Histone H2B monoubiquitination is essential for anther development.
Abstract
Histone H2B monoubiquitination (H2Bub1) is an important regulatory mechanism in eukaryotic gene transcription and is essential for normal plant development. However, the function of H2Bub1 in reproductive development remains elusive. Here, we report rice (Oryza sativa) HISTONE MONOUBIQUITINATION1 (OsHUB1) and OsHUB2, the homologs of Arabidopsis (Arabidopsis thaliana) HUB1 and HUB2 proteins, which function as E3 ligases in H2Bub1, are involved in late anther development in rice. oshub mutants exhibit abnormal tapetum development and aborted pollen in postmeiotic anthers. Knockout of OsHUB1 or OsHUB2 results in the loss of H2Bub1 and a reduction in the levels of dimethylated lysine-4 on histone 3 (H3K4me2). Anther transcriptome analysis revealed that several key tapetum degradation-related genes including OsC4, rice Cysteine Protease1 (OsCP1), and Undeveloped Tapetum1 (UDT1) were down-regulated in the mutants. Further, chromatin immunoprecipitation assays demonstrate that H2Bub1 directly targets OsC4, OsCP1, and UDT1 genes, and enrichment of H2Bub1 and H3K4me2 in the targets is consistent to some degree. Our studies suggest that histone H2B monoubiquitination, mediated by OsHUB1 and OsHUB2, is an important epigenetic modification that in concert with H3K4me2, modulates transcriptional regulation of anther development in rice.
The reversible monoubiquitination of histone H2B (H2Bub1) in chromatin is an important biochemical event in regulating important cellular processes through gene regulation in eukaryotes. H2Bub1 levels are dynamically regulated via deposition and removal of ubiquitin by specific enzymes. Three enzymes work coordinately to conjugate ubiquitin via its C-terminal residue to the side chain of a Lys residue of the substrate/acceptor protein (Hershko and Ciechanover, 1998). Firstly, ubiquitin is activated by an ATP-dependent reaction involving the ubiquitin-activating enzyme E1. It is then conjugated to the active-site Cys residue of the ubiquitin-conjugating enzyme E2 and finally transferred to the target Lys residue of the substrate protein by the ubiquitin-protein isopeptide ligase E3. Most organisms have only one E1 but typically dozens of different E2 and hundreds to thousands of different E3 enzymes, which provide the necessary variation for substrate specificity (Hua and Vierstra, 2011; Braun and Madhani, 2012). Identification and characterization of specific E3s involved in histone ubiquitination from a range of organisms are proving important in understanding the broader biological role of this important epigenetic modification in cellular and developmental regulation.
H2Bub1 plays a critical role in the growth of eukaryotes and is typically associated with transcriptional activation (Feng and Shen, 2014). In yeast (Saccharomyces cerevisiae), Drosophila spp., and mammal cells, H2B is ubiquitylated by the E3 ubiquitin ligase Brefeldin A-sensitivity protein1 (BRE1). The yeast bre1 mutant, which lacks ubiquitinated H2B, shows a developmental defect resulting in increased cell size (Hwang et al., 2003). Mutations in Drosophila spp. dBre1 are lethal due to blocked notch signaling (Bray et al., 2005). In mammals, the hBRE1 (RING FINGER PROTEIN20 [RNF20]/RNF40 complex functions as the E3 ligase (Kim et al., 2005; Zhu et al., 2005). hBRE1 (RNF20) acts as a putative tumor suppressor by selective regulation of subsets of genes with some being up-regulated and others down-regulated. Depletion of RNF20 results in a decrease in p53 expression and an increase in cell migration and tumorigenesis as well as an increase in expression of c-myc, a proto-oncogene (Shema et al., 2008). A tumor-suppressive role for RNF40 mediated through H2Bub1 has also been demonstrated in breast cancer cells (Prenzel et al., 2011).
In Arabidopsis (Arabidopsis thaliana), histone H2B is monoubiquitinated by two E3 ligases, HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 (Fleury et al., 2007; Liu et al., 2007; Cao et al., 2008). The hub1 and hub2 mutants display reduced seed dormancy correlated with decreased expression of several dormancy-related genes such as DELAY OF GERMINATION1 (Liu et al., 2007). During vegetative growth stages, hub mutants exhibit pale leaves, modified leaf shape, reduced rosette biomass, and inhibited root growth (Fleury et al., 2007). At the floral transition stage of development, hub1 and hub2 single mutants and hub1 hub2 double mutant lines exhibit an early flowering phenotype compared with the wild type (Cao et al., 2008; Gu et al., 2009; Xu et al., 2009). More recent studies have shown that several circadian clock genes are down-regulated in hub mutants and that their respective chromatin regions contain lower levels of H2Bub1 (Bourbousse et al., 2012; Himanen et al., 2012). Thus, H2Bub1 appears to be a mechanism that contributes to comparatively rapid regulation of plant growth responses to environment through expression modulation of some circadian clock genes. Histone H2B monoubiquitination is also involved in the regulation of cutin and wax composition in the Arabidopsis cuticle. Irregular epidermal cells and disorganized cuticle layers were observed in rosette leaves of hub1-6 and hub2-2 mutants (Ménard et al., 2014). In addition, H2Bub1 has been found to play a role in plant defense against pathogens. hub1 mutants show increased susceptibility to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola (Dhawan et al., 2009), and HUB1 and HUB2 have been shown to regulate the expression of R genes SUPPRESSOR OF npr1-1, CONSTITUTIVE1 and RESISTANCE TO PERONOSPORA PARASITICA4 via histone monoubiquitination at the R gene locus (Zou et al., 2014). Further, H2Bub1 mediated by HUB1/HUB2 is also involved in regulating the dynamics of microtubules to alter the defense response to Verticillium dahliae toxins in Arabidopsis (Hu et al., 2014).
H2Bub1 has also been found to be involved in reproductive development of eukaryotes. A yeast strain with specific impairment of H2Bub1 through mutation of RAD6, a major E2 conjugating enzyme, is found to be unable to form spores on account of meiotic prophase arrest (Robzyk et al., 2000). Another study has shown that male mice, deficient for HR6B/hRAD6B, are infertile due to abnormal spermatogenesis (Roest et al., 1996). Male mice deficient in the E3 ligase RNF8 are also sterile, and this correlates with a decrease in both H2Bub1 and H2Aub in elongating spermatids (Lu et al., 2010). Moreover, in Arabidopsis, mutations in the genes encoding the facilitates chromatin transcription (FACT) subunits Structure-Specific Recognition Protein1 and SPT16, which can interact synergistically with H2Bub1, result in various aberrant developmental phenotypes, which include reduced fertility (Lolas et al., 2010). A genetic analysis also showed that H2Bub1 acts on some phenotypes, including silique size, in the same pathway as FACT. UBIQUITIN-SPECIFIC PROTEASE26 can deubiquitinate H2Bub1 in Arabidopsis, and ubp26 mutants show increased level of H2Bub1 (Sridhar et al., 2007). The ubp26 mutants also show distinct reproductive developmental defects, including poor seed production, which may be the result of impaired pollen release from nondehiscent anthers and the lack of functional embryo sacs (Luo et al., 2008; Schmitz et al., 2009). These findings imply that H2Bub1 may play an important role in male reproductive development, though currently there is a paucity of information regarding the molecular basis of H2Bub1 involvement in this process.
Here, we identified OsHUB1 and OsHUB2 as two E3 ligases of H2Bub1 with a function in male reproductive development in rice (Oryza sativa). Loss of function of OsHUB1 and OsHUB2 results in abnormal H2Bub1 deposition and a reduction in histone H3 lysine-4 (H3K4) dimethylation. Many genes associated with development of the anther tapetum and pollen were found to be down-regulated in oshub1. We propose that during anther development, H2Bub1 is involved in regulating gene transcription, likely by promoting histone H3K4 dimethylation (H3K4me2) in the chromatin of specific genes.
RESULTS
Rice oshub1 Mutants Show Developmental Defects, Including a Partially Sterile Phenotype
Arabidopsis HUB1 and HUB2 have been found to play various roles in seed performance and plant development through mediating H2Bub1 modification (Feng and Shen, 2014), but nothing has been reported about their rice homologs. In rice, there are two homologous genes, OsHUB1 (Loc_Os04g46450) and OsHUB2 (Loc_Os10g41590). To analyze their function, two alleles of oshub1 (oshub1-1 and oshub1-2) and one allele of oshub2 were investigated. In oshub1-1, oshub1-2, and oshub2 mutant alleles, a transfer DNA (T-DNA) insertion was located in the 12th intron, second exon, and 17th exon, respectively (Supplemental Fig. S1, A and B), and no full-length transcripts were detected for any of the three alleles (Supplemental Fig. S1, C–E).
The three alleles of oshub1 and oshub2 showed quite similar phenotypes, including slightly early heading (data not shown), semidwarfism (Supplemental Fig. S2), and partial sterility of spikelets (Fig. 1, A and B). Seed set was 100%, 40%, 30%, and 50% in the wild type, oshub1-1, oshub1-2, and oshub2, respectively (Fig. 1C). When the oshub mutants were crossed with the wild type, all F1 plants displayed normal phenotypes, and the F2 plants had an approximate 3:1 ratio for phenotypic segregation (in oshub1-1, normal plants:mutant plants = 45:12 and χ2 = 0.474 for 3:1; in oshub1-2, normal plants:mutant plants = 62:19 and χ2 = 0.103 for 3:1; and in oshub2, normal plants:mutant plants = 53:19 and χ2 = 0.074 for 3:1), suggesting that these phenotypes segregated as monofactorial recessive alleles in linkage.
