Abstract
Deficiency of the Golgi N-glycan branching enzyme Mgat5 in mice promotes T cell hyperactivity, endocytosis of CTLA-4 and autoimmunity, including a spontaneous multiple sclerosis (MS)-like disease. Multiple genetic and environmental MS risk factors lower N-glycan branching in T cells. These include variants in interleukin-2 receptor-α (IL2RA), interleukin-7 receptor-α (IL7RA), and MGAT1, a Golgi branching enzyme upstream of MGAT5, as well as vitamin D3 deficiency and Golgi substrate metabolism. Here we describe linked intronic variants of MGAT5 that are associated with reduced N-glycan branching, CTLA-4 surface expression and MS (p = 5.79 × 10−9, n = 7,741), the latter additive with the MGAT1, IL2RA and IL7RA MS risk variants (p = 1.76 × 10−9, OR = 0.67−1.83, n = 3,518).
Keywords: Multiple sclerosis, N-glycosylation, MGAT5, MGAT1, Interleukin-7 receptor, Interleukin-2 receptor
1. Introduction
Multiple sclerosis (MS) is a complex trait disease, whereby multiple genetic and environmental factors combine to determine MS susceptibility (Steinman, 2001; Hafler et al., 2007; Smolders et al., 2008). The majority of genetic variants that increase disease risk appear to be present in loci involved with regulating the immune response, such as the major histocompatibility complex locus (HLA) and interleukin receptors IL2RA and IL7RA (Gregory et al., 2007; Lundmark et al., 2007; Schmidt et al., 2007; Maier et al., 2009a,b; International Multiple Sclerosis Genetics Consortium and Wellcome Trust Case Control Consortium, 2011). In addition to the genetic component, environmental factors such as exposure to sunlight are also significant determinants in MS risk (Smolders et al., 2008). The importance of the effects from the environment is most notably demonstrated in the case of monozygotic twins that are discordant for MS ~70% of the time despite virtually no differences in their genome, transcriptome and epigenetic profiles (Ebers et al., 1986; Baranzini et al., 2010). Evidence supporting the role of the environment in disease risk includes studies involving vitamin D3, where serum levels correlate with sunlight exposure, vary along the earth's longitudinal axis, and inversely correlate with MS susceptibility and severity (van der Mei et al., 2003; Soilu-Hanninen et al., 2005; Munger et al., 2006).
Understanding how genetic and environmental risk factors combine at the molecular level to promote loss of self-tolerance in MS is critical to identifying safe and efficacious therapies. In this regard, we recently reported that multiple genetic and environmental factors converge to disturb protein N-glycosylation of T cells in MS (Mkhikian et al., 2011). In mice, the Golgi N-glycan pathway is a critical modulator of T cell function and autoimmune disease (Grigorian et al., 2009). N-acetylglucosamine (GlcNAc) branched N-glycans attached to cell surface immuno-modulatory receptors are bound by a multimeric family of mammalian lectin proteins, called galectins (Dennis et al., 2009). This network of crosslinked galectins and glycoproteins form a molecular lattice on the cell surface that regulates surface concentration, lateral distribution, and signaling strength of the bound glycoproteins (Demetriou et al., 2001; Morgan et al., 2004; Chen et al., 2007; Grigorian et al., 2007; Lau et al., 2007; Lee et al., 2007; Grigorian and Demetriou, 2011). The Golgi enzymes Mgat1, 2, 4a/b, 5, along with metabolic production of their shared substrate UDP-GlcNAc, control the degree of N-glycan branching, the strength of the lattice and cell growth and differentiation (Grigorian et al., 2007; Lau et al., 2007; Grigorian et al., 2012a). For example, murine Mgat5 deficiency results in hyperactive TCR clustering and signaling, surface loss of the anti-autoimmune receptor CTLA-4, pro-inflammatory TH1 differentiation and development of spontaneous autoimmunity (Demetriou et al., 2001; Morgan et al., 2004; Lau et al., 2007; Grigorian et al., 2011). In the PL/J background, N-glycan branching deficiency promotes a spontaneous MS-like disease characterized by inflammatory demyelination and neurodegeneration (Lee et al., 2007). In MS, multiple genetic and environmental risk factors converge to disturb N-glycan branching by altering MGAT1 expression/activity (Mkhikian et al., 2011). These include genetic variants in MGAT1 (VAVT-T haplotype: rs7726005, rs2070924, rs2070925), IL2RA (rs2104286) and IL7RA (rs6897932), as well as environmental factors consisting of vitamin D3 and metabolic production of UDP-GlcNAc. These interact with a polymorphism in CTLA-4 (Thr17Ala, rs231775) that reduces the number of N-glycans attached to the protein by 50% to suppress surface expression of CTLA-4 in human T cell blasts (Anjos et al., 2002; Maurer et al., 2002). Supplementing cells or mice with the monosaccharide N-acetylglucosamine (GlcNAc) enhances UDP-GlcNAc production and N-glycan branching to reverse the T cell hyperactivity and/or autoimmunity induced by genetic deficiencies in N-glycan branching (Grigorian et al., 2007, 2011).
