Integration of gene interaction data across a model dicot and a monocot identifies conserved and distinct regulatory network modules involved in nitrogen use, enabling translational discoveries from models to crops.
Abstract
In this study, we used a cross-species network approach to uncover nitrogen (N)-regulated network modules conserved across a model and a crop species. By translating gene network knowledge from the data-rich model Arabidopsis (Arabidopsis thaliana) to a crop, rice (Oryza sativa), we identified evolutionarily conserved N-regulatory modules as targets for translational studies to improve N use efficiency in transgenic plants. To uncover such conserved N-regulatory network modules, we first generated an N-regulatory network based solely on rice transcriptome and gene interaction data. Next, we enhanced the network knowledge in the rice N-regulatory network using transcriptome and gene interaction data from Arabidopsis and new data from Arabidopsis and rice plants exposed to the same N treatment conditions. This cross-species network analysis uncovered a set of N-regulated transcription factors (TFs) predicted to target the same genes and network modules in both species. Supernode analysis of the TFs and their targets in these conserved network modules uncovered genes directly related to N use (e.g. N assimilation) and to other shared biological processes indirectly related to N. This cross-species network approach was validated with members of two TF families in the supernode network, BASIC-LEUCINE ZIPPER TRANSCRIPTION FACTOR1-TGA and HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING1 (HRS1)/HRS1 Homolog family, which have recently been experimentally validated to mediate the N response in Arabidopsis.
The goal of this study is to translate network knowledge from Arabidopsis (Arabidopsis thaliana), a data-rich model species, to enhance the identification of nitrogen (N)-regulatory networks in rice (Oryza sativa), one of the most important crops in the world. With a significantly smaller genome size than other cereals (approximately 430 Mb), the ability to perform genetic transformations (Hiei and Komari, 2008), and a finished genome sequence (International Rice Genome Sequencing Project, 2005), rice is an excellent monocot model for genetic, molecular, and genomic studies (Gale and Devos, 1998; Sasaki and Sederoff, 2003). In this study, we constructed N-regulatory gene networks in rice using network knowledge from Arabidopsis, a data-rich laboratory model for dicots. Thus, this cross-species network study exploits the best-characterized experimental models for dicot and monocot plants.
N is a rate-limiting element for plant growth. Rice plants absorb NH4+ at a higher rate than NO3– (Fried et al., 1965). Because NH4+ strongly inhibits NO3– uptake in agricultural soils where both NO3– and NH4+ are present (Kronzucker et al., 1999a), root NH4+ uptake may be favored as a result of the specific down-regulation of NO3– uptake systems (Kronzucker et al., 1999b). In rice, combinations of NO3– and NH4+ usually result in a greater vegetative growth than when either N form is supplied alone (Cramer and Lewis, 1993). Therefore, we designed our N treatment experiments in this study to include both NO3– and NH4+.
In previous studies of the Arabidopsis N response, we analyzed transcriptome data in the context of gene interactions to identify and validate N-regulated gene networks in planta (Gifford et al., 2008; Gutiérrez et al., 2008; Krouk et al., 2010). In this article, we compare the N-regulated genes and gene networks between Arabidopsis and rice. This cross-species network analysis provides a unique opportunity to examine the conservation and divergence of N-regulated networks in the context of monocot and dicot transcriptomes. As rice and Arabidopsis are highly divergent phylogenetically, any evolutionarily conserved networks should be of special importance.
Establishing the architecture of gene regulatory networks requires gathering information on transcription factors (TFs), their targets in the genome, and their corresponding binding sites in gene promoter regions. Generation of N-responsive transcriptome data from rice and Arabidopsis enabled us to identify conserved N-regulatory gene network modules shared between dicots and monocots. We analyzed the rice and Arabidopsis transcriptomes (using Affymetrix GeneChips) in response to N treatments in roots and shoots. The VirtualPlant software platform (Katari et al., 2010), which is operational for both Arabidopsis and rice, was used to perform much of the analysis, including homology mapping analysis and significance of overlap in gene lists using the Genesect tool (www.virtualplant.org).
The N-regulated gene network includes expression data generated in this study and metabolic and protein-protein interactions from publicly available rice data (Rohila et al., 2006, 2009; Ding et al., 2009; Gu et al., 2011; Dharmawardhana et al., 2013). Despite the fact that much of the genomic and systemic rice data have been generated over the past years, a lot of information is still missing. For example, Arabidopsis has much more experimental data with regard to cis-binding sites and protein-protein interaction. To fill these gaps in rice network knowledge, we integrated orthology-based Arabidopsis interaction data (Palaniswamy et al., 2006; Yilmaz et al., 2009; Gu et al., 2011; Ho et al., 2012) and searched for functional Arabidopsis cis-binding sites in rice, to identify N-regulatory network modules and biological processes (network biomodules) conserved between dicots and monocots.
An important issue in this analysis is orthology. Monocots and dicots are quite distantly related, with divergence estimation of 140 to 150 million years ago (Chaw et al., 2004). A naive and crude method for identifying putative orthologs is to use reverse BLAST hit thresholds: the putative orthologs must map to each other with a BLAST e-value less than some cutoff. The identification of putative orthologs between monocots and dicots is confounded by the presence of paralogs (homologous genes originating from gene duplication events). There are several algorithms, such as OrthoMCL (Fischer et al., 2011), that are designed to help distinguish an ortholog from a paralog by comparing sequences within species in addition to between species. However, even if these algorithms can detect true orthologs with greater specificity, there is always a possibility that different gene family members in each species take on the responsibility of responding to nutrients, like N. Here, we test and compare the performance of the reverse BLAST hit method and OrthoMCL in identifying genes and gene interactions whose functions are conserved across species. From here on, the cross-species gene mapping based on BLASTP will be referred to as homologs, and the matches based on OrthoMCL will be called orthologs.
Finally, this cross-species network study significantly contributes to two important areas: (1) studying N-regulated gene networks in rice, an important crop, and (2) identifying conserved and distinct N-regulatory hubs controlling network biomodules, which can be used to enhance translational discoveries between a model plant and crops. Our aim to identify N-regulated genes across a model dicot and a monocot crop, and to interpret it in a systems biology/network context, is essential to derive testable biological hypotheses. By applying network information, we can identify key regulators of these N-responsive gene networks and biomodules, which can be further manipulated to study N use efficiency in transgenic plants. This approach has the potential to enhance translational discoveries from Arabidopsis to a crop (rice) with the goal of improving plant N use efficiency, which will contribute to sustainable agricultural practices by diminishing the use of N fertilizers.
