Abstract
Female multiple mating (polyandry) is widespread across many animal taxa and indirect genetic benefits are a major evolutionary force favouring polyandry. An incentive for polyandry arises when multiple mating leads to sperm competition that disadvantages sperm from genetically inferior mates. A reduction in genetic quality is associated with costly selfish genetic elements (SGEs), and studies in invertebrates have shown that males bearing sex ratio distorting SGEs are worse sperm competitors than wild-type males. We used a vertebrate model species to test whether females can avoid an autosomal SGE, the t haplotype, through polyandry. The t haplotype in house mice exhibits strong drive in t heterozygous males by affecting spermatogenesis and is associated with homozygous in utero lethality. We used controlled matings to test the effect of the t haplotype on sperm competitiveness. Regardless of mating order, t heterozygous males sired only 11% of zygotes when competing against wild-type males, suggesting a very strong effect of the t haplotype on sperm quality. We provide, to our knowledge, the first substantial evidence that polyandry ameliorates the harmful effects of an autosomal SGE arising through genetic incompatibility. We discuss potential mechanisms in our study species and the broader implications for the benefits of polyandry.
Keywords: t haplotype, segregation distortion, polyandry, embryo viability, indirect benefits, genetic incompatibility
1. Introduction
When females mate with multiple males, sperm from different males compete for fertilization of the ova [1]. By inciting sperm competition, females may prolong male–male competition beyond pre-copulatory contest and bias fertilization towards males of high quality or compatibility [2], with major effects on sexual behaviour, sex allocation, social networks, sexually transmitted infections, population viability and speciation [3]. Despite the many demonstrations of direct and indirect (genetic) benefits of polyandry [2], there is still no real consensus on why polyandry is so ubiquitous in nature.
One possibly underappreciated benefit of polyandry is protection from costly selfish genetic elements (SGEs) driving through males [4]. SGEs are sequences that alter DNA replication in their own favour, increasing their representation in the subsequent generation (called drive or segregation distortion) at the cost of their homologous sequences and usually also of the rest of the genome [5]. SGEs that kill or interfere with gametes carrying the homologous gene or chromosome, called gamete killers [5], typically drive through males. This is presumably because male gametes are produced in excess so that destruction of gametes has a smaller effect on fertility in males than in females [6]. Driving elements can occur on sex chromosomes or on autosomes, but sex chromosome drive is expected to arise more easily than autosomal drive [7]. However, modifiers of sex chromosome drive are strongly selected for because mating with a driving male will result in a costly single sex brood [7]. Given the relative amount of information encoded on autosomes versus sex chromosomes, more genomic regions may be available in which novel autosomal drivers can evolve. In addition, autosomal drive is much less likely to be detected because of the lack of sex-biased broods [6]. Consequently, there has been a detection bias towards sex ratio distorting SGEs [6]. Indeed, autosomal drive has so far mainly been studied in model systems, such as mice (t haplotype [8]), and Drosophila (Segregation Distorter [9]). Thus, autosomal drive through males may be more common than observed, but the relative importance of autosomal versus sex chromosomal drive for evolution within the genome remains unclear. As whole genome scans become increasingly common, more SGEs are likely to be identified [10].
Male drivers can be expected to incur fitness disadvantages. Male-driving autosomal SGEs are associated with inferior genetic quality, the most extreme costs arising through recessive lethal mutations or sterility in homozygous carriers [11–13]. In heterozygous males, SGE bearing sperm harm their wild-type bearing counterparts and ensure the SGE's transmission to a large proportion of the offspring [5]. In SGE homozygous males however, sperm bearing homologous copies of the SGE can render each other dysfunctional, leading to strong fertility reduction or even sterility [14,15]. Despite strongly deleterious effects of reduced male fertility or homozygous lethality, autosomal SGEs can be maintained in populations through drive [16]. Females thus face the risk of mating with males of inferior genetic quality with negative effects on the number and genetic quality of their offspring.
When the drive mechanism involves killing or harming sperm not carrying the SGE during spermatogenesis, polyandry can be an effective means of avoiding carriers of SGEs because as a direct consequence of drive, these males have fewer viable or functional sperm [11,17–19]. Indeed, reduced sperm competitiveness of males carrying SGEs has been reported in stalk-eyed flies and several Drosophila species [20–23]. Further support comes from studies reporting associations between female remating rate and sex ratio distorting chromosomes across wild populations of Drosophila and stalk-eyed flies [24–26]. Empirical evidence for the effect of polyandry on autosomal SGEs is however very scarce [19]. Here, we investigated the influence of an autosomal SGE on postcopulatory sexual selection in a vertebrate.
The t haplotype in house mice is a very intensively studied SGE [5]. Typically, t haplotypes are inherited by 90% of the offspring of male carriers (denoted as +/t) and by 50% of offspring of female carriers, but t/t offspring perish in utero owing to recessive lethal mutations [27,28]. Immediate fitness costs associated with the t haplotype are thus related to genetic incompatibility: +/t females mated to +/t males have 40% smaller litters than when mated to +/+ males [28]. +/t females are predicted to avoid this strong cost of genetic incompatibility associated with fertilization by +/t males. There is ample empirical evidence that sexually receptive +/t females prefer the odour and the proximity of +/+ males over +/t males [29]. However, +/+ females might also benefit from avoiding fertilization by +/t males if the t haplotype also exhibits additive detrimental fitness effects, but the evidence so far is mixed (e.g. behavioural dominance: [30,31]).
