Abstract
Biological cells sense and respond to mechanical forces, but how such a mechanosensing process takes place in a nonlinear inhomogeneous fibrous matrix remains unknown. We show that cells in a fibrous matrix induce deformation fields that propagate over a longer range than predicted by linear elasticity. Synthetic, linear elastic hydrogels used in many mechanotransduction studies fail to capture this effect. We develop a nonlinear microstructural finite-element model for a fibre network to simulate localized deformations induced by cells. The model captures measured cell-induced matrix displacements from experiments and identifies an important mechanism for long-range cell mechanosensing: loss of compression stiffness owing to microbuckling of individual fibres. We show evidence that cells sense each other through the formation of localized intercellular bands of tensile deformations caused by this mechanism.
Keywords: fibrous matrix, buckling, three-dimensional traction force, cell mechanics
1. Introduction
Physical cues control cell behaviour through various mechanisms collectively referred to as mechanotransduction [1]. For example, the stiffness of a cell's environment controls cellular morphology, migration and development [2]. Equally important is the response of cells to direct physical forces either through cell–cell adhesions [3,4] or through the extracellular matrix [5–7]. Nearly all previous work on mechanotransduction has used synthetic, linear elastic gels [8]. The mechanical properties of physiological extracellular environment, however, deviate entirely from simple homogeneous linear elasticity. Natural fibrous matrices exhibit strain stiffening [9], tensile normal strains under shear loading [10], negative compressibility [11] and lower stiffness in compression than in tension [10]. These nonlinear properties of biological gels can have a dramatic effect on behaviours like cell spreading [12].
Various models have simulated nonlinearity of fibrous biological materials, but relatively few have considered local, non-uniform deformations in such nonlinear inhomogeneous materials [13,14]. Instead, nearly all previous studies have focused on homogeneous shearing [9,15–17] or uniaxial tension [11] of the bulk material. These studies of uniform deformations have revealed novel constitutive behaviour of fibrous materials, but they fail to simulate deformations similar to those applied by a cell. By contracting and changing shape, cells apply localized forces to their surroundings, resulting in inhomogeneous stress and deformation fields in the matrix. Given the lack of theoretical and experimental studies of cell–matrix interactions at the local scale, there remains a need to quantify cell-generated forces and displacements and to discern how cells respond to nonlinear properties of fibrous materials at the scale sensed by the cell.
Here we experimentally measure three-dimensional cell-induced matrix displacements and report two findings: (i) displacements decay much slower with distance from the cell than predicted by linear elasticity; (ii) multiple cells cause localized matrix densification and fibre alignment in tether-like bands joining them. We hypothesize that the mechanism responsible for these phenomena is loss of compression strength owing to microbuckling of individual fibres. To test this claim, we develop a microstructural finite-element (FE) network model of the fibrin matrix. Buckling of individual fibres is modelled by a loss of stiffness in compression for network elements. Our model agrees with previous experimental observations for fibrin, and it predicts both the slow decay of displacements and localization of intercellular tethers. Variants of the model without loss of stiffness in compression fail to predict these effects. The long range of cell-generated displacements and stresses, and the localization into intercellular tensile tethers, allow cells to sense each other and their surroundings over larger distances through a fibrous matrix than through homogeneous hydrogels with linear elastic behaviour. We show evidence that cells respond to localized tension by growing protrusions towards one another, guided by the dense aligned fibres in tethers. This points to fibre microbuckling as an important mechanism responsible for enhanced range of cell mechanosensing in fibrous matrix environments.
2. Results
2.1. Cell-induced matrix displacements
We motivate our model by first considering cell-induced displacements within a three-dimensional fibrous matrix during initial cell spreading. A cell seeded in a three-dimensional matrix initially applies tensile tractions to the fibres by undergoing uniform isotropic contraction while in an essentially spherical state. This suggests Eshelby's solution for a contracting spherical inclusion in a homogeneous, linear elastic, infinite medium [18], as a simple analytical model for cell-induced matrix deformation. In this solution, the displacement magnitude u = u(r) scales as u(r) ∼ r−2 with distance r from the cell centre. Stress components, e.g. the radial component σrr, scale as σrr ∼ r−3. A spreading, elongated, ellipsoidal cell with polarized alignment, applies tractions equivalent to equal and opposite forces at its poles, i.e. a dipole [19], in view of force equilibrium. Displacements owing to a dipole in a three-dimensional linear elastic continuum also scale as u ∼ r−2. One would thus expect displacements induced by a spreading cell in a three-dimensional matrix to scale similarly.
Using confocal microscopy and digital volume correlation [14,20,21], we measure displacements induced by isolated fibroblast cells embedded in a three-dimensional fibrin matrix. Displacements induced by the cells are largest near to the cell and decrease with distance from the cell (figure 1a). We quantify the rate at which displacements decay over distance by computing displacements along linear paths starting at the centre of cell and ending approximately 100 µm away (figure 1a, white line). Experimental data from multiple different cells are plotted on a logarithmic scale in figure 1b. Data are fit to the form u(r) = Ar−n. Here A and n are constants; n > 0 is a decay power. The larger the value of n, the faster the displacement u decays with distance r from the cell centre. Fits of the experimental data yield n = 0.52 (mean over data from six cells during multiple time points), indicating that displacements decay much slower than predicted by the linear elastic solution n = 2. The ratio of the RMS errors of fits to u ∼ r−0.52 and u ∼ r−2 is 0.14 ± 0.07 (mean ± s.d.); hence the scaling u ∼ r−0.52 describes cell-induced displacements in a fibrin matrix far better than the three-dimensional linear elastic scaling u ∼ r−2.
Figure 1.
