Abstract
Some venomous cone snails feed on small fishes using an immobilizing combination of synergistic venom peptides that target Kv and Nav channels. As part of this envenomation strategy, δ-conotoxins are potent ichtyotoxins that enhance Nav channel function. δ-Conotoxins belong to an ancient and widely distributed gene superfamily, but any evolutionary link from ancestral worm-eating cone snails to modern piscivorous species has not been elucidated. Here, we report the discovery of SuVIA, a potent vertebrate-active δ-conotoxin characterized from a vermivorous cone snail (Conus suturatus). SuVIA is equipotent at hNaV1.3, hNaV1.4 and hNaV1.6 with EC50s in the low nanomolar range. SuVIA also increased peak hNaV1.7 current by approximately 75% and shifted the voltage-dependence of activation to more hyperpolarized potentials from –15 mV to –25 mV, with little effect on the voltage-dependence of inactivation. Interestingly, the proximal venom gland expression and pain-inducing effect of SuVIA in mammals suggest that δ-conotoxins in vermivorous cone snails play a defensive role against higher order vertebrates. We propose that δ-conotoxins originally evolved in ancestral vermivorous cones to defend against larger predators including fishes have been repurposed to facilitate a shift to piscivorous behaviour, suggesting an unexpected underlying mechanism for this remarkable evolutionary transition.
Keywords: conotoxin, defence, predation, venom, molecular evolution
1. Introduction
Cone snails are highly specialized marine predators that use potent venom to subdue polychaete worms, molluscs and fishes [1]. The extremely fast prey immobilization achieved by some piscivorous species represents one of the most efficient and remarkable prey-capture strategies existing in nature [2]. The combination of toxins that cause this powerful immobilization include the κ- and δ-conotoxins found in predatory venom, which target Kv and Nav channels, respectively [2]. Through a cladistics approach and cDNA cloning, it was shown that all fish-hunting Conus species analysed produced at least one δ-conotoxin-like sequence, supporting an essential biological role for these secreted peptides [3]. In addition, δ-conotoxins belong to the widely distributed and ancestral plesiotypic gene superfamily O1, suggesting their early evolution in Conidae [4]. While ancestral cone snails preyed upon marine worms, the evolutionary path allowing a slow-moving, worm-eating (vermivorous) snail to switch to fish-hunting (piscivorous) behaviour has not been unravelled [5].
Previous studies have shown that lineage-specific dietary shifts in cone snails could be attributed to the asymmetric evolution of conotoxin loci, especially in the gene superfamily O1 [6]. Moreover, such differential expression of conotoxin gene superfamilies was also shown to allow the shift from a worm-based diet to fish-hunting in cones [7]. While these studies provide a mechanistic insight into the evolution and diversification of cone snail venom, they failed to identify a specific evolutionary path that drove the evolution of piscivory in cone snails. Unexpectedly, it was recently reported that cone snails can inject distinct venoms in response to predatory or defensive stimulus [8]. To this end, the venom gland is divided into two main parts, with the distal part producing conotoxins involved in prey capture, whereas the proximal section produces defensive conotoxins. These paradigm-shifting results suggested a novel theory for the origins of piscivory in cone snails, where toxins used to defend against large predators including fishes may have been repurposed, allowing the evolution of fish-hunting.
In this study, we report the discovery and characterization of SuVIA, a potent δ-conotoxin active on vertebrate Nav isolated from the venom of a vermivorous Conus species (Tesselliconus clade). Interrogation of the venom gland transcriptomes of other worm-hunting species uncovered a further 11 similar sequences, demonstrating the wide occurrence of δ-conotoxin-like sequences in worm-hunting species. Importantly, we demonstrate that SuVIA induces nocifensive or pain-like behaviours in mice when administered locally, and that this peptide is expressed in the proximal part of the venom gland, indicative of a specific defensive role. Our results provide, to our knowledge, the first evidence at the molecular level that δ-conotoxins originally evolved to defend against vertebrate predators such as large fishes, and that this pharmacological activity may have been repurposed to allow the evolution of fish-hunting behaviour, effectively turning predators into prey in a surprising evolutionary twist.
2. Material and methods
(a). Crude venom extraction and fractionation
The dissected venom obtained from three specimens of Conus suturatus, collected from the Great Barrier Reef, was used for extraction and fractionation. Dissection was performed on ice and the venom ducts were squeezed and the contents were collected in 1 ml of 0.1% formic acid, and lyophilised immediately. The total 30 mg crude venom was extracted by 30% acetonitrile/0.1% trifluoroacetic acid. Fractionation of crude venom was carried out through reversed-phase high-performance liquid chromatography (RP-HPLC) using Vydac 218TP-C18 (4.6 × 250 mm) and Thermo Hypersil-C18 (4.6 × 150 mm) columns fitted to a Shimadzu Prominence HPLC system with 0.043% TFA/90% acetonitrile (aq) as elution solvent B and 0.05% TFA (aq) as solvent A. A linear 1% B min−1 gradient was delivered to the column over 80 min. The eluent was monitored using a dual wavelength UV detector set to 214 and 280 nm and fractions were collected from the 214 nm trace.
(b). Activity-guided isolation of SuVIA
Activity-guided isolation of SuVIA was carried out using a FLIPR high-throughput Ca2+ assay as previously described [9,10]. Briefly, the human neuroblastoma cell line SH-SY5Y (ATCC) endogenously expressing hNaV1.2, hNaV1.3 and hNaV1.7 [10] was cultured in Roswell Park Memorial Institute (RPMI) medium containing 15% fetal bovine serum (FBS) and 2 mM l-glutamine and split every 3–4 days using 0.25% trypsin/EDTA (Life Technologies, Mulgrave, Victoria, Australia). Cells were plated at a density of 30 000–50 000 cells per well on black-walled 384 well imaging plates (Corning, Mornington, Vic., Australia) and cultured for 48 h at 37°C/5% CO2 before loading with Calcium 4 No Wash dye (Molecular Devices, Sunnyvale, CA, USA) for 30 min at 37°C. Changes in fluorescence (excitation 475–495, emission 515–575 nm) in response to addition of crude venom or venom fractions were measured every 1 s for 300 s using a FLIPRTetra fluorescent plate reader (Molecular Devices) and analysed using Screenworks 3.1.1.4 (Molecular Devices).