A recent genome-wide association study (GWAS) screening for variants that alter MS severity in Europeans identified two linked intronic MGAT5 polymorphisms as their top hits (rs4953911 and rs3814022) (Brynedal et al., 2010). Although this association was not fully replicated in other cohorts for disease severity, whether rs4953911 and rs3814022 are involved in MS susceptibility remains unknown (Esposito et al., 2011; International Multiple Sclerosis Genetics Consortium, 2011). For example, the largest GWAS of MS risk done to date did not utilize a SNP chip that contained these two MGAT5 SNPs (International Multiple Sclerosis Genetics Consortium and Wellcome Trust Case Control Consortium, 2011). In addition to disease risk, the ability of these SNPs to alter N-glycan branching and thus cell surface concentration of immuno-modulatory receptors have yet to be investigated. Here, we report that MGAT5 rs4953911 and rs3814022 are associated with reduced N-glycan branching and CTLA-4 surface expression as a consequence of decreased MGAT5 expression in blasting T cells, a phenotype rescued by metabolic supplementation of cells with GlcNAc. Moreover, we find that MGAT5 rs4953911 and rs3814022 associate withMS in two independent MS case-control cohorts. Risk of MS was more strongly associated with MGAT5 rs4953911 and as expected, combining this stronger variant with other known variants that also lower N-glycan branching, namely MGAT1 VAVT-T, IL2RA (rs2104286) and IL7RA (rs6897932), additively increased the risk of MS (Gregory et al., 2007; Lundmark et al., 2007; Maier et al., 2009a,b; Mkhikian et al., 2011). Our results provide additional evidence that defective N-glycosylation plays a critical role in MS and further support the potential of metabolic supplementation to the N-glycan pathway with GlcNAc as a therapeutic tool for MS treatment and risk management.
2. Materials and methods
2.1. Human subjects
All human subjects used for both T cell experiments and genetic correlation studies are Caucasian (non-Hispanic) and include both males and females. For T cell experiments, human whole blood was collected from healthy control individuals through the Research Blood Donor Program serviced by the Institute for Clinical & Translational Science (ICTS) at University of California, Irvine (UCI). For genetic correlation studies, we examined two independent Caucasian MS case-control cohorts, one from North America and one from Europe. Cohort 1 cases (n = 1201 total) consisted of unrelated, non-Hispanic Caucasians with clinically definite MS obtained at UCI (n = 82) or from across the United States by the Multiple Autoimmune Disease Genetics Consortium (n = 176, MADGC, www.madgc.org) and the Accelerated Cure Project for Multiple Sclerosis (n = 943, www.acceleratedcure.org). Cohort 1 controls (n = 2377 total) were unrelated, non-diseased, non-Hispanic Caucasians obtained at UCI (n = 119), which include individuals used for T cell experiments, and from across North America by North American Rheumatoid Arthritis Consortium (n = 192, NARAC, www.naracdata.org), the Type1 Diabetes Genetics Consortium (n = 1199, T1DGC, www.t1dgc.com), the Genetics of Kidneys in Diabetes (GoKinD) (n = 746, www.niddkrepository.org), and publically available datasets (n = 121) from the HapMap Project (www.hapmap.org) and 1000 Genomes Project (www.1000genomes.org). T1DGC and GoKinD DNA samples were obtained from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Cohort 2 data was largely obtained from publically available datasets. Cases (n = 1913 total) were European [Sweden (n = 988), France (n = 568) and Italy (n = 357)] MS subjects previously genotyped for MGAT5 rs4953911 and rs3814022 (Brynedal et al., 2010). Cohort 2 controls (n = 2338 total) were Europeans from the ‘British 1958 Birth Cohort’ (n = 1480) and T1DGC (n = 858). Both control cohorts were in Hardy Weinberg Equilibrium (HWE). The sample size for individual alleles reported in Tables 1 and 2 varies marginally from these numbers due to failed genotyping for that specific allele in a few cases and/or controls.
Table 1.
MGAT5 variants in multiple sclerosis.
| Variant frequency | p-value | OR [95% CI] | |||
|---|---|---|---|---|---|
| Control (n) | MS (n) | ||||
| MGAT5*T (rs4953911) | Cohort 1 | 0.302 (2364) | 0.357 (1183) | 7.77 × 10−7 | 1.31 [1.17–1.47] |
| Cohort 2 | 0.288 (2338) | 0.323 (1856) | 3.23 × 10−4 | 1.17 [1.07–1.29] | |
| Combined p-value: | 5.79 × 10−9 | ||||
| MGAT5*G (rs3814022) | Cohort 1 | 0.263 (2363) | 0.309 (1201) | 6.50 × 10−6 | 1.29 [1.15–1.45] |
| Cohort 2 | 0.254 (2331) | 0.286 (1897) | 4.21 × 10−4 | 1.18 [1.07–1.29] | |
| Combined p-value: | 5.67 × 10−8 | ||||
Reported are one-sided p-values computed using the Cochran–Armitage test of North American (cohort 1) and European (cohort) non-Hispanic Caucasian case-control cohorts. All individual control cohorts are in HWE. P-values were combined using Fishers method. OR=odds ratio. 95% CI = 95% confidence intervals.
Table 2.
MGAT5 with IL2RA, IL7RA and MGAT1 IVAVT-T in multiple sclerosis.
| Variables: | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| Controls | 2 | 62 | 281 | 727 | 817 | 387 | 68 |
| Cases | 0 | 21 | 95 | 328 | 427 | 242 | 61 |
| OR (x vs rest)c | – | 0.67 | 0.64 | 0.86 | 1.07 | 1.31 | 1.83 |
| P-valuea | 2.78 × 10−9 | ||||||
| OR (x + 1 vs x)d | 1.22 [1.14–1.30] | ||||||
| Multiple logistic regression p-valueb | 1.76 × 10−9 | ||||||
Analysis of Cohort 1 (North American non-Hispanic Caucasian case-control MS cohort).
