Abstract
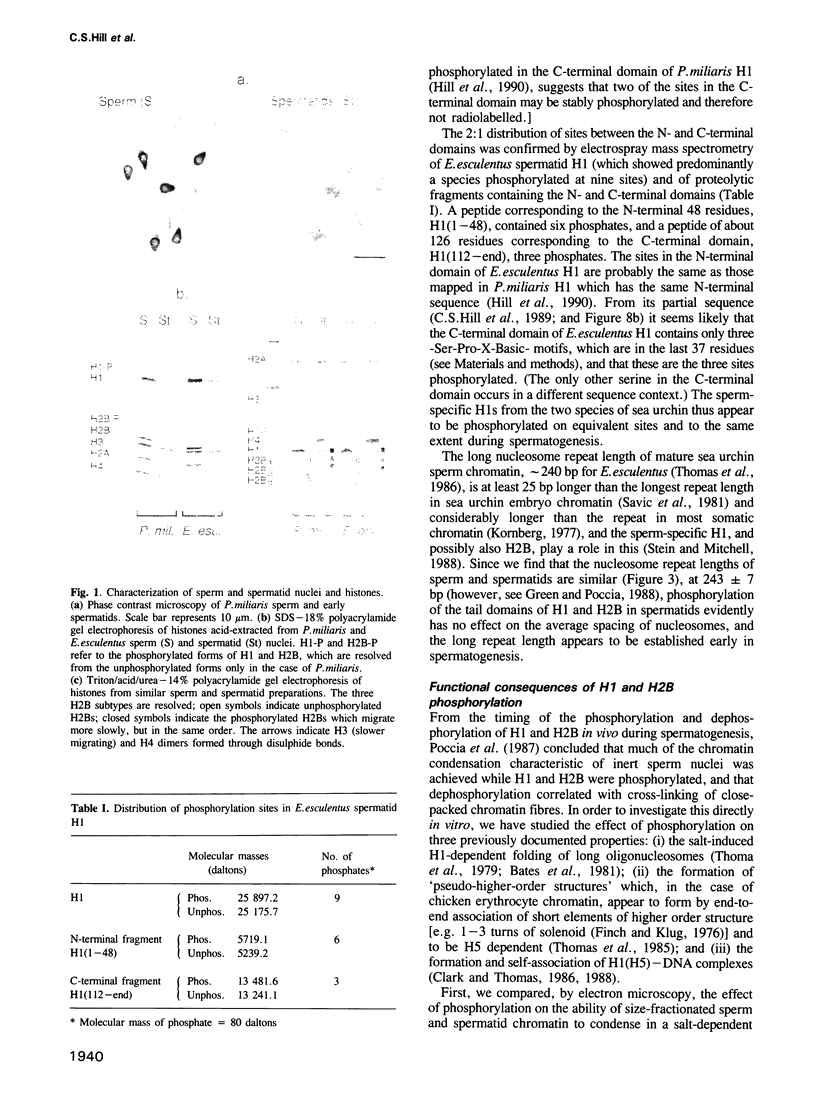
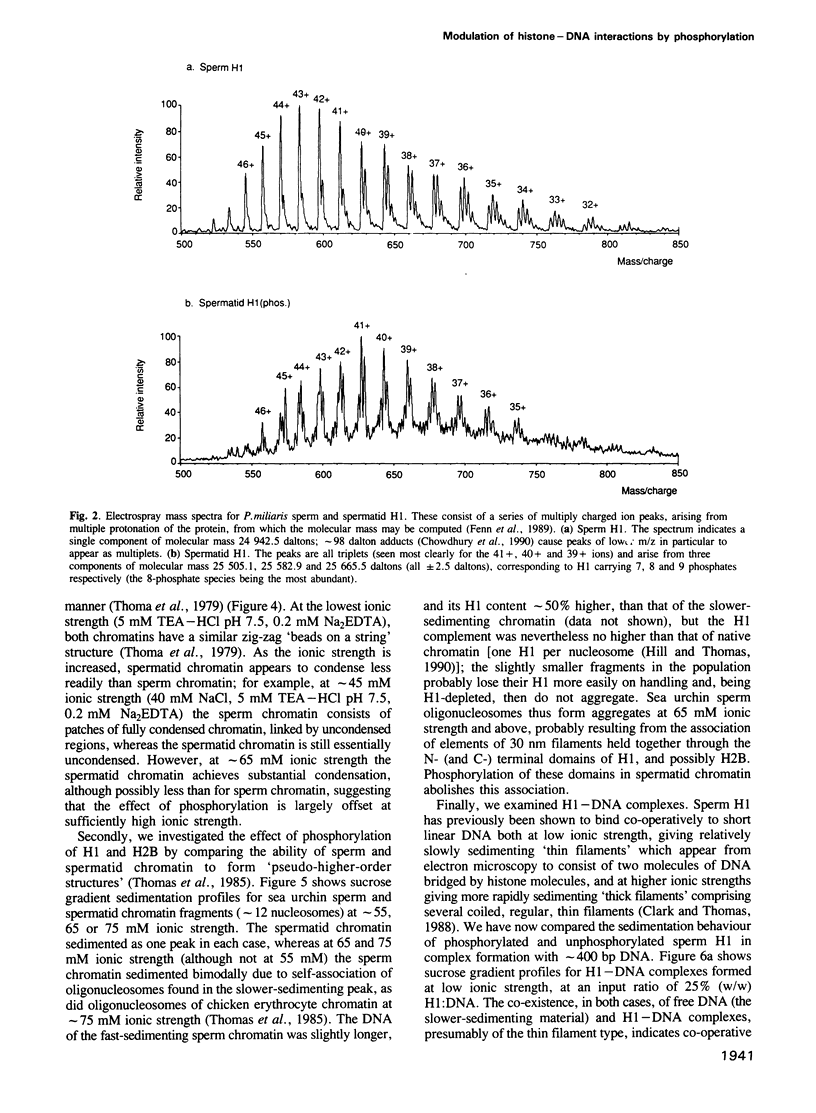
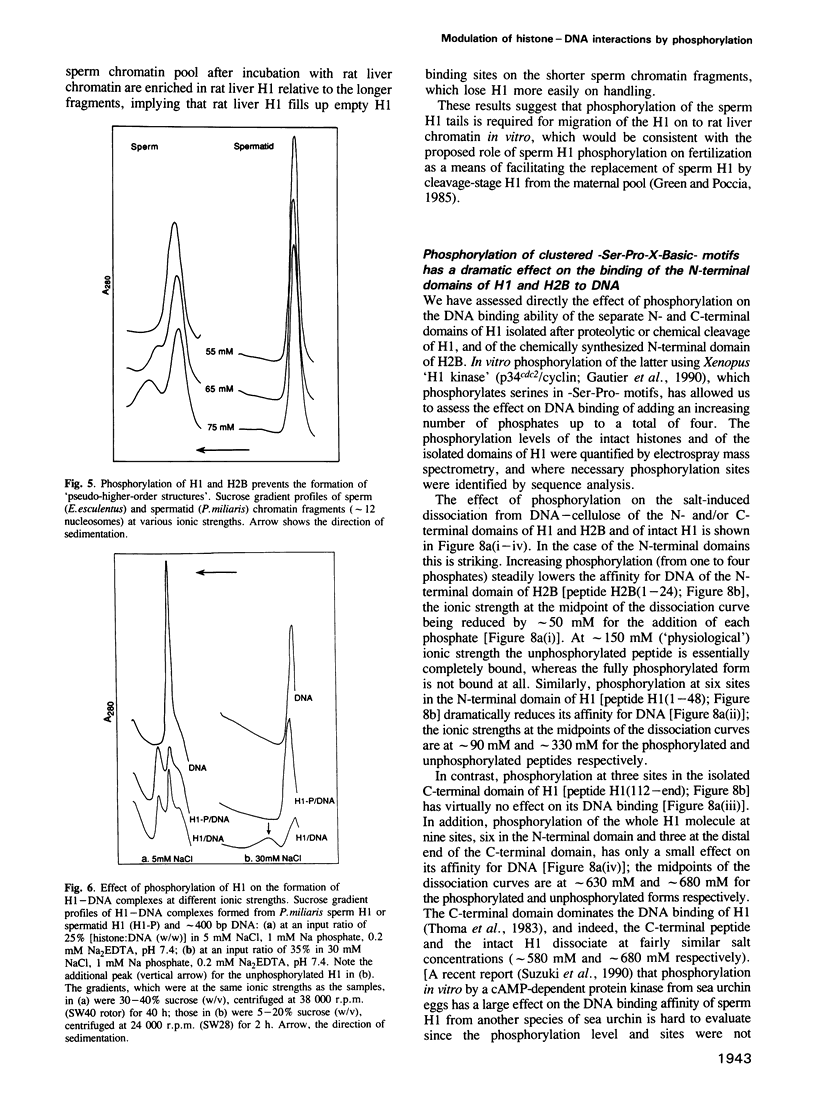
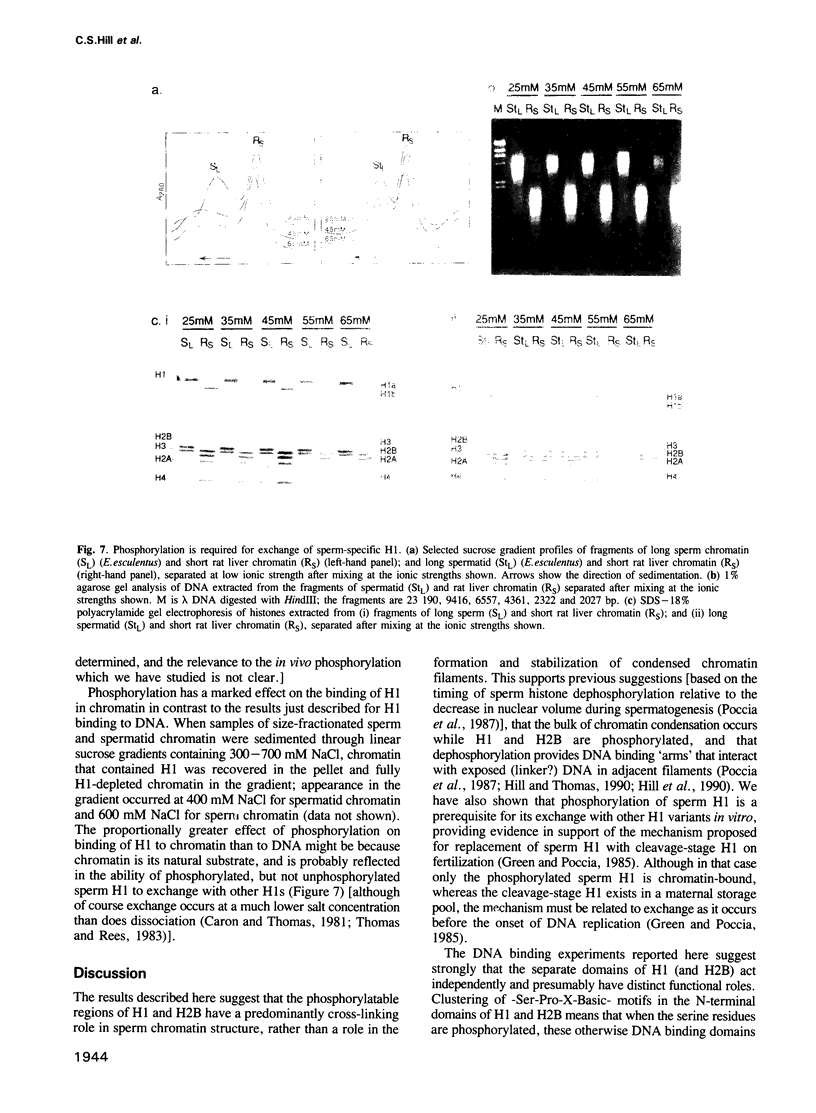
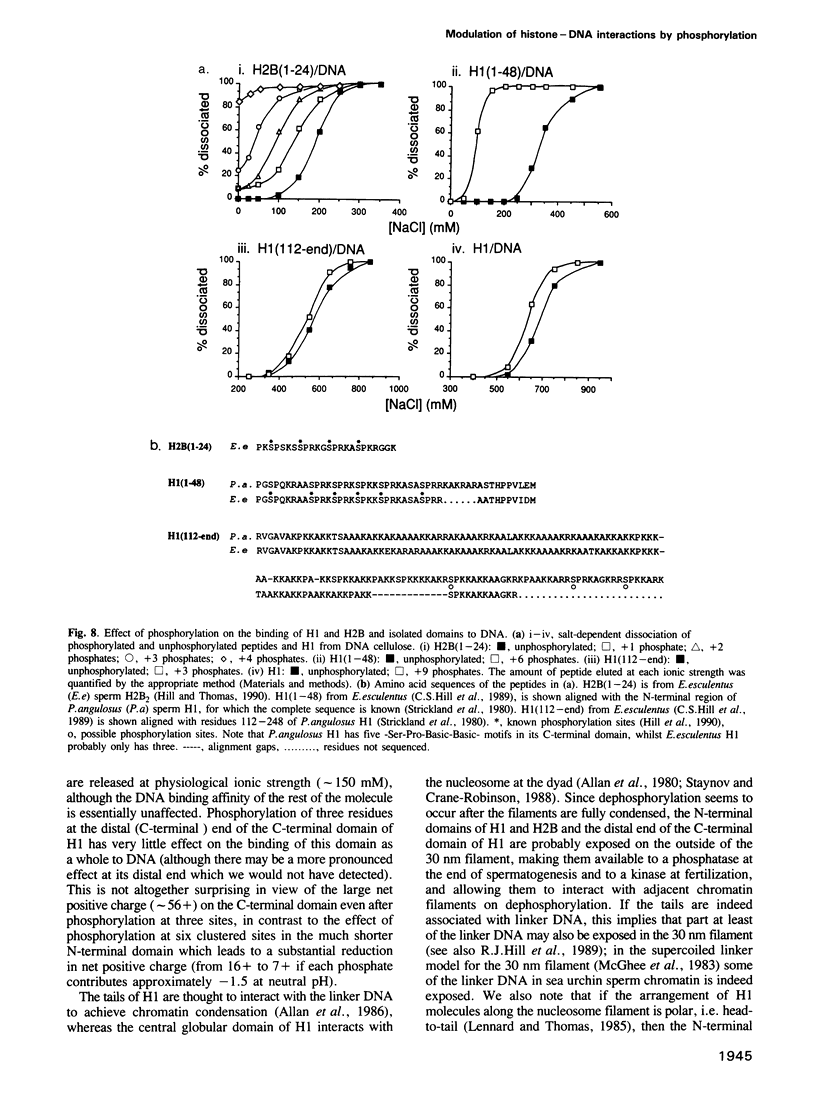
The sea urchin sperm-specific histones H1 and H2B are multiply phosphorylated in spermatids, dephosphorylated in the final stages of spermatogenesis to give mature sperm, and rephosphorylated upon fertilization. Phosphorylation in spermatids, and probably at fertilization, occurs at repeated -Ser-Pro-X-Basic-motifs in the distinctive N-terminal basic domains of both histones and at the end of the much longer C-terminal domain of H1. Here we identify the consequences of multiple phosphorylation through comparison of some physical and biochemical properties of spermatid (phosphorylated) and sperm (dephosphorylated) chromatin and histones. Study of the DNA binding properties of the intact histones and isolated basic domains suggests that phosphorylation at three dispersed sites in the C-terminal tail of H1 has little effect on its overall DNA binding affinity, whereas, strikingly, binding of the N-terminal domains of H2B and H1 is abolished by phosphorylation at four or six tandemly repeated sites respectively. Together with the relative timing of events in vivo, this suggests that phosphorylation/dephosphorylation of the N-terminal (and distal end of the C-terminal) tail of H1, and/or the N-terminal tail of H2B, effectively controls intermolecular interactions between adjacent chromatin filaments, and hence chromatin packing in the sperm nucleus.