The basis for the t haplotype's selfishness—arguably its main effect—is its impact on spermatogenesis. Drive in +/t males is due to an elaborate molecular mechanism resulting in abnormal flagellar function of +sperm, comparable to a ‘poison–antidote’ system [32]. This is predicted to have an effect on sperm competitiveness of +/t males through a numerical reduction of functional sperm. To achieve a drive of 90%, most +sperm in a +/t male's ejaculate are rendered dysfunctional, reducing the number of functional sperm by about 45%. Although +/t males have the same number of epididymal sperm as +/+ males, their sperm show reduced velocity and linearity and importantly, fewer sperm at the site of fertilization (reviewed in [33]). In monogamous matings, fertility of +/t males tends to be lower than that of +/+ males [28,31]. Thus, +/t males probably ejaculate fewer functional sperm. However, the effect on the inter-ejaculate competitive ability of the remaining functional sperm remains unknown. Indications for reduced sperm competitive abilities of +/t males are restricted to few studies using very small sample sizes [34] and which did not use controlled matings. Assuming a fair raffle model where the number of functional sperm corresponds to the number of tickets bought in a lottery [35], the predicted paternity share of +/t males is about 35% owing to the reduction in functional sperm numbers.
Here, we used many experimental matings to investigate: (i) sperm competitiveness of +/t males, (ii) fitness consequences for polyandrous females in the form of embryo viability, and (iii) whether +/t and +/+ males invest differentially into sperm production.
2. Material and methods
(a). Experimental animals
We used 90 male and 140 female laboratory-born house mice (Mus musculus domesticus), F1 to F3 descendants from a free-living population of wild house mice in Switzerland [36]. At every generation, we introduce mice from the free-living population into our breeding colony. Laboratory conditions were a reversed 14 L : 10 D cycle (lights on at 17.30) and a temperature of 22–24°C. Food (mouse and rat breeding diet, Provimi Kliba AG) and water were provided ad libitum, paper towels and cardboard served as enrichment and nest building material. Breeding pairs consisted of monogamously paired non-sibling +/+ males and +/t females, producing on average 50% +/t offspring. Offspring were weaned at 28 days after birth and kept in same sex sibling groups in Macrolon Type III cages (425 × 266 × 155 mm). We used +/t and +/+ males and females and diagnosed their t haplotype status before they entered the experiment. An ear punch taken at weaning was used for genotyping and individual marking. t haplotype status was diagnosed by PCR [28,37]. Male mice were separated latest at the onset of aggression between brothers and kept individually in Macrolon Type II cages (180 × 240 × 140 mm). The experimenter was blind with respect to the mice's t genotype during all procedures, including mating trials, female and male dissections, and video observations (see below).
(b). Sperm competition trials
For our experimental matings, we followed a protocol modified after [38]. Details on mating design and paternity assignment are given in the electronic supplementary material. Briefly, we conducted sperm competition trials using full brother pairs differing in t haplotype genotype by mating them to virgin +/+ and +/t females in cycling oestrous. By using full brothers, we largely controlled for potential effects of genetic background and maternal environment on sperm competitiveness. We conducted up to four trials to balance mating order (as there is first male precedence in house mice [38]) and female t genotype. During mating trials, pairs were checked every 1–1.5 h for copulatory plugs indicative of ejaculation [39]. Once a copulatory plug was detected, the female was added to the second male's cage and checked every 30–60 min until either a second copulatory plug was observed or until the beginning of the next dark phase. We confirmed and counted ejaculations using video recordings. To obtain unbiased estimates of paternity share (before t/t embryos are resorbed [28]), we sacrificed females 9 days (±1 day) post coitum using gradual CO2 filling in their home cage and dissected females to retrieve implanted embryos. We scored 12 microsatellites spread across 10 autosomes and assigned paternity using Cervus v. 3.0 [40].
(c). Embryo viability
To investigate fitness consequences for females, we assessed embryo viability based on developmental stage. At day 9, normal embryos have clearly visible somites and forelimb buds begin to form (Theiler Stages 13 or 14 [41]). During dissection, we recorded the number of implantation sites and the development stage of individual embryos. Embryos with normal morphological appearance were classified as viable, whereas embryos with arrested development (i.e. Theiler Stage 10 or earlier) as well as resorbed embryos were classified as inviable.
(d). Male reproductive organs
As we could not measure ejaculate size directly, we investigated potential t haplotype-associated differences in sperm production and storage by weighing testes and epididymides post-mortem.
(e). Statistical analyses
Sample sizes available for statistical analyses are summarized in the electronic supplementary material, table S1. Of the 140 females used for mating trials, 95 mated after an average of two trials (range 1–12). Seventeen females did not become pregnant, 15 did not mate with the second male and remating could not be unambiguously determined for a further six. Because of our focus on postcopulatory processes, trials without ejaculation by the second male were omitted from further analyses, except for analysis of the effect of +/t paternity share on embryo viability (see below). For 16 of the 57 remaining females, we were not able to unambiguously quantify the number of ejaculations. Thus, our final sample sizes were 41 females (320 out of 329 embryos genotyped) for the effect of ejaculation number on paternity share and 57 females (440 out of 453 embryos genotyped) for the other variables.
All statistical analyses were performed in R, v. 3.0.2 [42]. We analysed t paternity share with generalized linear mixed models (GLMMs), using the function glmer in lme4 [43]. The number of embryos sired by the +/t male was included as the dependent variable and the number of embryos genotyped for a given female as the binomial denominator. Mating order, female t genotype, the relative difference in body weight between the competing males and the difference in the number of ejaculations of the +/t versus +/+ male were fitted as fixed effects with biologically relevant two-way interactions. To avoid pseudo-replication, we included male pair as a random factor. We accounted for overdispersion by including an observation-level random effect and compared models based on the Akaike information criterion corrected for small sample sizes (AICc) using the dredge function in MuMIn [44]. To get estimates and confidence intervals (CIs), we back-transformed best model estimates from the logit to the original scale. We obtained approximate 95% confidence intervals by multiplying Student's t-values for our sample sizes by standard errors of the predicted values before back-transformation to the original scale [45].