One-sided p-value computed using the Cochran–Armitage test for trend with MGAT5*T (rs4953911), IL2RA*T (rs2104286) and IL7RA*C (rs6897932) coded as 0, 1 or 2 and MGAT1 IVAVT-T (rs7726005, rs2070924 and rs2070925) non-carriers coded as 0, carriers with IL2RA*T + IL7RA*Cb4 as 1 and carriers with IL2RA*T + IL7RA*C = 4 as −1.
P-value from the likelihood ratio test (4 degrees of freedom) from fitting a multiple logistic regression with 4 terms: MGAT5*T, MGAT1 IVAVT-T, IL2RA*T + IL7RA*C and the interaction between MGAT1 IVAVT-T and IL2RA*T + IL7RA*C. Variables were coded as 0, 1 or 2 for the number of risk alleles, except MGAT1 IVAVT-T which was coded as 0 or 1 for carrier status.
Odds ratio (OR) for carriers with X risk alleles versus carriers with other numbers of risk alleles.
OR for X + 1 versus X determined from the logistic regression coefficient.
2.2. T cell isolation and analysis
Human T cells from healthy control individuals (n = 11 total, 4 females, 7 males; ages 22–55, mean age = 33) were isolated from whole blood using the RosetteSep Human CD4+ T cell enrichment kit (StemCell Technologies) and stimulated with PMA (1 ng/ml) and ionomycin (125 ng/ml) for 5 days in culture. In GlcNAc supplementation experiments, 40 mM GlcNAc (Ultimate Glucosamine, Wellesley Therapeutics Inc.) and 10 mM uridine (Sigma) were added to cells in culture at Day 0. For flow cytometry, after normalizing for cell number, CD4+ T cells were stained with 2 ug/ml L-PHA-FITC (Vector Labs) to detect β1,6-GlcNAc branched N-glycans. T cells were also co-stained with CD4-PE-Cy5 and CD25-PE (eBioscience) to mark CD4+ T cell blasts. For CTLA-4 cell surface measurements, T cells were stained with anti-CTLA-4-PE or its respective PE-labeled isotype control (eBioscience), and final CTLA-4 values were adjusted for nonspecific binding by isotype subtraction. Fluorescence levels were measured by flow cytometry using BD's FACSCalibur flow cytometer. Post-acquisition analysis was performed using FlowJo software. For RNA analysis, CD4+ T cells were stimulated as above and harvested at the indicated time points. RNA was isolated using RNeasy Mini Kit (Qiagen) and used to generated cDNA using M-MLV Reverse Transcriptase (Invitrogen). Gene expression analysis was performed using real-time qPCR with TaqMan Gene Expression Assays (ABI) specific for humanMGAT5 (Hs00327733_s1) and beta-actin (Hs99999903_m1). The ΔΔCt method was used to calculate relative fold enrichment in MGAT5 RNA expression using beta-actin for normalization.
2.3. SNP analysis
Single nucleotide polymorphism (SNP) genotyping was performed using TaqMan SNP Genotyping Assays (ABI) as previously described (Mkhikian et al., 2011). TaqMan SNP genotyping assays utilized for MGAT5 SNPs rs4953911 and rs3814022 are C___3258342_10 and C___2163491_10, respectively. Genotyping assays for IL2RA (rs2104286) and IL7RA (rs6897932) are C__16095542_10 and C___2025977_10, respectively. The MGAT1 IVAVT-T haplotype (rs7726005, rs2070924 and rs2070925) was genotyped using two custom TaqMan primer/probe sets targeting rs7726005 and rs2070924 as previously described (Mkhikian et al., 2011). The two MGAT1 VT-T SNPs (rs2070924 and rs2070925) are 10nt apart and sequenom genotyping of 1387 subjects revealed 100% concordance. The genotyping success rate for all 7 SNPs examined was > 99% in all TaqMan genotyped cohorts except for MGAT5 rs4953911 in MS cohort 1 (> 98%). Concordance for duplicate genotyping was determined by re-genotyping all 6 SNPs and revealed 100% concordance (n = 735) for all SNPs except MGAT5 rs4953911 and rs3814022 (99.5%, n = 374 and n = 376, respectively).
2.4. Statistical analysis
Individual p-values for association studies in Table 1 for North American cohort 1 and European cohort 2 are one-sided and were computed using the Cochran–Armitage test, while combined p-values were generated using Fishers method. For genetic interaction studies in Table 2 for cohort 1, MGAT5*T (rs4953911), IL2RA*T (rs2104286) and IL7RA*C (rs6897932) were coded as 0, 1 or 2 for the number of risk alleles. In addition, MGAT1 IVAVT-T (rs7726005, rs2070924 and rs2070925) non-carriers were coded as 0, while carriers with less than four total risk alleles for IL2RA*T + IL7RA*C were coded as 1 and carriers with exactly four risk alleles for IL2RA*T + IL7RA*C as −1. The one-sided p-value for this trend analysis was computed using the Cochran–Armitage test. Multiple logistic regression analysis was performed using the following four terms: MGAT5*T, MGAT1 IVAVT-T, IL2RA*T + IL7RA*C and the interaction between MGAT1 IVAVT-T and IL2RA*T + IL7RA*C. Alleles were coded as 0, 1 or 2 for the number of risk alleles, except for MGAT1 IVAVT-T which was coded as 0 or 1 for carrier status. From this fitting, a likelihood ratio test with four degrees of freedom was used to generate a multiple logistic regression p-value. Odds ratios for carriers with varying numbers of risk alleles were determined from the logistic regression coefficient.