(c). Molecular mass characterization and sequence determination
A pure active fraction containing SuVIA was analysed and its molecular mass determined on an AB Sciex 4700 TOF-TOF Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA). The plate was calibrated using Calmix (4700 Proteomics analyzer calibration mixture) from Applied Biosystems. All spectra were acquired in reflector mode and 20 spectra of 50 laser shots were accumulated based on defined acceptance parameters and adequate signal intensity in the 800–5000 m z−1 range. The amino acid sequence of SuVIA was determined by automated Edman degradation using an Applied Biosystems 494 Procise Protein sequencing system (Australian Proteome Analysis Facility). The sample was dissolved in 10% ACN/90% 25 mM NH4HCO3 and reduced using DTT (25 mM) at 56°C for 0.5 h. Alkylation followed by addition of iodoacetamide (55 mM) at room temperature for 0.5 h. The sample mixture was purified using RP-HPLC on a Zorbax 300SB-C18 (3 × 150 mm) column. The target peptide was loaded onto a precycled, Biobrene-treated disc and subjected to up to 35 cycles of Edman N-terminal sequencing. The ambiguity of the last amino acid (residue 27) at the C-terminus was clarified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) sequencing applied to the Glu-C digested peptide segments using an AB Sciex QStar system. The reduction and alkylation for LC-MS/MS sequencing were carried out using the fast triethylphosphine/iodoethanol protocol [11] and the dried reaction mixture was reconstituted directly in a Glu-C solution and digested overnight at 37°C.
(d). FLIPR membrane potential assay
The pharmacological activity of SuVIA was assessed using a FLIPRTetra fluorescent membrane potential assay as previously described [12]. Briefly, HEK293 cells expressing hNaV1.3, hNaV1.4, hNaV1.6 and hNaV1.7 (Scottish Biomedical, Glasgow, Scotland) were cultured under selection with blasticidin, G418 or zeocin in minimal essential medium (MEM; Sigma-Aldrich, Castle Hill, NSW, Australia) supplemented with l-glutamine and 10% FBS (Bovogen Biologicals, France). Cells were passaged every 3–5 days using TrypLE Express (Life Technologies), seeded at a density of 10 000–15 000 cells per well on black-walled 384 well imaging plates (Corning) and cultured for 48 h at 37°C/5% CO2. Cells were then loaded for 30 min at 37°C with red membrane potential dye (Molecular Devices) diluted according to the manufacturer's instructions in physiological salt solution (composition (in millimolar): NaCl (140), glucose (11.5), KCl (5.9), MgCl2 (1.4), NaH2PO4 (1.2), NaHCO3 (5), CaCl2 (1.8), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (10), pH 7.4). To assess activation of NaV subtypes by SuVIA, a two-addition protocol (for each addition: read interval 1 s, number of reads 300) measuring changes in fluorescence (excitation 510–545 nm, emission 565–625 nm) in response to addition of varying concentrations of SuVIA and an EC10 concentration of the site 2 toxin veratridine (10–20 µM) was used. Potentiation of veratridine-induced membrane potential changes was analysed using Screenworks 3.1.1.4 (Molecular Devices) and expressed relative to control veratridine (70 µM) responses.
(e). Electrophysiology
Whole-cell patch-clamp experiments were conducted using the QPatch (16-channel) automated electrophysiology platform using 16 well planar patch chip plates (QPlate) with a patch hole diameter of 1 µm and resistance of 2 ± 0.02 MΩ. HEK293 expressing human NaV1.7 were cultured as mentioned in the above FLIPR assay. On the day of the assay cells were 70% confluent before they were harvested using Detachin (Bio-Scientific, Kirrawee, NSW, Australia) and resuspended in Ex-Cell ACF CHO medium containing 25 mM HEPES (Sigma-Aldrich). Approximately 7 × 106 cells were transferred to the QPatch (Sophion Biolin Scientific, North Brunswick, NJ, USA) and washed once in extracellular solution (1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, 3 mM KCl, 30 mM NaCl, 0.1 mM CdCl2 and 130 mM TEA-Cl. The pH was adjusted to 7.3 with NaOH. The osmolarity was adjusted to 320 mOsm with sucrose) before being applied to the QPlate.