The proportion of viable embryos was analysed in analogy to t paternity share, using 57 polyandrous females (446 out of 453 embryos classified for viability) and 15 monandrous females (122 out of 124 embryos classified for viability). The delay between mating and dissection did not have an effect on embryo viability and was not included in subsequent models. To test for a benefit of a reduction in +/t paternity share on embryo viability, +/t male paternity share, the female's genotype and an interaction between +/t male paternity share and female genotype were included as fixed effects. Female body weight was included as an additional fixed effect and male pair was included as a random effect.
We analysed testes and epididymides weights with linear models and log-transformed organ and body weight to achieve normality of residuals. Full models included t genotype, body weight and its interaction as fixed effects. We selected the minimal adequate model using stepwise backwards model selection based on log-likelihood.
3. Results
(a). +/t paternity share
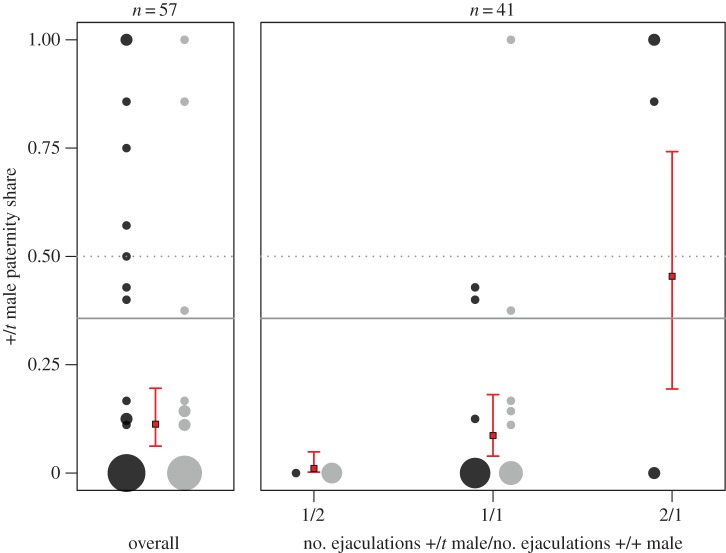
Female t genotype was not retained during model selection for +/t paternity share analysis and females were hence pooled. In 57 trials of polyandrous females, +/t males sired only 57 of the 440 embryos genotyped (12.9%). The GLMM including mating order and the relative weight difference between males performed best as indicated by the lowest AICc value. Here, mating order (z =−4.11, n = 55, p < 0.001) and body weight difference (z = 4.04, n = 55, p < 0.001) had significant effects on paternity share, but +/+ and +/t males did not differ in body weight (ANOVA, F1,74 = 0.12, p = 0.731). When mating first, +/t males sired 21.7% of the offspring as opposed to 4.7% when mating second. The model prediction for mean +/t male paternity share was 11.3% (approx. 95% CI 6.2–19.6%; left chart in figure 1). This strongly differs from the null hypothesis of equal paternity share between +/t and +/+ males (dashed grey line in figure 1; z =−4.33, n = 55, p < 0.001). Notably, the upper confidence level of the +/t paternity share was also well below the adjusted null hypothesis, predicted by the reduction in the number of functional sperm through drive. With 90% drive by the t allele (previously measured in [28]), the majority of +sperm are rendered dysfunctional and are not competitive against other males' sperm (predicted +/t paternity share 35%, solid grey line in figure 1). We obtained an estimate of male drive from 37 embryos sired by a +/t male mated to a +/+ female. Thirty-one out of 37 (84%) embryos paternally inherited the t haplotype, not significantly different from 90% (χ21 = 1.59, p = 0.208). In the reciprocal cross, 60 out of 125 (48%) embryos maternally inherited the t, not different from Mendelian segregation (χ21 = 0.2, p = 0.655).
Figure 1.
Paternity share of +/t males in sperm competition with +/+ males. Shown are overall paternity share (left chart) and paternity share as a function of the number of ejaculations (right chart). The surface area of grey circles is proportional to the number of observations. Colours of circles represent mating order, with dark grey for trials in which the +/t male was first to mate. Mating order did not have a significant effect on paternity share when accounting for the number of ejaculations and is included for illustrational purposes only. Squares and bars represent mean and approximate 95% confidence interval estimates. The grey dotted line shows equal paternity share for +/+ and +/t males and the grey solid line represents the prediction based on a numerical reduction in functional sperm through drive (see main text). (Online version in colour.)
(b). Ejaculation frequency
During video analysis, we found that males ejaculated twice between two cage checks in some of the trials. Thus, in our second model selection approach, we included only trials for which we knew the exact number of ejaculations by both males. The model including only the difference in number of ejaculations between competitors received strongest AICc support. An additional ejaculation by the +/t male enhanced his paternity share to 45% (right chart in figure 1; GLMM: z = 3.895, n = 41, p < 0.001). In 11 out of 41 trials, the first male to mate ejaculated twice, whereas the second male ejaculated twice in only 1 out of 41 trials. Thus, when accounting for the number of ejaculations, neither mating order nor body weight had a significant effect on paternity share. Ejaculation number was independent of male t status, with five +/t males and seven +/+ males ejaculating twice (figure 1; χ21 = 0.05, p = 0.818).