3. Results
3.1. Downregulation of N-glycan branching and surface CTLA-4 by MGAT5 genetic variants
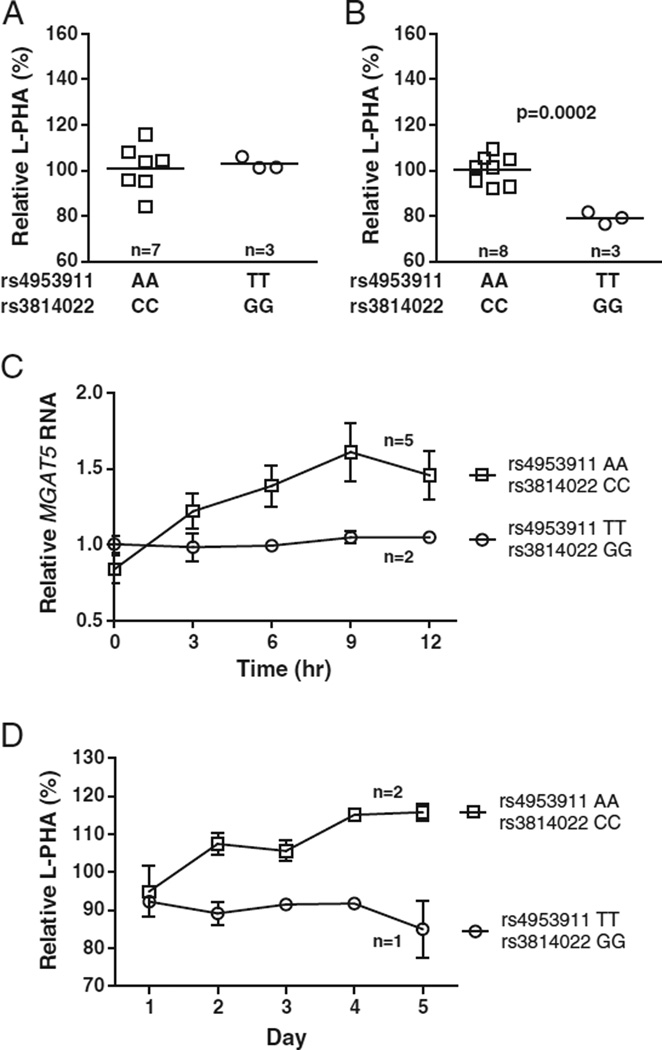
MGAT5 rs4953911 and rs3814022 are in strong linkage disequilibrium (Lewontin's D' = 0.98 and Pearson's correlation coefficient r = 0.90), and both reside within intron 2 of the MGAT5 gene. Direct assessment of the biological effect of intronic SNPs is difficult; therefore we correlated changes in MGAT5-induced β1,6- GlcNAc branching in CD4+ T cells isolated from healthy donors containing the MGAT5 risk variants or their common allele counterparts. For the sake of simplicity and to provide the clearest comparison, we analyzed cells from individuals who were homozygous for both rs4953911 and rs3814022, or homozygous for their common alleles. In addition, to minimize variability, we also matched cells for the other MS variants known to alter N-glycan branching, namely MGAT1 VAVT-T, IL2RA (rs2104286) and IL7RA (rs6897932) (Mkhikian et al., 2011). Cell surface β1,6-GlcNAc branching was assessed by flow cytometry with L-PHA (Phaseolus vulgaris, leukoagglutinin), a plant lectin that binds specifically to β1,6-GlcNAc-branched N-glycans and serves as a quantitative marker of N-glycan branching (Cummings and Kornfeld, 1982; Demetriou et al., 2001). In non-activated, resting T cells, very little difference in L-PHA binding was seen on average between rs4953911/rs3814022 (TT/GG) homozygotes and the common allele homozygotes (AA/CC) (Fig. 1A). In contrast, activated T cell blasts homozygous for rs4953911/rs3814022 displayed significant decreases in L-PHA binding over the homozygous controls (~21% average decrease, p = 0.0002) (Fig. 1B). Consistent with this decrease in N-glycan branching, MGAT5 RNA levels are also lower in activated T cells homozygous for the MGAT5 variants when compared to the common alleles, with the differences peaking at nine hours post stimulation (~35% average decrease) (Fig. 1C). This suggests that rs4953911/rs3814022 SNPs have a role in modulating N-glycan branching in T cells in the activated state, and this modulation likely occurs via effects on MGAT5 RNA expression levels.
Fig. 1.
MGAT5 rs4953911/rs3814022 reduce GlcNAc branching and MGAT5 RNA in activated T cells. (A) Scatterplot of L-PHA binding by flow cytometry in non-activated, resting human T cells. Purified CD4+ T cells homozygous for rs4953911/rs3814022 (TT/GG) or their common allele control counterparts (AA/CC) were rested 5 days in culture before analysis for surface levels of GlcNAc branching. Each data point (square or circle) represents the average of triplicate L-PHA measurements for each individual. Relative L-PHA values were generated by normalization to the average of the control group (set at 100%). The horizontal line across each data group represents the average L-PHA value of that entire group. (B) Scatterplot of L-PHA binding in activated CD4+ CD25+ T cell blasts. Purified CD4+ T cells were activated with PMA (1 ng/ml) and ionomycin (125 ng/ml) for 5 days in culture. L-PHA binding measurements and analysis were performed as in (A). (C) Time course analysis of MGAT5 RNA levels in activated CD4+ T cells. CD4+ T cells were stimulated as in (B) for 0, 3, 6, 9, and 12 h before harvesting for analysis. Each line represents combined average MGAT5 RNA values for the specified group. Relative MGAT5 RNA values were generated by normalization to the average value of rs4953911/rs3814022 homozygotes within each hour (set at 1). (D) Time course analysis of L-PHA binding changes in CD4+ CD25+ T cell blasts. Activated cells were stimulated as in (B) and harvested each day for a total of 5 days. L-PHA binding measurements and analysis were performed as in (A). Each line represents combined average branching values for the specified group. Relative L-PHA values were generated by normalization to a control individual at Day 1 (set at 100%). For all panels, n is the total number of unique individuals analyzed per group. Error bars represent stand error of the mean (SEM). P-values were generated using one-tailed unpaired t-tests.