Cells were positioned in the QPlate by the robotic fluid handler and the whole-cell parameters were set as follows: positioning pressure −60 mbar, minimum seal resistance 0.1 GΩ, holding pressure −20 mbar and holding potential −100 mV. The whole-cell configuration was formed using an internal solution containing 140 mM CsF, 1 mM EGTA and 10 mM HEPES. The pH was adjusted to 7.3 using CsOH. The extracellular solution contained 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, 3 mM KCl, 30 mM NaCl, 0.1 mM CdCl2 and 130 mM TEA-Cl. The pH was adjusted to 7.3 with NaOH. Whole-cell currents were filtered at 5 kHz and acquired at 25 kHz. Following establishment of the whole-cell configuration, cells were held at −80 mV and hNav1.7 currents were produced following a pre-pulse of −100 mV for 50 ms and a step to −20 mV for 50 ms before returning to a holding potential of −80 mV. This pulse protocol was applied every 30 s for 5 min. Current (I)–voltage (V) relationships were constructed with a holding potential of −80 mV followed by a pre-pulse of −100 mV for 50 ms and a series of 50 ms step pulses that ranged from −80 to 50 mV in 5 mV increments before returning to a holding potential of −80 mV. To investigate the effects of SuVIA on fast inactivation, cells were clamped at a holding membrane potential of −90 mV before a series pre-pulses of 500 ms ranging from −120 to −10 mV in 10 mV increments. A test pulse of −20 mV was then applied to determine the fraction of inactivated ion channels. The voltage protocol used to investigate ramp current consisted of a 50 ms pre-pulse of −100 mV from a holding potential of −80 mV, followed by a voltage ramp from −100 to 20 mV over 50 ms. Current (Inorm) plotted for the IV curve and fast inactivation analysis was normalized using the following equation: vInorm = I/Imax, where I is the current measured and Imax is peak current. Conductance (G) plotted for the GV curve was normalized using the following equation: G = I/(Vmemb − Vrev), where Vmemb is the membrane potential and Vrev is the reversal potential. Normalized GV and voltage of inactivation curves were fitted using the following Boltzmann distribution equation: y = 1/{1 + exp [(V50 − Vmemb)/k]}, where y is normalized current, V50 is the potential at half maximal current and k is the slope factor.
(f). Transcriptomic analysis of vermivorous cone snails
The venom gland transcriptome was obtained for the following worm-hunting cone snail species: Conus varius, Conus frigidus, Conus miliaris, Conus sanguinolentus and Conus ebraeus. Total RNA extraction, mRNA purification and 454 pyrosequencing were carried out as described previously [4]. The raw reads were uploaded in our in-house searchable database and δ-conotoxin-like sequences were retrieved using ConoSorter [13], aligned with Multalin program [14] and edited in Jalview [15].
(g). In vivo nocifensive behaviour
Male C57BL/6 mice were housed with a 12 L : 12 D cycle at 21°C with free access to water and standard rodent chow. To assess the behavioural effect induced by SuVIA, nocifensive responses after intraplantar administration were assessed as previously described [16]. Animals were anesthetised using 3% isofluane and SuVIA (50 nM in sterile saline) was administered by shallow subcutaneous injection into the left hind paw in a total volume of 20 μl. Nocifensive behaviour was quantified by a blinded observer from video recordings.
(h). Data analysis
GraphPad Prism (v. 4.00, San Diego, California) was used to fit a 4-parameter Hill equation with variable Hill slope to the data. All data, unless otherwise stated, are expressed as the mean ± standard error of the mean (s.e.m.).
3. Results
(a). Isolation of a novel excitatory conotoxin from Conus suturatus venom
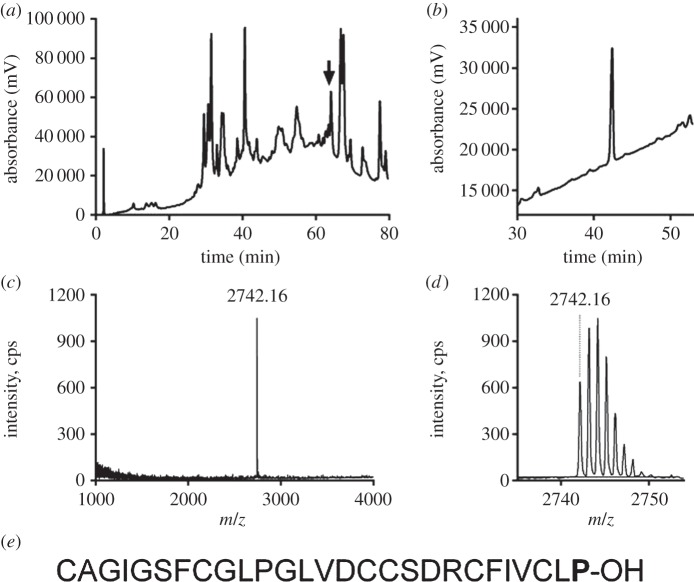
Crude venom isolated from C. suturatus elicited increases in intracellular Ca2+ that were associated with oscillations in SH-SY5Y cells and potentiated veratridine-induced Ca2+ responses which are mediated through endogenously expressed hNaV1.2, hNaV1.3 and hNaV1.7 [10]. Using assay-guided fractionation, we found that activity was associated with a single peak eluting at 63% B that was dominated by a single monoisotopic mass of 2741.16 Da (M+H, 2742.16) (figure 1a–d). The N-terminal Edman degradation of the corresponding peptide yielded 26 residues with the following sequence CAGIGSFCGLPGLVDCCSDRCFIVCL and a calculated monoisotopic molecular mass of 2644.10 Da. The missing last residue was determined as proline after GluC-digestion and MS/MS fragmentation of the C-terminal piece of the peptide (RC(EtO)FIVC(EtO)LP). Thus, the full amino acid sequence of the active fraction was determined as CAGIGSFCGLPGLVDCCSDRCFIVCLP-OH using a combination of Edman degradation and MS/MS (figure 1e). The detected mass of 2742.16 Da (M + H) corresponded to the calculated mass of 2741.157 Da.
Figure 1.
Isolation and sequence characterization of SuVIA. (a) Fractionation of the venom extract from C. suturatus using a Thermo Hypercil-C18 4.6 × 150 mm column eluted with a linear gradient from 0 to 80% of buffer B, over 80 min at 1 ml min−1. The arrow indicated where SuVIA eluted. (b) The peak indicated by the arrow was further characterized on a Thermo Hypersil-C18 2.1 × 150 mm column using a linear gradient from 20 to 80% of buffer B, over 60 min at 0.3 ml min−1. (c,d) Single monoisotopic mass of 2742.16 Da (M + H) was detected using 4700 MALDI-TOF MS. (e) The amino acid sequence of the SuVIA was obtained by Edman degradation and tandem mass spectrometry. The calculated monoisotopic mass of 2741.157 Da matched the above-detected mass.