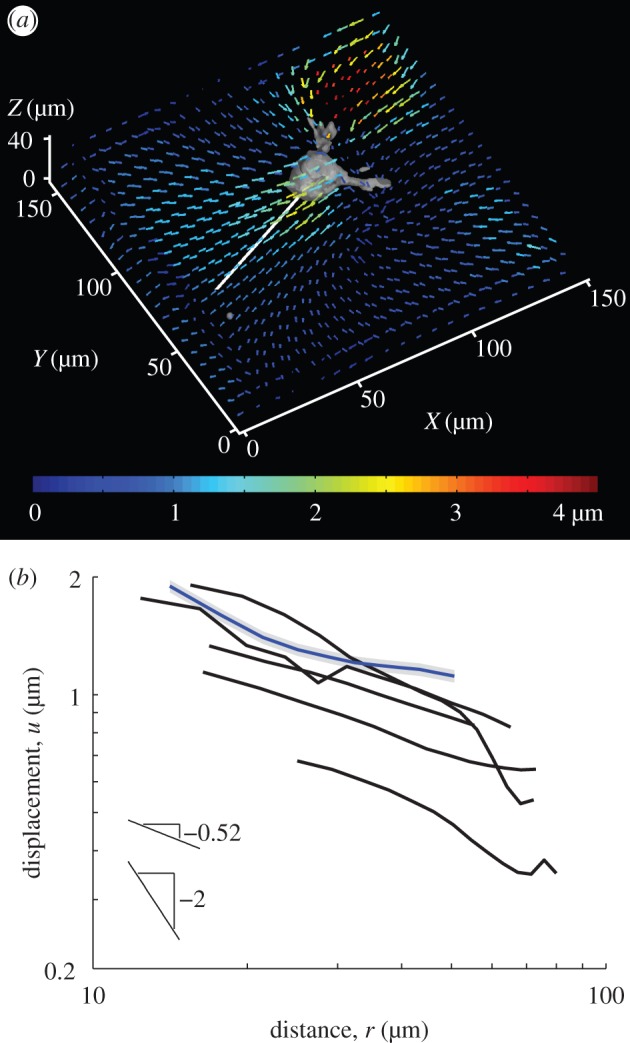
Experimentally measured displacements induced by isolated cells embedded within a three-dimensional fibrous matrix. (a) The coloured quivers plot three-dimensional matrix displacement vectors applied by a cell to a three-dimensional fibrin matrix. Paths (white) are chosen proceeding outward from the cell body. (b) Displacement magnitudes along the paths are averaged for multiple time points and plotted. Each curve is for a different cell. The blue curve shows displacements for the cell in (a). The grey shading behind the blue curve shows typical error of the displacement measurement after averaging. Data used to generate these curves is in the electronic supplementary material.
A striking difference between fibrin networks and homogeneous gel matrices is the phenomenon of buckling of individual fibres under compression (e.g. [22, fig. 5]). This is directly responsible for the decreased ability of fibrin networks to sustain compressive stresses. Each fibre has very low resistance to bending, much like a flexible string [23]. If one pulls at the ends of a string, it resists tension. If one pushes the ends of a string towards each other, the string bends easily without resisting compression (i.e. it buckles), and this buckling can change the mechanical response of a network [24]. Fibrin exhibits a larger stiffness in tension than compression [10,25] owing to buckling of individual fibres under compression [15,22,26,27]. Is this nonlinearity responsible for the discord between the observed displacement scaling and the prediction based on a linear elastic matrix assumption? While it may be possible to address this via a continuum model for a material with lower stiffness in compression [28], here we present a simple theoretical continuum argument, which we will investigate in detail using a discrete model. As the cell exerts radial contractile traction forces, the stress tensor in the matrix has a tensile (positive) radial component in three-dimensional spherical coordinates, and two contractile (negative) hoop (angular) components. Assuming the individual fibres of the fibrin matrix buckle under a small compressive load, the contractile hoop components of the stress tensor are small and can be neglected. This assumption reduces the radial equilibrium equation [29] to
| 2.1 |
Solving equation (2.1) gives 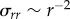 . Thus, stress owing to cell contraction is transmitted over a longer range than under the scaling
. Thus, stress owing to cell contraction is transmitted over a longer range than under the scaling 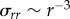 predicted by linear elasticity. Assuming piecewise linear stress–strain relations with zero stiffness in compression, σrr is proportional to the radial strain du(r)/dr which gives u ∼ r−1 (coupling between σrr and the hoop strains γθθ, γϕϕ vanishes due to hyperplastic reciprocity: ∂σrr/∂γθθ = ∂σθθ/∂γrr = 0 as σθθ = 0 in the compressive regime. For more details on a hyperelastic material model that leads to equation (2.1) as a special case, see [28].) The scaling from this simple analysis, u ∼ r−1, points towards displacements that propagate over a longer range than the three-dimensional linear elastic scaling, u ∼ r−2. Furthermore, the scaling from the theoretical analysis is closer to the experimentally observed scaling, u ∼ r−0.52 than to the linear elastic one. This plausibility argument shows the right trend, but ignores the inhomogeneous and discrete nature of the fibrin network. To account for these factors, we turn to a microstructural network model.
predicted by linear elasticity. Assuming piecewise linear stress–strain relations with zero stiffness in compression, σrr is proportional to the radial strain du(r)/dr which gives u ∼ r−1 (coupling between σrr and the hoop strains γθθ, γϕϕ vanishes due to hyperplastic reciprocity: ∂σrr/∂γθθ = ∂σθθ/∂γrr = 0 as σθθ = 0 in the compressive regime. For more details on a hyperelastic material model that leads to equation (2.1) as a special case, see [28].) The scaling from this simple analysis, u ∼ r−1, points towards displacements that propagate over a longer range than the three-dimensional linear elastic scaling, u ∼ r−2. Furthermore, the scaling from the theoretical analysis is closer to the experimentally observed scaling, u ∼ r−0.52 than to the linear elastic one. This plausibility argument shows the right trend, but ignores the inhomogeneous and discrete nature of the fibrin network. To account for these factors, we turn to a microstructural network model.
2.2. Model
We develop a FE-based microstructural model consisting of a two-/three-dimensional network of linear elements representing fibres. This model expands on one that we have recently developed [14]. Each element undergoes uniaxial tension/compression and rotates with no resistance. We model buckling of fibres as a loss of stiffness in compression in the stress–strain relation of individual elements. This agrees qualitatively with observed behaviour in similar systems [30]. In the context of our model, ‘microbuckling’ will refer to elements obeying a stress–strain relation where the stiffness (slope) under compression is smaller than the stiffness under tension (figure 2a, blue line). In the following simulations, we use a ratio of stiffness in compression to stiffness in tension ρ = 0.1. While the choice of ρ = 0.1 is arbitrary, we find that any positive ratio of stiffnesses ρ significantly less than unity yields similar results. By contrast, ‘no microbuckling’ will refer to elements with ρ = 1, i.e. elements with a linear stress–strain relation without a reduced compression stiffness. For most simulations, networks comprise elements with a bilinear stress–strain relationship (figure 2a, different slopes in tension and compression). We will also account for the possibility of entropic elasticity by employing a wormlike chain-type (WLC) stress–strain relationship [9,23], where the stiffness increases with strain in tension (figure 2b). The elements connect an array of nodes as in figure 2c. Randomness is added to nodal positions to simulate the random array of fibres of different lengths typical of a fibrous network (figure 2d).