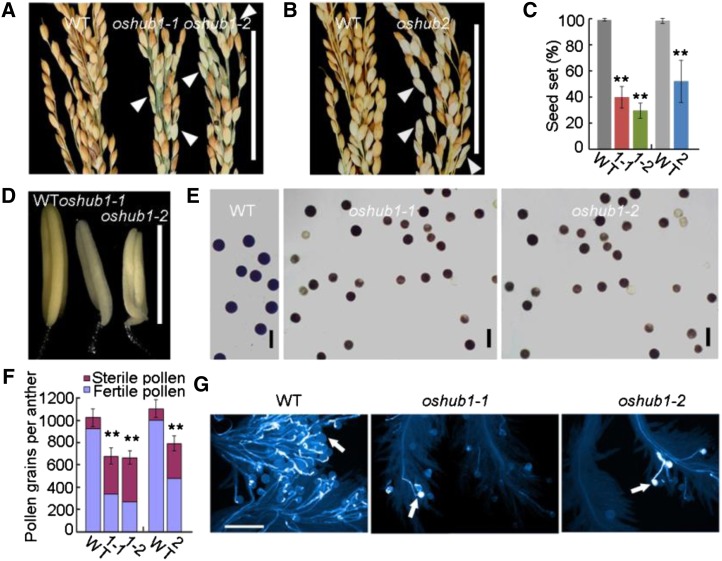
Figure 1.
Phenotypic analysis of the wild type (WT) and oshub1 and oshub2 mutants. A, Comparison of the percentage seed set of the wild type (left), oshub1-1 (middle), and oshub1-2 (right). B, Comparison of the percentage seed set of the wild type (left) and oshub2 (right). Arrowheads in A and B indicate sterile spikelets. C, Seed set of wild-type and oshub1-1 (1-1), oshub1-2 (1-2), and oshub2 (2) mutant plants. Seed set was calculated as the proportion of fertile spikelets to all spikelets for each plant at maturity. Data are means ± sd (n = 20). D and E, Comparison of anther phenotype before anthesis (D) and I2-KI-stained pollens (E) of the wild type and oshub1 mutants. F, The number of sterile and fertile pollen grains per anther as determined by I2-KI staining of wild-type, oshub1-1 (1-1), oshub1-2 (1-2), and oshub2 (2) materials. Data are means ± sd of total pollen grains (n = 5). G, Comparison of aniline blue-stained pollen grains on stigmas for the wild type and oshub1 mutants. Arrows indicate the pollen grains with pollen tubes. Student’s t test was used to analyze significant differences between the wild type and mutants (*P < 0.05; and **P < 0.01). Bars = 5 cm (A and B), 2 mm (D), 50 µm (E), and 200 µm (G).
Because flowering time and semidwarfism phenotypes have already been well studied in Arabidopsis (Fleury et al., 2007; Cao et al., 2008), the focus in this study was detailed characterization of the partial-sterile phenotype of oshub1 and oshub2 mutants. The oshub1 mutants produced flowers having the full complement of floral organs, with the lemma, palea, and pistil developing normally (data not shown); however, mutants failed to generate normal anthers. Compared with wild-type anthers, mutant anthers were smaller and paler (Fig. 1D), and the anther length in oshub1-1 and oshub1-2 lines was 89% and 84% of that of the wild type, respectively. An iodine-potassium iodide (I2-KI) staining assay was utilized to estimate pollen viability and this indicated that viability of the wild type was 95%, but only 50% and 40% in oshub1-1and oshub1-2, respectively (Fig. 1, E and F).
As 50% pollen viability should be enough to effect good seed set (Zhou et al., 2011), there are likely additional aspects to the mutant phenotype that explain the observed partial spikelet sterility. The total number of pollen grains per anther was counted. Wild-type plants had 1,024 ± 92 pollen grains per anther, while the oshub1-1 and oshub1-2 mutants had 679 ± 89 and 665 ± 84 pollen grains per anther, respectively. Thus, in total, oshub1-1 and oshub1-2 pollen viability was just 37% and 29% that of the wild type (Fig. 1F). Furthermore, 2 h after anthesis, the number of pollen grains shed onto the stigmas of mutants is markedly reduced compared with the wild type (Fig. 1G). Analysis of the oshub2 mutant was found to closely mirror that of oshub1 having reduced pollen number and pollen viability (Fig. 1F). Together, these data suggest that the partial reproductive sterile phenotype observed in oshub mutants is caused by defects in the anther developmental program and pollen formation.
OsHUB1 and OsHUB2 Have Effects on Late Anther Development
To further characterize the morphological defects in the oshub1 and oshub2 mutants, transverse sections of anthers were examined. No significant differences were detected between the wild type and the mutant anthers from the early developmental stages through to stage 9 (Fig. 2), by which time meiosis would have completed. Meiosis in the mutant lines also appeared to be normal, as revealed by karbol fuchsin staining analysis (Supplemental Fig. S3). In subsequent developmental stages, mutant lines had obviously abnormal anthers. In wild-type plants at stage 10, tapetal cells had become more degenerated and had deeply stained cytoplasm. The adjacent middle layer was thin and barely visible, and the wild-type microspores appeared round and vacuolated (Fig. 2B). By contrast, mutant lines exhibited a thicker tapetum with enhanced staining, and the microspores had irregular appearance (Fig. 2, F and J). At stage 11 of pollen development in the wild type, the endothecium and middle layer degenerate, and the tapetal cells became differentiated and degenerated. Typical falcate pollen grains were also formed at this stage following the first mitotic division of microspores (Fig. 2C). However, the mutant lines showed delayed degradation of the anther middle layer and endothecium, and tapetal cells appeared lightly stained, with evidence of collapse into the anther locule and cellular degeneration. In addition, the pollen grains of mutant lines remained highly vacuolated and exine deposition was absent (Fig. 2, G and K). By the mature pollen stage, the tapetum and middle layer had completely degenerated in the mutants, similar to wild-type anthers (Fig. 2, D, H, and L). Wild-type anther locules at this stage were rounded and began to dehisce. By contrast, mutant locules were narrower and longer and were largely indehiscent. At this stage, mature wild-type pollen grains were typically round and stained deeply with toluidine blue, indicating that the pollen is full of storage materials (Fig. 2D). However, by contrast, few pollen grains from mutant lines were normally developed. The majority of grains were irregularly shaped, stained weakly with toluidine blue, and eventually collapsed (Fig. 2, H and L). These observations suggest that oshub1-1, oshub1-2 lines have developmental defects in anther wall layers during late phases of stamen and that these defects impact microspore formation and subsequent release.
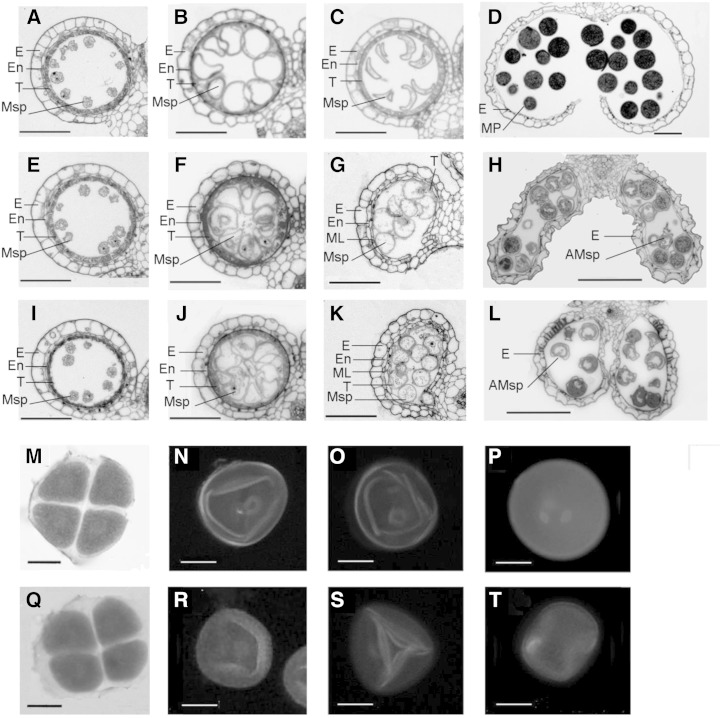
Figure 2.
Comparison of male gametogenesis in wild-type and oshub1 plants. A to D and M to P show the wild type. E to H, I to L, and Q to T show the oshub1-1, oshub1-2, and oshub1-1 mutant, respectively. E, Epidermis; En, endothecium; ML, middle layer; T, tapetum; Msp, microspore; MP, mature pollen; AMsp, aborted microspore. A, E, and I, Cross section of anthers at stage 9. B, F, and J, Cross section of anthers at stage 10. C, G, and K, Cross section of anthers at stage 11. D, H, and L, Anthers at stage 13. M and Q, N and R, O and S, and P and T show microspores at stage 8b, stage 10, stage 11, and stage 12, respectively. N to P and R to T are DAPI stained to show nuclei. M and Q are karbol fuchsin stained to show tetrad. Bars = 100 µm (A–L) and 10 µm (M–T).
To ascertain whether gametogenesis was affected in oshub1-1, 4′,6-diamidino-2-phenylindole (DAPI) staining was used to image the nuclei of developing pollen in mutant and wild-type plants. oshub1-1 meiocytes were found to be normal and were able to undergo normal meiosis to form tetrad, similar to wild-type plants (Fig. 2, M and Q). During postmeiotic developmental stages, wild-type microspores proceeded through a first and second mitotic division to form two sperm nuclei and one vegetative nucleus (Fig. 2, N–P). By contrast, in the mutant lines, approximately 60% of microspores were irregularly shaped and appeared to lack nuclei (Fig. 2, R–T). These results strongly suggest that OsHUB1 and OsHUB2 are required for normal postmeiotic pollen development.