A BLAST search using this sequence as a query revealed that Bt6.4, a superfamily O1 conotoxin isolated from the vermivorous Conus betulinus, was the closest homologue to this 27 residue long peptide (electronic supplementary material, figure S1), with 96% sequence identity (Asp > Gly difference). However, except for Bt6.4, SuVIA appears more closely related to piscivorous δ-conotoxins than other worm- or mollusc-hunting cone sequences (electronic supplementary material, figure S2). Therefore, as SuVIA is rich in hydrophobic residues (approx. 60%) and activates NaV channels, features reminiscent of δ-conotoxins, this novel peptide isolated from C. suturatus was named δ-SuVIA.
(b). Transcriptomic analyses
To date, pharmacological characterization of δ-conotoxins active at mammalian Nav has only been reported for peptides isolated from the venom of fish-hunting cone snails, although δ-conotoxin-like sequences have been isolated from mollusc- and worm-hunting species (electronic supplementary material, table S3), with those from molluscivorous species found to modulate molluscan Nav. However, the role and biological activity of δ-conotoxin-like sequences from vermivorous species remains unknown. To investigate how widely distributed these δ-conotoxins are in various worm-hunting cone snail species, we interrogated the venom gland transcriptome of five common species from four different clades, namely C. ebraeus and C. miliaris (Virroconus), C. frigidus (Virgiconus), C. sanguinolentus (Lividoconus) and C. varius (Strategoconus). A total of 11 new δ-conotoxin-like sequences were retrieved based on sequence homology (electronic supplementary material, figure S4), including four from C. varius (Va6.1, Va6.2, Va6.3 and Va6.4), three from C. miliaris (Mi6.1, Mi6.2 and Mi6.3), two from C. sanguinolentus (Sa6.1 and Sa6.2) and one each from C. frigidus (Fr6.1) and C. ebraeus (Eb6.1). While all species expressed sequences similar to SuVIA, C. frigidus, C. sanguinolentus and C. miliaris also produced variants with an extended C-terminus. Overall, our result demonstrate that δ-conotoxin-like sequences are broadly distributed in worm-hunting cone snails, including several species that belong to different phylogenetic clades, supporting an important role in prey capture and/or defence.
(c). Pharmacological and electrophysiological activities of SuVIA
We sought to assess the pharmacological activity of SuVIA across the mammalian neuronal and skeletal muscle NaV isoforms NaV1.3, NaV1.4, NaV1.6 and NaV1.7, as activity at these targets could induce significant behavioural responses in higher order vertebrates. SuVIA potentiated veratridine-induced responses in a concentration-dependent manner, with approximately equipotent activity at hNaV1.3 (EC50 3.98 ± 0.97 nM), hNaV1.4 (EC50 4.99 ± 0.92 nM), hNaV1.6 (EC50 1.27 ± 0.56 nM) and hNaV1.7 (EC50 2.42 ± 0.12 nM) (figure 2).
Figure 2.
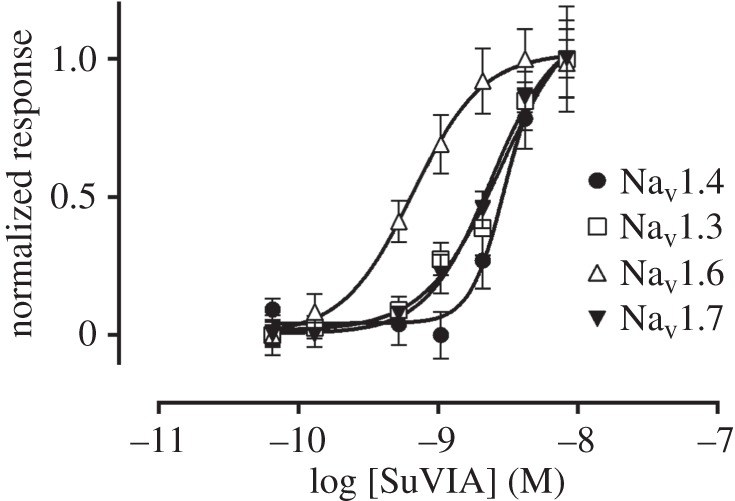
Native SuVIA activates hNaV1.3, hNaV1.4, hNaV1.6 and hNaV1.7. The activity of SuVIA at human sodium channel isoforms NaV1.3, NaV1.4, NaV1.6 and NaV1.7 was assessed using a fluorescent membrane potential assay in stably transfected HEK293 cells. SuVIA was approximately equipotent at hNaV1.3 (EC50 3.98 ± 0.97 nM), hNaV1.4 (EC50 4.99 ± 0.92 nM), hNaV1.6 (EC50 1.27 ± 0.56 nM) and hNaV1.7 (EC50 2.42 ± 0.12 nM). Data are expressed as mean ± s.e.m. from n = 3 wells and is representative of three independent experiments.
