Abstract
Summary: Metabolic network mapping is a widely used approach for integration of metabolomic experimental results with biological domain knowledge. However, current approaches can be limited by biochemical domain or pathway knowledge which results in sparse disconnected graphs for real world metabolomic experiments. MetaMapR integrates enzymatic transformations with metabolite structural similarity, mass spectral similarity and empirical associations to generate richly connected metabolic networks. This open source, web-based or desktop software, written in the R programming language, leverages KEGG and PubChem databases to derive associations between metabolites even in cases where biochemical domain or molecular annotations are unknown. Network calculation is enhanced through an interface to the Chemical Translation System, which allows metabolite identifier translation between >200 common biochemical databases. Analysis results are presented as interactive visualizations or can be exported as high-quality graphics and numerical tables which can be imported into common network analysis and visualization tools.
Availability and Implementation: Freely available at http://dgrapov.github.io/MetaMapR/. Requires R and a modern web browser. Installation instructions, tutorials and application examples are available at http://dgrapov.github.io/MetaMapR/.
Contact: ofiehn@ucdavis.edu
1 Introduction
Metabolomic experiments contain both high-dimensional and complex biological, chemical and analytical information. Mass spectrometry based analyses can generate measurements for many hundreds to thousands of small molecules. In addition to compounds with identified biological roles many measurements may only contain mass spectral or empirical information. Analysis of metabolomic data in the context of biological domain knowledge (e.g. enzymatic precursor to product relationships) is a well-established approach for metabolic network generation (Gao et al., 2010). However, real world metabolomic experiments can measure a wide range of biochemical domains [for example (Grapov et al., 2012)] for which direct biochemical intermediates may be absent or unknown, leading to sparse disconnected biochemical representations. Inclusion of non-measured metabolites in the reconstructed metabolic networks using tools like Metscape (Karnovsky et al., 2012) can help overcome this issue, but requires calculation of minimum spanning trees, which can still fail to associate metabolites lacking biochemical domain knowledge (e.g. complex lipids). Barupal et al. (2012) recently showed in their tool Metamapp that structural similarity information can be used to enhance enzymatic transformation networks and fill in the gaps between missing biochemical intermediates or domains. However, neither Metamapp nor Metscape directly calculate structural similarity, mass spectral similarity nor empirical relationships, and lack standalone interactive network visualization and threshold tuning interfaces featured in MetaMapR. In addition to biochemical transformations and structural similarity, MetaMapR also incorporates mass spectral similarity and empirical correlation information. The combination of these four orthogonal measures of molecular association provides a robust framework for generating richly connected biochemical representations which can combine molecules with unknown biochemistry, unknown structures and integrate non-metabolomic data (genomic, proteomic, clinical) into the reconstructed metabolic networks.
2 Methods
MetaMapR (http://dgrapov.github.io/MetaMapR/) is implemented in the R programming language (http://cran.us.r-project.org/) and requires the R package Shiny (http://www.rstudio.com/) and a modern web browser (Chrome, Firefox, IE10, Safari, etc). Internet connection is required for calculation of biochemical (KEGG, http://www.genome.jp/kegg/) and structural similarity network (PubChem, https://pubchem.ncbi.nlm.nih.gov/) relationships.
The user interface is implemented using the Twitter Bootstrap front-end (http://getbootstrap.com/2.3.2/) and enhanced by custom CSS, HTML and JavaScript. Interactive networks are created using the D3.js (http://d3js.org/) JavaScript library and the R package d3Networks. Networks can be exported as scalable vector graphic or portable network graphic formats. Alternatively, network edge list and node attributes can be exported as a comma separated value (.csv) files which can be extended using other third party software such as Cytoscape (Shannon et al., 2003). This licensed (GPLv3) cross-platform (windows, OSX and linux) software can be deployed locally or as a hosted web application using the Shiny server (https://github.com/rstudio/shiny-server). Download and installation instructions can be found at https://github.com/dgrapov/MetaMapR.
2.1 Features
Data can be uploaded as comma separated values (.csv) or other delimited formats through the application paste field. Accepted metabolite identifiers include synonyms or one of over 200 common biological database identifiers (see Identifier Translation). Mass spectra can be uploaded as mass-to-charge and intensity pair strings (e.g. “m/z1:intensity1 m/z2:intensity2”). Measured metabolite concentrations, peak areas/heights, intensities or other experimental data can be used to calculate empirical correlation relationships.