RESULTS
Equilibrating N Treatment Conditions for Arabidopsis and Rice
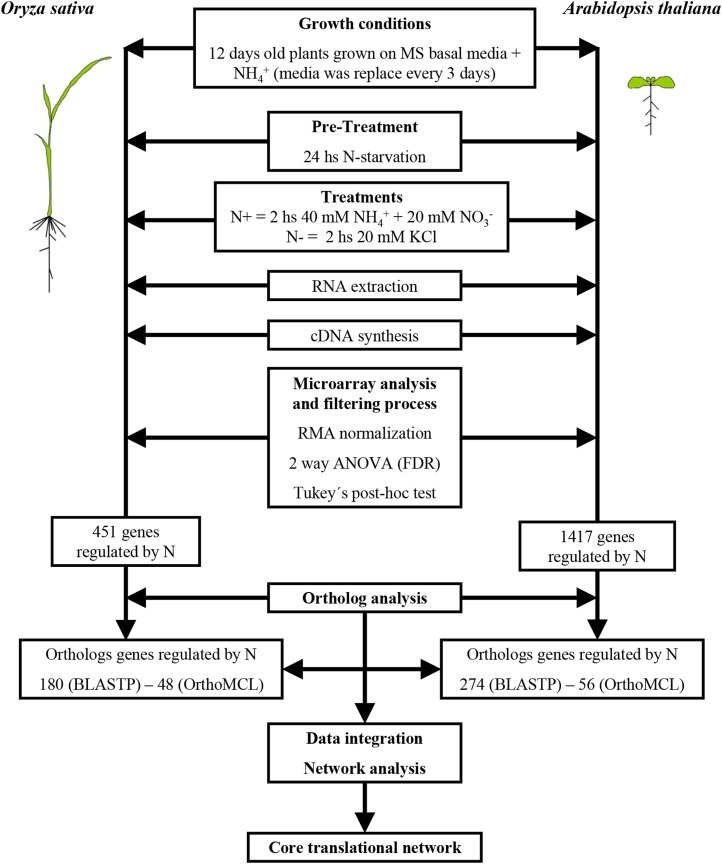
The goal of this study is to identify conserved N response networks in two species by comparison. Thus, in our study, we made our N treatments and growth conditions of rice and Arabidopsis as comparable as possible. We adapted our hydroponic system for Arabidopsis (Gifford et al., 2008) to grow and treat rice seedlings, with only the plant roots submerged in liquid medium. For plants with minimal seed reserves such as Arabidopsis, an external N supply is required to allow plant growth and development. By contrast, rice can grow for longer periods using N nutrients stored in their seeds. In order to equilibrate the growth conditions of these two species, and to eliminate the seed-nutrient effect during N treatment, the nutritive rice seed tissue was dissected away from the rice seedlings once the cotyledon and roots emerged, and only the germinated embryo was placed in the hydroponic system. For both species, the N source during this initial growth phase contained 0.5 mm ammonium succinate, which was renewed every 2 to 3 d with fresh medium to avoid NH4+ depletion due to different consuming rates between species. This growth on a low level of an N source (ammonium) was the background in which to observe the effects of transient treatments with nitrate (Wang et al., 2000, 2004) and/or high ammonium. As the N regulation of gene expression is largely dependent on carbon (C) resource provision in Arabidopsis (Krouk et al., 2009), 0.5% (w/v) Suc was included in the growth medium as a constant nutrient to eliminate C signaling effects during transient N treatments. After 12 d, plants were N starved for 24 h. Finally, at the start of their light cycle, plants were N treated for 2 h with a combination of NO3– (40 mm) and NH4+ (20 mm), the amount of N in Murashige and Skoog (MS) medium (Murashige and Skoog, 1962; for details, see “Materials and Methods”). Shoot and root RNA samples were hybridized to the Arabidopsis ATH1 and Rice Genome Arrays from Affymetrix to evaluate changes in global gene expression (see “Materials and Methods”) in response to N treatments. The normalized microarray data for each species have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38102.
The Effect of N Treatment on Genome-Wide Expression in Rice
Our first aim was to identify N-regulated genes and study their response in rice shoots and roots. Following robust microarray analysis normalization, two-way ANOVA with false discovery rate (FDR) correction, and filtering of transcriptome data using a 1.5-fold cutoff (described in “Materials and Methods”; Fig. 1), we found a set of 451 genes in rice that were significantly regulated in rice by N treatment (Table I). In rice shoots, 103 genes were N induced, and 39 genes were repressed in response to N treatment. In rice roots, 234 genes were N induced, while 106 genes were repressed in N-treated samples, compared with control treatments (Table I; for a complete list of regulated genes, see Supplemental Table S1; for organ-specific gene responses, see Supplemental Fig. S1). Rice roots appear to have a much larger response in terms of the number of genes, which has also been observed previously in Arabidopsis (Wang et al., 2003). Additionally, these results from the rice microarray data were confirmed by reverse transcription-quantitative PCR (RT-qPCR) for a number of selected genes (Supplemental Fig. S2).
Figure 1.
Schematic diagram of the experimental and data-mining approach used in this study. Briefly, rice and Arabidopsis plants were grown for 12 d before a 2-h treatment with N at a final concentration of 20 mm KNO3 and 20 mm NH4NO3 versus KCl control (see “Materials and Methods”). Genome-wide analysis using Affymetrix chips was used to quantify mRNA levels. Modeling of microarray data, using ANOVA, homology/orthology, and network analysis (detailed in “Materials and Methods”), was used to identify a core translational N-regulatory network shared between rice and Arabidopsis.
Table I. Number of N-regulated genes in rice and Arabidopsis.
Percentages of regulated genes for each type of regulation are in parentheses.
| Genes | Roots |
Shoots |
||
|---|---|---|---|---|
| Induced | Repressed | Induced | Repressed | |
| Rice, 451 genes | 234 (51.8%) | 106 (23.5%) | 103 (22.8%) | 39 (8.6%) |
| Arabidopsis, 1,417 genes | 757 (53.4%) | 424 (29.9%) | 166 (11.7%) | 184 (12.9%) |
The 451 N-regulated rice genes included genes involved in nitrate uptake and metabolism, sugar biosynthesis, and ammonium assimilation, among others (Table II). Specifically, some of the genes in these groups are involved in producing reductants for nitrite uptake and also include enzymes of the pentose phosphate pathway, which generates the NADPH necessary for N assimilation (Table II). We also observed N induction of a gene that encodes the pentose phosphate enzymes in both tissues: Glucose-6-Phosphate Dehydrogenase (G6PDH) (LOC_Os07g22350). Such genes involved in C metabolism are related to the production of energy for nitrate or nitrite reduction. These types of genes have also been identified previously as N responsive in Arabidopsis (Wang et al., 2003).
Table II. Selected rice genes regulated by N in shoots and roots (for details, see “Materials and Methods”).
For the full list of genes, see Supplemental Table S1. The fold change of N-response genes was calculated as the ratio between N and KCl expression values. The P value cutoff is 0.05 or less, and the fold change is 1.5-fold or greater (shown are log2 values; the fold change cutoff log2 for 1.5 = 0.585). NC, No change.
| Gene Identifier | Gene Description | Log2 Ratio |
|
|---|---|---|---|
| Root | Shoot | ||
| Nitrate uptake | |||
| LOC_Os02g02170 | High-affinity nitrate transporter, putative, expressed | −0.93 | NC |
| LOC_Os02g38230 | Component of high-affinity nitrate transporter, putative, expressed | 1.66 | NC |
| Nitrate/nitrite assimilation | |||
| LOC_Os02g53130 | Nitrate reductase, putative, expressed | 2.15 | 3.32 |
| LOC_Os01g25484 | Ferredoxin-nitrite reductase, chloroplast precursor, putative, expressed | 2.37 | 2.67 |
| Ferredoxin reduction | |||
| LOC_Os01g64120 | Ferredoxin6, chloroplast precursor, putative, expressed | 3.28 | 4.22 |
| LOC_Os05g37140 | Ferredoxin6 | 1.33 | 0.89 |
| LOC_Os03g57120 | Ferredoxin-NADP reductase, root isozyme, chloroplast precursor, putative, expressed | 1.55 | 1.95 |
| LOC_Os04g44650 | Ferredoxin-thioredoxin reductase | 0.67 | 0.95 |
| Pentose phosphate pathway | |||
| LOC_Os08g08840 | Glc-6-P/phosphate translocator2, chloroplast precursor, putative, expressed | 0.84 | NC |
| LOC_Os07g22350 | Glc-6-P 1-dehydrogenase2, chloroplast precursor, putative, expressed | 1.62 | 1.07 |
| Ammonium assimilation | |||
| LOC_Os01g48960 | Glu synthase, chloroplast precursor, putative, expressed | 1.17 | NC |
| LOC_Os04g56400 | Gln synthetase, chloroplast precursor, putative, expressed | 1.09 | 0.71 |
Finally, rice genes involved in ammonium assimilation were found to respond to N treatments in our study (Table II). NADH-GOGAT (LOC_Os01g48960) was N induced in rice roots, while Gln synthetase (LOC_Os04g56400) was found to be N regulated (1.09- and 0.71-fold change, respectively) in both roots and shoots (Table II). The complete list of N-regulated genes in rice is shown in Supplemental Table S1.
Genome-Wide Effects of N Treatment in Arabidopsis
Arabidopsis seedlings were N treated as described above for rice (for details, see “Materials and Methods”; Fig. 1), and following RNA extraction, gene responses to N treatments were analyzed using microarrays. Following normalization, two-way ANOVA, FDR correction, and filtering for 1.5-fold change, 1,417 Arabidopsis genes were identified to be N responsive compared with the control treatment. In Arabidopsis shoots, 166 genes were N induced and 184 genes were repressed in response to N treatments. In Arabidopsis roots, 757 genes were N induced and 424 genes were repressed (Table I; for a complete list of regulated genes, see Supplemental Table S2). The N-regulated genes in Arabidopsis included genes involved in nitrate uptake and metabolism, genes in the pentose phosphate pathway, and ammonium assimilation, among others (Table III).
Table III. Selected Arabidopsis genes regulated by N in shoot and/or roots (for details, see “Materials and Methods”).