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986 Feb 20;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Hill C. S., Martin S. R., Thomas J. O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988 Jan;7(1):69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Differences in the binding of H1 variants to DNA. Cooperativity and linker-length related distribution. Eur J Biochem. 1988 Dec 1;178(1):225–233. doi: 10.1111/j.1432-1033.1988.tb14447.x. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989 Oct 6;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Minshull J., Lohka M., Glotzer M., Hunt T., Maller J. L. Cyclin is a component of maturation-promoting factor from Xenopus. Cell. 1990 Feb 9;60(3):487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Interaction of sperm histone variants and linker DNA during spermiogenesis in the sea urchin. Biochemistry. 1988 Jan 26;27(2):619–625. doi: 10.1021/bi00402a019. [DOI] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Phosphorylation of sea urchin sperm H1 and H2B histones precedes chromatin decondensation and H1 exchange during pronuclear formation. Dev Biol. 1985 Mar;108(1):235–245. doi: 10.1016/0012-1606(85)90026-0. [DOI] [PubMed] [Google Scholar]
- Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978 Mar;84(1):1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Martin S. R., Thomas J. O. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. doi: 10.1002/j.1460-2075.1989.tb08398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Packman L. C., Thomas J. O. Phosphorylation at clustered -Ser-Pro-X-Lys/Arg- motifs in sperm-specific