(c). Embryo viability
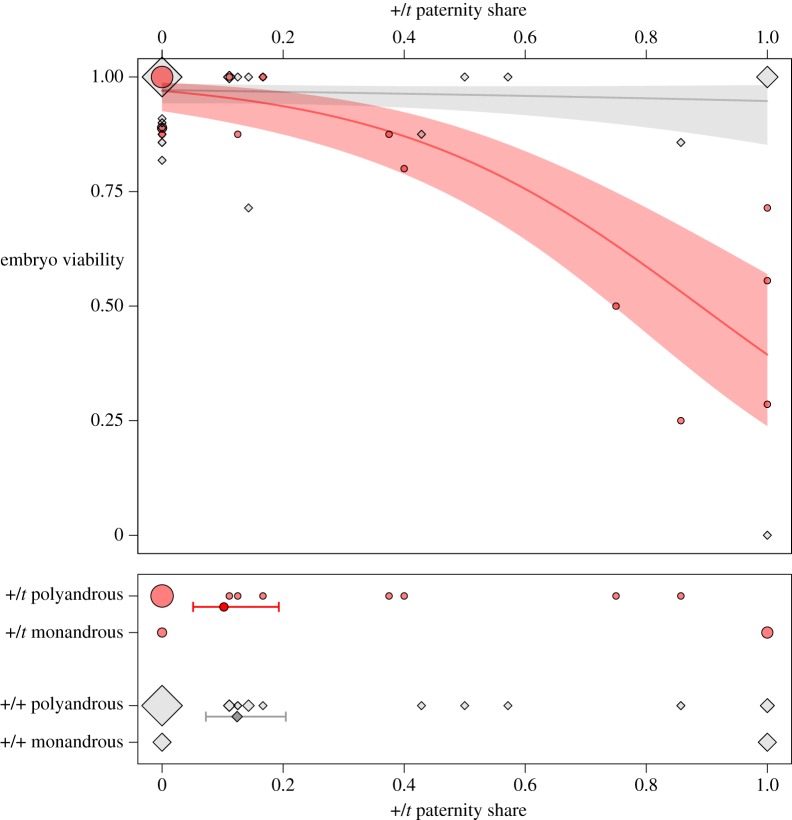
The model best explaining embryo viability included the interaction between +/t male paternity share and female genotype as well as female body weight. Thus, the proportion of viable embryos was significantly influenced by the interaction between +/t paternity share and female genotype, i.e. +/t females had a lower proportion of viable embryos when +/t paternity share increased (top chart in figure 2; GLMM: z = 3.59, n = 70, p < 0.001). Indeed, all 18 embryos that had the t/t genotype were inviable. By contrast, only 8 out of 152 (5.3%) of the +/t embryos and 15 out of 373 (4.0%) of the +/+ embryos were inviable, respectively. Female body weight at the time of mating had a positive effect on embryo viability (GLMM: z = 2.38, n = 70, p = 0.017) but body weight did not differ between +/+ and +/t females (ANOVA, F1,68 = 0.035, p = 0.853) or between monandrous and polyandrous females (F1,68 = 0.71, p = 0.401).
Figure 2.
+/t paternity share in polyandrous and monandrous females (bottom chart) and consequences for embryo viability (top chart). Monandrous females were mated to either a +/t or a +/+ male (+/t male paternity share 1 and 0, respectively), whereas polyandrous females mated with both a +/t and a +/+ male. Colours and shapes indicate female genotype (+/+ in lighter grey diamonds, +/t in darker grey circles). The surface area of diamonds and circles is proportional to the number of observations. Mean and approximate 95% confidence interval estimates are indicated by points and bars (bottom chart) and lines and shaded areas (top chart), respectively. (Online version in colour.)
(d). Male reproductive organs
Both testis and epididymis weight correlated positively with body weight, but there were no differences between +/+ and +/t males in body weight (see § +/t paternity share). Epididymis weight was slightly more strongly correlated to body weight (F1,74 = 3.99, p < 0.001, R2 = 0.12) than was testis weight (F1,74 = 2.93, p = 0.005, R2 = 0.10). Neither organ showed an association with t genotype (testes: F2,73 = −0.68, p = 0.502; epididymis: F2,73 = 0.32, p = 0.750).
4. Discussion
We show that the t haplotype in house mice is associated with a strong disadvantage in postcopulatory competition. +/t males sired dramatically fewer offspring than their +/+ brothers, regardless of mating order. This paternity share was significantly lower than the adjusted null hypothesis (35%), which accounts for the effect of drive on the number of functional sperm in a +/t male's ejaculate. We further show that this severely reduced paternity share results in an immediate benefit for polyandrous +/t females by reducing costly t-associated genetically incompatible fertilizations.
(a). Postcopulatory competition
In sperm competition against +/+ males, +/t males sired only 11% of a female's implanted embryos. Notably, t paternity share was even lower than predicted from the number of functional sperm in a raffle model. If t haplotype drive is achieved by harming +sperm alone, then the 90% drive observed in our study population should reflect a decrease in the number of functional sperm by about 45%, providing an adjusted null hypothesis of about 35% t paternity share. The upper level of the approximate CI (20%) was well below this prediction. This suggests that not only does drive harm +sperm [33], but also damages t sperm in +/t males. The ‘poison–antidote’ mechanism favouring t sperm within a +/t male's ejaculate (see [32] for details) thus appears to be imperfect insofar as it results in a strong between-ejaculate disadvantage when a +/t ejaculate competes against a +/+ ejaculate. Thus, the t haplotype's ‘antidote’ does not appear to provide full protection from the t haplotype's own ‘poisonous’ effect. Previous experimental evidence for a +/t male sperm competition disadvantage has been very scarce. Using artificial insemination of eight +/+ females with equal sperm numbers from +/+ and +/t males, Olds-Clarke & Peitz [34] inferred that the t haplotype was transmitted to 22% (5 out of 23) of the fetuses. This is a broad proxy of the +/t male's paternity share, because assignment depended on the tailless phenotype (genotype T/t) traditionally used for t haplotype detection. Consequently, paternity could be assigned only to offspring that inherited the t from their father and the tailless mutation T (brachyury) from their mother. Thus, accurate phenotypic paternity estimation relied on strong male drive, Mendelian inheritance of T in females and random fusion of the t and T gametes. Given these limitations and the small sample size associated with a large standard error, the authors were unable to conclude whether +/t paternity success was lower than expected from drive (the adjusted null hypothesis). Other studies suggesting a sperm competition disadvantage for +/t males based their estimate of paternity share on low numbers of multiply sired litters [46,47]. Apart from being based on very few litters, these estimates are prone to a biased estimation of +/t male sperm competitiveness, as litters resulting from multiple mating but with exclusive paternity for one male would not have been included. In our mating trials, ejaculation by both males resulted in multiple paternity in only 17 out of 57 litters (29.8%) which is remarkably similar to estimates of multiple paternity from wild populations [28,48,49]. If we had only analysed multiply sired litters, we would have overestimated +/t paternity share by a factor of almost three at 31.5%. Using controlled matings, we were able to overcome major limitations of previous studies and thus, to our knowledge, we provide the first comprehensive estimate of +/t male disadvantage in postcopulatory competition.