Figure 2.
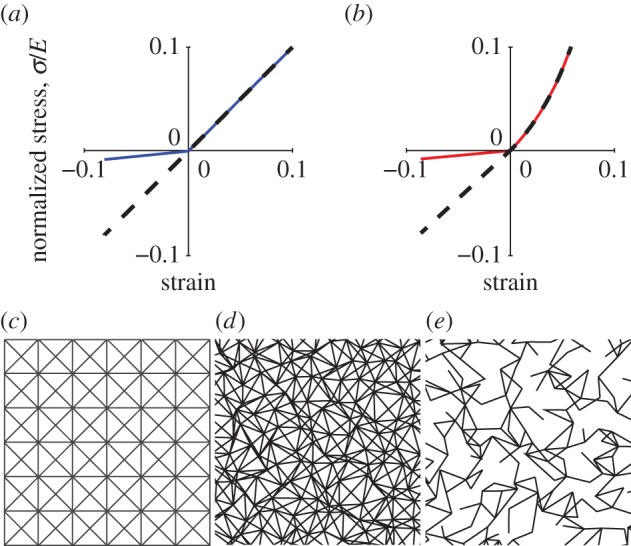
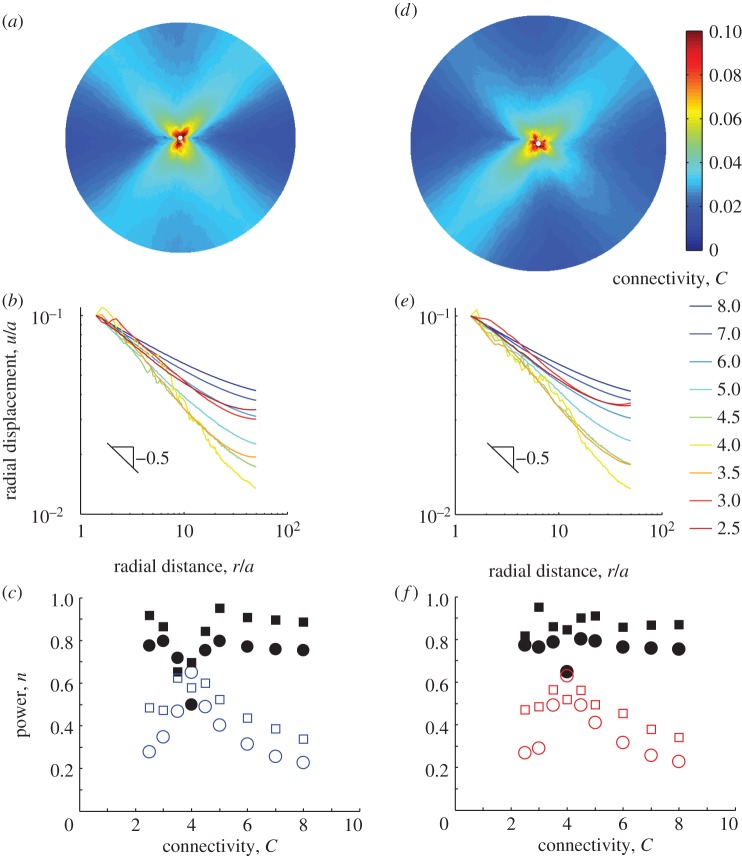
Finite-element model network details. (a) Stress–strain curves for bilinear model. Stress σ is normalized by Young's modulus E. Dashed black: linear without microbuckling (ρ = 1); solid blue: bilinear with microbuckling (ρ = 0.1). (b) Normalized stress–strain curves for the strain-stiffening model, which exhibits WLC-like behaviour in tension. For this model ρ is defined as the slope upon approaching the origin from the left divided by the slope upon approaching the origin from the right. A continuous slope at the origin (ρ = 1) was used to simulate non-buckling elements (dashed black line) and a discontinuous slope (ρ = 0.1) was used to simulate microbuckling elements (solid red line). (c) Network array. (d) Randomized network, C = 8. (e) Network with reduced connectivity, C = 3.
Another important aspect of actual fibrin networks is their low connectivity, or coordination number C, i.e. the average number of fibres meeting at a node. The network of figure 2c,d has C = 8, while actual fibrin often has a typical value of C = 3 [31]. This is below the critical value for rigidity, C = 6 or 4 for three- and two-dimensional networks, respectively. As a result, fibrin is typically a ‘floppy’ network, and this affects its mechanical properties [31]. To obtain a model network with lower connectivity (such as C = 3 in figure 2e), we removed elements at random from the original C = 8 network of figure 2d. As in [31], deleted elements were replaced by weak elements, whose stiffness was six orders of magnitude less than that of the deleted ones; this ensured stability of numerical calculations.
In contrast to previous models that focus on the macroscale behaviour of a fibre network [15–17], we simulate the inhomogeneous, localized displacements induced in a fibrin matrix by an embedded cell. We begin with two-dimensional FE simulations where the cell is modelled as a contracting circle. The matrix occupies the region a < r < b, where r is distance from the cell centre; here a is the cell radius, and b/a = 50. The outside boundary r = b is free (a zero traction boundary condition is imposed). The cell boundary r = a undergoes a radial contractile displacement u(a) = −0.1a. Simulations were performed for different connectivities in the interval 2.5 ≤ C ≤ 8 for bilinear element networks with microbuckling and without. The displacement magnitude u was computed (figure 3a), averaged around the circular region (figure 3b; electronic supplementary material, figure S1a), and fit against distance from the centre of the circular region r to u = Ar−n for the constants A and n. Results, n plotted versus connectivity C, are shown in figure 3c. In general, the decay power n for networks with microbuckling (figure 3c, open circles) is substantially lower, by at least 0.4, than for networks without microbuckling of fibres (figure 3c, black circles). This is true for a wide range of connectivities, with an exception near the critical value C = 4; for these values n ≈ 0.6 in both types of networks. We observe larger spatial inhomogeneities of displacement at the scale of individual fibres in networks with C = 4 than in those with both subcritical and supercritical connectivity (figure 3b; electronic supplementary material, figure S2). These fluctuations are due to the change in phase from subcritical to supercritical connectivity as detailed elsewhere [31,32]. For the case without microbuckling (i.e. with linear stress–strain relation) as individual elements have linear stress–strain behaviour, we compare the displacement with the linear elastic two-dimensional solution u = Ar−n + Brn for the constants A, B and n. Except near C = 4, we find n = 0.89 ± 0.04 (mean ± standard deviation, essentially independent of C over all connectivities). This value of n = 0.89 is close to the two-dimensional linear elastic solution n = 1. Connectivity does not appear to play a major role in displacement decay except near the critical value. We find no change in these conclusions when the zero traction boundary condition is replaced by a zero displacement condition fixing the external boundary (see electronic supplementary material, figure S3). Thus, we conclude microbuckling is crucial for the slow decay of displacements.