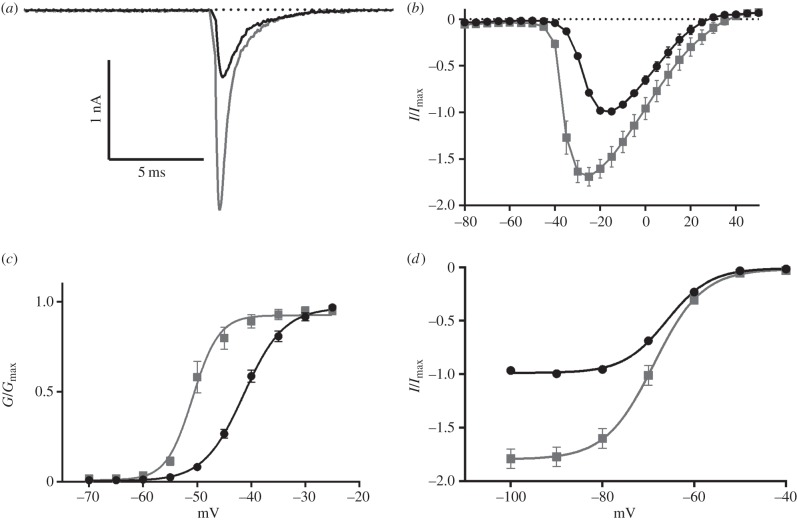
Next, we evaluated the electrophysiological properties including peak current, ramp current and the voltage of activation and inactivation in HEK293 cells expressing human NaV1.7 in the absence and presence of SuVIA (5 nM). SuVIA increased the peak hNaV1.7 current by approximately 75 ± 10.18% and shifted the voltage-dependence of activation to more hyperpolarized potentials from –15 mV to –25 mV (figure 3a,b). The V50 of activation was similarly shifted to more hyperpolarized potentials with cells exposed to SuVIA recording a V50 of –50.85 ± 0.44 mV, a decrease of 9.47 ± 0.52 mV compared with untreated cells (figure 3c). SuVIA also slightly shifted the V50 of inactivation by –2.83 ± 1.01 mV (p < 0.05; figure 3d), which in combination with effects on activation and peak current, results in dramatically enhanced hNaV1.7 current. Indeed, ramp currents were also significantly increased by 168.5 ± 31.6% and the membrane potential at which peak ramp current was observed was shifted to more hyperpolarized potentials (from –21.58 ± 1.0 to –25.97 ± 0.53 mV) in the presence of SuVIA (electronic supplementary material, figure S5). Surprisingly, in contrast to conotoxins from piscivorous cone snails, SuVIA had little effect on hNaV inactivation.
Figure 3.
SuVIA (5 nM) (grey) alters the electrophysiological properties of human NaV1.7 (black). (a) A representative trace of hNaV1.7 showing an increase in peak current induced by SuVIA. (b) Current–voltage relationship in the presence and absence of SuVIA. Peak current is increased and the voltage of activation of hNaV1.7 is shifted by 10 mV to more hyperpolarized potentials in the presence of SuVIA. (c) The conductance—voltage relationship is shifted towards more hyperpolarized potentials by SuVIA. Control V50: −41.38 ± 0.28 mV, SuVIA V50: −50.85 ± 0.44 mV. (d) The voltage of inactivation is slightly altered by SuVIA although peak current at hyperpolarized potentials is increased. Control V50: −65.98 ± 0.4 mV, SuVIA V50: −68.81 ± 0.93 mV. Data are expressed as mean ± s.e.m.; n = 8–9.
(d). A defensive role for SuVIA?
Our recent discovery that cone snails can produce different venoms in response to predatory or defensive stimulus prompted the investigation of the putative role of δ-like conotoxins from vermivorous species. Distal venom peptides are associated with prey capture, whereas proximal conotoxins are found almost exclusively in the defensive venom. To determine the regional distribution of SuVIA in the venom duct, extracts from the proximal and distal parts of C. suturatus venom duct were subjected to LC-MS analysis. The LC-MS traces show a clear pattern of expression along the duct, with SuVIA, being much more abundant (greater than threefold) in the proximal than the distal section (figure 4a,b). Thus, the regional distribution of SuVIA is consistent with a defensive rather than a prey-capture role in vermivorous species (figure 4c). To determine its deterrent activity in higher order vertebrates, we assessed the effect of SuVIA after shallow subcutaneous injection into the foot pad of mice. This approach was chosen to mimic the sting of a cone snail, and directly exposes peripheral sensory nerve endings to test compounds. Consistent with a putative defensive role, robust nocifensive or pain-like behaviours were elicited by intraplantar administration of SuVIA (50 nM), including increased licking of the injected hind paw compared with control (figure 5). Thus, the pharmacological activity of SuVIA, its distribution along the venom duct and its physiological effects are consistent with a role in deterring potential vertebrate predators.
Figure 4.
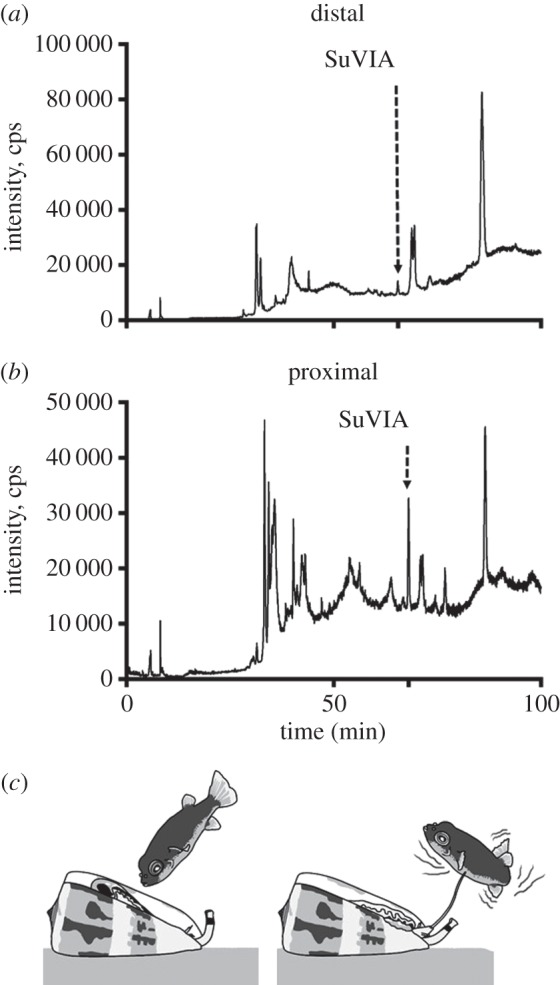
Regional expression of SuVIA in the venom duct and its proposed defensive role. LC-MS trace of the distal venom extract (a) shows a low expression level of SuVIA, whereas it is abundant in the proximal extract (b). (c) Based on its proximal expression profile and its mode of action probably inducing pain, we propose a defensive role for δ-conotoxins to deter potential vertebrate predators such as fishes.