(b). +/t male ejaculate features
If sperm competition is the main explanation for the drastically reduced paternity share of +/t males, what sperm features might be causing this effect? While motile sperm from +/t males are hyperactivated sooner and show a faster initial rate of fertilization in vitro, their velocity and linearity are reduced (reviewed in [33]). This results in a lower number of progressive sperm, reducing the number of sperm reaching the site of fertilization in vivo [50]. These t-associated sperm motility features might relate to the paternity pattern found here. Our initial analysis suggested a first male benefit consistent with previous findings in mice [38]. However, closer inspection revealed that differences in the number of ejaculations between competing males were responsible for this order effect (figure 1). Thus, the absence of an order effect when accounting for the number of ejaculations was surprising. This suggests that +/t males ejaculate sperm that fail to benefit from the mating order typically favoured in this species (first male).
As an alternative to intrinsic sperm motility differences between +/+ and +/t males, sperm viability and motility of +/t males may be influenced by the seminal fluids of wild-type males in sperm competition. In the stalk-eyed fly Cyrtodiopsis whitei that harbours a sex chromosome driver, the seminal fluid of wild-type males incapacitates sperm from drive males, strongly reducing their fertilization success [51].
(c). Ejaculate allocation and female choice
Alternative explanations for the observed low paternity share other than intrinsic differences in sperm competitiveness between +/t and +/+ males are: (i) differential sperm investment depending on male genotype, and (ii) female choice.
(i) Males might employ different strategies for gaining paternity, such as differential investment into sperm production and differential ejaculate allocation. Here, the investigated organs involved in sperm production and sperm storage did not differ in size between +/+ and +/t males. This finding has to be interpreted with caution, as the intra-specific correlation between testis/epididymis weight and sperm production may be weak, and cryptic differences in testicular efficiency may remain undetected when looking at simple weight measurements [52]. However, in support of our findings, previous studies of congenic +/+ and +/t males consistently found no differences in the number of stored sperm [33]. The paternity outcome may also be attributed to differences in ejaculate allocation. Our finding that the number of ejaculations affects +/t male paternity share supports ejaculate allocation as a means by which males can affect the outcome of sperm competition. However, +/t males were not more likely to ejaculate twice than wild-type males. In conclusion, given the strong effect of male genotype on paternity share and the significant effect of the number of ejaculations on paternity outcome, we deem it unlikely that comparably minor differences in sperm production or ejaculate investment are responsible for the low +/t paternity share in our experiment.
(ii) Females are known to discriminate between males and to show pre-copulatory mating preferences [53]. In a series of experiments testing olfactory and social female preference, +/t females preferred +/+ males over +/t males, while +/+ females showed no preference [29]. A small paternity bias consistent with mate choice for genetic compatibility has also been found in a wild population [28]. A recent study where females had free access to a +/t and +/+ male found paternity share to be lower for +/t than +/+ males, but was unable to distinguish between pre- and postcopulatory processes [54]. Here, we measured the paternity outcome only when females received ejaculations by both males, thus the only avenue for female choice would be cryptic [55]. In previous studies, transmission of the t haplotype was lower than expected in crosses in which +/t males were mated to +/t rather than +/+ females [28,56], possibly indicating that females may be able to select genetically compatible sperm for fertilization. Although we cannot distinguish between sperm competition and cryptic female choice, we found no direct evidence for discrimination compatible with cryptic female choice for genetic compatibility, as female genotype did not affect the paternity outcome.
(d). Fitness consequences for females
Owing to strong male drive and t homozygote lethality, +/t females mated monandrously to +/t males have much smaller litters than +/t females mated to +/+ males because many offspring from the former mating cross have the lethal genotype t/t ([31,57], this study). Here, we confirm that early embryo lethality in +/t females is a direct consequence of t homozygosity, as all detected t/t embryos were inviable. The proportion of viable embryos decreased with +/t male paternity share in +/t females but not in +/+ females. This has important implications for +/t females. By mating with more than one male, females can increase the probability of fertilization by a genetically compatible +/+ male. This appears to be a direct consequence of incompatible +/t males having a strong disadvantage in sperm competition. Lorch & Chao [18] formally modelled selection for female multiple mating in the presence of fitness reducing mates. They concluded that multiple mating is only favoured when female fitness is a concave-down function of the proportion of costly mates, i.e. females mating with a costly and a non-costly male have less than half their offspring sired by the costly male [18]. We show that the female fitness function is indeed strongly concave-down (figure 2) and thus that female multiple mating can be selectively favoured by the presence of the t haplotype. Compared to randomly mating monandrous females with an average +/t paternity share of 50%, polyandrous females reduce the +/t paternity share (to the left in figure 2) with a positive effect on embryo viability (moving upwards in the top chart of figure 2). When focusing on the immediate negative consequences as we did here, only +/t females benefit from polyandry. However, in natural populations, polyandrous +/+ females could also benefit from avoiding fertilization by +/t males by avoiding maternal investment into sons that are bad sperm competitors [58]. Similarly, females that invest into +/t daughters that face a risk of reduced reproductive success through genetic incompatibility are likely to have lower long-term fitness. Thus, all females potentially benefit from avoiding +/t males, but the magnitude of this benefit will depend on the genotype-specific benefits and the cost of polyandry [54].