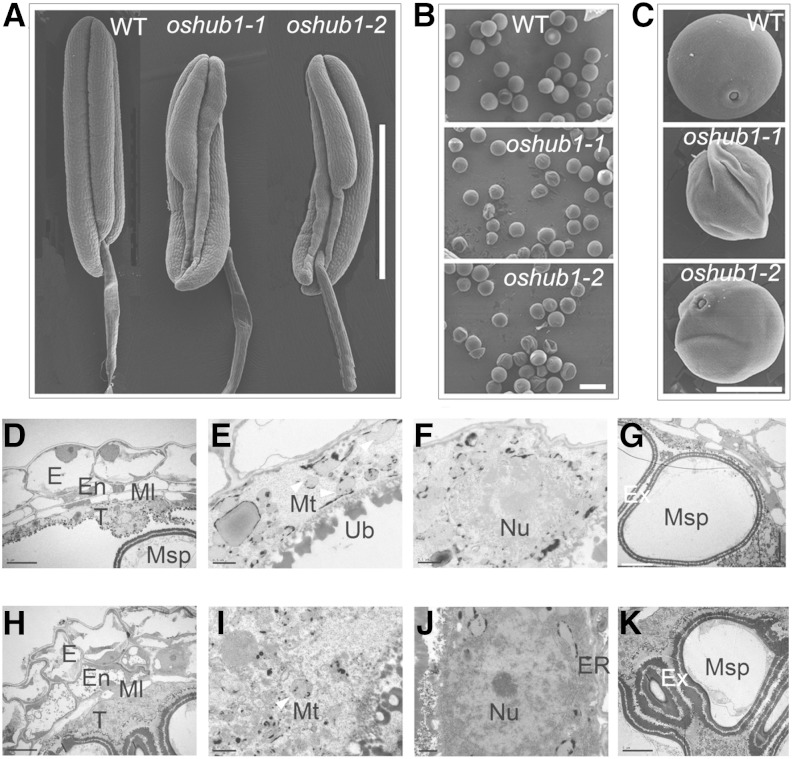
To gain more detailed insight into the oshub1 anther abnormalities, scanning electron microscopy was used. Wild-type anthers were found to be full and regular, whereas mutant anthers were shriveled in the basal sections of the locules (Fig. 3A). Mature wild-type pollen was round and plump, whereas a proportion of the mutant pollen had an irregular and shrunken morphology (Fig. 3, B and C). The percentage of abnormal pollen was 50% and 60% in oshub1-1 and oshub1-2, respectively. Transmission electron microscopy was used to analyze the abnormal tapetal layer and microspore of oshub1 mutants. At stage 10, the wild-type tapetal cytoplasm was condensed (Fig. 3D), with swollen mitochondria and nucleus, which showed a diffuse cellular organization, associated with programmed cell death (Fig. 3, E and F). By contrast, oshub1-1 tapetal layer was less condensed, and organelles were clearly defined, particularly mitochondria, nucleus, and endoplasmic reticulum, with integrated membrane and normal appearance, suggesting the delayed cell death process in the tapetum (Fig. 3, H–J). Moreover, compared with the wild-type microspore at stage 10, the oshub1-1 microspore developed a thick and abnormal exine, resulting in irregular shape (Fig. 3, G and K). These observations suggest that oshub1 mutant undergoes delayed tapetal degeneration, which affects microspore development.
Figure 3.
Scanning electron microscopy and transmission electron microscopy of wild-type (WT) and oshub1 anthers. A and B, Comparison of anthers before anthesis (A) and mature pollen grains (B) derived from the wild type and oshub1 mutants. C, Higher magnification of pollen grains from wild-type and oshub1 mutant plants. D to G, The cross section of wild-type anthers at stage 10 showing anther wall layers (D), mitochondria (E) and nucleus (F) in tapetal layer, and microspore (G). H to K, The cross section of oshub1-1 anthers at stage 10 showing anther wall layers (H), mitochondria (I) and nucleus (J) in tapetal layer, and microspore (K). Mitochondria is marked by the white arrowhead in E and I. E, Epidermis; En, endothecium; ER, endoplasmic reticulum; Ex, exine; Ml, middle layer; Msp, microspore; Mt, mitochondria; Nu, nucleus; T, tapetum; Ub, ubisch body. Bars = 1 mm (A), 50 µm (B), 20 µm (C), 5 µm (D, H, G, and K), and 0.5 µm (E, F, I, and J).
Defect in H2Bub1 Reduces H3K4me2 Level in oshub1 and oshub2 Mutant Plants
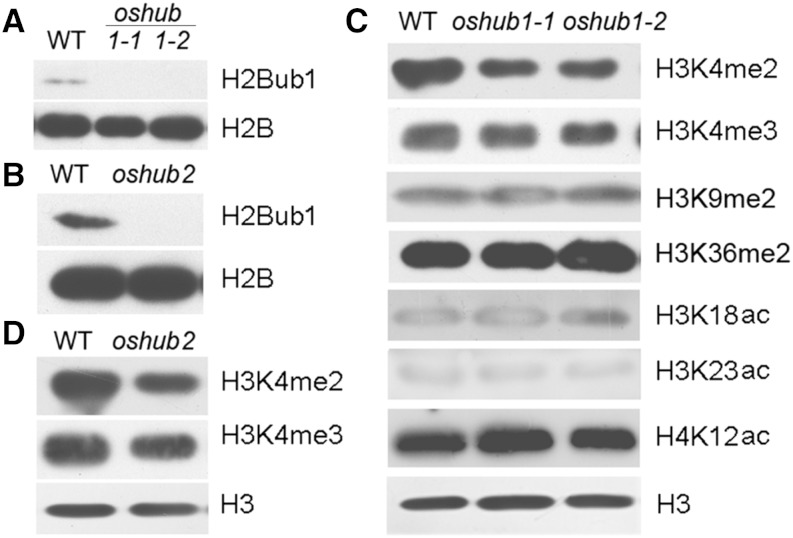
As OsHUB1 is the putative E3 ligase that directs histone H2B monoubiquitination, it infers that H2Bub1 will be lost in the oshub1 mutant. To verify this hypothesis, an anti-H2Bub1 antibody was used to analyze the presence of H2Bub1 by immunoblotting. As shown in Figure 4A, H2Bub1 was not detected in the oshub1 mutant at a global level. In addition, there was also a loss of H2Bub1 in the oshub2 mutant line (Fig. 4B). These data indicate that OsHUBs are required for H2Bub1 in rice. Many studies have indicated that presence of H2Bub1 influences other histone modifications (Hwang et al., 2003; Zhu et al., 2005). Thus, the interaction between H2Bub1 and H3 methylation, H3 acetylation, or H4 acetylation was examined. Our data showed that loss of H2Bub1 leads to decreased global levels of H3K4me2 in oshub1 and oshub2 mutant plants compared with the wild type (Fig. 4, C and D); however, no significant differences in the genome-wide level of H3K4me3, H3K9me2, H3K36me2, histone H3 lysine-18 acetylation (H3K18ac), H3K23ac, and H4K12ac modification were detected (Fig. 4C). This indicates that H2Bub1 is necessary for the enhancement of H3K4me2 in rice.
Figure 4.
Global histone modification in the wild type (WT) and oshub mutants. A and B, Analysis of H2B monoubiquitination in the wild type and mutants using an anti-H2Bub1 antibody. H2B was used as a loading control. C, Detection of H3 methylation and H3 and H4 acetylation on a genome-wide scale in the wild type and oshub1 mutants. D, Analysis of H3K4 di- and trimethylation in the wild type and oshub2 mutant. H3 was used as loading control in C and D.
Physical Interactions between OsHUB1 and OsHUB2
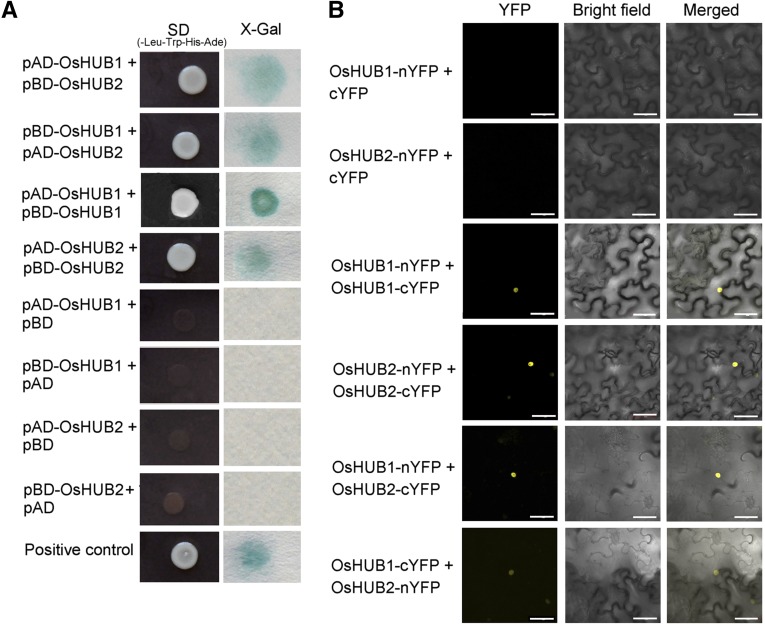
The oshub1 and oshub2 phenotypic analysis indicated that both OsHUB1 and OsHUB2 are necessary for rice spikelet fertility. In addition to sharing a partially sterile phenotype, oshub1 and oshub2 showed other similarities, including early heading time and a semidwarf plant architecture. This data are consistent with the hypothesis that the two proteins may function in the same complex, as has previously been demonstrated for their Arabidopsis homologs, HUB1 and HUB2 (Cao et al., 2008). Thus, testing for an interaction between OsHUB1 and OsHUB2 was performed by utilizing yeast two-hybrid and bimolecular fluorescence complementation assays. Results from these studies revealed that OsHUB1 interacts with OsHUB2 and also that each of the two proteins can interact with itself (Fig. 5, A and B). Consistent with nucleus localization of OsHUB1 (Supplemental Fig. S4), the interactions were localized to the nucleus (Fig. 5B), which indicates that the proteins function in histone modification.
Figure 5.