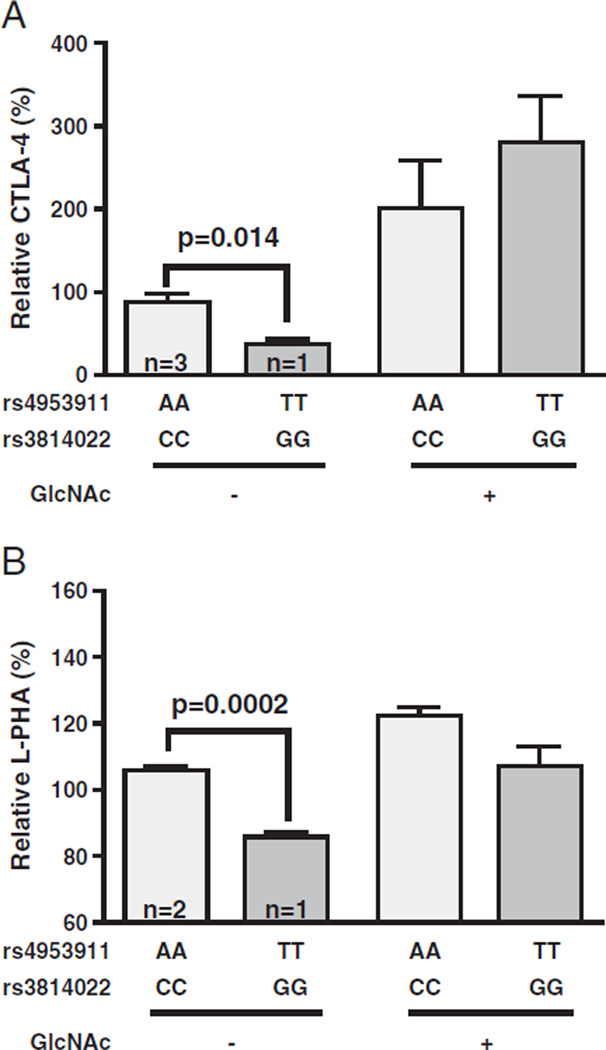
We then investigated more closely the changes in N-glycosylation on a day-by-day basis for the MGAT5 variants. Alterations in L-PHA binding associated with rs4953911/rs3814022 did not peak until Days 4–5 following stimulation (~20–27% average decrease) (Fig. 1D). This is the period of the T cell growth cycle when up-regulation of anti-autoimmune CTLA-4 at the cell surface is required to induce growth arrest, a phenotype regulated by N-glycan branching (Grigorian et al., 2007; Lau et al., 2007). Thus, we investigated the possibility of these MGAT5 variants altering CTLA-4 surface levels in individuals matched for the CTLA-4 Thr17Ala allele (rs231775), which is known to alter the number of N-glycans attached to CTLA-4 (Anjos et al., 2002). Indeed, rs4953911/rs3814022 homozygosity (TT/GG) was also associated with a reduction in CTLA-4 surface expression in 5-day old T cell blasts compared to average values for its common allele control counterparts (AA/CC) (~58% average decrease, p = 0.014) (Fig. 2A). Supplementing T cells homozygous for the MGAT5 variants with GlcNAc restored L-PHA binding (Fig. 2B) and CTLA-4 surface expression (Fig. 2A) to that of control homozygotes, confirming that the observed differences resulted from alterations in branching. These results are consistent with the MGAT5 SNPs playing a role in MS severity and in line with published reports which demonstrated murine Mgat5 deficiency lowers CTLA-4 surface expression and promotes MS-like disease in mice (Demetriou et al., 2001; Lau et al., 2007; Brynedal et al., 2010).
Fig. 2.
MGAT5 rs4953911/rs3814022 downregulate CTLA-4 surface expression. Bar graphs of surface CTLA-4 positive cells (A) or L-PHA binding levels (B) in individuals homozygous for the common MGAT5 alleles (AA and CC for rs4953911 and rs3814022, respectively) or the average values from one individual homozygous for both variant alleles (TT and GG, respectively) are shown. CD4+ T cells were activated as in Fig. 1B, with or without treatment with 40 mM GlcNAc and 10 mM uridine. Cell surface expression of CTLA-4 or L-PHA binding on CD4+ T cell blasts were measured by flow cytometry. Relative L-PHA values and relative percentage of cells positive for CTLA-4 surface expression were calculated by normalization to control individuals (set at 100%). In addition, CTLA-4 values were adjusted by subtracting for their respective isotype control. n = 3 and n = 2 for the number of unique control individuals in panels (A) and (B), respectively. Error bars represent standard error of the mean (SEM). P-values were generated using one-tailed unpaired t-tests of triplicate staining.