The fold change of N-response genes was calculated as the ratio between the N and KCl expression values. The P value cutoff is 0.05 or less, and the fold change is 1.5-fold or greater (shown are log2 values; the fold change cutoff log2 for 1.5 = 0.585). NC, No change.
| Gene Identifier | Gene Description | Log2 Ratio |
|
|---|---|---|---|
| Roots | Shoots | ||
| Nitrate uptake | |||
| At1g69850 | Nitrate transporter1:2; calcium ion binding/transporter (NRT1:2) | −0.62 | −1.04 |
| At5g50200 | Nitrate transmembrane transporters (NRT3:1) | 1.79 | 2.04 |
| At1g08090 | High-affinity nitrate transporter2.1 (NRT2:1) | 2.76 | 2.35 |
| Nitrate/nitrite assimilation | |||
| At1g37130 | Nitrate reductase2 (NIA2) | NC | 2.65 |
| At1g77760 | Nitrate reductase1 (NIA1) | 3.69 | 5.85 |
| At2gl5620 | Nitrite reductase; ferredoxin-nitrate reductase (NIR1) | 3.33 | 6.33 |
| Ferredoxin reduction | |||
| At2g27510 | Ferredoxin3; electron carrier (ATFD3) | 1.52 | 3.16 |
| At4g05390 | Root FNR1; oxidoreductase (ATRFNR1) | 2.49 | 3.99 |
| At1g30510 | Root FNR2; oxidoreductase (ATRFNR) | 2.75 | 4.43 |
| Pentose phosphate pathway | |||
| At1g24280 | Glc-6-P dehydrogenase3 (G6PD3) | 3.34 | 4.78 |
| At5g13110 | Glc-6-P dehydrogenase2 (G6PD2) | 1.93 | 2.99 |
| Ammonium assimilation | |||
| At5g35630 | Gln synthetase2 (GS-GLN2) | 1.14 | NC |
| At5g16570 | Gln synthetase1;4 (GLN1;4) | −1.20 | NC |
| At5g53460 | NADH-dependent Glu synthase1 (GLT1) | 1.38 | 2.40 |
| Glu biosynthesis/degradation | |||
| At1g51720 | Glu dehydrogenase, putative | 1.14 | NC |
| At5g07440 | Glu dehydrogenase2 (GDH2) | 1.76 | NC |
As observed for rice, the majority of N-regulated genes in Arabidopsis are root specific (also found previously [Wang et al., 2004]). For example, 75% of genes were uniquely N regulated in Arabidopsis roots versus shoots, while only 16% of N-regulated genes were expressed exclusively in shoots (Supplemental Fig. S3). Several known Arabidopsis N-induced genes were also responsive to our treatments with ammonium nitrate, including NIA1, NIA2, NIR, NRT2:1, NRT1:2, NRT3:1, FERREDOXIN3, G6PD2, G6PD3, GLT1, ASN2, and GDH2, among others (Table III; for a complete list, see Supplemental Table S2; Wang et al., 2003; Krouk et al., 2010). Additionally, our microarray data were confirmed by RT-qPCR results in a number of selected Arabidopsis genes (Supplemental Fig. S4).
To determine whether the overlap between the rice and Arabidopsis N-responsive genes was significant, a permutation test was performed. A total of 1,417 genes were selected randomly from Arabidopsis genes present on the Affymetrix chip, and 451 rice genes were selected randomly from genes present on the rice Affymetrix chip. Using BLASTP homology, the overlap was measured in terms of rice and Arabidopsis genes. This was done 10,000 times, and then the number of times the overlap was greater than or equal to the observed was counted. The overlap obtained from random sampling was never greater than or equal to the observed, making P < 0.0001. These results suggest that, despite the difference in the number of responsive genes, rice and Arabidopsis respond very similarly to the N treatments provided.
Network Analysis Identifies Conserved Genes Involved in N Signaling in Rice
It is known that the expression of many TFs is regulated by NO3–. However, to date, only a few such NO3–-regulated TFs have been shown to be involved in NO3– signaling in Arabidopsis (Alvarez et al., 2014; Medici et al., 2015; for review, see Castaings et al., 2011).
Creation of a Rice-Arabidopsis N-Regulatory Network
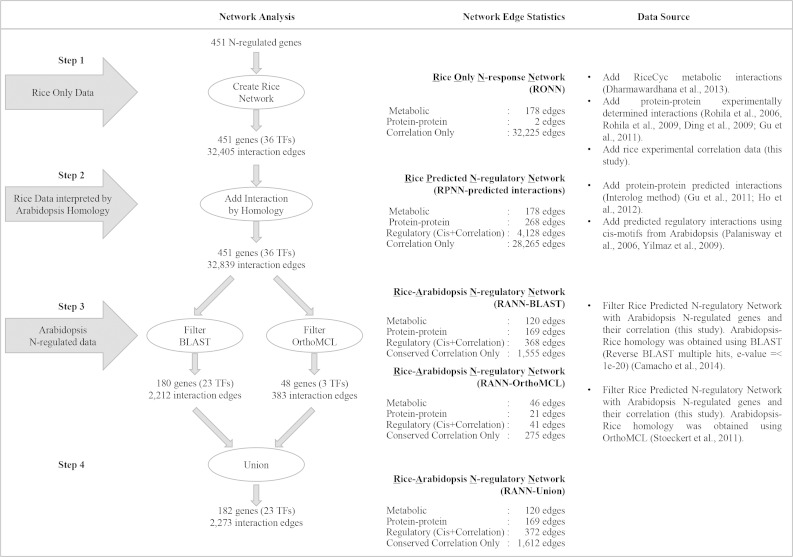
To identify novel TFs that may play a global role in an N-regulatory network, we performed network analysis that exploited our microarray data sets from Arabidopsis and rice (Fig. 2). We generated a network using the limited knowledge of known rice interactions and then, to enrich the existing network in rice, we introduced predicted interaction data based on homology to the large amount of Arabidopsis network knowledge. For this purpose, we started our network analysis by creating a rice-only nitrogen-response network (RONN; Fig. 2, step 1). In step 1, we used the rice experimental data generated in our study by looking at significant correlations among N-regulated rice genes (Pearson correlation coefficient with a P cutoff of 0.05), metabolic pathways from RiceCyc (Dharmawardhana et al., 2013), and experimentally determined protein-protein interactions in rice (Rohila et al., 2006, 2009; Ding et al., 2009; Gu et al., 2011) for this network creation (for details, see “Materials and Methods”). This rice-only analysis resulted in a network of 451 N-regulated genes, with 36 TFs and 32,405 interactions among them (Fig. 2, RONN).
Figure 2.
Work flow of the network analysis of N-regulated genes differentially expressed in rice resulting in RANN-Union. The input was 451 rice N-regulated genes. In each of the three steps, we introduced rice and Arabidopsis data in order to identify RANN-Union, which includes N-regulated genes and network modules conserved between rice and Arabidopsis (for details, see “Materials and Methods”).
Next, in step 2 (Fig. 2), predicted protein-protein interactions in rice and cis-binding site information from Arabidopsis were added to the RONN. This generated a new predictive network: the rice predicted N-regulatory network (RPNN; for details, see “Materials and Methods”). The RPNN included rice predicted regulatory interactions obtained from cis-binding site data in Arabidopsis and TF family information in rice from PlantTFDB (Jin et al., 2014; for details, see “Materials and Methods”). In the RPNN, predicted regulatory edges are defined by the presence of a cis-binding site and a significant correlation between a TF and a target. In this analysis, 3,960 of the 32,225 correlation edges also contain cis-binding information, thus recategorizing them as regulatory edges. In the case where the target of one TF (e.g. TF1) is another TF (e.g. TF2), there is a possibility that TF1 is a target of TF2 (and vice versa), in which case one correlation edge between two TFs is converted to two regulatory edges. There are 168 such TF1-TF2 correlation edges, thus increasing the number of regulatory edges from 3,960 to 4,128 (Fig. 2, RPNN). The RPNN-predicted interactions network had the same number of genes as the RONN; however, the addition of predicted protein-protein interactions along with regulatory data increases the total number of interactions to 32,839 in the RPNN-predicted interactions network (Fig. 2).
Next, we were interested in further filtering the RPNN to identify the N-regulatory genes and network modules whose regulation is conserved across two species, Arabidopsis and rice. To this end, in step 3 (Fig. 2), we introduced the Arabidopsis experimental data of N-responsive genes generated in our study into the RPNN-predicted interactions network. We approached this research question using two different orthology methods (BLASTP and OrthoMCL) to obtain two different rice-Arabidopsis N-regulated networks (RANNs; RANN-BLAST and RANN-OrthoMCL, respectively). Both RANN-BLAST and RANN-OrthoMCL contain only rice genes where the rice gene and its putative ortholog in Arabidopsis is N regulated in our experimental conditions. Additionally, the correlation and regulatory edges between these conserved N-regulatory genes also had to be conserved (Fig. 2; for details, see “Materials and Methods”).