Identifier Translation can be optionally used to map user metabolite names or identifiers to KEGG or PubChem CIDs required to calculate biochemical and chemical similarity networks. Translations are accomplished using CTSgetR (https://github.com/dgrapov/CTSgetR), an R interface to the Chemical Translation System (http://cts.fiehnlab.ucdavis.edu/).
Biochemical Reaction Networks are generated based on the KEGG RPAIR (http://www.genome.jp/kegg/reaction/, ftp://ftp.genome.jp/pub/db/rclass/rpair) substrate-product pair reaction database. User supplied metabolite names or database identifiers are optionally translated to KEGG identifiers which are then used to query for biochemical substrate-product relationships using a curated lookup table based on over 14 000 biochemical reactions in the KEGG Database (Kanehisa et al., 2014).
Structural Similarity Networks are determined based on similarities between PubChem Substructure Fingerprints (ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt). The R package Chemminer is used to generate molecular fingerprints using the PubChem Power User Gateway (PUG). Molecular fingerprints in the form of ordered lists of binary bits defining presence or absence of physical properties (e.g. element type, functional group, nearest neighbors) are used to calculate structural similarities. Pairwise similarities are calculated based on the Tanimoto similarity between two bit vectors (Willett et al., 1998). Similarity scores are bound between 0 and 1, where a score of 0 or 1 defines no or complete overlap in structural properties between two molecules.
Spectral Similarity Networks are calculated based on pairwise similarities between mass spectra. Cosine correlations are calculated between molecular features’ mass spectra which are encoded as mass-to-charge ratio (m/z) and intensity pairs. The results are bounded between 0 and 1, with zero defining no and 1 complete correlation between two mass spectra. Generated mass spectral similarity networks can be optimized based on control of the cosine correlation threshold for edge acceptance, limit of total edges per object, and object-specific control of edge acceptance (e.g. limiting connections to only show annotated to unknown relationships).
Empirical Dependency Networks are calculated (Langfelder and Horvath, 2008) based on the parametric Pearson and biweight correlations or non-parametric Spearman correlations between measured metabolite values (e.g. concentration, peak intensity, etc.) for any or all samples. Measures of significance or P-values and the false discovery rate (FDR) adjusted P-values can be used to alter the statistical confidence of the correlation networks. For example a correlation network based monotonic linear relationships between metabolites which is robust to outliers and mitigates spurious false discoveries can be calculated using Spearman correlations with edge acceptance at FDR P-values < 0.05.
3 Results and Discussion
MetaMapR is implemented using the shiny R package, a tool for building browser-based applications, which can be deployed on the desktop using a modern web browser or can be hosted as a standalone web-application using the shiny-server (https://github.com/rstudio/shiny-server).
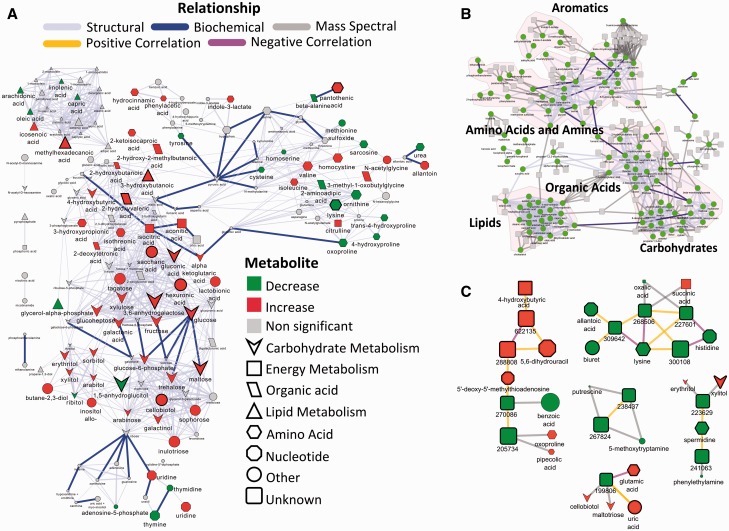
A variety of MetaMapR applications are described at (http://dgrapov.github.io/MetaMapR/) including the analysis of type 1 diabetes-dependent (T1D) biochemical changes in NOD mice (Grapov et al., 2014) and metabolic changes in lung cancer in humans (Wikoff et al., 2015). The study by Grapov et. al. compared metabolic profiles of animals progressing to T1D (progressor) to those maintaining normoglycemic control (non-progressor), to identify age and gender independent T1D-associated biochemical perturbations in over 470 plasma metabolites measured by gas-chromatography time-of-flight mass spectrometry (GC–TOF–MS). A biochemical reaction and structural similarity metabolic network (Fig. 1 A) was calculated to show key biochemical alterations in progressor compared with non-progressors. Due to the limitation of the KEGG database description of the directionality of biochemical transformations, ‘the terms “reversible” and “irreversible” do not necessarily reflect biochemical properties of each reaction’ (http://www.genome.jp/kegg/xml/docs/) and because MetaMapR uniquely combines biochemical relationships, many of which are reversible given appropriate conditions, with structural similarity, mass spectral similarity and correlations, all of which lack directionality; the current implementation of MetaMapR treats all enzymatic relationships as undirected. The network structural similarity threshold was set to Tanimoto score > 0.7, which maintains non-overlapping network modularity for the biochemical classes described in Figure 1A. A more detailed description of structural similarity threshold selection can be found in Barupal et. al. (Barupal et al., 2012).