histones H1 and H2B. EMBO J. 1990 Mar;9(3):805–813. doi: 10.1002/j.1460-2075.1990.tb08177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Hill R. J., Watt F., Wilson C. M., Fifis T., Underwood P. A., Tribbick G., Geysen H. M., Thomas J. O. Bands, interbands and puffs in native Drosophila polytene chromosomes are recognized by a monoclonal antibody to an epitope in the carboxy-terminal tail of histone H1. Chromosoma. 1989 Dec;98(6):411–421. doi: 10.1007/BF00292786. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lennard A. C., Thomas J. O. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985 Dec 16;4(13A):3455–3462. doi: 10.1002/j.1460-2075.1985.tb04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion C., Martinage A., Tirard A., Roux B., Daune M., Mazen A. Histone phosphorylation in native chromatin induces local structural changes as probed by electric birefringence. J Mol Biol. 1985 Nov 20;186(2):367–379. doi: 10.1016/0022-2836(85)90111-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Meijer L., Arion D., Golsteyn R., Pines J., Brizuela L., Hunt T., Beach D. Cyclin is a component of the sea urchin egg M-phase specific histone H1 kinase. EMBO J. 1989 Aug;8(8):2275–2282. doi: 10.1002/j.1460-2075.1989.tb08353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Kirschner M. W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989 May 25;339(6222):275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Poccia D. L., Simpson M. V., Green G. R. Transitions in histone variants during sea urchin spermatogenesis. Dev Biol. 1987 Jun;121(2):445–453. doi: 10.1016/0012-1606(87)90181-3. [DOI] [PubMed] [Google Scholar]