Figure 3.
Long-range propagation of displacements is due to microbuckling. (a) Inhomogeneous forces, like those applied by a cell, are modelled by a circle of radius a contracting in a circular region with radius b = 50a. Contours of normalized displacement u/a are shown here for the case of the bilinear model (figure 2a) with microbuckling (ρ = 0.1) and C = 3. For a cell of radius 10 µm, the applied displacement u/a = 0.1 would correspond to 1 µm. (b) Displacements are averaged around a circle of radius r about the centre of the model and plotted for simulations that used different connectivities ranging from C = 2.5 to C = 8. All curves show long-range propagation of displacements with slopes ≈−0.5. At the critical connectivity, C = 4, displacements exhibit spatial inhomogeneities, resulting in fluctuations. (c) Decay power n versus connectivity C. Circles show fits to u = Ar−n; squares show fits to u = Ar−n + Brn. Solid black symbols represent fibres that do not buckle (ρ = 1); open symbols represent fibres that do buckle (ρ = 0.1). Most powers n for the case of microbuckling ρ = 0.1 are ≈0.5, in agreement with the slope of −0.5 observed in (b). The value of n ≈ 0.5 indicates displacement propagate over a longer range than predicted by linear elasticity, for which n = 1 in two dimensions. Simulations are repeated for the strain-stiffening WLC-type relationship (figure 2b). (d) Contours of displacement u/a for the strain-stiffening relationship with microbuckling (ρ = 0.1). (e) Averaged displacements and (f) decay powers n for the strain-stiffening relationship. As in (c), circles show fits to u = Ar−n; squares show fits to u = Ar−n + Brn. Solid black symbols represent fibres that do not buckle (ρ = 1); open symbols represent fibres that do buckle (ρ = 0.1).
The long range of cell-induced displacements has been previously attributed to strain stiffening [12], but this has been disputed [33]. We observe that the experiments of [12] were performed on fibrin, which exhibits microbuckling. Also, Rudnicki et al. [33] provide evidence against strain stiffening as the underlying mechanism, but do not seem to propose an alternative. To help settle this, we repeated our simulations with elements whose stress–strain curve is of WLC type and stiffens in tension (figure 2b). Two versions of stiffening WLC stress–strain curves were compared. A curve whose slope is continuous at zero strain and increases smoothly in tension models a tension-stiffening material that does not undergo microbuckling (black dashed line, figure 2b). The alternative stress–strain curve has a discontinuous slope at zero strain (10 times smaller than the tangent stiffness for small tensile strain). It models microbuckling (red solid line, figure 2b) combined with tension stiffening. In all cases, values of the decay exponent n from fits for WLC networks (figure 3d–f; electronic supplementary material, figure S1c) agreed well with fits for bilinear networks of the same connectivity and same (buckling or non-buckling) type (figure 3c). This provides strong evidence that the tension-stiffening nonlinearity in the absence of microbuckling is not the cause of the slow displacement decay that we observe.
Until now, we have considered round cell geometries, which do not capture the elongated shape of spread cells. For an anisotropic cell contracting along its long axis, an ellipsoid more accurately captures the cell's shape. For this geometry, linear elasticity predicts that displacements far from the cell scale as 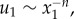 where n = 2 in three dimensions, n = 1 in two dimensions; x1 is the distance along the major axis from the centre of the ellipsoid (or ellipse in two dimensions) and u1 is the displacement in the x1 direction [18]. To compare with the linear elastic solution, we placed in our fibrous network model an ellipse with a ratio of semi-major and semi-minor axes a1/a2 = 4. As with the contracting circle, the matrix occupied a circular region of radius b = 50a with the nodes on the boundary r = b free and a defined for the ellipse as
where n = 2 in three dimensions, n = 1 in two dimensions; x1 is the distance along the major axis from the centre of the ellipsoid (or ellipse in two dimensions) and u1 is the displacement in the x1 direction [18]. To compare with the linear elastic solution, we placed in our fibrous network model an ellipse with a ratio of semi-major and semi-minor axes a1/a2 = 4. As with the contracting circle, the matrix occupied a circular region of radius b = 50a with the nodes on the boundary r = b free and a defined for the ellipse as 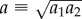 . Contractile displacements were applied on the boundary of the ellipse, with non-zero component u1(x1) = −0.1a(x1/a1) (along the long axis of the ellipse). This is equivalent to subjecting the ellipse to a negative uniaxial strain along the ellipse's long axis. The largest magnitude of contractile displacement is |u1(a1)| = 0.1a (at the ellipse tip), the same value as for the contracting circle. Displacements along the axis of the ellipse (figure 4a) appear to scale similarly to the displacements induced by the contracting circle (figure 3b). Indeed, the fittings to
. Contractile displacements were applied on the boundary of the ellipse, with non-zero component u1(x1) = −0.1a(x1/a1) (along the long axis of the ellipse). This is equivalent to subjecting the ellipse to a negative uniaxial strain along the ellipse's long axis. The largest magnitude of contractile displacement is |u1(a1)| = 0.1a (at the ellipse tip), the same value as for the contracting circle. Displacements along the axis of the ellipse (figure 4a) appear to scale similarly to the displacements induced by the contracting circle (figure 3b). Indeed, the fittings to 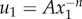 show decay powers n that are significantly smaller for networks with microbuckling (figure 4b, ρ = 0.1) than without (figure 4b, ρ = 1; electronic supplementary material, figure S1b). Like the contracting circle, the ellipse exhibits an exception at the critical connectivity C = 4. The trend shown in figures 3 and 4 is clear: microbuckling results in cell-induced displacements that propagate over a longer range than predicted by linear elasticity for both a contracting circle and a contractile ellipse.
show decay powers n that are significantly smaller for networks with microbuckling (figure 4b, ρ = 0.1) than without (figure 4b, ρ = 1; electronic supplementary material, figure S1b). Like the contracting circle, the ellipse exhibits an exception at the critical connectivity C = 4. The trend shown in figures 3 and 4 is clear: microbuckling results in cell-induced displacements that propagate over a longer range than predicted by linear elasticity for both a contracting circle and a contractile ellipse.