RANN-BLAST comprised 180 rice N-regulated genes, of which 23 are TFs. By contrast, RANN-OrthoMCL had only 48 rice N-regulated genes, of which three genes are TFs. It is not surprising that RANN-OrthoMCL is smaller than RANN-BLAST, since OrthoMCL differentiates between orthologs and paralogs. It is important to note that out of 48 genes from RANN-OrthoMCL, only two additional genes were present uniquely in RANN-OrthoMCL and not in RANN-BLAST. These genes comprise a glycoprotein, LOC_Os10g41250, and a protein of unknown function, LOC_Os05g46340. As discussed below, we identified validated gene interactions using RANN-BLAST that would have been missed had we only used RANN-OrthoMCL. Therefore, a union of the two conserved cross-species networks, RANN-BLAST and RANN-OrthoMCL, was performed to generate the rice-Arabidopsis N-regulatory network (RANN-Union), which contains 182 rice N-regulated genes, of which 23 genes are TFs (Fig. 2, step 4).
Of the 182 genes in RANN-Union (Fig. 2, step 4), some of the genes are known to be directly involved in N assimilation, such as nitrate transporters, nitrate and nitrite reductase, Gln synthetase, and Glu synthase, among others (for the complete list of regulated genes, see Supplemental Table S3). RANN-Union also contains ferredoxin reductase genes (LOC_Os03g57120, LOC_Os05g37140, and LOC_Os01g64120) whose encoded proteins are indirectly involved in nitrite reduction by providing reducing power, as shown in Arabidopsis (Wang et al., 2000). Additionally, LOC_Os03g57120 is orthologous to ATRFNR1 in Arabidopsis (At4g05390; based on BLASTP and OrthoMCL), which has also been shown previously to be involved in supplying reduced ferredoxin for nitrate assimilation (Hanke et al., 2005). In addition, two calcineurin B-like-interacting protein kinases (CIPKs) are present in the group of 182 N-regulated genes in RANN-Union. LOC_Os03g03510 has Arabidopsis CIPK23 as its ortholog (based on OrthoMCL and BLASTP), while LOC_Os03g22050 is homolog to Arabidopsis CIPK23 based only on BLASTP (but not OrthoMCL). Interestingly, CIPK23 has been identified as an NO3−-inducible protein kinase (Castaings et al., 2011). Additionally, both rice CIPK loci (LOC_Os03g22050 and LOC_Os03g03510) are homologous to Sucrose Nonfermenting1 (SNF1)-related protein kinase (KIN11) and to Mitogen-activated protein kinases/Extracellular signal-regulated kinases kinase kinase (MEKK1) based on BLASTP but not OrthoMCL. KIN11 is an Snf1-related kinase proposed to be part of an energy-sensing mechanism in Arabidopsis (Baena-González et al., 2007) and also found to be related to N assimilation (Gutiérrez et al., 2008). Also, MEKK1 is involved in Glu signaling in root tips of Arabidopsis (Forde, 2014). Moreover, LATERAL ORGAN BOUNDARY DOMAIN39 (LOC_Os03g41330), a TF present in RANN-Union, was found to be regulated at the transcriptional level by NO3− and involved in N signaling in Arabidopsis (Rubin et al., 2009).
To study how TF connectivity changed throughout the network analysis, and to identify putative regulators that control the expression of conserved network modules, the TFs N regulated in these networks were ranked based on their hubbiness, the number of regulatory connections (Table IV). As mentioned previously, the number of connections found for TFs in the RPNN-predicted interactions (Fig. 2, step 2) decreased when the network was filtered with Arabidopsis N-regulatory genes and their correlations (Fig. 2, step 3). The TF with the highest number of connections in RANN-Union is LOC_Os03g55590 (Table IV), a gene that belongs to the G2-like transcription factor family and subgroup HHO (for HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING1 [HRS1] homolog). The HHO family has another member conserved in RANN-Union, LOC_Os07g02800. A naive assumption of the network analysis is that the TF with the most connections has the most influential regulatory role. In previous studies, we have used the ranking of TF hubbiness to identify candidates for follow-up mutational studies in which they were validated (Gutiérrez et al., 2008). To test the influence of orthology data, we investigated whether the rank of TFs based on hubbiness changed from the RPNN-predicted interactions network to the final RANN-Union using the Wilcoxon test. A P value of 1.423e-08 indicates that the connectivity rank presented in Table IV changed significantly through the network generation steps shown in Figure 2.
Table IV. TFs in RANN-Union.
For each step in network construction (Fig. 2), TFs were rank based on their number of connections in the network.
| Rice Gene Locus | Rice Gene Description | No. of Directed Connections |
|||
|---|---|---|---|---|---|
| RPNN-Predicted Interactions | RANN-BLAST | RANN-OrthoMCL | RANN-Union | ||
| LOC_Os03g55590 | DNA-binding protein, putative, expressed | 180 | 45 | 17 | 46 |
| LOC_Os01g53260 | OsWRKY23: superfamily of rice TFs having WRKY and zinc finger domains, expressed | 162 | 41 | 41 | |
| LOC_Os01g64000 | Abscisic acid response element-binding factor, putative, expressed | 138 | 35 | 35 | |
| LOC_Os01g06640 | DNA-binding protein, putative, expressed | 191 | 31 | 31 | |
| LOC_Os07g02800 | Myb-like DNA-binding domain, SHAQKYF class family protein, expressed | 150 | 30 | 14 | 30 |
| LOC_Os01g43650 | OsWRKY11: superfamily of rice TFs having WRKY and zinc finger domains, expressed | 166 | 26 | 10 | 29 |
| LOC_Os11g47870 | Chitin-inducible gibberellin-responsive protein2, putative, expressed | 163 | 27 | 27 | |
| LOC_Os09g35030 | sbCBF6, putative, expressed | 229 | 16 | 16 | |
| LOC_Os09g25070 | OsWRKY62: superfamily of rice TFs having WRKY and zinc finger domains, expressed | 170 | 14 | 14 | |
| LOC_Os01g34060 | DNA-binding protein, putative, expressed | 99 | 12 | 12 | |
| LOC_Os04g42950 | DNA-binding protein, putative, expressed | 96 | 12 | 12 | |
| LOC_Os09g32260 | Arabidopsis NAC079 (ANAC079)/ANAC080, putative, expressed | 129 | 12 | 12 | |
| LOC_Os11g08210 | NAC domain-containing protein71, putative, expressed | 99 | 11 | 11 | |
| LOC_Os02g15340 | NAC domain-containing protein76, putative, expressed | 67 | 10 | 10 | |
| LOC_Os04g55970 | DNA-binding protein, putative, expressed | 143 | 10 | 10 | |
| LOC_Os12g10660 | Salt tolerance-like protein, putative, expressed | 50 | 10 | 10 | |
| LOC_Os03g04310 | DNA-binding protein, putative, expressed | 84 | 7 | 7 | |
| LOC_Os10g42130 | ANAC071, putative, expressed | 78 | 6 | 6 | |
| LOC_Os01g64020 | TF Histone Gene Binding Protein-1b, putative, expressed | 219 | 5 | 5 | |
| LOC_Os08g42550 | AP2 domain-containing protein, expressed | 61 | 5 | 5 | |
| LOC_Os06g41100 | TGA10 TF, putative, expressed | 119 | 2 | 2 | |
| LOC_Os05g37170 | TF TGA6, putative, expressed | 90 | 1 | 1 | |
| LOC_Os02g06910 | Auxin response factor6, putative, expressed | 163 | |||
| LOC_Os03g21710 | WRKY DNA-binding domain-containing protein, expressed | 79 | |||
| LOC_Os03g47730 | Knotted1-interacting protein, putative, expressed | 61 | |||
| LOC_Os03g55220 | Helix-loop-helix DNA-binding protein, putative, expressed | 175 | |||
| LOC_Os04g56990 | Transcription factor, putative, expressed | 293 | |||
| LOC_Os05g20930 | Transcriptional regulator RABBIT EARS, putative, expressed | 195 | |||
| LOC_Os05g38140 | Basic helix-loop-helix (bHLH) TF, putative, expressed | 79 | |||
| LOC_Os06g07030 | Dehydration-responsive element-binding protein, putative, expressed | 101 | |||
| LOC_Os09g26420 | Ethylene response factor, putative, expressed | 69 | |||
| LOC_Os09g36160 | SHORT INTERNODES, putative, expressed | 30 | |||
Creation of an Arabidopsis-Rice N-Regulatory Network
Considering that there is more information available in Arabidopsis than in rice, we performed a similar network analysis as in Figure 2, but now using Arabidopsis N-regulated data as the starting point (Supplemental Fig. S5). We filtered the Arabidopsis network with rice experimental data generated in our study using BLASTP and OrthoMCL (Supplemental Fig. S5). The resulting Arabidopsis-rice N-regulatory network (ARNN-Union) has 276 genes. By definition, the identities of the genes from ARNN-Union (276 genes; Supplemental Fig. S5) are equal to those of RANN-Union (182 genes; Fig. 2). The number of genes is different, however, because in most cases rice genes have more than one N-regulated ortholog in Arabidopsis. Following this rationale, ARNN-Union contains 34 TFs (Supplemental Fig. S5), while RANN-Union contains only 23 TFs (Fig. 2; for a list of ARNN-Union TFs, see Supplemental Table S4). We also studied how TF connectivity changed throughout the steps of the network analysis in Supplemental Figure S5 by ranking TFs based on the number of regulatory connections (Supplemental Table S4). In the top five highly ranked TFs of ARNN-Union (Supplemental Table S4), we found three members of the HRS1/HHO family and TGA1, which were each validated to be involved in the N response in Arabidopsis (Alvarez et al., 2014; Medici et al., 2015), in addition to WRKY28, a novel finding of our study. We also investigated if the rank of TFs based on connectivity changed from the Arabidopsis-only N-regulatory network to the final ARNN-Union, again using a Wilcoxon test. P = 1.391e-10 denotes that the connectivity rank of TFs (e.g. numbers of connections) in Supplemental Table S4 changed significantly through the network generation process used in Supplemental Figure S5.