3.2. Association of MGAT5 variants with MS risk
MGAT5 rs4953911 and rs3814022 were first identified in a case-only MS study (n = 1040) screening for polymorphisms associated with disease severity, with reported p-values of 1.5 × 10−7 and 2.8 × 10−6, respectively (Brynedal et al., 2010). However, our previous work demonstrating a link between decreases in N-glycan branching and risk of MS in humans and MS-like disease in mice suggests that MGAT5 rs4953911 and rs3814022 should also increase the risk of MS (Demetriou et al., 2001; Lee et al., 2007; Mkhikian et al., 2011). To investigate this hypothesis, we genotyped both polymorphisms in a non-Hispanic Caucasian cohort of 1201 MS cases and 2377 controls from North America (cohort 1). MGAT5 rs4953911 and rs3814022 both associated with MS, with p-values of 7.77 × 10−7 and 6.50 × 10−6, respectively (Table 1). The two alleles are in high LD and halotype analysis gave p = 4.52 × 10−6 for the rare haplotype, providing no additional information over the point analysis. Association of the two variants was replicated in a second independent cohort consisting of European MS cases (n = 1913) previously genotyped for both MGAT5 SNPs (Brynedal et al., 2010) and separate European controls from the ‘1958 British Birth Cohort’ and the Type-1 Diabetes Genetics consortium (n = 2338). P-values of 3.23 × 10−4 and 4.21 × 10−4 for rs4953911 and rs3814022were observed in this cohort, respectively. Combining p-values from the two cohorts produced genome-wide significance for rs4953911 (p = 5.79 × 10−9) and near genome-wide significance for rs3814022 (p = 5.67 × 10−8).
3.3. Additive genetic risk of MGAT5 SNPs with MGAT1, IL2RA and IL7RA variants in MS
Next we examined whether combining the stronger associated MS risk allele rs4953911 with other known variants that also lower N-glycan branching and promote MS increases disease risk beyond what is seen with the individual variants. The MGAT1 VAVT-T haplotype (rs7726005, rs2070924, rs2070925), IL2RA*T (rs2104286) and IL7RA*G (rs6897932) all lower N-glycan branching and show point association with MS (Mkhikian et al., 2011). However, IL2RA*T and IL7RA*C reduce N-glycan branching by downregulating MGAT1 mRNA; whereas the MGAT1 IVAVT-T haplotype enhances MGAT1 mRNA expression yet lowers N-glycan branching by sequestering the shared substrate UDP-GlcNAc from MGAT5. These opposing effects on MGAT1 mRNA cancel each other out and result in enhanced N-glycan branching and reduced risk of MS when MGAT1 VAVT-T, IL2RA*T and IL7RA*G are combined. To account for this interaction in a trend analysis, IL2RA*T and IL7RA*C were both coded as 0, 1 or 2 for the number of risk alleles, while MGAT1 IVAVT-T non-carriers were coded 0, carriers with IL2RA*T + IL7RA*C < 4 as 1, and carriers with IL2RA*T + IL7RA*C = 4 as −1. MGAT5*T (rs4953911) was also encoded as 0, 1 and 2, as this variant is expected to reduce N-glycan branching irrespective of whether MGAT1 is increased or decreased. A trend analysis of our North American MS cohort (cohort 1) with this coding gives p = 2.78 × 10−9 (one-sided Cochran–Armitage test, Table 2). The odds ratio (OR) increases from 0.67 for individuals with 1 variant to 1.83 for those with 6 variants. Similarly, using the logistic regression coefficient to estimate the OR predicts an OR = 1.22 for each additional variant added.
We also fit a multiple logistic regression with four terms: MGAT5*T (rs4953911), MGAT1 IVAVT-T, IL2RA*T + IL7RA*C and the interaction between MGAT1 IVAVT-T and IL2RA*T + IL7RA*C. In this analysis, MGAT1 IVAVT-T was coded 0 or 1 for carrier status, while the others were coded as 0, 1 or 2 for the number of risk alleles. For cohort 1, the p-value based on the likelihood ratio test with four degrees of freedom is 1.76 × 10−9. Importantly, the p-value of the three terms without MGAT5*T (rs4953911) is 1.87 × 10−5 and comparing the two models indicates that MGAT5*T (rs4953911) is still significant (p = 2.59 × 10−6) after taking the other three terms into consideration. Thus, MGAT5*T (rs4953911) provides additional information for MS risk. For cohort 2, data for MGAT1 IVAVT-T, IL2RA*T and IL7RA*C are not publicly available for the MS cases and therefore could not be analyzed with MGAT5*T (rs4953911) for combined effects. However, the results in cohort 1 are consistent with additive effects of MGAT5 rs4953911 and the other known MS risk alleles that also lower N-glycan branching.
4. Discussion
Elucidating the many factors which drive autoimmune diseases such as MS remains a challenging prospect due to the complexities of various interacting pathways. In the case of MS, the crosstalk between genetics and the environment dictates the need for multiple factors to be considered at once in order to correctly assess their true impact on disease risk and activity (Mkhikian et al., 2011; Grigorian et al., 2012b). In this report, we have shown that the MGAT5 intronic variants rs4953911 and rs3814022 correlate with lower N-glycan branching, reduced surface CTLA-4 in human CD4+ T cell blasts, and associate with MS. In addition, the stronger MS risk allele rs4953911 additively combines with other known MS risk alleles that also lower N-glycan branching (i.e. MGAT1 VAVT-T, IL2RA, and IL7RA) to further enhance disease risk. Consistent with association in MS, we have recently reported family based association of MGAT5 rs4953911 and rs3814022 with Type 1 diabetes (5.8 × 10−3 and p = 1.5 × 10−3, respectively) via analysis of 2395 Caucasian families from the Type 1 Diabetes Genetics Consortium (Yu et al., in press). MGAT5 RNA levels are lower in rs4953911/rs3814022 homozygotes, suggesting these SNPs may cause a change in splicing efficiency and/or transcriptional regulation leading to a defect in MGAT5 expression and thus decreased N-glycan branching. However, strong linkage disequilibrium with another polymorphism that serves as the primary functional variant cannot be excluded.