Self-interaction and pairwise interaction of OsHUB1 and OsHUB2. A, Physical interactions between OsHUB1 and OsHUB2 in the yeast two-hybrid assay. pACT2 is the pray vector (AD), and pAS2 is the bait vector (BD). Yeast stains cotransformed with OsHUB1 and OsHUB2, OsHUB1 and OsHUB1, and OsHUB2 and OsHUB2 were assayed for growth on selective medium (-Leu, -Trp, -His, and -Ade; left) and β-galactosidase activity (right). Self-interaction of HUB1 (Cao et al., 2008) was used as a positive control (pAD-HUB1 + pBD-HUB1). Cotransformations of pAD-OsHUB1 with pBD, pBD-OsHUB1 with pAD, pAD-OsHUB2 with BD, and pBD-OsHUB2 with pAD are included as negative controls. B, Bimolecular fluorescence complementation assay showing the (self-)interaction of OsHUB1 and OsHUB2 in vivo. OsHUB1 and OsHUB2 were transiently coexpressed or were coexpressed with the vector alone in Nicotiana benthamiana leaf cells. Bars = 50 µm. SD, Synthetically defined medium; X-Gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid; YFP, yellow fluorescent protein.
OsHUB1 Is Ubiquitously Expressed
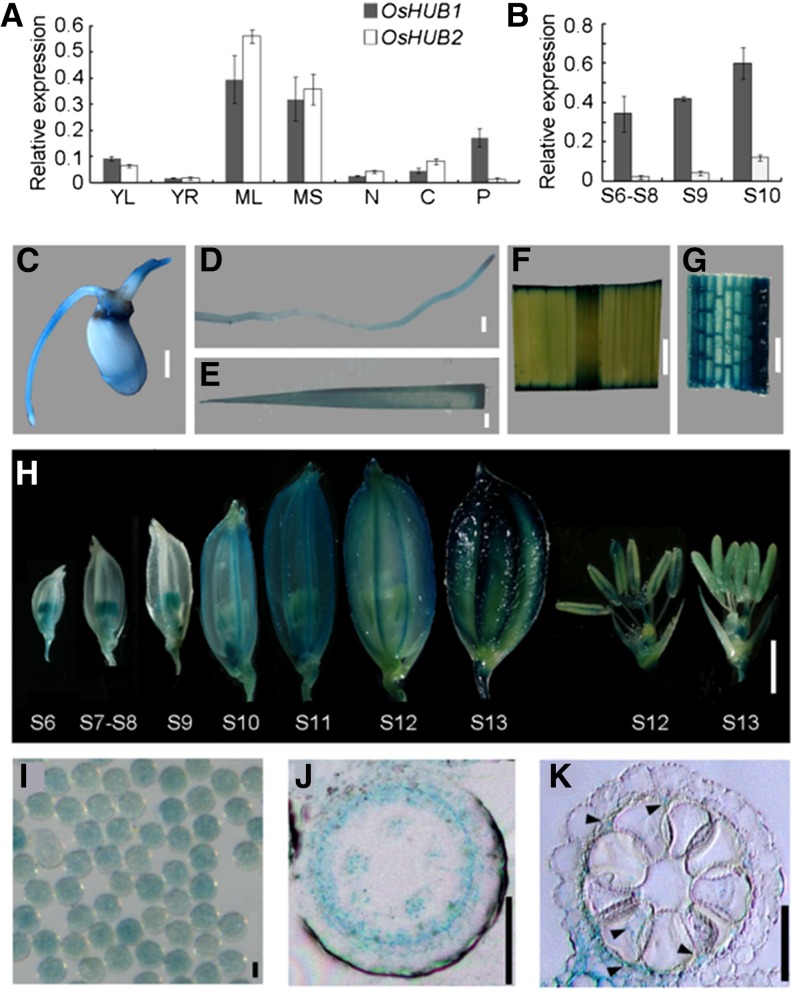
Quantitative reverse transcription (qRT)-PCR revealed that OsHUB1 and OsHUB2 have a ubiquitous expression profile; transcripts were detected in young leaves and roots, mature leaves, and mature sheath, culm, and panicle tissues (Fig. 6A). During anther development, both OsHUB1 and OsHUB2 had higher expression levels at stage 10 compared with stages 6 to 8 and stage 9, although OsHUB2 exhibited a lower expression level than that of OsHUB1 (Fig. 6B). Consistent with the qRT-PCR results, a GUS reporter construct driven by the OsHUB1 promoter (a 2.0-kb sequence upstream of the ATG start codon) showed that OsHUB1 has high expression in germinating seeds (Fig. 6C), young roots (Fig. 6D), young leaves (Fig. 6E), mature leaves (Fig. 6F), leaf sheathes (Fig. 6G), spikelets from various developmental stages (Fig. 6H), and mature pollen grains (Fig. 6I). In addition, OsHUB1 was preferentially expressed in anthers during early spikelet development, and later, GUS activity was detected in the whole spikelet. Further transverse section analysis of GUS-stained locules indicated that OsHUB1 is highly expressed in the tapetum (Fig. 6, J and K). These data suggest that OsHUB1 likely plays a role in tapetal cells during anther development.
Figure 6.
Expression pattern of OsHUB1 and OsHUB2. A and B, qRT-PCR analysis of expression profiles for OsHUB1 and OsHUB2. S6 to S8 and S9 and S10 represent anther development at stages 6 to 8 and stages 9 and 10, respectively. ACTIN1 was used as internal control. Error bars indicate sd of three independent samples. YL, Young leaf; YR, young root; ML, mature leaf; MS, mature sheath; N, node; C, culm; P, panicle. C to H, Histochemical analysis of OsHUB1 expression in a germinating seed (C), young root (D), young leaf (E), mature leaf (F), leaf sheath (G), spikelet at various stages (H), and mature pollen grains (I) from transgenic plants expressing GUS driven by the OsHUB1 promoter. S6 to S13 refer to stages 6 to 13. J and K, GUS staining in a transverse section of a stage-8 (J) and a stage-10 (K) anther locule. Arrowheads in K show the GUS signal. Bars = 2 mm (C–H) and 20 µm (I–K).
Transcriptome Analysis of Wild-Type and oshub1-1 Anthers
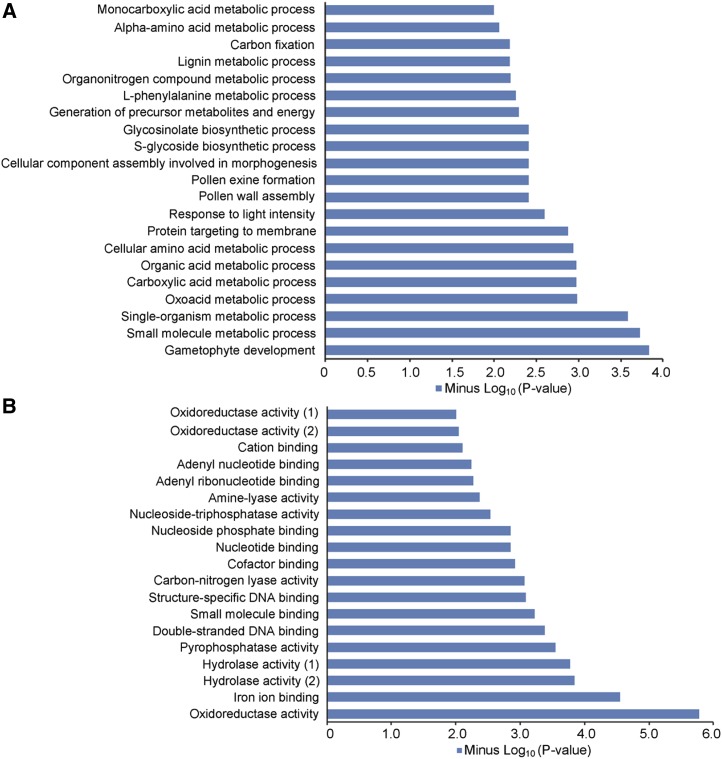
H2Bub1 has been recognized as a critical epigenetic marker associated with transcriptional activation in eukaryotes (Zhang, 2003). We showed that H2Bub1 is undetectable in oshub1 mutant plants (Fig. 4A). Therefore, mRNA-sequencing (RNA-seq) analysis of wild-type and oshub1-1 anthers at stage 10 was performed to identify genes with altered expression levels. In the oshub1-1 mutant, 1,843 down-regulated and 1,067 up-regulated genes were detected based on a false discovery rate (FDR) cutoff of 0.001 (FDR ≤ 0.001) and a greater than 2-fold (log2 fold ≥ 1) expression difference, indicating that a large proportion of genes are positively regulated by H2Bub1. To understand the biological roles of the differentially expressed genes (DEGs) in the anther, we conducted Gene Ontology (GO) enrichment analysis for biological process and molecular function. The most significantly enriched GO term for biological process was gametophyte development (Fig. 7A), and for molecular function, it was oxidoreductase activity (Fig. 7B). We also performed GO analysis of down-regulated genes and up-regulated genes separately. The most significantly enriched categories between the down-regulated genes and the up-regulated ones are distinctive, with pollen formation for the down-regulated genes and lignin metabolic process for the up-regulated genes, respectively (Supplemental Fig. S5), indicating that down-regulated genes might have more effects on pollen development. Some genes previously identified as being critical for anther development were found to be down-regulated in the oshub1-1 RNA-seq data (Table I).
Figure 7.
Biological process and molecular function by GO analysis of genes that were up-regulated and down-regulated at least 2-fold in oshub1-1. A, Biological process indicated by GO analysis. B, Molecular function indicated by GO analysis. Hydrolase activity (1) refers to hydrolase activity acting on acid anhydrides in phosphorus-containing anhydrides. Hydrolase activity (2) refers to hydrolase activity acting on acid anhydrides. Oxidoreductase activity (1) refers to oxidoreductase activity acting on single donors with incorporation of molecular oxygen. Oxidoreductase activity (2) refers to oxidoreductase activity acting on NAD(P)H.