Supernode Analysis of RANN-Union
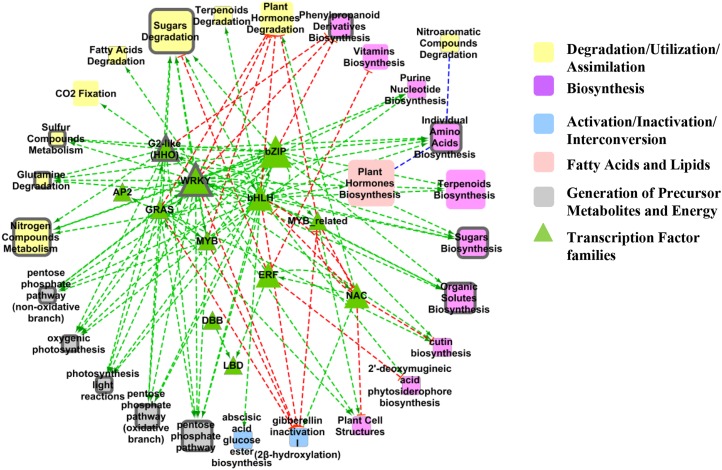
The supernode analysis groups genes with the same biological processes, functional terms, and annotations into a single node whose size is proportional to the number of genes in the supernode. To gain an understanding of how the conserved genes were connected to each other when categorized with plant metabolic network pathway information, a supernode network analysis was performed using TF families (PlantTFDB; Jin et al., 2014) and OryzaCyc pathway associations (OryzaCyc version 1.0; Dharmawardhana et al., 2013) for the 182 genes in RANN-Union (Fig. 2). The resulting supernode network of RANN-Union identified several well-represented TF families highly connected to major metabolic pathways (Fig. 3). The supernode network analysis also revealed that the TF families with the highest number of members in this network are basic-leucine zipper (bZIP) and WRKY.
Figure 3.
Supernode network analysis created from the 182 genes of RANN-Union. Individual nodes were clustered based on PlantCyc pathways and TF family classification to form supernodes. Genes that do not belong to either of the two classifications are not shown here. Triangles represent TF families, and squares represent PlantCyc pathways (Plant Metabolic Network, 2013). The size of the nodes is proportional to the number of genes within that particular category (from 1 to 5). Nodes are connected by TF-target (red dashed lines = predicted negative correlation; green dashed lines = predicted positive correlation) and predicted protein-protein (blue dashed lines) interactions. All nodes are present in RANN-BLAST. Nodes circled in thick gray lines are also present in RANN-OrthoMCL.
RANN-Union top TF hubs include four members of the bZIP TF family in rice (LOC_Os05g37170, LOC_Os01g64020, LOC_Os06g41100, and LOC_Os01g64000). Homologs of these family members have been validated to be involved in N responses in Arabidopsis (Gutiérrez et al., 2008; Hanson et al., 2008; Jonassen et al., 2009; Obertello et al., 2010; Para et al., 2014; Fig. 3; Table IV). Three members of the bZIP TF family belong to the subfamily TGA, which was recently indicated to be involved in N regulation (see below; Alvarez et al., 2014). The supernode network analysis also shows that the TF families bZIP, bHLH, WRKY, and G2-like (HHO) are involved in the N regulation of genes related to N compound metabolism, which contains genes involved in the N assimilation pathway.
OsWRKY23, the second in the rank of most connected TFs in RANN-Union (Table IV), is homologous to Arabidopsis WRKY75 (At5g13080) based on BLASTP only and has been shown to be related to phosphate acquisition (Devaiah et al., 2007). Also, OsWRKY23 is orthologous to Arabidopsis WRKY28 (At4g18170) based on BLASTP and OrthoMCL, which has been shown to be involved in the activation of salicylic acid biosynthesis (van Verk et al., 2011).
Two Predicted TF Families Conserved in RANN-Union Are Biologically Validated
Among the list of 23 TFs present in RANN-Union, we found two TF families whose roles in N signaling have been experimentally validated. We first investigated the HHO/HRS1 family. This TF family has two N-regulated members in rice and four homologs in Arabidopsis (Supplemental Fig. S6). To gain insight into the HHO/HRS family and their conserved N regulation, we performed a phylogenetic analysis and found that the common N-responsive members of the HHO family from rice and Arabidopsis fall in the same clade (Supplemental Fig. S6). The phylogenetic tree was built by ClustalW alignment and the maximum likelihood method. This group of HHO family members present in the same clade is also orthologous to each other using either OrthoMCL or BLASTP (Supplemental Fig. S6). This result is an in silico validation of our cross-species network approach. Also, it was recently validated that two members of this TF family, HRS1 and HHO1, have important roles in integrating nitrate signaling in the Arabidopsis root (Medici et al., 2015).
Based on supernode analysis, the bZIP family has 20 connections to biological processes, making it the third most highly connected TF family in RANN-Union. The three N-regulated rice TGA family members (LOC_Os01g64020, LOC_Os05g37170, and LOC_Os06g41100) are putative homologs to the four N-regulated Arabidopsis TGA family members: At1g22070 (TGA3), At1g77920 (TGA7), At5g10030 (TGA4), and At5g65210 (TGA1; Supplemental Fig. S7). Based on our supernode network analysis, discussed above, these TFs have connections with biosynthesis and degradation/utilization and assimilation metabolic pathway processes (Fig. 3). We performed a phylogenetic tree analysis using all TGA family members in Arabidopsis and rice identified by BLASTP. The phylogenetic tree (Supplemental Fig. S7) shows that the rice and Arabidopsis N-regulated members of the TGA family are paralogs, as confirmed by OrthoMCL. As shown in Supplemental Figure S7, all N-regulated TGA family members in each species were identified by homology based on BLASTP. However, it is important to point out that two of the members of the TGA TF family identified in RANN-BLAST (TGA1 and TGA4) were recently validated as important regulatory components of the nitrate response in Arabidopsis (Alvarez et al., 2014). We also observed a significant overlap (P = 0.008) between the validated targets identified in planta in the tga1/4 double mutant (data available from Alvarez el al., 2014) and the predicted targets from RANN-Union analysis (analysis done using the Genesect tool in VirtualPlant; www.virtualplant.org). These TGA1/TGA4 targets identified in our analysis and validated in planta include two proteins that have been shown to be involved in N signaling. These TGA1 targets include HRS1, a TF involved in N signaling as mentioned earlier (Medici et al., 2015), and CIPK3, one of several kinases identified to have a role in N signaling (Hu et al., 2009). The last gene present in this intersect set of validated HRS1 targets in RANN is a proteasome subunit, a potential gene hypothesized to be involved in N regulation (Regulatory Particle 5b), a potential new hypothesis for N signaling via the proteasome that our analysis has uncovered. Thus, the conservation of function across rice and Arabidopsis indicated the role of the TGA family in the N response. It is noteworthy that this prediction, which is also supported by recent experimental data (Alvarez et al., 2014), would have been missed if we relied only on orthology based on OrthoMCL. Importantly, our cross-species network analysis has also opened new hypotheses for testing about N-regulatory mechanisms in plants.