The identification and characterization of MGAT5 (rs4953911/rs3814022) bring the number of independent MS genetic and environmental risk factors demonstrated to dysregulate N-glycosylation and CTLA-4 surface expression in T cell blasts to seven. These are MGAT5 (rs4953911, rs3814022), MGAT1 IVAVT-T (rs7726005, rs2070924, rs2070925), IL2RA (rs2104286), IL7RA (rs6897932), CTLA-4 (rs231775), vitamin D3 deficiency and metabolic production of UDP-GlcNAc (Mkhikian et al., 2011). The largest GWAS of MS risk to date recently increased the number of MS associated loci to > 50 (International Multiple Sclerosis Genetics Consortium and Wellcome Trust Case Control Consortium, 2011). Based on the biology of the nearest gene, many of these are likely to also dysregulate N-glycosylation. For example, two loci are in/near vitamin D3 metabolic enzymes (CYP27B1, CYP24A1), while another is in/near the IL-7 cytokine gene. Moreover, many genomic regions were near genes that control TCR signal strength, a critical determinant of N-glycan branching and thus CTLA-4 surface retention in T cell blasts (Demetriou et al., 2001; Lau et al., 2007; Chen et al., 2009). These include CD37, CD40, CD58, CD80, CD86, CLECL1, CBLB, MALT1, TAGAP and MAPK1. The othermajor category of loci identified contained genes for cytokines and their signaling machinery, including IL12A, IL12B IL12RB1, IL22RA2, IRF8, TNFRSF1A, TNFRSF14, TNFSF14, TYK2 and STAT3.We are currently examining these pathways and it appears that a number of these may also regulate N-glycan branching. With further analysis, a significant proportion of the polymorphisms associated with MS may directly or indirectly regulate N-glycosylation to affect T cell growth and differentiation. Such a result is supported by our data in mice, where Mgat5 and N-glycan branching deficiency induce a spontaneous MS-like disease (Demetriou et al., 2001; Lee et al., 2007). To our knowledge, this has not been demonstrated for any other MS-associated gene outside of HLA-TCR double transgenic mice (Madsen et al., 1999). Finally, continued expansion of the number of MS associated loci known to dysregulate N-glycosylation will further validate the potential use of GlcNAc as a therapeutic tool for the treatment and risk management of MS.
Acknowledgments
The research was supported by the National Institutes of Health R01AI082266 to M.D. through the National Institute of Allergy and Infectious Disease as well as through a Collaborative Multiple Sclerosis Research Center Award to M.D. H.M. was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number F30HL108451. The research was also supported by the Institute for Clinical and Translational Science at UC Irvine through Clinical and Translational Science Award from the NIH. The authors thank the National Institute of Diabetes and Digestive and Kidney Diseases for providing DNA samples.
References
- Anjos S, Nguyen A, Ounissi-Benkalha H, Tessier MC, Polychronakos C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J. Biol. Chem. 2002;277:46478–46486. doi: 10.1074/jbc.M206894200. [DOI] [PubMed] [Google Scholar]
- Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, May GD, Woodward JE, Caillier SJ, McElroy JP, Gomez R, Pando MJ, Clendenen LE, Ganusova EE, Schilkey FD, Ramaraj T, Khan OA, Huntley JJ, Luo S, Kwok PY, Wu TD, Schroth GP, Oksenberg JR, Hauser SL, Kingsmore SF. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynedal B, Wojcik J, Esposito F, Debailleul V, Yaouanq J, Martinelli-Boneschi F, Edan G, Comi G, Hillert J, Abderrahim H. MGAT5 alters the severity of multiple sclerosis. J. Neuroimmunol. 2010;220:120–124. doi: 10.1016/j.jneuroim.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Chen IJ, Chen HL, Demetriou M. Lateral compartmentalization of T cell receptor versus CD45 by galectin-N-glycan binding and microfilaments coordinate basal and activation signaling. J. Biol. Chem. 2007;282:35361–35372. doi: 10.1074/jbc.M706923200. [DOI] [PubMed] [Google Scholar]
- Chen HL, Li CF, Grigorian A, Tian W, Demetriou M. T cell receptor signaling co-regulates multiple Golgi genes to enhance N-glycan branching. J. Biol. Chem. 2009;284:32454–32461. doi: 10.1074/jbc.M109.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD, Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J. Biol. Chem. 1982;257:11230–11234. [PubMed] [Google Scholar]
- Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebers GC, Bulman DE, Sadovnick AD, Paty DW, Warren S, Hader W, Murray TJ, Seland TP, Duquette P, Grey T, et al. A population-based study of multiple sclerosis in twins. N. Engl. J. Med. 1986;315:1638–1642. doi: 10.1056/NEJM198612253152603. [DOI] [PubMed] [Google Scholar]
- Esposito F, Wojcik J, Rodegher M, Radaelli M, Moiola L, Ghezzi A, Capra R, Brambilla P, Sorosina M, Giacalone G, Martinelli V, Comi G, Abderrahim H, Martinelli Boneschi F. MGAT5 and disease severity in progressive multiple sclerosis. J. Neuroimmunol. 2011;230:143–147. doi: 10.1016/j.jneuroim.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, Lincoln R, McCauley JL, Sawcer SJ, Compston DA, Dubois B, Hauser SL, Garcia-Blanco MA, Pericak-Vance MA, Haines JL. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- Grigorian A, Demetriou M. Mgat5 deficiency in T cells and experimental autoimmune encephalomyelitis. ISRN Neurol. 2011:374314. doi: 10.5402/2011/374314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorian A, Lee SU, Tian W, Chen IJ, Gao G, Mendelsohn R, Dennis JW, Demetriou M. Control of T Cell-mediated autoimmunity by metabolite flux to N-glycan biosynthesis. J. Biol. Chem. 2007;282:20027–20035. doi: 10.1074/jbc.M701890200. [DOI] [PubMed] [Google Scholar]