Fig. 1.
Mapped metabolic networks combining a variety of edge combinations available in MetaMapR. Metabolomic networks can be generated based on (A) biochemical substrate-product and structural similarity relationships (Grapov et al., 2014), which can also display (B) mass spectral similarity information or combined with C) empirical correlations. BinBase identifiers (Fiehn et al., 2005) are reported for unknowns. Node size represents the fold change in metabolite levels relative controls
T1D was associated with large scale metabolic perturbations in plasma metabolites including increases in the majority of carbohydrates (red downward arrows), and a decrease in the structurally similar but not directly biochemically related 1,5-anhydroglucitol. Dietary derived 1,5-anhydroglucitol (bottom left) is an established marker of glucose control (Kim and Park, 2013), the levels in which drop in response to competition with increasing glucose for reabsorption in the kidneys. Networks in Figure 1 were calculated in MetaMapR, exported to Cytoscape (Shannon et al., 2003) and further enhanced by mapping various empirical and domain knowledge-based variables to the network node attributes, the process of which is described in detail elsewhere (Grapov et al., 2014). Mass spectral information can be used to extend the analysis of biochemical and structural similarity relationships to molecules without structural annotation (unknowns; Fig. 1B). Mass spectral similarity network analysis has been previously used to link structurally unknown features with known molecules (Watrous et al., 2012). Mass spectral similarity is defined based on the cosine of the angle between two or more mass spectra represented as vectors (cosine correlation) which was set to > 0.7 for Figure 1B and 1C. We suggest that the user considers tuning the threshold for mass spectral similarity based on their needs (Stein and Scott, 1994).
MetaMapR uniquely combines molecular biochemical and structural information (Fig. 1A) with mass spectral similarity (Fig. 1B) and correlation based associations (Fig. 1C). The combination of orthogonal information can help link structurally unknown metabolites (Fig. 1C, rounded rectangles) to other identified species. MetaMapR is freely available open source software which includes ongoing efforts to integrate the analysis of gene-metabolite and protein-metabolite biochemical information, calculation of Gaussian graphical Markov metabolomic networks (GGM) and an enhanced dynamic network mapping interface.
Acknowledgements
We acknowledge the exceptional work of the R Development Core Team, Shiny and authors of R community contributed packages.
Funding
This project was funded by the National Institutes of Health NIH, 1 U24 DK097154 for the West Coast Metabolomics Center (OF, DG) and NIH P20 HL113452 (OF).
Conflict of Interest: none declared.
References
- Barupal D.K., et al. (2012) MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics, 13, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O., et al. (2005) Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. In: Ludäscher B., Raschid L. (eds.) Data Integration in the Life Sciences. Springer, Berlin, pp. 224–239. [Google Scholar]
- Gao J., et al. (2010) Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics, 26, 971–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapov D., et al. (2012) Type 2 diabetes associated changes in the plasma non-esterified fatty acids, oxylipins and endocannabinoids. PloS one, 7, e48852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapov D., et al. (2014) Diabetes associated metabolomic perturbations in NOD mice. Metabolomics, 11, 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., et al. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res., 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A., et al. (2012) Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics, 28, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.J., Park C.Y. (2013) 1,5-Anhydroglucitol in diabetes mellitus. Endocrine, 43, 33–40. [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S., Scott D. (1994) Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Spectrom., 5, 859–866. [DOI] [PubMed] [Google Scholar]
- Watrous J., et al. (2012) Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA, 109, E1743–E1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W., et al. (2015) Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prevent. Res, 8, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett P., et al. (1998) Chemical similarity searching. J. Chem. Inf. Comput. Sci., 38, 983–996. [Google Scholar]