(e). Polyandry and the t frequency paradox
The t frequency in natural populations is typically dramatically lower than predicted by theory (the t frequency paradox; for a review, see [59]). As polyandry rates in natural house mouse populations are considerable [28,48,49], and females show high remating rates in the laboratory [60], our results strongly suggest that polyandry is likely to answer this long-standing puzzle in evolutionary genetics. Using a high rate of polyandry and a low sperm competitiveness of +/t males, a modelling approach showed that polyandry alone could account for the t frequency decline observed in the wild population from which our mice were derived [47]. Polyandry might positively correlate with population density in wild populations, because females have more mating opportunities [49], which may account for the fact that t frequencies are typically much lower in large than in small populations [59].
(f). Selfish genetic elements and polyandry
We found that an autosomal SGE has a strong impact on sperm competitiveness in house mice. Our results suggest that not only can polyandry prevent the spread of autosomal drive, but that polyandry is so effective at preventing fertilization by SGE bearing sperm, that even moderate costs to females associated with a driver could cause the evolution of increased polyandry. In modelling scenarios for sex-chromosome-linked male drive, Holman et al. [61] found that polyandry can evolve as an effective response to sperm competition disadvantaged drive if there are additional costs to drive homozygotes.
Our results are in agreement with findings in other species bearing SGEs driving through males. Sex chromosome drivers in several plant and invertebrate species are associated with reduced competitive ability of male gametes, with 20–40% paternity share when averaged across mating order [20–23,62,63]. Similar disadvantages in sperm competition have been found in studies investigating B chromosomes and cytoplasmic incompatibility inducing Wolbachia [57,64,65]. Moreover, in response to a sex ratio biasing SGE, Drosophila pseudoobscura populations evolved higher remating rates and shorter remating latency after only 10 generations of experimental evolution [66]. Here, we show that autosomal drive is associated with an extreme disadvantage in sperm competition in a mammal. Thus, our findings generalize the notion that male drivers cause a disadvantage in sperm competition [19].
Undetected autosomal drive that manipulates spermatogenesis could be common and is likely to incur fitness costs [5,6]. If fitness costs of SGEs arise solely from genetic incompatibility, polyandry is not predicted to evolve even if SGE males have reduced sperm competitiveness [57]. This is because the frequency at which females encounter incompatible mates determines the benefit of polyandry, which cannot offset even mild costs of polyandry when SGE carriers are rare [4,57]. However, if male carriers of SGEs are costly to all females, e.g. owing to reduced fertility, polyandry can readily evolve if SGE-carrying males are disadvantaged in sperm competition [4,18]. Thus, it is possible that polyandry may have evolved, or may persist, in a wide range of species due to its benefits in resisting SGEs.
Acknowledgements
We thank Jari Garbely for genotyping, Gabi Stichel for animal husbandry and Barbara König for support. We also thank Renée Firman, Andri Manser, Tom Price, Laura Travers and two anonymous reviewers for helpful feedback on earlier versions of this manuscript. Mollie Brooks, Manuela Ferrari, Andri Manser and Erik Postma advised on statistical issues.
Ethics
Experimental procedures received ethics approval by the veterinary office of the canton Zurich and were conducted in accordance with Swiss law.
Data accessibility
Data are archived in the Dryad data repository: http://dx.doi.org/10.5061/dryad.m2f45.
Authors' contributions
A.S. participated in the design of the study, carried out the experiment and the statistical analyses, and drafted the manuscript. A.L. conceived of the study and helped draft the manuscript. Both authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This study was supported by the Swiss National Science Foundation grant no. 138389.
References
- 1.Parker GA. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567. ( 10.1111/j.1469-185X.1970.tb01176.x) [DOI] [Google Scholar]
- 2.Jennions MD, Petrie M. 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. Camb. Phil. Soc. 75, 21–64. ( 10.1017/S0006323199005423) [DOI] [PubMed] [Google Scholar]
- 3.Pizzari T, Wedell N. 2013. The polyandry revolution. Phil. Trans. R. Soc. B 368, 20120041 ( 10.1098/rstb.2012.0041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedell N. 2013. The dynamic relationship between polyandry and selfish genetic elements. Phil. Trans. R. Soc. B 368, 20120049 ( 10.1098/rstb.2012.0049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt A, Trivers R. 2006. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Harvard University Press. [Google Scholar]
- 6.Taylor DR, Ingvarsson PK. 2003. Common features of segregation distortion in plants and animals. Genetica 117, 27–35. ( 10.1023/A:1022308414864) [DOI] [PubMed] [Google Scholar]
- 7.Hurst L, Pomiankowski A. 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics 128, 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silver LM. 1985. Mouse t haplotypes. Annu. Rev. Genet. 19, 179–208. ( 10.1146/annurev.ge.19.120185.001143) [DOI] [PubMed] [Google Scholar]