Table I. A selection of genes involved in anther development with altered expression in oshub1-1 anthers as identified by RNA-seq analysis.
| Gene Locus | Gene Annotation | Gene Name | Fold Change | P Value |
|---|---|---|---|---|
| log2(mutant/wild type) | ||||
| Os08g43290 | Lipid transfer protein precursor | OsC4 | −1.0 | 0 |
| Os04g57490 | Cys protease | OsCP1 | −1.2 | 2.58E–09 |
| Os03g07140 | Fatty acyl-CoA reductase | DPW | −1.4 | 0 |
| Os03g07250 | Cytochrome P450 | CYP704B2 | −1.1 | 0 |
| Os01g10504 | MADS-box family protein | MADS3 | −1.7 | 1.44E–26 |
| Os04g37570 | Aspartic protease | AP37 | −2.1 | 6.63E–190 |
| Os04g51070 | Basic Helix-Loop-Helix protein | EAT1 | −1.6 | 7.00E–46 |
| Os06g40550 | ATP-Binding Cassette2 type transporter | PDA1 | −1.1 | 3.70E–104 |
| Os08g03682 | Cytochrome P450 | CYP703A3 | −1.2 | 0 |
| Os09g27620 | Plant Homedomain-finger domain containing protein | PTC1 | −1.4 | 7.88E–24 |
| Os06g08290 | MYB family transcription factor | Anther Indehiscence1 | −1.2 | 6.85E–06 |
| Os04g52320 | Pectin lyase family protein | Unnamed | −1.2 | 3.58E–153 |
| Os04g24530 | 4-Coumarate-CoA ligase | Unnamed | −1.6 | 3.33E–50 |
| Os04g39470 | MYB family transcription factor | OsMYB103 | −1.4 | 4.90E–29 |
| Os10g34360 | Stilbene synthase | YY2 | −1.4 | 0 |
| Os01g49650 | Lipid transfer protein precursor | Unnamed | −1.14 | 7.60E–157 |
| Os07g22850 | Chalcone and stilbene synthases | Unnamed | −1.61 | 4.34E–281 |
| Os07g37090 | Ribosome inactivating protein | RA39 | −1.3 | 2.43E–186 |
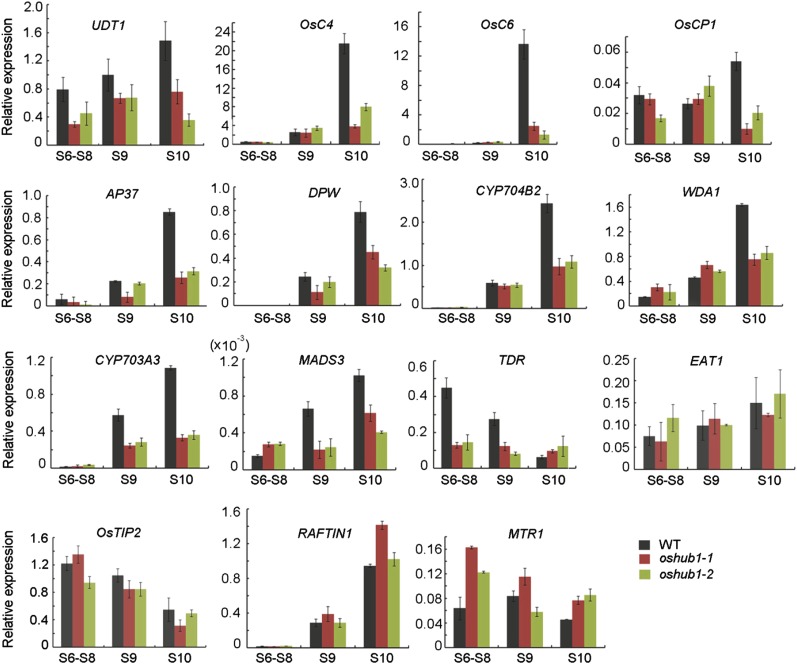
Furthermore, by utilizing qRT-PCR, we verified the expression pattern of a selection of genes identified as having changed expression in oshub1 (Table I). Anthers from three developmental stages (stages 6–8, stage 9, and stage 10) were used in the analysis (Fig. 8). These sequences included aspartic protease encoding gene (AP37; Niu et al., 2013), Cys protease1 gene (OsCP1; Lee et al., 2004), lipid transfer protein gene (OsC4; Tsuchiya et al., 1994), Cytochrome P450 704B2 gene (CYP704B2; Li et al., 2010), CYP703A3 (Yang et al., 2014), Defective Pollen Wall (DPW; Shi et al., 2011), Eternal Tapetum1 (EAT1; Niu et al., 2013), and floral homeotic C-class gene (MADS3; Hu et al., 2011). The expression pattern of these genes was largely consistent with the RNA-seq data set, with the exception of EAT1 (Fig. 8). The expression of EAT1 was down-regulated in the oshub1-1 RNA-seq data, but it showed no difference compared to the wild type by qRT-PCR analysis. In addition, the expression of a selection of genes that appeared to have no change in expression according to the RNA-seq data was also checked by qRT-PCR (Fig. 8). These were the lipid transfer protein gene (OsC6; Zhang et al., 2010), Undeveloped tapetum1 (UDT1; Jung et al., 2005), Wax-Deficient Anther1 (WDA1; Jung et al., 2006), Tapetum Degeneration Retardation (TDR; Li et al., 2006), TDR INTERACTING PROTEIN2 (OsTIP2; Fu et al., 2014), MICROSPORE AND TAPETUM REGULATOR1 (MTR1; Tan et al., 2012), and RAFTIN1 (Wang et al., 2003). It was found that OsC6, UDT1, and WDA1 were down-regulated in the oshub1 mutant at the vacuolated pollen stage, while TDR, TIP2, MTR1, and RAFTIN1 showed no significant change.
Figure 8.
qRT-PCR expression analysis of genes reported to be involved in rice anther development in wild-type (WT) and oshub1 plants. S6 to S8 and S9 and S10 represent stages 6 to 8 and stages 9 and 10 of anther development, respectively. For RNA extraction, anthers were used. ACTIN1 was used as the internal control. Each data point is the average of three biological repeats, and error bars indicate sd.
Expression of meiosis-related genes in oshub1 mutants was largely unaffected. For example, PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1; Nonomura et al., 2004), PAIR2 (Nonomura et al., 2006), PAIR3 (Yuan et al., 2009), MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1; Nonomura et al., 2007), UDP-glucose pyrophosphorylase1 (UGP1; Chen et al., 2007), OsRAD21-4 (Zhang et al., 2006), GAMYB (Aya et al., 2009), and MER3 (Wang et al., 2009) displayed no significant difference in expression from those in the wild type, with only MER3 (Wang et al., 2009) having decreased expression in oshub1 (Supplemental Fig. S6). This suggests that meiotic division of the microspore mother cell may not be affected in the oshub1 mutant. In addition, the chromosome behavior of male meiocytes from the wild type and oshub1-1 was examined. This revealed that oshub1-1 plants complete a normal meiotic program producing free microspores indistinguishable from those in wild-type plants (Supplemental Fig. S3). These data further support the thesis that oshub1-1 defects are restricted to postmeiotic anther development stages.
Levels of H2B Monoubiquitination Are Altered in Tapetum Development-Related Genes
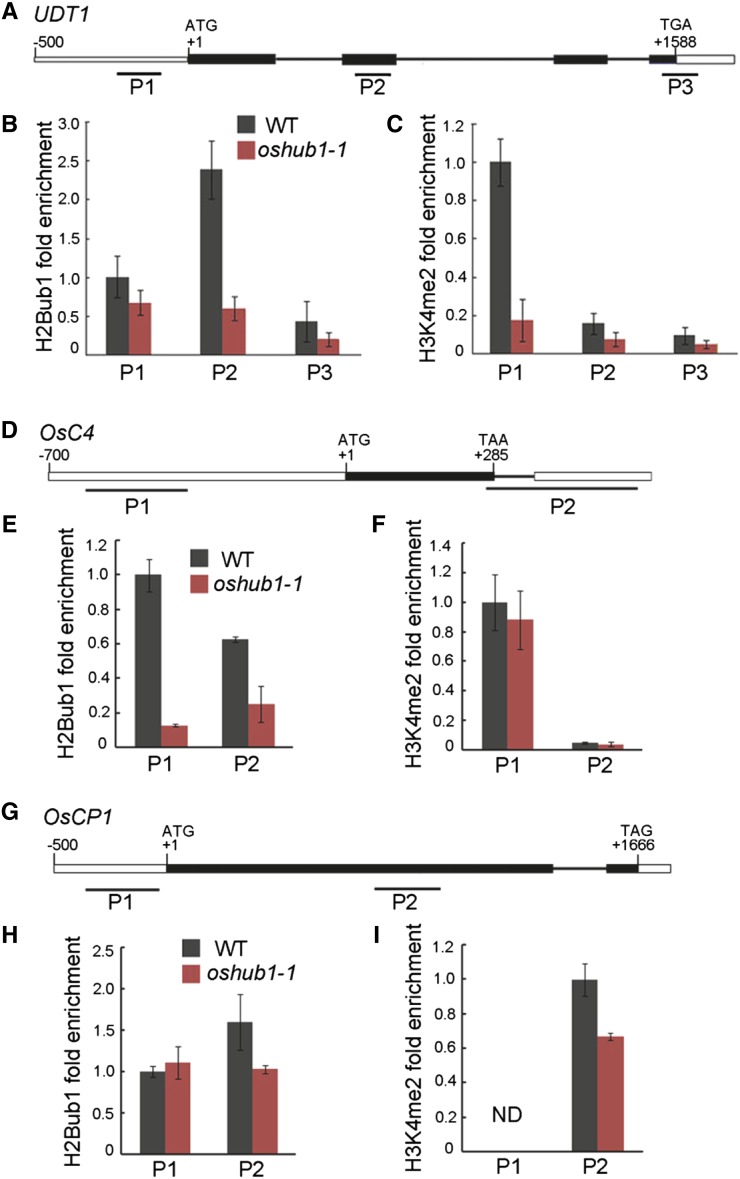
As the expression of genes known to be involved in late development of both the tapetum and pollen was down-regulated with the loss of H2Bub1, we next examined whether the abundance of H2Bub1 in their chromatin was altered in oshub1. A chromatin immunoprecipitation (ChIP) assay was performed with an anti-H2Bub1 antibody against chromatin derived from rice spikelets at stage 10 of development for both the wild type and oshub1-1 line. Comparative qRT-PCR was then used to check for H2Bub1 enrichment in the chromatin of down-regulated genes. Results revealed that in the oshub1-1 mutant, H2Bub1 is significantly reduced at the second exon of UDT1 (Fig. 9, A and B), at the promoter and gene body region of OsC4 (Fig. 9, D and E), and at the first exon of OsCP1 (Fig. 9, G and H). Thus, H2B monoubiquitination in the chromatin of these three genes is dependent on OsHUB1. However, either no or only minor modification of H2Bub1 was detected in the other down-regulated genes (data not shown). It is possible that OsC4, OsCP1, and UDT1 are the direct target genes of H2Bub1.