- Savić A., Richman P., Williamson P., Poccia D. Alterations in chromatin structure during early sea urchin embryogenesis. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3706–3710. doi: 10.1073/pnas.78.6.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M. V., Poccia D. Sea urchin testicular cells evaluated by fluorescence microscopy of unfixed tissue. Gamete Res. 1987 Jun;17(2):131–144. doi: 10.1002/mrd.1120170205. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z., Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988 Dec 1;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. J Mol Biol. 1988 Oct 20;203(4):1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- Strickland W. N., Strickland M., Brandt W. F., Von Holt C., Lehmann A., Wittmann-Liebold B. The primary structure of histone H1 from sperm of the sea urchin Parechinus angulosus. 2. Sequence of the C-terminal CNBr peptide and the entire primary structure. Eur J Biochem. 1980 Mar;104(2):567–578. doi: 10.1111/j.1432-1033.1980.tb04460.x. [DOI] [PubMed] [Google Scholar]
- Sung M. T., Freedlender E. F. Sites of in vivo phosphorylation of histone H5. Biochemistry. 1978 May 16;17(10):1884–1890. doi: 10.1021/bi00603a013. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Sohma H., Yazawa M., Yagi K., Ebashi S. Histone H1 kinase specific to the SPKK motif. J Biochem. 1990 Sep;108(3):356–364. doi: 10.1093/oxfordjournals.jbchem.a123206. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Losa R., Koller T. Involvement of the domains of histones H1 and H5 in the structural organization of soluble chromatin. J Mol Biol. 1983 Jul 5;167(3):619–640. doi: 10.1016/s0022-2836(83)80102-8. [DOI] [PubMed] [Google Scholar]