- 9.Sandler L, Hiraizumi Y, Sandler I. 1959. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of segregation-distortion. Genetics 44, 233–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casellas J, et al. 2012. Genome scans for transmission ratio distortion regions in mice. Genetics 191, 247–259. ( 10.1534/genetics.111.135988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeh JA, Zeh DW. 1996. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. Biol. Sci. 263, 1711–1717. ( 10.1098/rspb.1996.0250) [DOI] [Google Scholar]
- 12.Tregenza T, Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 9, 1013–1027. ( 10.1046/j.1365-294x.2000.00964.x) [DOI] [PubMed] [Google Scholar]
- 13.Hartl D, Hiraizumi Y, Crow J. 1967. Evidence for sperm dysfunction as the mechanism of segregation distortion in Drosophila melanogaster. Genetics 58, 2240–2245. ( 10.1073/pnas.58.6.2240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyon M. 1986. Male sterility of the mouse t-complex is due to homozygosity of the distorter genes. Cell 44, 357–363. ( 10.1016/0092-8674(86)90770-1) [DOI] [PubMed] [Google Scholar]
- 15.Hartl D. 1969. Dysfunctional sperm production in Drosophila melanogaster males homozygous for the segregation distorter elements. Proc. Natl Acad. Sci. USA 63, 782–789. ( 10.1073/pnas.63.3.782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl DL. 1970. A mathematical model for recessive lethal segregation distorters with differential viabilities in the sexes. Genetics 66, 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haig D, Bergstrom C. 1995. Multiple mating, sperm competition and meiotic drive. J. Evol. Biol. 8, 265–282. ( 10.1046/j.1420-9101.1995.8030265.x) [DOI] [Google Scholar]
- 18.Lorch PD, Chao L. 2003. Selection for multiple mating in females due to mates that reduce female fitness. Behav. Ecol. 14, 679–686. ( 10.1093/beheco/arg045) [DOI] [Google Scholar]
- 19.Price TA, Wedell N. 2008. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica 132, 295–307. ( 10.1007/s10709-007-9173-2) [DOI] [PubMed] [Google Scholar]
- 20.Wu C-I. 1983. Virility deficiency and the sex-ratio trait in Drosophila pseudoobscura. I. Sperm displacement and sexual selection. Genetics 105, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson GS, Fry CL. 2001. Meiotic drive alters sperm competitive ability in stalk-eyed flies. Proc. Biol. Sci. 268, 2559–2564. ( 10.1098/rspb.2001.1831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atlan A, Joly D, Capillon C, Montchamp-Moreau C. 2004. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J. Evol. Biol. 17, 744–751. ( 10.1111/j.1420-9101.2004.00737.x) [DOI] [PubMed] [Google Scholar]
- 23.Price TA, Bretman AJ, Avent TD, Snook RR, Hurst GDD, Wedell N. 2008. Sex ratio distorter reduces sperm competitive ability in an insect. Evolution 62, 1644–1652. ( 10.1111/j.1558-5646.2008.00386.x) [DOI] [PubMed] [Google Scholar]
- 24.Pinzone CA, Dyer KA. 2013. Association of polyandry and sex-ratio drive prevalence in natural populations of Drosophila neotestacea. Proc. R. Soc. B 280, 20131397 ( 10.1098/rspb.2013.1397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price TA, Bretman A, Gradilla AC, Reger J, Taylor ML, Giraldo-Perez P, Campbell A, Hurst GDD, Wedell N. 2014. Does polyandry control population sex ratio via regulation of a selfish gene? Proc. Biol. Sci. 281, 20133259 ( 10.1098/rspb.2013.3259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson GS, Swallow JG, Christensen SJ, Madden K. 2003. Phylogeography of sex ratio and multiple mating in stalk-eyed flies from southeast Asia. Genetica 117, 37–46. ( 10.1023/A:1022360531703) [DOI] [PubMed] [Google Scholar]
- 27.Klein J, Sipos P, Figueroa F. 1984. Polymorphism of t-complex genes in European wild mice. Genet. Res. 44, 39–46. ( 10.1017/S0016672300026239) [DOI] [Google Scholar]
- 28.Lindholm AK, Musolf K, Weidt A, König B. 2013. Mate choice for genetic compatibility in the house mouse. Ecol. Evol. 3, 1231–1247. ( 10.1002/ece3.534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenington S, Coopersmith C, Williams J. 1992. Genetic basis of mating preferences in wild house mice. Am. Zool. 32, 40–47. ( 10.1093/icb/32.1.40) [DOI] [Google Scholar]
- 30.Lenington S, Drickamer LC, Robinson AS, Erhart M. 1996. Genetic basis for male aggression and survivorship in wild house mice (Mus domesticus). Aggress. Behav. 22, 135–145. () [DOI] [Google Scholar]
- 31.Carroll LS, Meagher S, Morrison L, Penn DJ, Potts WK. 2004. Fitness effects of a selfish gene (the Mus t complex) are revealed in an ecological context. Evolution 58, 1318–1328. ( 10.1111/j.0014-3820.2004.tb01710.x) [DOI] [PubMed] [Google Scholar]
- 32.Herrmann BG, Bauer H. 2012. The mouse t-haplotype: a selfish chromosome: genetics, molecular mechanism, and evolution. In Evolution of the house mouse (eds Macholán M, Baird SJE, Munclinger P, Piálek J.), pp. 297–314. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 33.Olds-Clarke P. 1997. Models for male infertility: the t haplotypes. Rev. Reprod. 2, 157–164. ( 10.1530/ror.0.0020157) [DOI] [PubMed] [Google Scholar]
- 34.Olds-Clarke P, Peitz B. 1985. Fertility of sperm from t/+ mice: evidence that +-bearing sperm are dysfunctional. Genet. Res. 47, 49–52. ( 10.1017/S0016672300024502) [DOI] [PubMed] [Google Scholar]
- 35.Parker GA. 1990. Sperm competition games: raffles and roles. Proc. R. Soc. Lond. B 242, 120–126. ( 10.1098/rspb.1990.0114) [DOI] [Google Scholar]