Figure 9.
Evaluation of H2B monoubiquitination and H3K4 dimethylation levels in UDT1, OsC4, and OsCP1 chromatin. A, The structure of UDT1. B and C, ChIP-qPCR analysis to determine the level of uH2B (B) and H3K4me2 (C) in UDT1 chromatin. D, The structure of OsC4. E and F, ChIP-qPCR analysis to determine the level of the uH2B (E) and H3K4me2 (F) in OsC4 chromatin. G, The structure of OsCP1. H and I, ChIP-qPCR analysis to determine the level of uH2B (H) and H3K4me2 (I) in OsCP1 chromatin. The start codon ATG and stop codon TGA (or TAA or TAG) are indicated. +1 indicates the translation initiation point. Boxes and lines represent exons and introns, respectively. P1 to P3 represent regions covered by the primers used to assess the level of uH2B and H3K4me2 by qPCR following ChIP. Data were normalized to the input chromatin, the P1 region in the wild type (WT) was set to be 1 in B, C, E, F, and H, and the P2 region in the wild type was set to be 1 in I. This experiment was repeated three times with independent samples. Data are means ± se. ND, Not detected.
H2Bub1 Is Required for H3K4me2 in the Chromatin of OsCP1 and UDT1
We showed that H2Bub1 is necessary for the enhancement of H3K4me2 at a genome-wide level (Fig. 4, C and D). To further investigate whether H2Bub1 has an effect on gene-specific H3K4me2 in the chromatin of H2Bub1 target genes, ChIP-quantitative PCR (qPCR) was performed using an anti-H3K4me2 antibody. Compared with the wild type, the level of H3K4me2 was found to be significantly decreased in the promoter region of UDT1 chromatin and the gene body region of OsCP1 chromatin, respectively, in the oshub1-1 mutant (Fig. 9, C and I). In the chromatin of OsC4, levels of H3K4me2 were not significantly different between wild-type and oshub1-1 plants (Fig. 9F). These results indicate that H2Bub1 is required for H3K4 dimethylation in the chromatin of OsCP1 and UDT1.
DISCUSSION
H2Bub1 Mediated by OsHUB1 and OsHUB2 Enhances H3K4me2 Deposition at a Genome-wide Level
H2Bub1 is a feature of eukaryotic chromatin being detected from yeast to humans and plants (Zhang, 2003; Shilatifard, 2006; Liu et al., 2007; Zhang et al., 2007; Weake and Workman, 2008). Many E3 ligases directed at H2Bub1 synthesis have been characterized, shedding light on the biological function of H2Bub1, including Bre1 (Hwang et al., 2003), dBre1 (Bray et al., 2005), RNF20/RNF40 (Kim et al., 2005; Zhu et al., 2005), and HUB1/HUB2 (Fleury et al., 2007; Liu et al., 2007). Here, we report the identification of two rice E3 ligases, OsHUB1/OsHUB2, and further prove that they are also involved in production of H2Bub1 in vivo. Western blotting using an H2Bub1 antibody revealed that the rice loss-of-function mutants are devoid of ubiquitinated H2B (Fig. 4, A and B), implying that rice OsHUB1 and OsHUB2 function as E3 ligases directed at histone H2B ubiquitination, similar to their Arabidopsis homologs. It is also likely that OsHUB1 and OsHUB2 form a heterotetrameric E3 ligase complex, as shown by their mutual interactions (Fig. 5). These observations suggest that H2Bub1 is a well-conserved cell biological event in higher eukaryotes.
H2Bub1 is associated with the transcribed regions of highly expressed genes (Minsky et al., 2008) and could regulate transcription indirectly through trans-tail histone crosstalk with H3K4me2/H3K4me3, H3K79me3, and H3K36me2/H3K36me3 (Shilatifard, 2006; Weake and Workman, 2008; Berr et al., 2011; Braun and Madhani, 2012). In yeast, ubiquitination of H2B (Lys-123) mediated by Rad6 and Bre1 is a prerequisite for methylation of H3K4 and H3K79 (Robzyk et al., 2000; Sun and Allis, 2002; Hwang et al., 2003; Wood et al., 2003). Interestingly, deubiquitination of H2B-K123ub is required for histone H3-K36 methylation (Henry et al., 2003). In human cells, overexpression or RNA interference-mediated down-regulation of the RNF20/RNF40 complex consistently gives rise to changed levels of monomethyl-H3K4, trimethyl-H3K4, and dimethyl-H3K79 (Zhu et al., 2005). It has also been reported that reduction of H2B ubiquitination in chickens results in substantial reductions of H3K4me3 and H3K79me2 levels, a minor reduction in H3K4me2, and slightly reduced H3K9acK14ac. However, in Arabidopsis, no obvious differences were found in the genome-wide levels of H3K4me3, H3K4me2, H3K4me1, H3K36me2, or H3K9me2 between wild-type and hub1, hub2, or ubiquitin carrier protein1 (ubc1) ubc2 mutant plants (Cao et al., 2008). In our study, absence of H2Bub1 in oshub1 and oshub2 mutants was only associated with substantially decreased H3K4me2 (Fig. 4, C and D), suggesting that H2Bub1 influences histone methylation variably between species.
Phylogenetic analysis of BREs and HUBs involved in H2Bub1 shows that BREs in yeast, Drosophila spp. and humans belong to one clade and BRE1/HUB homologs in plants fall into another (Supplemental Fig. S7), indicating that the mechanism of H2B monoubiquitination, mediated by BRE1 or HUB1 homologs, may be different between plants and other organisms. Interestingly, phylogenetic analysis based on either HUB1 or HUB2 sequences places dicotyledonous and monocotyledonous plants in separate groups. Again, this opens up the possibility that distinctive pathways operate to regulate H2Bub1 levels and thus plant growth and development, in monocot and dicot plant lineages. In support of this, H2Bub1 mediated by OsHUB1/OsHUB2 in rice can regulate H3K4me2 at a global level (Fig. 4, C and D), but H2Bub1 mediated by HUB1/HUB2 in Arabidopsis does not (Cao et al., 2008). H2Bub1 mediated by yeast BRE1 and human RNF20/RNF40 is a prerequisite for H3K4 and H3K79 methylation (Hwang et al., 2003; Kim et al., 2005), but this does not seem to be the case in plants. It is possible that the proteins downstream of BRE1/HUB bring about the diversity in histone methylation. H2B ubiquitylation has been shown to directly stimulate Dot1/hDot1L-mediated H3K79 methylation (Dover et al., 2002; McGinty et al., 2008) and SET1/hSET1 complex-mediated H3K4 di- and trimethylation (Shilatifard, 2006; Kim et al., 2009) in yeast and human cells. But nothing is known about the proteins mediating the crosslink between H2Bub1 and H3 Lys methylation in plant.
OsHUB1 and OsHUB2 Are Involved in Late Anther Development
Loss of function of HUB1/HUB2 enzymes that are involved in H2Bub1 has been reported to lead to many developmental defects in Arabidopsis, such as altered leaf morphology, seed dormancy, and flowering time (Fleury et al., 2007; Liu et al., 2007; Cao et al., 2008). Here, we analyzed rice plants harboring mutations in the genes encoding E3 ligases OsHUB1 and OsHUB2 to get an insight into their role in growth and development of a model monocotyledonous plant. oshub1-1, oshub1-2, and oshub2 mutant lines had a variety of phenotypes, including earlier heading and semidwarfism, features also observed in Arabidopsis hub mutants (Fleury et al., 2007; Liu et al., 2007; Cao et al., 2008), thus pointing to conserved physiological and biochemical functions of HUBs in plants.
We further reported that mutation of OsHUB1 and OsHUB2 resulted in striking defects in anther development and pollen formation, features not reported in Arabidopsis hub1 and hub2 mutants (Feng and Shen, 2014). Several lines of evidence presented here support the hypothesis that H2Bub1, mediated by OsHUB1/OsHUB2, plays a key role in late anther development. Firstly, loss-of-function mutations of OsHUBs results in altered stamen morphology, with anthers being shorter and having abnormal wall layers and significant levels of aborted pollen (Figs. 1–3). Secondly, RNA-seq data and gene expression profile analysis revealed that the expression of many anther development-related genes changes in oshub1 mutants. Furthermore, GO analysis of these genes indicate that they have a broad range of molecular and cellular functions, including oxidoreduction activity, DNA binding, and hydrolase activities, which are highly likely to significantly impact normal anther development if deregulated (Figs. 7 and 8; Table I). Thirdly, knockout of OsHUB1 and OsHUB2 led to loss of H2Bub1 in vivo (Fig. 4, A and B). Taken together, these data prove that H2Bub1 mediated by OsHUB1 and OsHUB2 is an essential histone modification involved in anther and pollen development through regulating gene expression profiles.