Figure 4.
Simulated displacements due to an elongated cell. An ellipse with a ratio of semi-major to semi-minor axes a1/a2 = 4 is simulated contracting along its long axis in a circular region of radius b = 50a where 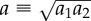 . (a) Displacements along the major axis u1 are plotted against distance along the axis from the centre of the ellipse x1 for connectivities ranging from C = 2.5 to C = 8. As in figure 3, fluctuations are observed for connectivites near the critical value C = 4. (b) Displacements far from the cell (i.e. for x1/a > 5) are fit to
. (a) Displacements along the major axis u1 are plotted against distance along the axis from the centre of the ellipse x1 for connectivities ranging from C = 2.5 to C = 8. As in figure 3, fluctuations are observed for connectivites near the critical value C = 4. (b) Displacements far from the cell (i.e. for x1/a > 5) are fit to 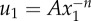 . Solid black circles represent decay powers n for fibres that do not buckle (ρ = 1); open blue circles represent n for fibres that do buckle (ρ = 0.1). For simulations with buckling, except near the critical connectivity C = 4, decay powers are smaller than the linear elastic solution n = 1 and smaller than simulations without buckling.
. Solid black circles represent decay powers n for fibres that do not buckle (ρ = 1); open blue circles represent n for fibres that do buckle (ρ = 0.1). For simulations with buckling, except near the critical connectivity C = 4, decay powers are smaller than the linear elastic solution n = 1 and smaller than simulations without buckling.
We also performed three-dimensional simulations (contracting spherical cell), with similar conclusions. We recall that the three-dimensional linear elastic solution predicts u ∼ r−2. The theoretical argument based on equation (2.1) gives u ∼ r−1, while a fit to our experiments yields u ∼ r−0.52. For three-dimensional networks (microbuckling bilinear elements) with C = 14, a fit to u = Ar−n gives n = 0.67. For C = 3.5 (below the critical value for rigidity C = 6), we found n = 0.82 (electronic supplementary material, figure S4). These results combine to show that microbuckling of fibres is the key mechanism responsible for the longer range of cell-induced deformations in a fibrin matrix.
2.3. Tethers
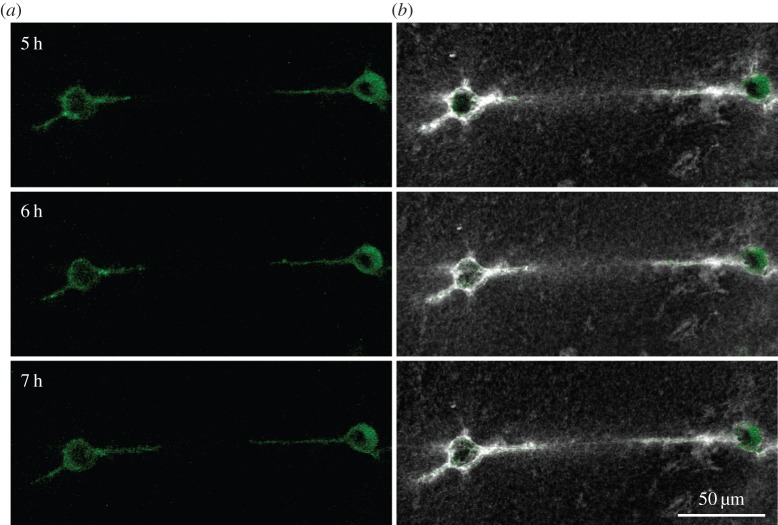
Can cells exploit the long propagation range of matrix deformations they themselves induce for sensing the presence of other cells? We use confocal microscopy to visualize both the matrix and multiple fibroblast cells embedded in it. We observe that cells whose distance from each other is of the order of 10 cell diameters are connected to each other by linear bands consisting of aligned and densely packed matrix fibres (figure 5). Within these ‘tethers' fibres appear to be in tension in the direction joining the cells. These tethers also occur between multicellular explants in a fibrous matrix [34,35], but the mechanism for their formation remains unknown. The tethers extend far beyond a single cell's protrusion (figure 5). Matrix remodelling by degradation or deposition cannot be responsible for alignment at such a large distance from the cell. This leads us to examine the hypothesis that tethers form owing to tensile forces.
Figure 5.
Pairs of cells spread toward one another along tethers. (a) The cells (green), and (b) the cells (green) with the matrix (grey/white) at the same time point. The cells apply tensile force to the fibrin matrix resulting in matrix tethers connecting the cells (white). These tethers have a high density of matrix fibres, as apparent by the bright fluorescent signal in the space between the cells. The cells then spread along these tethers. Times are hours after the cells were seeded in the fibrin matrix.
A previous model has shown that a point force in a fibrous medium induces forces which propagate through tether-like paths [36]. The point force loading of this previous model was not intended to simulate forces owing to cells, which maintain force equilibrium while pulling on the matrix. To investigate the physical mechanism of tether formation, we used our FE network model with microbuckling to simulate a pair of contracting cells. A symmetric boundary condition is imposed at the bottom of a square region containing a circle of radius a (the other boundaries are free of applied tractions). By symmetry this is equivalent to a pair of identical contracting circular cells in the fibrous matrix. We apply an isotropic inward radial displacement of 0.1a to the circular region. For a cell with a radius of a = 10 µm, this value of 0.1a corresponds to 1 µm, in agreement with the experimental data (figure 1b).
A different model [37] requires cell displacements nearly an order of magnitude higher than the experimentally observed value of 1 µm in order to predict appreciable interaction between cells. The simulated tensile strains in our network occur almost entirely in the band between the two cells, along aligned linear paths formed by elements in tension (figure 6a). Compressive strains localize perpendicular to these tensile tethers (figure 6b). Owing to low compression stiffness (microbuckling), the magnitude of compressive strain in elements roughly perpendicular to the tether is more than twice the magnitude of the tensile strain. Thus within the tether, the trace of the strain tensor, or the volumetric strain, is negative, consistent with the observation that matrix fibre density increases between pairs of cells (figure 5). When simulating networks without microbuckling, we found no such tethers forming; instead, tensile strains had a nearly radially symmetric distribution around each cell (electronic supplementary material, figure S5). Thus, we conclude that localization of matrix deformation caused by multiple cells in the tensile, tether-like regions joining those cells occurs because of microbuckling of fibres normal to the tethers.
Figure 6.