Figure 5.
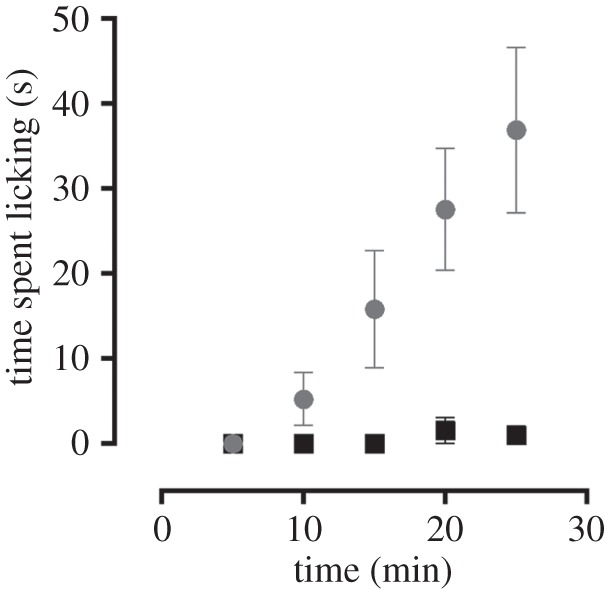
Nocifensive behaviour elicited by intraplantar administration of SuVIA. Shallow subcutaneous injection of SuVIA (grey; 50 nM, 20 µl) into the hindpaw of C57BL/6 mice elicited nocifensive behaviour evidenced by increased licking of the affected hind paw compared with saline (black).
4. Discussion
Cone snails are marine predators that have evolved an envenomation strategy facilitating predation on worms (vermivorous), molluscs (molluscivorous) or fishes (piscivorous) using a cocktail of venom peptides that target a range of receptors and ion channels crucial for normal physiological function [1]. Recent estimates suggest that each species of cone snail produces hundreds to thousands of conotoxins, the majority of which remain pharmacologically uncharacterized [4,17]. Not surprisingly, many conotoxins affect action potential initiation and propagation, including the μ- and μO-conotoxins that block the pore or alter NaV channel gating, [1,18–20], the δ-conotoxins that delay NaV inactivation by binding to site 6, and the ι-conotoxins that enhance NaV channel opening [21–23]. While inhibition or enhanced activation of NaV can cause paralysis, cardiac arrhythmias or epilepsy in the case of central NaV, the precise role of conopeptides targeting NaV channels in prey capture and/or defence is unclear. In piscivorous cone snails such as Conus purpurascens or Conus ermineus, δ-conotoxins are thought to be essential for the rapid immobilization of their fast moving prey [2], and this view is supported by the apparent paucity of δ-conotoxin sequences in venom of molluscivorous or vermivorous cone snails. When injected in fishes, δ-conotoxins elicit rapid swimming with twisted motions, quivering fins and a ‘lockjaw extended mouth’ syndrome, effects that could also deter larger predators [24]. In this study, we characterize a vermivorous δ-conotoxin (δ-SuVIA) and reveal another 11 novel δ-conotoxin-like sequences from other vermivorous cone snails.
SuVIA possesses all the hallmarks of typical δ-conotoxins, with a cysteine framework VI and a highly hydrophobic sequence. Remarkably for a peptide isolated from a vermivorous species, SuVIA was active at low nanomolar concentrations at mammalian NaV isoforms, including hNaV1.3, hNaV1.4, hNaV1.6 and hNaV1.7. Most invertebrates possess only a single NaV channel gene (NaV1), possibly two (NaV2) [25], and the primary sequence appears relatively divergent from vertebrate channels, especially near the inactivation gate (IVS3–S4). Furthermore, as the few annelid NaV channels that have been cloned to date generally show low sequence identity with vertebrate isoforms (33–44%), it suggests that cross-species activity is unlikely [26]. While the binding site of SuVIA remains to be determined, its divergent sequence, NaV subtype-selectivity and mechanism of action shows little overlap with the δ-conotoxins from piscivorous cone snails (electronic supplementary material, table S3). Thus, the newly described δ-conotoxin SuVIA from the venom of C. suturatus, as well as 11 related peptides from other vermivorous Conus, represent a potentially novel set of conotoxins that will provide further insight into the pharmacology of NaV channels and the evolution of cone snails.
Of the nine known mammalian NaV isoforms (NaV1.1–NaV1.9), NaV1.1, NaV1.2 and NaV1.3 are found predominantly in the central nervous system, while NaV1.7, NaV1.8 and NaV1.9 play a dominant role in peripheral sensory neurons and transmission of nociceptive signals. By contrast, NaV1.4 is expressed in skeletal muscle, NaV1.5 is crucial for the cardiac action potential and NaV1.6 is expressed at nodes of Ranvier of myelinated neurons where it is involved in saltatory action potential conduction [27]. Furthermore, loss of function mutations in the NaV1.7 gene are known to underlie congenital indifference to pain in humans, whereas gain of function mutations produce erythromelalgia and paroxysmal extreme pain disorder, conditions characterized by episodes of uncontrollable pain [28,29]. Thus, activation of NaV1.7 by SuVIA is likely to produce a painful sensation and could participate in this broadly evolved defensive mechanism. Consistent with such a role, the increase in paw licking which we observed after intraplantar administration of SuVIA indicates that δ-conotoxins can indeed produce nocifensive effects. As SuVIA is expressed in the proximal section of the venom gland of C. suturatus, it is thus likely that δ-conotoxins play a defensive role in vermivorous Conidae.