In addition, many studies have shown that chromatin modifications are involved in plant anther and pollen development (He et al., 2011; Thorstensen et al., 2011). Histone deacetylation mediated by Histone Deacetylase19 in Arabidopsis was found to be involved in plant reproductive development. Loss-of-function hda19 plants displayed abnormal flowers, reduced male and female fertility, and smaller siliques (Tian and Chen, 2001; Tian et al., 2005; Zhou et al., 2005). Histone H3 methyltransferase ASH1 HOMOLOG2 is required for ovule and anther development in Arabidopsis by modifying H3K36 trimethylation (Grini et al., 2009). The increase in bonsai methylation1 mutants induced a variety of developmental phenotypes such including abnormal pollen, which depend on methylation of histone H3 at Lys-9 (Saze et al., 2008). Arabidopsis SET DOMAIN GROUP2 mediates global H3K4me3 deposition and is responsible for male and/or female gametophyte development (Berr et al., 2010). Here, we report that H2Bub1 has a marked effect on anther development in rice, indicating that a complicated histone modification regulatory network is involved in this important process. However, our results did not indicate that H2Bub1 mediated by OsHUB1 and OsHUB2 is required for H3K36 and H3K9 methylation (Fig. 4). So far, little is known about the crosstalk between histone modifications associated with anther and pollen development.
Our study indicates that H2Bub1 in rice has both conserved and species-specific functions compared with Arabidopsis. Abnormal anther development is a feature of oshub1 and oshub2 mutant lines in rice, but this is not the case for Arabidopsis hub1 and hub2 mutants (Feng and Shen, 2014), which may be due to some fundamental differences in the regulation of male reproductive development between rice and Arabidopsis. For example, loss-of-function mutations of the rice H3K9 demethylase JMJ706 led to severe abnormalities in spikelet development affecting floral morphology and organ number (Sun and Zhou, 2008), but by contrast, mutants of its Arabidopsis homologs (EARLY FLOWERING6) result in early flowering, with no apparent infertility phenotype (Noh et al., 2004).
H2Bub1 Regulates Transcription of Tapetal Development-Related Genes Coordinately with H3K4me2
H2Bub1 can modulate chromatin structure and DNA accessibility to regulate diverse DNA-dependent processes such as gene transcription (Zhang, 2003; Shilatifard, 2006; Pinder et al., 2013). RNA interference against human RNF20/RNF40 leads to reduced H2Bub1 and subsequent repression of HOX gene expression (Zhu et al., 2005). The absence of H2Bub1 in Arabidopsis hub1 mutants also results in reduced expression of seed dormancy-related genes (Liu et al., 2007). A genome-wide analysis revealed that in Arabidopsis, H2Bub1 is associated with active genes distributed throughout the genome (Roudier et al., 2011). In this study, we showed that reduced levels of H2Bub1 in the rice oshub1 mutant causes widespread changes in gene expression, with nearly 2,000 genes identified as being down-regulated, many of which are known to be involved in anther and pollen development (Table I). CYP703A3 (Aya et al., 2009), CYP704B2 (Li et al., 2010), OsC4 (Tsuchiya et al., 1994), OsC6 (Zhang et al., 2010), and DPW (Shi et al., 2011) are involved in late stages of tapetal development, including lipid metabolism and transport (Fu et al., 2014). OsCP1 (Lee et al., 2004) and AP37 (Li et al., 2011; Niu et al., 2013) promote tapetal programmed cell death and pollen formation. In addition, PERSISTENT TAPETAL CELL1 (PTC1; Li et al., 2011), UDT1 (Jung et al., 2005), and MADS3 (Hu et al., 2011) regulate tapetal programmed cell death during anther development. The development, differentiation, and degradation of tapetal cells is a highly regulated process that works in coordination with microspore development. In the study, we found that enlarged tapetum occurs at stage 10 and the microspores also appeared abnormal compared with the wild type (Figs. 2 and 3). We speculate that down-regulation of tapetum-related genes in oshub1 could be associated with the reduced fertility phenotype. The results indicate that the defect in H2Bub1 leads to the repression of genes involved in late tapetum development delaying degeneration and thus affecting pollen development.
In our study, we confirmed that three genes having reduced expression in oshub1 (UDT1, OsCP1, and OsC4) are direct targets of H2Bub1 (Fig. 9). Rice UDT1 is required for the differentiation of secondary parietal cells into mature tapetal cells and plays an essential role in maintaining the tapetal developmental program (Jung et al., 2005). During the process of late anther development, it is possible that reduced UDT1 expression in the oshub1 mutant could lead to defects in tapetal development. OsCP1 is a Cys protease gene that, when mutated, causes defects in pollen development (Lee et al., 2004). Thus, a reduction OsCP1 transcript levels in oshub1 may also affect pollen formation. OsC4, a lipid transfer protein, and the related lipid transfer protein OsC6 were both down-regulated in oshub1. Both are proposed to play a role in the development of lipidic orbicules and pollen exine through transporting lipophilic materials from tapetal cytoplasm to the locule for pollen wall development, and thus their concomitant down-regulation likely explains the pollen degeneration phenotype of oshub1 (Tsuchiya et al., 1994; Zhang et al., 2010; Ji et al., 2013). Collectively, H2Bub1 mediated by OsHUB1/OsHUB2 regulates late tapetum development and the transport of lipophilic materials from tapetum to pollen through directly regulating the expression of UDT1, OsCP1, and OsC4.
As discussed previously, H2Bub1 mediated by OsHUB1 and OsHUB2 can enhance H3K4me2 at a genome-wide level (Fig. 4). H3K4me2 is not only enriched in genomic regions surrounding transcriptional start sites (Zhang et al., 2009), but also found within gene body regions, which is proposed to permit refinement of tissue-specific gene expression (Pekowska et al., 2010). In our study, OsCP1 was found to have high levels of H3K4me2 within the gene body region, and UDT1 had high H3K4me2 in the promoter region (Fig. 9). UDT1 is preferentially expressed in anthers at early developmental stages (Jung et al., 2005). OsCP1 is mainly expressed in the anther, especially in the locule region, including the pollen and tapetum (Lee et al., 2004). Therefore, we speculate that H2Bub1, mediated by OsHUB1 and OsHUB2, is directly involved in the transcriptional regulation of anther and pollen development-related genes coordinately with H3K4me2 as a mechanism refining tissue specificity expression patterns.
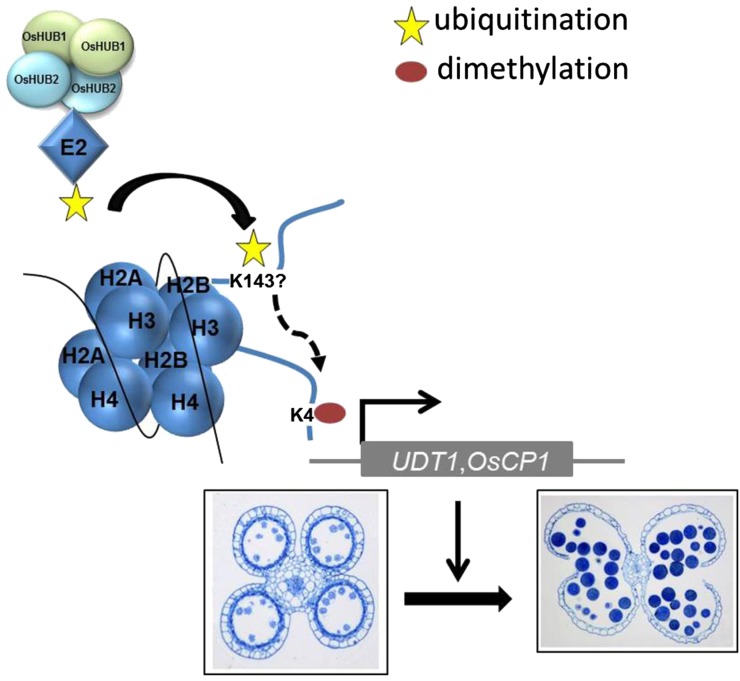
In summary, this work has uncovered a previously unknown role of histone H2B monoubiquitination mediated by OsHUB1 and OsHUB2 in the regulation of male reproductive development in plants, as depicted in a working model (Fig. 10). In this model, OsHUB1 and OsHUB2 may form a heterotetrameric complex to function as an E3 ligase in the process of histone H2B monoubiquitination. H2Bub1, once produced, can then regulate the expression of many genes during anther development, such as CP1, OsC4, and UDT1, and cooperates with a mechanism for H3K4me2 that refines control of gene transcription. Thus, H2Bub1, mediated by OsHUB1 and OsHUB2 is essential for the reproductive development in rice. This finding may provide an epigenetic molecular approach for the rational manipulation of pollen fertility to improve crop yields.
Figure 10.
A proposed model for H2B monoubiquitination mediated by OsHUB1 and OsHUB2 during anther development in rice. In this model, OsHUB1 and OsHUB2 form a heterotetramer and recruit E2s to UDT1 and OsCP1 chromatin, leading to transfer of an ubiquitin molecule to H2B possibly at Lys-143. H2Bub1 formation enhances H3K4me2, together promoting transcription initiation. The fine-tuning of gene expression of UDT1/OsCP1 is important for late anther development of rice.
MATERIALS AND METHODS
Plant Materials and Growth Condition
oshub1-1 (PFG_1B-08922.L), oshub1-2 (PFG-1B_24518.L), and oshub2 (PFG_2A-00426.R) T-DNA insertion mutants produced in Dongjin and Hwayoung background, respectively, were obtained from the Rice Functional Genomics Express Database (Jeong et al., 2002). All rice (Oryza sativa) plants for phenotype characterization were grown in Beijing from June to October.
qRT-PCR
For tissue expression profile, total RNA was isolated using Trizol reagent (Invitrogen) as described by the supplier from rice tissues: young root, young leaf, mature leaf, mature sheath, node, culm, panicle, and panicles at different stages. For RNA-seq data verification, total RNA was isolated from anthers at meiotic stage, microspore stage, and vacuolated stage. The stages of developing anthers were classified according to spikelet length in the following categories (Feng et al., 2001; Li et al., 2006; Zhang et al., 2011): meiosis stage (stages 6–8) anthers within 1- to 3-mm spikelets, young microspore stage (stage 9) anthers within 3- to 5-mm spikelets, vacuolated pollen stage (stage 10) anthers within 5- to 7-mm spikelets, and mature pollen stage (stages 12–13) anthers within 7- to 8-mm spikelets. After treatment with DNase I (Takara, 2270A), 1.0 µg of RNA was used to synthesize the oligo(dT)-primed first strand. All the primers for qRT-PCR are listed in Supplemental Table S1. qRT-PCR analysis was performed using SYBR Green Real-Time PCR Master Mix (TOYOBO, QPK-201) on a Eppendorf Realplex detection system.