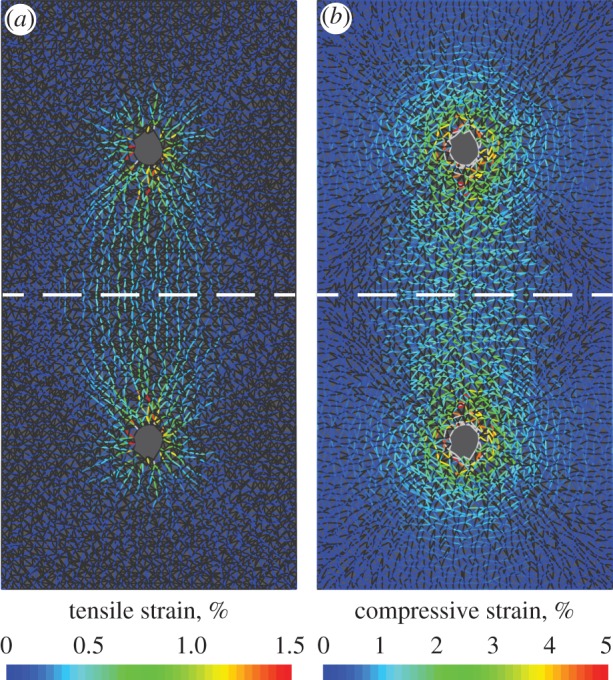
Fibre alignment and densification provide a mechanism for long-range cell mechanosensing. Mechanical interactions between cells are simulated using the FE model with a contracting circle and a symmetric boundary (dashed line). Plots show tensile (a) and compressive (b) strains within fibres. Fibres under tension (a) form intercellular tethers. Compressed fibres (b) are roughly perpendicular to tensile ones. The strains below the dashed line are the reflection of the strains above the dashed line.
3. Discussion
We have shown that cells embedded within a fibrin matrix exert forces that cause matrix displacements to propagate over a longer range than predicted by linear elasticity. The long-range propagation of displacements has been previously observed for cells on a flat, two-dimensional fibrous substrate [12] and for multicellular constructs in a three-dimensional matrix [38]. Our observations, first reported in ref. [14], confirm this result for single cells in a three-dimensional system. Here, we further quantify the spatial decay of displacements by fitting to a power law resulting in displacements scaling as 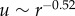 . While the propagation of displacements over a long range is now apparent, the precise mechanism is still unclear. Recent studies have argued for [12] and against [33] the hypothesis that long-range propagation of displacements results from strain stiffening. When we included strain stiffening in the behaviour of fibres, but suppressed compression weakening owing to buckling, long-range propagation was not observed in simulations of our model. Thus, we conclude that fibre buckling—rather than strain stiffening in tension—explains the long-range propagation of displacements observed in the experiments.
. While the propagation of displacements over a long range is now apparent, the precise mechanism is still unclear. Recent studies have argued for [12] and against [33] the hypothesis that long-range propagation of displacements results from strain stiffening. When we included strain stiffening in the behaviour of fibres, but suppressed compression weakening owing to buckling, long-range propagation was not observed in simulations of our model. Thus, we conclude that fibre buckling—rather than strain stiffening in tension—explains the long-range propagation of displacements observed in the experiments.
To simulate buckling, we used a model that does not resist changes in angle between the elements. Previous work [32] has pointed to bending as an important mechanism that controls the mechanical response of fibrous materials. However, the model of Broedersz et al. [32] does not allow for buckling or even bending of individual fibres. Instead, it models bending by penalizing changes in angle between initially collinear elements that meet at a node. Moreover, even for models that include bending, forces are dominated not by transverse bending displacements but by axial ones [36]. To address here the question of microbuckling, we focus on these axial displacements. We assume fibres buckle immediately after a compressive load is applied, i.e. we assume the fibre buckling load (equivalently the buckling strain) is equal to zero. Is this assumption reasonable? A typical fibrin fibre with a length of 1 µm, diameter of 0.2 µm, persistence length of 40 µm [23] and Young's modulus of 15 MPa [39] will buckle at a compressive strain of approximately 4 × 10−4%. This value is small compared with typical strains experienced in the matrix (approx. 1%), so our choice of setting the transition point between different stiffnesses at the onset of compression (vanishing buckling load) is justified.
Besides long-range propagation of displacements, fibrous materials exhibit what is termed in [11] a ‘negative compressibility’ in uniaxial tension, i.e. a negative ratio of the trace of the stress tensor and the trace of the strain tensor during a uniaxial tension experiment. Fibrous materials also exhibit tensile normal stresses under prescribed shear deformation [10]. This is in essence equivalent to negative normal (compressive) strains when the material is subjected to external tangential forces (prescribed shear stress), but not constrained to expand or contract. To test whether our fibre model is consistent with these experimental observations, we simulated homogeneous uniaxial tension. We found that when fibres buckle, the model exhibits negative compressibility in tension. In addition, under applied tangential forces equivalent to an external shear stress, the model responded with negative normal strains in shear. When microbuckling is removed from the model, neither of the aforementioned behaviours occurs (electronic supplementary material, figure S6). Thus, our model with fibre microbuckling is consistent with previous experimental work on fibrin [10,11] and collagen [26]. Certainly fibrous materials exhibit nonlinear behaviours besides microbuckling in compression, but our model points to microbuckling as being both consistent with previous experimental work and of major importance to the mechanical response of fibrous materials.
Together, our simulations and experiments reveal that microbuckling of fibrin enables cells to induce displacements that follow linear, tether-like paths that lead to other cells. These displacements propagate over a dramatically longer range than in a linear material. A remaining question of biological relevance is whether cells physically respond to the formation of tethers. In our experiments, we have observed pairs of cells forming pointed protrusions along these tethers and subsequently growing toward one another by several cell diameters (figure 5), sometimes eventually joining two cells (electronic supplementary material, figure S7). A different model indicated that elongated cells initially pointed toward one another may sense displacements induced by their neighbours [37], but it did not answer the question of how cells break their initial spherical symmetry to spread toward one another as observed in our experiments. Our model, which we present here and have described previously [14], suggests a mechanism whereby cells can sense one another during the initially spherical state. Even if each cell is initially spherical and contracts isotropically, the tether formation mechanism that we describe results in greater tension and fibre density that is highly polarized in the direction of neighbouring cells (figure 5). Both tension and fibre density may provide a directional signal: by growing protrusions along the direction of the tethers, cells have a higher chance of approaching one another. The fact that cells change shape and grow along such tethers supports the hypothesis that they use this very same mechanism to sense and even approach their neighbours. We expect future work to further clarify how cells sense the mechanical properties of fibrous materials and how we can better design artificial cell culture platforms to better control cellular response to forces within the extracellular matrix.