- 36.König B, Lindholm AK. 2012. The complex social environment of female house mice (Mus domesticus). In Evolution of the house mouse (eds Macholán M, Baird SJE, Munclinger P, Piálek J.), pp. 114–134. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 37.Schimenti J, Hammer M. 1990. Rapid identification of mouse t haplotypes by PCR polymorphism (PCRP). Mouse Genome 87, 108. [Google Scholar]
- 38.Firman RC, Simmons LW. 2008. Polyandry, sperm competition, and reproductive success in mice. Behav. Ecol. 19, 695–702. ( 10.1093/beheco/arm158) [DOI] [Google Scholar]
- 39.Rugh R. 1968. The mouse: its reproduction and development. Minneapolis, MN: Burgess Publishing Company. [Google Scholar]
- 40.Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. ( 10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 41.Theiler K. 1989. The house mouse: atlas of embryonic development. New York, NY: Springer. [Google Scholar]
- 42.R Core Team. 2014. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 43.Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: linear mixed-effects models using eigen and S4. R package v. 1.0-6 See http://CRAN.R-project.org/package=lme4. [Google Scholar]
- 44.Bartoń K. 2013. MuMIn: multi-model inference. R package v. 1.9.13 See http://CRAN.R-project.org/package=MuMIn. [Google Scholar]
- 45.Crawley MJ. 2007. The R book. Chichester, UK: John Wiley and Sons. [Google Scholar]
- 46.Ardlie KG, Silver LM. 1996. Low frequency of mouse t haplotypes in wild populations is not explained by modifiers of meiotic drive. Genetics 144, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manser A, Lindholm AK, König B, Bagheri HC. 2011. Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evolution 65, 2435–2447. ( 10.1111/j.1558-5646.2011.01336.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firman RC, Simmons LW. 2008. The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J. Evol. Biol. 21, 1524–1533. ( 10.1111/j.1420-9101.2008.01612.x) [DOI] [PubMed] [Google Scholar]
- 49.Dean MD, Ardlie KG, Nachman MW. 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol. Ecol. 15, 4141–4151. ( 10.1111/j.1365-294X.2006.03068.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tessler S, Olds-Clarke P. 1981. Male genotype influences sperm transport in female mice. Biol. Reprod. 24, 806–813. ( 10.1095/biolreprod24.4.806) [DOI] [PubMed] [Google Scholar]
- 51.Fry CL, Wilkinson GS. 2004. Sperm survival in female stalk-eyed flies depends on seminal fluid and meiotic drive. Evolution 58, 1622–1626. ( 10.1111/j.0014-3820.2004.tb01743.x) [DOI] [PubMed] [Google Scholar]
- 52.Firman RC, Garcia-Gonzalez F, Thyer E, Wheeler S, Yamin Z, Yuan M, Simmons LW. 2015. Evolutionary change in testes tissue composition among experimental populations of house mice. Evolution 69, 848–855. ( 10.1111/evo.12603) [DOI] [PubMed] [Google Scholar]
- 53.Andersson M. 1994. Sexual selection. Princeton, NJ: Princeton University Press. [Google Scholar]
- 54.Manser A, König B, Lindholm AK. 2014. Female house mice avoid fertilization by t haplotype incompatible males in a mate choice experiment. J. Evol. Biol. 28, 54–64. ( 10.1111/jeb.12525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eberhard WG. 1996. Female control: sexual selection by cryptic female choice. Princeton, NJ: Princeton University Press. [Google Scholar]
- 56.Bateman N. 1960. Selective fertilization at the T-locus of the mouse. Genet. Res. 1, 226–238. ( 10.1017/S0016672300000215) [DOI] [Google Scholar]
- 57.Champion de Crespigny FE, Hurst LD, Wedell N. 2008. Do Wolbachia-associated incompatibilities promote polyandry? Evolution 62, 107–122. ( 10.1111/j.1558-5646.2007.00274.x) [DOI] [PubMed] [Google Scholar]
- 58.Keller L, Reeve H. 1995. Why do females mate with multiple mates? The sexually selected sperm hypothesis. Adv. Study Behav. 24, 291–315. ( 10.1016/s0065-3454(08)60397-6) [DOI] [Google Scholar]
- 59.Ardlie KG. 1998. Putting the brake on drive: meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 14, 189–193. ( 10.1016/S0168-9525(98)01455-3) [DOI] [PubMed] [Google Scholar]
- 60.Rolland C, MacDonald D, de Fraipont M, Berdoy M. 2003. Free female choice in house mice: leaving best for last. Behaviour 140, 1371–1388. ( 10.1163/156853903771980639) [DOI] [Google Scholar]
- 61.Holman L, Price TA, Wedell N, Kokko H. 2015. Coevolutionary dynamics of polyandry and sex-linked meiotic drive. Evolution 69, 709–720. ( 10.1111/evo.12595) [DOI] [PubMed] [Google Scholar]
- 62.Taylor D, Saur M, Adams E. 1999. Pollen performance and sex-ratio evolution in a dioecious plant. Evolution 53, 1028–1036. ( 10.2307/2640808) [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson GS, Johns PM, Kelleher ES, Muscedere ML, Lorsong A. 2006. Fitness effects of X chromosome drive in the stalk-eyed fly, Cyrtodiopsis dalmanni. J. Evol. Biol. 19, 1851–1860. ( 10.1111/j.1420-9101.2006.01169.x) [DOI] [PubMed] [Google Scholar]
- 64.Champion de Crespigny FE, Wedell N. 2006. Wolbachia infection reduces sperm competitive ability in an insect. Proc. R. Soc. B 273, 1455–1458. ( 10.1098/rspb.2006.3478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beukeboom LW. 1994. Phenotypic fitness effects of the selfish B chromosome, paternal sex ratio (PSR) in the parasitic wasp Nasonia vitripennis. Evol. Ecol. 8, 1–24. ( 10.1007/BF01237662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price TA, Hodgson DJ, Lewis Z, Hurst GDD, Wedell N. 2008. Selfish genetic elements promote polyandry in a fly. Science 322, 1241–1243. ( 10.1126/science.1163766) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are archived in the Dryad data repository: http://dx.doi.org/10.5061/dryad.m2f45.