DISCUSSION
This study provides a novel analysis of N-regulated gene networks conserved across two highly divergent species: rice (a monocot) and Arabidopsis (a dicot). Despite their large phylogenetic distance, our analysis revealed a set of N-regulated genes, TFs, and network modules conserved in rice and Arabidopsis exposed to the same N-treatment conditions. Our analysis shows a statistically significant overlap, indicating that rice and Arabidopsis respond very similarly to the N treatments. The list of genes regulated by N treatments in rice includes many of the known nitrate/ammonium-regulated genes previously identified in Arabidopsis, including genes known to respond to nitrate (Nitrate Reductase, Nitrite Reductase, Ferredoxin, Ferredoxin-Dependent NADPH Reductase, and G6PDH). These results are not surprising in hindsight, given that the former are important to reduce the plant’s risk of nitrite toxicity. Selected genes from the N-responsive lists were corroborated by RT-qPCR analysis. One of the important aspects of this genomic analysis is that the N treatments performed on rice and Arabidopsis were comparable, so that the gene responses could be directly compared. Genome profiling revealed that 1.32% of the rice genome is regulated in response to N treatment while 6.76% of the Arabidopsis genome responds to N treatment, and in both cases, roots were more sensitive to N than shoots. The result of the permutation test, which was performed to determine whether the overlap between the rice and Arabidopsis N-responsive genes was significant, suggests that, despite the difference in number of N-responsive genes, rice and Arabidopsis respond very similarly to N treatment.
The rice genome size is more than three times that of Arabidopsis and is estimated to have significantly more genes (Yu et al., 2005). According to that estimate, we would have expected more N-regulated genes in rice; however, the difference in the total number of N-regulated genes between the species might be mainly due to the fact that the N treatment used here affects these two plants differently. In support of that notion, it has long been known that rice can form natural associations with endophytic diazotrophs, which are responsible for supplying the plants with fixed N, increasing plant height, root length, and dry matter production. In rice and maize (Zea mays), associative N fixation can supply 20% to 25% of total N requirements (Santi et al., 2013). The experiments performed here were done in a sterile environment, so the difference in the number of N-regulated genes might be due to the fact that the N-response pathway in rice needs the bacterial association to be completely active.
The N-signaling network has gained new levels of complexity in recent years and is as yet far from being completely understood (Vidal et al., 2010; Castaings et al., 2011; Bargmann et al., 2013; Medici et al., 2015). In addition, it is an open question how well gene networks derived from model dicots, such as Arabidopsis, might faithfully reconstruct pathways in a monocot, such as rice.
Our hypothesis was that the conserved network nodes (genes) and edges (interactions) among species would provide an initial framework to understand the complex functional genomic and genetic knowledge of N-regulatory networks. To address this, we generated a gene expression network based on coexpression, homologs based on BLASTP, and orthologs based on OrthoMCL to reveal conserved coexpression relationships between rice and Arabidopsis. Our results suggest that using BLASTP homology produced a more complete core N-regulatory network between rice and Arabidopsis compared with OrthoMCL alone. When we use OrthoMCL to distinguish between orthologs and paralogs, we lose promising candidates from the network. For example, if we only used OrthoMCL to obtain orthology information, we would have missed the TGA family members and their interaction to regulate N-responsive biological processes. From the phylogenetic analysis, it is clear that TGA family members evolved in their function so much that different members of the family have taken on the responsibility to be N responsive in each species. Since it is well accepted that different members of the TF family bind to the same binding site, this hypothesis is quite reasonable. As described in “Results,” our predicted TGA1 and TGA4 target genes from RANN-Union overlap significantly with published and biologically validated in planta data in Arabidopsis (Alvarez et al., 2014).
In this cross-species network approach, we used known rice annotation and experimental data to generate RONN (Fig. 2, step 1), to which we added known Arabidopsis annotation data (Fig. 2, step 2), and subsequently filtered it with the Arabidopsis N-treatment experimental data generated in this study (Fig. 2, step 3). This analysis identified a core N-regulatory network conserved between rice and Arabidopsis (RANN-Union). This cross-species network analysis enabled us to identify conserved N-regulated genes, network modules, TFs, and biological processes related to this essential nutrient. The list of potential N-responsive genes in rice was considerably reduced when we integrated our experimental data from Arabidopsis (Fig. 2, step 3). In addition, the supernode network analysis allowed us to visualize how N-responsive biological processes, such as N compound metabolism and sugar biosynthesis, are related to each other and which TF families regulate them. The presence of metabolic pathways related to sugar metabolism and amino acid biosynthesis is important in this context, since the production of reduced carbon is necessary to produce both the energy and carbon skeletons required for the incorporation of inorganic N into amino acids.
By starting with the experimental data from the model plant Arabidopsis and subsequently filtering it with the rice experimental data generated in this study, we uncovered a subset of conserved TFs potentially involved in N regulation. However, compared with the N-regulated network information already known in Arabidopsis, we conclude that, while we did not significantly improve our knowledge of Arabidopsis interactions by integrating rice data, we did identify a smaller evolutionarily conserved network. On the other hand, when we started with rice experimental data and then added predicted network knowledge inferred from Arabidopsis, subsequently introducing Arabidopsis experimental data, we significantly improved our network connections and identified TF-target connections that have been experimentally validated in Arabidopsis. To summarize, using Arabidopsis network knowledge, including gene interactions and experimental data, highly refined our rice networks, enabling us to identify potential master TFs involved in the N response, some of which have been biologically validated in Arabidopsis by independent experiments (e.g. members of the TGA and HHO TF members).
In plants, transcriptional regulation is mediated by a large number of TFs controlling the expression of tens or hundreds of target genes in various signal transduction cascades. Interestingly, a recent transcriptome data analysis supports our predictions for the TFs controlling the core N-regulatory network uncovered in our analysis. Specifically, Canales et al. (2014) integrated publicly available root microarray data under contrasting nitrate conditions and concluded that the most represented TFs families are AP2/ERF, MYB, bZIP, and bHLH. In our study, the TFs regulated by N treatment were ordered by their network connectivity, under the premise that highly connected genes are more likely to be involved in biological processes. These TF families are also present in our supernode analysis based on RANN-Union. Additionally, our supernode analysis also revealed the G2-like (HHO) family in rice, based on orthology to Arabidopsis, as one of the most highly connected TF families. In addition, there is recent experimental validation of several members of the HHO family being involved in the N response in Arabidopsis (Medici et al., 2015). Another highly connected TF family obtained from the supernode analysis was the TGA family, three members of which were N regulated and conserved in RANN-BLAST but not in RANN-OrthoMCL. With these results, we conclude that it is important to consider homologs based on BLASTP for the retrieval of conserved network modules. We further validated RANN-Union by determining that our predicted targets of TGA1/4 significantly overlap (P = 0.008) with validated targets identified in planta in the tga1/4 double mutant (Alvarez et al., 2014). Thus, this novel finding of TFs implicated in the N regulation of genes and network modules, conserved in both rice and Arabidopsis according to our predicted network, is strongly supported by the experimental study of tga1 and tga4 mutants (Alvarez et al., 2014).
Finally, our study addresses a major challenge of translational research, which is to transfer network knowledge from data-rich model species, such as Arabidopsis, to data-poor crop species, such as rice. The results presented here describe the transfer of network knowledge from Arabidopsis to crops (Fig. 2, steps 2 and 3) and how it can help develop effective and sustainable biotechnological solutions to enhance N acquisition by plants in natural or agricultural environments. Proper plant N nutrition in the environment will not only improve production but will also contribute to sustainable agricultural practices by diminishing the use of N fertilizers, thus reducing greenhouse gases, stratospheric ozone, acid rain, and nitrate pollution of surface and ground water.
