. ( 10.1038/nrmicro3241) [DOI] [PubMed] [Google Scholar]
- 65.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26, 335–340. ( 10.1016/j.tig.2010.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bataillon T, Zhang T, Kassen R. 2011. Cost of adaptation and fitness effects of beneficial mutations in Pseudomonas fluorescens. Genetics 189, 939–949. ( 10.1534/genetics.111.130468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 8, 172 ( 10.1186/1471-2105-8-172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samson JE, Moineau S. 2013. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 4, 347–368. ( 10.1146/annurev-food-030212-182541) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full dataset is available on Dryad (http://dx.doi.org/10.5061/dryad.11r07).