- Thomas J. O. Chemical radiolabeling of lysines that interact strongly with DNA in chromatin. Methods Enzymol. 1989;170:369–385. doi: 10.1016/0076-6879(89)70057-4. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Butler P. J. Salt-induced folding of sea urchin sperm chromatin. Eur J Biochem. 1986 Jan 15;154(2):343–348. doi: 10.1111/j.1432-1033.1986.tb09403.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Rees C. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem. 1983 Jul 15;134(1):109–115. doi: 10.1111/j.1432-1033.1983.tb07538.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Pearson E. C. Histone H5 promotes the association of condensed chromatin fragments to give pseudo-higher-order structures. Eur J Biochem. 1985 Feb 15;147(1):143–151. doi: 10.1111/j.1432-1033.1985.tb08730.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Wagner T. E., Hartford J. B., Serra M., Vandegrift V., Sung M. T. Phosphorylation and dephosphorylation of histone (V (H5): controlled condensation of avian erythrocyte chromatin. Appendix: Phosphorylation and dephosphorylation of histone H5. II. Circular dichroic studies. Biochemistry. 1977 Jan 25;16(2):286–290. doi: 10.1021/bi00621a020. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter M., Braun W. Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton-proton distances. J Mol Biol. 1984 Dec 15;180(3):715–740. doi: 10.1016/0022-2836(84)90034-2. [DOI] [PubMed] [Google Scholar]