4. Material and methods
4.1. Cell culture and matrix preparation
3T3 fibroblast cells stably expressing a green fluorescent protein–actin fusion protein were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum and 1 × non-essential amino acids. Fibrin was fluorescently labelled by mixing fibrinogen (Omrix Biopharmaceuticals, Israel) and 546 Alexa Fluor (Life Technologies, Carlsbad, CA, USA) for 1 h before filtering with a HiTrap desalting column (GE Healthcare, Milwaukee, WI, USA). Cell–fibrin constructs were created by suspending the cells in 20 U ml−1 thrombin solution (Omrix), mixing with 5 mg ml−1 labelled fibrinogen solution and placing on a #1.5 coverslip.
4.2. Microscopy and cell-induced matrix displacements
Within 1 h of seeding, cell–matrix constructs were transferred to a custom built 5% CO2, 37°C microscope enclosure. Imaging was performed with a Swept Field confocal microscope using a 40 × NA 1.15 water immersion objective (Nikon Instruments, Melville, NY, USA). Volume stacks of the cells and fibrin matrix were captured every 15 min over time periods of several hours.
Three-dimensional matrix displacements were computed directly from the images of the labelled fibrin using digital volume correlation [20] with the initial volume stack (before cell spreading) taken as a reference for the correlation. The digital volume correlation software, written in Matlab (The Mathworks), is freely available online [40]. Propagation of cell-induced matrix displacements was quantified by computing displacement magnitudes along multiple linear paths propagating outward from the centre of each initially rounded cell. To reduce errors caused by inhomogeneities within the matrix, displacements were averaged over approximately three different paths and over approximately 10 time points for each cell. After averaging, the standard deviation of the noise level was found to be 0.04 µm.
4.3. Microstructural model
The microstructural model was developed in the FE software Abaqus v. 6.10 (Dassault Systemes, Waltham, MA). Rod elements supporting tension and compression but not bending were connected as shown in figure 2c. Elements were randomly deleted to reduce the network connectivity. Removed elements were replaced by elements with stiffness 6 orders of magnitude smaller than the original elements. The choice of using weak elements with stiffness six orders of magnitude smaller than the original elements came after a series of convergence studies showed that further reduction in the stiffness of the weak elements had no effect on the displacements. Under tension, both a linear (figure 2a) and a strain-stiffening WLC relationship were investigated (figure 2b). Under compression a linear stress–strain relationship was used with slope given by ρ times the slope at small tensile strains with ρ = 0.1 for microbuckling and ρ = 1 for no buckling. Strains within each element (as plotted in figure 5; electronic supplementary material, figure S5) were computed by taking the natural logarithm of the stretch ratio, defined as the final element length divided by the initial element length. The three-dimensional model used element connectivity as shown in electronic supplementary material, figure S4 and the bilinear stress–strain relationship. Uniaxial tension was simulated in a square region by applying displacements on the top side, a symmetric boundary on the bottom side, and traction free boundaries on the right and left sides. Shear loading was simulated by applying horizontal displacements to the top of a thin rectangular region (aspect ratio 1/10) with a fixed bottom boundary and traction free conditions on the right and left. Apparent strains were computed by numerically computing the displacement gradients using a linear fitting. Effective Poisson's ratio was defined as the opposite of the ratio of apparent strains in the transverse and axial directions.
Supplementary Material
Competing interests
We have no competing interests.
Authors' contributions
J.N. and A.L. performed the experiments. J.N. performed the simulations. J.N. and P.R. wrote the manuscript. All authors discussed the results and gave approval for publication.
Funding
This work was funded in part by a grant from the National Science Foundation (Division of Materials Research No. 0520565) through the Center for the Science and Engineering of Materials at the California Institute of Technology, and in part, by National Science Foundation grant no. DMR-1206121. J.N. was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE-1144469.
References
- 1.Vogel V, Sheetz M. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275. ( 10.1038/nrm1890) [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang Y-L. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143. ( 10.1126/science.1116995) [DOI] [PubMed] [Google Scholar]
- 3.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. 2009. Physical forces during collective cell migration. Nat. Phys. 5, 426–430. ( 10.1038/nphys1269) [DOI] [Google Scholar]
- 4.Notbohm J, Kim J-H, Asthagiri AR, Ravichandran G. 2012. Three-dimensional analysis of the effect of epidermal growth factor on cell-cell adhesion in epithelial cell clusters. Biophys. J. 102, 1323–1330. ( 10.1016/j.bpj.2012.02.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo C-M, Wang H-B, Dembo M, Wang Y-L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144–152. ( 10.1016/S0006-3495(00)76279-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhart-King CA, Dembo M, Hammer DA. 2008. Cell-cell mechanical communication through compliant substrates. Biophys. J. 95, 6044–6051. ( 10.1529/biophysj.107.127662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang X, Bajaj P, Bashir R, Saif TA. 2011. How far cardiac cells can see each other mechanically. Soft Matt. 7, 6151–6158. ( 10.1039/c0sm01453b) [DOI] [Google Scholar]
- 8.Kandow CE, Georges PC, Janmey PA, Beningo KA. 2007. In Cell mechanics, vol. 83 (eds Wang Y-L, Discher DE.). New York, NY: Academic Press. [Google Scholar]
- 9.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. 2005. Nonlinear elasticity in biological gels. Nature 435, 191–194. ( 10.1038/nature03521) [DOI] [PubMed] [Google Scholar]
- 10.Janmey PA, McCormick ME, Rammensee S, Leight JL, Georges PC, MacKintosh FC. 2007. Negative normal stress in semiflexible biopolymer gels. Nat. Mater. 6, 48–51. ( 10.1038/nmat1810) [DOI] [PubMed] [Google Scholar]