Recently, a comprehensive molecular phylogeny of Conidae including 320 species provided a useful canvas to study the evolution of specific traits such as diet type, biogeographic origin and toxin diversity [30]. Interestingly, the major venom peptides expressed in a given species were found to tightly correlate with the clade this particular species belongs to [30]. For instance, in the ubiquitous α-conotoxin family (nicotinic antagonists), four of seven subfamilies is found predominantly in vermivorous species, whereas three of five subfamilies occurs exclusively in piscivorous species. However, the O1 superfamily appears as an exception, as highly similar δ-conotoxins are found in unrelated phylogenetic clades and independently of the diet type, supporting its early evolution in ancestral cone snails [4]. The vermivorous species studied here belong to the Tesselliconus (C. suturatus), Virroconus (C. ebraeus and C. miliaris), Virgiconus (C. frigidus), Lividoconus (C. sanguinolentus) and Strategoconus (C. varius) phylogenetic clades. Interestingly, a similar δ-conotoxin isolated from another vermivorous, Conus tessulatus (Tesseliconus clade, closely related to C. suturatus), has been reported very recently [31]. The mode of action of TsVIA is reminiscent of SuVIA, but rather than a defensive weapon, the authors suggest that this peptide is used by vermivorous cone snails to deter fish competitors and to allow direct predation on fishes. However, a predatory use seems unlikely as we show that δ-conotoxins are produced in the proximal part of the gland, which is consistent with a defensive weapon. Therefore, it will be interesting to search for additional δ-conotoxins in other clades of vermivorous cone snails in order to define how broadly this defensive strategy has occurred.
In conclusion, our results suggest that vertebrate-active δ-conotoxins contribute to defence rather than predation in vermivorous snails. Extending our recently developed hypothesis that defensive venoms in vermivorous cone snails are repurposed for mollusc- and fish-hunting [8], we propose that defensive δ-conotoxins were originally used by ancestral worm-hunting cone snails to protect against threats such as cephalopod and fish predation, and have been repurposed for fish-hunting in piscivorous Conidae. This finding has important implications for the diversification of diet in cone snails, with defensive δ-conotoxins providing a molecular basis facilitating the shift from a diet of worms to fishes.
Supplementary Material
Acknowledgement
The authors thank Valentin Dutertre for drawing panel (c) of figure 4.
Ethics
Ethical approval for experiments involving animals was obtained from the University of Queensland Institutional Animal Ethics Committee. All experiments were conducted in accordance with the Animal Care and Protection Regulation Qld (2012), the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 8th edition (2013) and the International Association for the Study of Pain Guidelines for the Use of Animals in Research.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
A.-H.J. carried out HPLC, MALDI-MS, LC-MS/MS experiments and drafted the manuscript; M.R.I. contributed to high-throughput pharmacological characterization; M.C.I. carried out electrophysiological characterization; J.J.S. contributed to HPLC experiments and peptide isolation; R.J.L. and P.F.A. contributed reagents, participated in the design of the study and drafted the manuscript; I.V. contributed to activity-guided isolation, high-throughput pharmacological characterization, in vivo characterization, participated in the design of the study and drafted the manuscript; S.D. carried out transcriptomic and phylogenetic analysis, participated in the design of the study and drafted the manuscript.
Competing interests
We declare we have no competing interests.
Funding
I.V. was supported by an ARC Future Fellowship (FT130101215); S.D. was supported by an UQ postdoctoral fellowship; R.J.L. was supported by an NHMRC Principal Research Fellowship and P.F.A. and R.J.L. were supported by a NHMRC Program grant (569927).
References
- 1.Lewis RJ, Dutertre S, Vetter I, Christie MJ. 2012. Conus venom peptide pharmacology. Pharmacol. Rev. 64, 259–298. ( 10.1124/pr.111.005322) [DOI] [PubMed] [Google Scholar]
- 2.Terlau H, Shon KJ, Grilley M, Stocker M, Stuhmer W, Olivera BM. 1996. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature 381, 148–151. ( 10.1038/381148a0) [DOI] [PubMed] [Google Scholar]
- 3.Bulaj G, DeLaCruz R, Azimi-Zonooz A, West P, Watkins M, Yoshikami D, Olivera BM. 2001. δ-conotoxin structure/function through a cladistic analysis. Biochemistry 40, 13 201–13 208. ( 10.1021/bi010683a) [DOI] [PubMed] [Google Scholar]
- 4.Dutertre S, Jin AH, Kaas Q, Jones A, Alewood PF, Lewis RJ. 2013. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteomic. 12, 312–329. ( 10.1074/mcp.M112.021469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imperial J, et al. 2007. Using chemistry to reconstruct evolution: on the origins of fish-hunting in venomous cone snails. Proc. Am. Phil. Soc. 151, 185–200. [Google Scholar]
- 6.Duda TF., Jr 2008. Differentiation of venoms of predatory marine gastropods: divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J. Mol. Evol. 67, 315–321. ( 10.1007/s00239-008-9155-8) [DOI] [PubMed] [Google Scholar]