MATERIALS AND METHODS
Plant Growth and Treatment Conditions
Rice seeds (Oryza sativa japonica) were kindly provided by Dale Bumpers of the National Rice Research Center. Seeds were surface sterilized in 70% (v/v) ethanol for 3 min, followed by commercial hydrogen peroxide for 30 min with gentle agitation, and washed with distilled water. Seeds were sown onto 1× MS basal salts (custom made; GIBCO) with 0.5 mm ammonium succinate, 3 mm Suc, and 0.8% (w/v) BactoAgar at pH 5.5 for 3 d in dark conditions at 27°C. Following germination, embryos with developed root systems and aerial tissues were dissected from the rest of the seed using a sterile blade and transferred to a hydroponic system (Phytatray II; Sigma-Aldrich) containing basal MS salts (custom made; GIBCO) with 0.5 mm ammonium succinate and 3 mm Suc at pH 5.5. Fresh medium was replaced every 3 d to maintain a steady nutritional state and optimal pH levels. After 12 d under long-day (16 h of light/8 h of dark) growth conditions, at a light intensity of 180 μE s–1 m–2 and 27°C, plants were transferred to fresh medium containing only custom basal MS salts for 24 h prior to treatment. On day 13, plants were transiently treated for 2 h at the start of their light cycle by adding N at a final concentration of 20 mm KNO3 and 20 mm NH4NO3. Control plants were treated with KCl at a final concentration of 20 mm. After treatment, roots and shoots were harvested separately using a blade, immediately submerged into liquid N, and stored at −80°C prior to RNA extraction.
Arabidopsis (Arabidopsis thaliana) seeds were placed in the dark for 2 d at 4°C to synchronize germination. Seeds were surface sterilized and then transferred to a hydroponic system (Phytatray I; Sigma-Aldrich) containing the same medium described for rice (pH 5.7). Growth conditions were the same as in rice, except that plants were under 50 μE s–1 m–2 light intensity at 22°C. N starvation and treatments were as described above.
RNA Isolation and RT-qPCR Analysis
RNA was isolated from roots and shoots with TRIzol reagent following the manufacturer’s protocols (Invitrogen Life Technologies). Standard manufacturer’s protocols were used to reverse transcribe total RNA (1–2 μg) to one-strand complementary DNA using ThermoScript reverse transcriptase (Invitrogen). Real-time quantitative PCR measurements were obtained for a set of selected genes using gene-specific primers (Supplemental Table S5) and LightCycler FastStart DNA Master SYBR Green (Roche Diagnostics). Expression levels of the tested genes were normalized to expression levels of the actin or clathrin gene as described (Obertello et al., 2010).
Microarray Experiments and Analysis
Complementary DNA synthesis, array hybridization, and normalization of the signal intensities were performed according to the instructions provided by Affymetrix. Affymetrix Arabidopsis ATH1 Genome Array and Rice Genome Array were used for the respective species. The Affymetrix microarray expression data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38102.
Gene expression values were transformed by taking the logarithm to base 2 (log2) of the ratio of N at a final concentration of 20 mm KNO3 and 20 mm NH4NO3 (experimental state) over KCl treatment (control state) to yield the magnitude of the deviations in up- and down-regulated genes symmetrically (log2 value of the ratio of 1-fold is 0). Data normalization was performed using the robust microarray analysis method in the Bioconductor package in the R statistical environment.
Two-way ANOVA was performed using a custom-made function in R to identify probes that were differentially expressed following N treatment. The P values for the model were then corrected for multiple hypotheses testing using FDR correction at 5% (Benjamini and Hochberg, 1995). The probes passing the cutoff (P ≤ 0.05) for the model, and N treatment or interaction of N treatment and tissue, were deemed significant. Tukey’s honestly significant difference posthoc analysis was performed on significant probes to determine the tissue specificity of N regulation at P ≤ 0.05 and 1.5-fold or greater change (log2 of 1.5 is 0.585). Probes mapping to more than one gene were disregarded. Finally, for the cases of multiple probe sets representing the same gene, the assumption was that the expression levels should be up-regulated or down-regulated in all the probes representing the gene. Expression levels were combined for those that passed this criterion. A set of 451 N-regulated genes differentially expressed in rice and 1,417 N-regulated genes differentially expressed in Arabidopsis was obtained.
For both species, Pearson correlation coefficient was calculated for probes that passed the two-way ANOVA and FDR correction. Specifically, the Pearson correlation coefficient was computed between different pairs of probe sets using the mean value of their expression data across the replicates using a custom script in R. Correlation was calculated separately for root genes and shoot genes in both species, and the corresponding correlation edges were labeled accordingly.
Orthology Analysis
Sequence and annotation data for the rice genome were downloaded from The Institute for Genomic Research Rice Genome Annotation Database, version 6.1 (http://rice.plantbiology.msu.edu/). Similarly, data for the Arabidopsis genome were obtained from The Arabidopsis Information Resource Web site, version 10 (Lamesch et al., 2012). Homologous N-regulated genes between rice and Arabidopsis were obtained using Reverse BLAST (Camacho et al., 2009) with e ≤ 1e-20, thereby allowing for multiple orthologous gene hits. Orthology was determined using the data provided on the OrthoMCL Web site (Fischer et al., 2011).
Network Analysis and Data Retrieval
For the gene network analysis (Fig. 2), rice network interaction data were obtained as follows. For RONN (Fig. 2, step 1), metabolic interactions were obtained from RiceCyc, Gramene Pathways (Dharmawardhana et al., 2013), and experimentally determined protein-protein interactions were obtained from the PRIN database (Gu et al., 2011) and the Rice kinase database (Rohila et al., 2006, 2009; Ding et al., 2009). For RPNN-predicted interactions (Fig. 2, step 2), computationally predicted protein-protein interactions were obtained from the PRIN database (Gu et al., 2011) and the Rice journal database (Ho et al., 2012).
Additionally, for RPNN-predicted interactions (Fig. 2, step 2), regulatory interactions were predicted between a TF and its putative target. TF family membership in rice was obtained from PlantTFDB (Jin et al., 2014), and cis-regulatory motifs were obtained from AGRIS (Palaniswamy et al., 2006). The upstream promoter sequences (1 kb) in rice were retrieved from RAP-DB (http://rapdb.dna.affrc.go.jp/). The cis-motifs in promoter regions were searched using the DNA pattern-matching tool from the RSA Tools Plants server with default parameters (van Helden, 2003). HRS1-HHO family member targets were predicted similarly, and cis-motifs for the TF family members were obtained from Medici et al. (2015).
For RANN-BLAST and RANN-OrthoMCL (Fig. 2, step 3), a correlation edge was considered as a conserved correlation edge when the correlation between the N-regulated gene pair in rice was supported by a significant correlation edge between its respective Arabidopsis N-regulated orthologous gene pair, with correct directionality (both correlation edges [in each species] were either both positive or both negative) and tissue specificity (both correlation edges [in each species] were either both root correlation edge or both shoot correlation edge).
Network Construction
In step 1 (Fig. 2), the 451 rice N-regulated genes were queried against the metabolic and experimentally determined protein-protein interaction databases, and all the significant correlation edges between them (P ≤ 0.05) were used to generate RONN. Querying against the predicted protein-protein interactions databases in step 2 (Fig. 2) further enriched this network. Additionally, the predicted regulatory interactions, obtained using cis-motifs from Arabidopsis, were restricted to those TF-target gene pairs where the two were also significantly correlated (P ≤ 0.05). The resulting network, RPNN predicted for step 2 (Fig. 2), had 451 rice genes with 36 TFs and a total of 32,839 interactions between them.
The RPNN-predicted interactions network has reduced numbers of correlation-only edges compared with RONN, because adding cis-motif information to the network resulted in some of the correlation-only edges being reassigned as regulatory edges. This also increased the total number of regulatory (4,128) edges and correlation-only (28,265) edges in the network to 32,393 edges from 32,225 correlation-only edges (Fig. 2). The 168 additional edges were the result of added directionality of regulation, accounting for cases where one TF (TF1) was targeting and was being targeted by another TF (TF2) in the network (Fig. 2).
In step 3 (Fig. 2), Arabidopsis N-regulated experimental correlation data were introduced using BLASTP and OrthoMCL, and individual networks were generated for each method following a similar work flow. Briefly, in both methods, the rice experimental correlation data were filtered with Arabidopsis correlation data, inferred in rice using orthology, to yield conserved correlation edges. If the significant correlation edge between the N-regulated gene pair in rice was also supported by a significant correlation edge between its respective Arabidopsis N-regulated orthologous gene pair, then it was considered a conserved correlation edge. The resulting networks for step 3 (Fig. 2), RANN-BLAST and RANN-OrthoMCL, comprised a total of 180 N-regulated rice genes with 2,212 total interactions and 48 N-regulated rice genes with 383 total interactions, respectively.