- Grigorian A, Torossian S, Demetriou M. T-cell growth, cell surface organization, and the galectin–glycoprotein lattice. Immunol. Rev. 2009;230:232–246. doi: 10.1111/j.1600-065X.2009.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorian A, Araujo L, Naidu NN, Place D, Choudhury B, Demetriou M. N-acetylglucosamine inhibits T-helper 1 (Th1)/T-helper 17 (Th17) responses and treats experimental autoimmune encephalomyelitis. J. Biol. Chem. 2011;286:40133–40141. doi: 10.1074/jbc.M111.277814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorian A, Mkhikian H, Demetriou M. Interleukin-2, Interleukin-7, T cell-mediated autoimmunity, and N-glycosylation. Ann. N. Y. Acad. Sci. 2012a;1253:49–57. doi: 10.1111/j.1749-6632.2011.06391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorian A, Mkhikian H, Li CF, Newton BL, Zhou RW, Demetriou M. Pathogenesis of multiple sclerosis via environmental and genetic dysregulation of N-glycosylation. Semin. Immunopathol. 2012b;34:415–424. doi: 10.1007/s00281-012-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genome wide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Genome-wide association study of severity in multiple sclerosis. Genes Immun. 2011;12:615–625. doi: 10.1038/gene.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Lee SU, Grigorian A, Pawling J, Chen IJ, Gao G, Mozaffar T, McKerlie C, Demetriou M. N-glycan processing deficiency promotes spontaneous inflammatory demyelination and neurodegeneration. J. Biol. Chem. 2007;282:33725–33734. doi: 10.1074/jbc.M704839200. [DOI] [PubMed] [Google Scholar]
- Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallstrom E, Khademi M, Oturai A, Ryder LP, Saarela J, Harbo HF, Celius EG, Salter H, Olsson T, Hillert J. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- Madsen LS, Andersson EC, Jansson L, Krogsgaard M, Andersen CB, Engberg J, Strominger JL, Svejgaard A, Hjorth JP, Holmdahl R, Wucherpfennig KW, Fugger L. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat. Genet. 1999;23:343–347. doi: 10.1038/15525. [DOI] [PubMed] [Google Scholar]
- Maier LM, Anderson DE, Severson CA, Baecher-Allan C, Healy B, Liu DV, Wittrup KD, De Jager PL, Hafler DA. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J. Immunol. 2009a;182:1541–1547. doi: 10.4049/jimmunol.182.3.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, Clark PM, Healy B, Walker N, Aubin C, Oksenberg JR, Hauser SL, Compston A, Sawcer S, De Jager PL, Wicker LS, Todd JA, Hafler DA. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009b;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M, Loserth S, Kolb-Maurer A, Ponath A, Wiese S, Kruse N, Rieckmann P. A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (CTLA4) gene (exon 1 + 49) alters T-cell activation. Immunogenetics. 2002;54:1–8. doi: 10.1007/s00251-002-0429-9. [DOI] [PubMed] [Google Scholar]
- Mkhikian H, Grigorian A, Li CF, Chen HL, Newton B, Zhou RW, Beeton C, Torossian S, Tatarian GG, Lee SU, Lau K, Walker E, Siminovitch KA, Chandy KG, Yu Z, Dennis JW, Demetriou M. Genetics and the environment converge to dysregulate N-glycosylation in multiple sclerosis. Nat. Commun. 2011;2:334. doi: 10.1038/ncomms1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Gao G, Pawling J, Dennis JW, Demetriou M, Li B. N-acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J. Immunol. 2004;173:7200–7208. doi: 10.4049/jimmunol.173.12.7200. [DOI] [PubMed] [Google Scholar]
- Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–2838. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am. J. Epidemiol. 2007;165:1097–1109. doi: 10.1093/aje/kwk118. [DOI] [PubMed] [Google Scholar]
- Smolders J, Damoiseaux J, Menheere P, Hupperts R. Vitamin D as an immune modulator in multiple sclerosis, a review. J. Neuroimmunol. 2008;194:7–17. doi: 10.1016/j.jneuroim.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Soilu-Hanninen M, Airas L, Mononen I, Heikkila A, Viljanen M, Hanninen A. 25-Hydroxyvitamin D levels in serum at the onset of multiple sclerosis. Mult. Scler. 2005;11:266–271. doi: 10.1191/1352458505ms1157oa. [DOI] [PubMed] [Google Scholar]
- Steinman L. Multiple sclerosis: a two-stage disease. Nat. Immunol. 2001;2:762–764. doi: 10.1038/ni0901-762. [DOI] [PubMed] [Google Scholar]
- van der Mei IA, Ponsonby AL, Dwyer T, Blizzard L, Simmons R, Taylor BV, Butzkueven H, Kilpatrick T. Past exposure to sun, skin phenotype, and risk of multiple sclerosis: case-control study. BMJ. 2003;327:316. doi: 10.1136/bmj.327.7410.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Gillen D, Li CF, Demetriou M. Incorporating parental information into family-based association tests. Biostatistics. doi: 10.1093/biostatistics/kxs048. in press. http://dx.doi.org/10.1093/biostatistics/kxs048. [DOI] [PMC free article] [PubMed] [Google Scholar]