- 7.Duda TF, Jr, Palumbi SR. 2004. Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc. Biol. Sci. 271, 1165–1174. ( 10.1098/rspb.2004.2708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutertre S, et al. 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 5, 3521 ( 10.1038/ncomms4521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vetter I, Dekan Z, Knapp O, Adams DJ, Alewood PF, Lewis RJ. 2012. Isolation, characterization and total regioselective synthesis of the novel µO-conotoxin MfVIA from Conus magnificus that targets voltage-gated sodium channels. Biochem. Pharmacol. 84, 540–548. ( 10.1016/j.bcp.2012.05.008) [DOI] [PubMed] [Google Scholar]
- 10.Vetter I, Mozar CA, Durek T, Wingerd JS, Alewood PF, Christie MJ, Lewis RJ. 2012. Characterisation of Nav types endogenously expressed in human SH-SY5Y neuroblastoma cells. Biochem. Pharmacol. 83, 1562–1571. ( 10.1016/j.bcp.2012.02.022) [DOI] [PubMed] [Google Scholar]
- 11.Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD. 2004. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal. Biochem. 333, 174–181. ( 10.1016/j.ab.2004.04.013) [DOI] [PubMed] [Google Scholar]
- 12.Durek T, Vetter I, Wang CI, Motin L, Knapp O, Adams DJ, Lewis RJ, Alewood PF. 2013. Chemical engineering and structural and pharmacological characterization of the α-scorpion toxin OD1. ACS Chem. Biol. 8, 1215–1222. ( 10.1021/cb400012k) [DOI] [PubMed] [Google Scholar]
- 13.Lavergne V, Dutertre S, Jin AH, Lewis RJ, Taft RJ, Alewood PF. 2013. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genomic. 14, 708 ( 10.1186/1471-2164-14-708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10 881–10 890. ( 10.1093/nar/16.22.10881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2---a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. ( 10.1093/bioinformatics/btp033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deuis JR, Zimmermann K, Romanovsky AA, Possani LD, Cabot PJ, Lewis RJ, Vetter I. 2013. An animal model of oxaliplatin-induced cold allodynia reveals a crucial role for Nav1.6 in peripheral pain pathways. Pain. 154, 1749–1757. ( 10.1016/j.pain.2013.05.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis J, Jones A, Lewis RJ. 2009. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 30, 1222–1227. ( 10.1016/j.peptides.2009.03.019) [DOI] [PubMed] [Google Scholar]
- 18.Leipold E, DeBie H, Zorn S, Borges A, Olivera BM, Terlau H, Heinemann SH. 2007. µO conotoxins inhibit NaV channels by interfering with their voltage sensors in domain-2. Channels. 1, 253–262. ( 10.4161/chan.4847) [DOI] [PubMed] [Google Scholar]
- 19.Hui K, Lipkind G, Fozzard HA, French RJ. 2002. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by µ-conotoxin. J. Gen. Physiol. 119, 45–54. ( 10.1085/jgp.119.1.45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li RA, et al. 2003. Molecular basis of isoform-specific µ-conotoxin block of cardiac, skeletal muscle, and brain Na+ channels. J. Biol. Chem. 278, 8717–8724. ( 10.1074/jbc.M210882200) [DOI] [PubMed] [Google Scholar]
- 21.Leipold E, Hansel A, Olivera BM, Terlau H, Heinemann SH. 2005. Molecular interaction of δ-conotoxins with voltage-gated sodium channels. FEBS Lett. 579, 3881–3884. ( 10.1016/j.febslet.2005.05.077) [DOI] [PubMed] [Google Scholar]
- 22.Fiedler B, Zhang MM, Buczek O, Azam L, Bulaj G, Norton RS, Olivera BM, Yoshikami D. 2008. Specificity, affinity and efficacy of ι-conotoxin RXIA, an agonist of voltage-gated sodium channels NaV1.2, 1.6 and 1.7. Biochem. Pharmacol. 75, 2334–2344. ( 10.1016/j.bcp.2008.03.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Liu J, Pi C, Zeng X, Zhou M, Jiang X, Chen S, Ren Z, Xu A. 2009. Identification of a novel M-superfamily conotoxin with the ability to enhance tetrodotoxin sensitive sodium currents. Arch. Toxicol. 83, 925–932. ( 10.1007/s00204-009-0453-8) [DOI] [PubMed] [Google Scholar]
- 24.Shon KJ, Hasson A, Spira ME, Cruz LJ, Gray WR, Olivera BM. 1994. δ-conotoxin GmVIA, a novel peptide from the venom of Conus gloriamaris. Biochemistry 33, 11 420–11 425. ( 10.1021/bi00204a003) [DOI] [PubMed] [Google Scholar]
- 25.Zakon HH. 2012. Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proc. Natl Acad. Sci. USA 109(Suppl. 1), 10 619–10 625. ( 10.1073/pnas.1201884109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackshaw SE, Henderson LP, Malek J, Porter DM, Gross RH, Angstadt JD, Levasseur SM, Maue RA. 2003. Single-cell analysis reveals cell-specific patterns of expression of a family of putative voltage-gated sodium channel genes in the leech. J. Neurobiol. 55, 355–371. ( 10.1002/neu.10214) [DOI] [PubMed] [Google Scholar]
- 27.Wingerd JS, Vetter I, Lewis RJ. 2012. Voltage-gated sodium channels as therapeutic targets. In Therapeutic targets: modulation, inhibition and activation (ed. LMBaML), pp. 63–122. Hoboken, NJ: John Wiley and Sons. [Google Scholar]
- 28.Fischer TZ, Waxman SG. 2010. Familial pain syndromes from mutations of the NaV1.7 sodium channel. Ann. NY Acad. Sci. 1184, 196–207. ( 10.1111/j.1749-6632.2009.05110.x) [DOI] [PubMed] [Google Scholar]
- 29.Cox JJ, et al. 2006. An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898. ( 10.1038/nature05413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puillandre N, Bouchet P, Duda TF, Jr, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. 2014. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol. Phylogenet. Evol. 78, 290–303. ( 10.1016/j.ympev.2014.05.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aman JW, et al. 2015. Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc. Natl Acad. Sci. USA 112, 5087–5092. ( 10.1073/pnas.1424435112) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.