- 11.Brown AEX, Litvinov RI, Discher DE, Purohit PK, Weisel JW. 2009. Multiscale mechanics of fibrin polymer: gel stretching with protein unfolding and loss of water. Science 325, 741–744. ( 10.1126/science.1172484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winer JP, Oake S, Janmey PA. 2009. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS ONE 4, e6382 ( 10.1371/journal.pone.0006382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shokef Y, Safran SA. 2012. Scaling laws for the response of nonlinear elastic media with implications for cell mechanics. Phys. Rev. Lett. 108, 178103 ( 10.1103/PhysRevLett.108.178103) [DOI] [PubMed] [Google Scholar]
- 14.Notbohm J. 2013. Dynamics of cell–matrix mechanical interactions in three dimensions. PhD thesis, California Institute of Technology. [Google Scholar]
- 15.Conti E, MacKintosh FC. 2009. Cross-linked networks of stiff filaments exhibit negative normal stress. Phys. Rev. Lett. 102, 088102 ( 10.1103/PhysRevLett.102.088102) [DOI] [PubMed] [Google Scholar]
- 16.Onck PR, Koeman T, Van Dillen T, Van der Giessen E. 2005. Alternative explanation of stiffening in cross-linked semiflexible networks. Phys. Rev. Lett. 95, 178102 ( 10.1103/PhysRevLett.95.178102) [DOI] [PubMed] [Google Scholar]
- 17.Heussinger C, Frey E. 2006. Stiff polymers, foams, and fiber networks. Phys. Rev. Lett. 96, 017802 ( 10.1103/PhysRevLett.96.017802) [DOI] [PubMed] [Google Scholar]
- 18.Eshelby JD. 1959. The elastic field outside an ellipsoidal inclusion. Proc. R. Soc. Lond. A 252, 561–569. ( 10.1098/rspa.1959.0173) [DOI] [Google Scholar]
- 19.Schwarz US, Safran SA. 2002. Elastic interactions of cells. Phys. Rev. Lett. 88, 048102 ( 10.1103/PhysRevLett.88.048102) [DOI] [PubMed] [Google Scholar]
- 20.Franck C, Hong S, Maskarinec SA, Tirrell DA, Ravichandran G. 2007. Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp. Mech. 47, 427–438. ( 10.1007/s11340-007-9037-9) [DOI] [Google Scholar]
- 21.Lesman A, Notbohm J, Tirrell DA, Ravichandran G. 2014. Contractile forces regulate cell division in three-dimensional environments. J. Cell Biol. 205, 155–162. ( 10.1083/jcb.201309029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim OV, Litvinov RI, Weisel JW, Alber MS. 2014. Structural basis for the nonlinear mechanics of fibrin networks under compression. Biomaterials 35, 6739–6749. ( 10.1016/j.biomaterials.2014.04.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piechocka IK, Bacabac RG, Potters M, MacKintosh FC, Koenderink GH. 2010. Structural hierarchy governs fibrin gel mechanics. Biophys. J. 98, 2281–2289. ( 10.1016/j.bpj.2010.01.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bischofs IB, Klein F, Lehnert D, Bastmeyer M, Schwarz US. 2008. Filamentous network mechanics and active contractility determine cell and tissue shape. Biophys. J. 95, 3488–3496. ( 10.1529/biophysj.108.134296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stylianopoulos T, Barocas VH. 2007. Volume-averaging theory for the study of the mechanics of collagen networks. Comput. Method Appl. Mech. Eng. 196, 2981–2990. ( 10.1016/j.cma.2006.06.019) [DOI] [Google Scholar]
- 26.Vader D, Kabla A, Weitz D, Mahadevan L. 2009. Strain-induced alignment in collagen gels. PLoS ONE 4, e5902 ( 10.1371/journal.pone.0005902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Münster S, Jawerth LM, Leslie BA, Weitz JI, Fabry B, Weitz DA. 2013. Strain history dependence of the nonlinear stress response of fibrin and collagen networks. Proc. Natl Acad. Sci. USA 110, 12 197–12 202. ( 10.1073/pnas.1222787110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosakis P, Notbohm J, Ravichandran G. 2014. A model for compression-weakening materials and the elastic fields due to contractile cells. (http://arxiv.org/abs/1412.2612)
- 29.Love AEH. 2013. A treatise on the mathematical theory of elasticity. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 30.Lakes R, Rosakis P, Ruina A. 1993. Microbuckling instability in elastomeric cellular solids. J. Mat. Sci. 28, 4667–4672. ( 10.1007/BF00414256) [DOI] [Google Scholar]
- 31.Wyart M, Liang H, Kabla A, Mahadevan L. 2008. Elasticity of floppy and stiff random networks. Phys. Rev. Lett. 101, 215501 ( 10.1103/PhysRevLett.101.215501) [DOI] [PubMed] [Google Scholar]
- 32.Broedersz CP, Mao X, Lubensky TC, MacKintosh FC. 2011. Criticality and isostaticity in fibre networks. Nat. Phys. 7, 983–988. ( 10.1038/nphys2127) [DOI] [Google Scholar]
- 33.Rudnicki MS, Cirka HA, Aghvami M, Sander EA, Wen Q, Billiar KL. 2013. Nonlinear strain stiffening is not sufficient to explain how far cells can feel on fibrous protein gels. Biophys. J. 105, 11–20. ( 10.1016/j.bpj.2013.05.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stopak D, Harris AK. 1982. Connective-tissue morphogenesis by fibroblast traction. 1. Tissue culture observations. Dev. Biol. 90, 383–398. ( 10.1016/0012-1606(82)90388-8) [DOI] [PubMed] [Google Scholar]
- 35.Sawhney RK, Howard J. 2002. Slow local movements of collagen fibers by fibroblasts drive the rapid global self-organization of collagen gels. J. Cell. Biol. 157, 1083–1091. ( 10.1083/jcb.200203069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heussinger C, Frey E. 2007. Force distributions and force chains in random stiff fiber networks. Eur. Phys. J. E. 24, 47–53. ( 10.1140/epje/i2007-10209-1) [DOI] [PubMed] [Google Scholar]
- 37.Abhilash AS, Baker BM, Trappmann B, Chen CS, Shenoy VB. 2014. Remodeling of fibrous extracellular matrices by contractile cells: predictions from discrete fiber network simulations. Biophys. J. 107, 1829–1840. ( 10.1016/j.bpj.2014.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gjorevski N, Nelson CM. 2012. Mapping of mechanical strains and stresses around quiescent engineered three-dimensional epithelial tissues. Biophys. J. 103, 152–162. ( 10.1016/j.bpj.2012.05.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collet JP, Shuman H, Ledger RE, Lee S, Weisel JW. 2005. The elasticity of an individual fibrin fiber in a clot. Proc. Natl Acad. Sci. USA 102, 9133–9137. ( 10.1073/pnas.0504120102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bar-Kochba E, Toyjanova J, Andrews E, Kim K-S, Franck C. 2015. A fast iterative digital volume correlation algorithm for large deformations. Exp. Mech. 55, 261–274. ( 10.1007/s11340-014-9874-2) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.