Finally, the two networks, RANN-BLAST and RANN-OrthoMCL, were merged in step 4 to yield RANN-Union, which had 182 N-regulated rice genes and 2,273 total interactions between them.
Network Visualization and Analysis
All network visualizations were created using Cytoscape (version 2.8.3) software (Shannon et al., 2003). Custom-made script was used to analyze the total number of direct targets for a TF for each regulatory network. The summarized results for the analysis across all networks are presented in Table IV. The Wilcoxon signed-rank test was used in R to validate that the change in the number of direct targets for the TFs is significant across the network generation process (Hollander and Wolfe, 2014).
Supernode Network Analysis
The supernode analysis merges the individual nodes (genes) into a single node, its size proportional to the number of nodes merged, based on the classification system selected. The TF families (Plant TFDB; Jin et al., 2014) and PlantCyc (OryzaCyc version 1.0; Plant Metabolic Network, 2013) pathways were the two major classification groupings used for our purposes, with level 3 subclass hierarchical classification (Fig. 3). The individual gene pair interactions were merged appropriately for the supernodes and were similar interaction types to those present in the gene network analysis.
Phylogenetic Analysis
The sequences coding for G2-like (HHO) and TGA proteins were retrieved from the AGRIS (http://arabidopsis.med.ohio-state.edu/) database and from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The alignment of the full-length amino acid sequences was performed in ClustalW using standard settings. The phylogeny reconstruction was inferred by using the maximum likelihood method. The bootstrap values were obtained based on 500 replicates. Phylogenetic analysis was conducted in MEGA5 software (Tamura et al., 2011).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Rice N-regulated gene lists compared using the Sungear tool.
Supplemental Figure S2. Quantification of mRNA levels of rice N-regulated genes.
Supplemental Figure S3. Arabidopsis N-regulated gene lists compared using the Sungear tool.
Supplemental Figure S4. Quantification of mRNA levels of Arabidopsis N-regulated genes.
Supplemental Figure S5. The workflow of the analysis of N-regulated genes differentially expressed in rice resulting in Arabidopsis-rice N-regulatory network (ARNN-Union).
Supplemental Figure S6. Arabidopsis and rice HRS1/HHO transcription factor family phylogenetic tree built by ClustalW alignment and maximum likelihood method.
Supplemental Figure S7. Arabidopsis and rice TGA transcription factor family phylogenetic tree built by ClustalW alignment and maximum likelihood method.
Supplemental Table S1. Genes regulated by nitrogen in rice shoots and roots.
Supplemental Table S2. Genes regulated by nitrogen in roots and shoots of Arabidopsis.
Supplemental Table S3. List of 182 genes in the Rice-Arabidopsis N-regulatory network (RANN-Union).
Supplemental Table S4. List of the transcription factors in the Arabidopsis-rice N-regulatory network (ARNN-Union).
Supplemental Table S5. Quantitative real-time PCR primers used in this study.
Supplementary Material
Acknowledgments
We thank Dan Tranchina for assistance with the statistical analysis of microarray data in the early stages of this study.
Glossary
- N
nitrogen
- TF
transcription factor
- MS
Murashige and Skoog
- C
carbon
- FDR
false discovery rate
- RT-qPCR
reverse transcription-quantitative PCR
- RONN
rice-only nitrogen-response network
- RPNN
rice predicted nitrogen-regulatory network
- RANN
rice-Arabidopsis nitrogen-regulated network
- RANN-Union
rice-Arabidopsis nitrogen-regulatory network
- ARNN-Union
Arabidopsis-rice nitrogen-regulatory network
- bHLH
basic helix-loop-helix
Footnotes
This work was supported by the National Science Foundation (Arabidopsis 2010 Genome grant no. MCB–0929338 to G.M.C., Database Activities grant no. DBI–0445666 to G.M.C. and M.S.K., and Integrative Organismal Systems grant no. IOS–0922738 to S.S.) and by the Consejo Nacional de Investigaciones Científicas y Técnicas (to M.O.).
Articles can be viewed without a subscription.
References
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L, et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Marshall-Colon A, Efroni I, Ruffel S, Birnbaum KD, Coruzzi GM, Krouk G (2013) TARGET: a transient transformation system for genome-wide transcription factor target discovery. Mol Plant 6: 978–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc 57: 289–300 [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA (2014) Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Marchive C, Meyer C, Krapp A (2011) Nitrogen signalling in Arabidopsis: how to obtain insights into a complex signalling network. J Exp Bot 62: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441 [DOI] [PubMed] [Google Scholar]
- Cramer M, Lewis OA (1993) The influence of nitrate and ammonium nutrition on the growth of wheat (Triticum aestivum) and maize (Zea mays) plants. Ann Bot (Lond) 72: 359–365 [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana P, Ren L, Amarasinghe V, Monaco M, Thomason J, Ravenscroft D, McCouch S, Ware D, Jaiswal P (2013) A genome scale metabolic network for rice and accompanying analysis of tryptophan, auxin and serotonin biosynthesis regulation under biotic stress. Rice (N Y) 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, Zheng X, Kanrar S, Stevenson RA, Dardick C, et al. (2009) A rice kinase-protein interaction map. Plant Physiol 149: 1478–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ Jr (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. (2014) Glutamate signalling in roots. J Exp Bot 65: 779–787 [DOI] [PubMed] [Google Scholar]
- Fried M, Zsoldos F, Vose PB, Shatokhin IL (1965) Characterizing the NO3 and NH4 uptake process of rice roots by use of 15N labelled NH4NO3. Physiol Plant 18: 313–321 [Google Scholar]
- Gale MD, Devos KM (1998) Plant comparative genetics after 10 years. Science 282: 656–659 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez R, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zhu P, Jiao Y, Meng Y, Chen M (2011) PRIN: a predicted rice interactome network. BMC Bioinformatics 12: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke GUYT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ 28: 1146–1157 [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3: 824–834 [DOI] [PubMed] [Google Scholar]
- Ho CL, Wu Y, Shen HB, Provart NJ, Geisler M (2012) A predicted protein interactome for rice. Rice (N Y) 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M, Wolfe D (2014) Nonparametric Statistical Methods. Wiley, New York [Google Scholar]
- Hu HC, Wang YY, Tsay YF (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–D1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Sévin DC, Lillo C (2009) The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J Plant Physiol 166: 2071–2076 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Glass AD, Yaeesh Siddiqi M (1999a) Inhibition of nitrate uptake by ammonium in barley: analysis of component fluxes. Plant Physiol 120: 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Siddiqi MY, Glass AD, Kirk GJ (1999b) Nitrate-ammonium synergism in rice: a subcellular flux analysis. Plant Physiol 119: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM (2010) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Tranchina D, Lejay L, Cruikshank AA, Shasha D, Coruzzi GM, Gutiérrez RA (2009) A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLoS Comput Biol 5: e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G (2015) AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun 6: 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM (2010) Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst Biol 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Li Y, Marshall-Colón A, Varala K, Francoeur NJ, Moran TM, Edwards MB, Hackley C, Bargmann BO, Birnbaum KD, et al. (2014) Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc Natl Acad Sci USA 111: 10371–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant Metabolic Network (2013) Plant Metabolic Network (PMN). http://pmn.plantcyc.org/ORYZA/class-tree?object=Pathways, on www.plantcyc.org (July 2013) [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Fujii H, Gribskov M, Kanrar S, et al. (2009) Protein-protein interactions of tandem affinity purified protein kinases from rice. PLoS ONE 4: e6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X, Gribskov M, Kanrar S, et al. (2006) Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J 46: 1–13 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi C, Bogusz D, Franche C (2013) Biological nitrogen fixation in non-legume plants. Ann Bot (Lond) 111: 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sederoff RR (2003) Genome studies and molecular genetics: the rice genome and comparative genomics of higher plants. Curr Opin Plant Biol 6: 97–100 [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J. (2003) Regulatory sequence analysis tools. Nucleic Acids Res 31: 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verk MC, Bol JF, Linthorst HJ (2011) WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutierrez RA (2010) Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley Interdiscip Rev Syst Biol Med 2: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama MY Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E (2009) GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol 149: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al. (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.