Histochemical GUS Assays
The 2-kb promoter of OsHUB1 (2,000 bp upstream of ATG) was fused with GUS and transformed into wild-type plants, and T2 lines were subjected to histochemical analysis. Freshly collected samples from transgenic plants expressing the promoter-GUS fusion were put in the staining solution under vacuum conditions for 15 min, followed by incubation at 37°C overnight. After staining, samples were rinsed in 70% (v/v) ethanol for 1 h before a photo was taken with a Nikon 80i digital camera.
Yeast Two Hybrid
The yeast (Saccharomyces cerevisiae) two-hybrid assay was performed using the Gal4 system. The coding regions of the genes tested were cloned into two vectors (pAS2 and pACT2) using Gateway technology (Invitrogen) to generate bait and prey vector. The two constructs were cotransformed into yeast strain AH109. Transformation, yeast growth, and β-galactosidase assays were performed as described in the Clontech Yeast Protocols Handbook.
Bimolecular Fluorescence Complementation
The bimolecular fluorescence complementation vectors pSPYNE173 and pSPYCE(M) were used (Waadt et al., 2008). Agrobacterium tumefaciens-mediated transient expression in Nicotiana benthamiana was performed as described by Waadt et al. (2008). The fluorescence was detected by a confocal microscope (Olympus FV1000MPE). The yellow fluorescent protein fluorescence was excited by a 514-nm laser and captured at 523 to 600 nm.
Evaluation of Pollen and Spikelet Fertility
To estimate the level of pollen viability, three spikelets were collected from each plant shortly before anthesis and fixed in 70% (v/v) ethanol. Three anthers were sampled at random from each spikelet. One hundred to 200 pollen grains were stained with 1% (w/v) I2-KI solution and observed with a Nikon 80i microscope. Spikelet fertility was calculated as the proportion of fertile spikelets to all spikelets of 10 plants at mature stage.
Scanning Electron Microscopy Analysis
Rice anthers were fixed in 45% (v/v) ethanol, 5% (v/v) acetic acid, and 1.9% (v/v) formaldehyde for 24 h. Samples were critical-point dried, sputter coated with gold in an E-100 ion sputter, and observed with a scanning electron microscope (Hitachi S-4800).
Semithin Transverse Section
Spikelets of different developmental stages were collected and fixed in a solution containing 45% (v/v) ethanol, 5% (v/v) acetic acid, and 1.9% (v/v) formaldehyde for 24 h at room temperature and dehydrated through an ethanol series. The samples were embedded into Spurr’s resin (SPI-CHEM) and polymerized at 60°C for 24 h. Transverse sections of 1.0 µm were cut with an ultramicrotome (Leica Ultracut R) and stained with 0.1% (w/v) toluidine blue O (Merck). Images were captured using a Nikon 80i microscope, merged, and enhanced using Photoshop CS (Adobe).
DAPI Staining
Spikelets at postmeiotic anther stage were fixed with ethanol:acetic acid (3:1), washed with 70% (v/v) ethanol, and stored at 4°C until observation. Then, the microspores were squeezed out to the slide and stained with 5 µL of DAPI stain solution (1 µg mL–1 DAPI in 10 mm phosphate-buffered saline buffer). After being covered with a cover glass, the nuclei were examined under the fluorescence microscope (Leica, DM5000B).
H2B Monoubiquitination, Histone Methylation, and Acetylation Assay
Histone was isolated from 2-week-old rice seedlings of the wild type, oshub1-1, oshub1-2, and oshub2, which were grown in a light chamber under a 14-h-light/10-h-dark cycle at 28°C. Histone extraction was performed as described (Charron et al., 2009). The proteins were then separated by 12% (w/v) SDS-PAGE and transferred to polyvinylidene fluoride membranes probed with anti-H2Bub1 (anti-H2B as loading control), anti-H3K4me2, anti-H3K4me3, anti-H3K9me2, anti-H3K36me2, anti-H3K18ac, anti-H3K23ac, and anti-H4K12ac (anti-H3 as loading control). The primary antibodies were purchased from Millipore Biotechnology, except for anti-H2B, which was purchased from Upstate Biotechnology.
RNA-seq Experiment and Data Analysis
Total RNA was prepared from wild-type and oshub1-1 anthers at stage 10. The RNA samples from two independent materials were sequenced by Beijing Genomics Institute, as described previously (Wang et al., 2013). The raw reads were filtered for adaptors and low-quality reads. Filtered reads were then mapped to the reference rice genome sequence using SOAPaligner/SOAP2 (Li et al., 2009). No more than two mismatches were allowed in the alignment. We applied the NOISeq method (Tarazona et al., 2011) to screen for DEGs between oshub1-1 and the wild type. The NOISeq method maintains good true positive and false positive rates when increasing sequencing depth. Also, NOISeq models the noise distribution from the actual data, so it can better adapt to the size of the data set, and is more effective in controlling the FDR. Genes satisfying the criteria fold change ≥ 2 and FDR ≤ 0.001 were considered as DEGs. GO annotation was obtained by BLAST (with parameters -p blastx -e 1e-5 -m 7) of coding sequences against the National Center for Biotechnology Information nonredundant database and then to the terms by the use of Blast2GO with default parameters. We used a hypergeometric test to find significantly enriched GO terms in DEGs comparing to the genome background. GO terms with P < 0.01 were presented as –log10 (P value) in Figure 7.
ChIP-qPCR
ChIP procedure of histone/DNA was performed as described by Bowler et al. (2004) using spikelets at stage 10. Anti-H2Bub1 and anti-H3K4me2 antibodies and salmon sperm DNA/protein A agarose were purchased from Millipore. The untreated sonicated chromatin was reversely cross linked and used as the input DNA control. Immunoprecipitation with antibody-specific immune antiserum and without any serum was performed as the reference. The amounts of immunoprecipitated DNA were assayed by qPCR using the same conditions of qRT-PCR.
Observation of Pollen Grains on the Stigma
The examination of pollen grains on the stigma was performed as described previously (Zhou et al., 2011). Briefly, the spikelets were collected 2 h after anthesis and fixed in formaldehyde-acetic acid solution (45% [v/v] ethanol, 5% [v/v] acetic acid, and 1.9% [v/v] formaldehyde) for 24 h at room temperature. The spikelets were then processed through an ethanol series and washed with distilled water. The spikelets were incubated in 10 m sodium hydroxide for 8 min at 56°C and then washed with distilled water and stained with 0.1% (w/v) aniline blue (Fluka). Finally, the samples were observed under a fluorescence microscope (Leica DM5000B).
Pollen Meiotic Chromosome Observation
The young spikelets (1.0–3.0 mm in length) at meiosis stage were fixed with ethanol:glacial acetic acid (3:1) for 24 h at room temperature, washed with 95% (v/v) ethanol twice, and stored in 70% (v/v) ethanol at 4°C until observation. The anthers were placed in a drop of improved karbol fuchsin, dissected and crushed with a needle, and examined under a Nikon 80i microscope. Images were enhanced with Photoshop CS.
Phylogenetic Analysis
The full amino acid sequences of OsHUB1 and OsHUB2 and the related proteins identified via BLAST search were aligned with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). A phylogenetic tree was constructed with MEGA software (version 4.0) using the maximum-likelihood method with the following parameters: Poisson model, pairwise deletion and, 1,000-replicates bootstrap (Tamura et al., 2007).
Sequence data from this article can be found in the rice database under the following accession numbers: OsHUB1 (Os04g46450), OsHUB2 (Os10g41590), UDT1 (Os07g36460), OsC4 (Os08g43290), OsC6 (Os11g37280), OsCP1 (Os04g57490), AP37 (Os04g37570), DPW (Os03g07140), CYP704B2 (Os03g07250), CYP703A3 (Os08g03682), MADS3 (Os01g10504), EAT1 (Os04g51070), TIP2 (Os01g18870), PTC1 (Os09g27620), Postmeiotic Deficient Anther1 (Os06g40550), PAIR1 (Os03g01590), PAIR2 (Os09g0506800), PAIR3 (Os10g26560), MEL1 (Os03g0800200), UGP1 (DQ395328), OsRAD21-4 (Os05g50410), GAMYB (Os01g59660), OsRAFTIN1 (Os08g38810), MTR1 (Os02g28970), and ACTIN1 (Os03g50890).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Scheme of OsHUB1 and OsHUB2 and identification of their respective T-DNA insertion mutations.
Supplemental Figure S2. Phenotypic comparison of wild-type and mutant plants at harvest stage.
Supplemental Figure S3. Analysis of meiosis in the anthers of wild-type and oshub1 plant lines.
Supplemental Figure S4. Subcellular localization of OsHUB1.
Supplemental Figure S5. Biological process by GO analysis of genes that were down-regulated and up-regulated at least 2-fold in oshub1-1.
Supplemental Figure S6. qRT-PCR analysis of the expression of rice meiotic division-related genes in the wild type and oshub1 mutants.
Supplemental Figure S7. Phylogenetic tree of OsHUB1 and its homologs.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Gynheung An (Postech Biotech Center) for providing oshub1and oshub2 seeds, Jingquan Li and Zengjuan Fu (Institute of Botany, Chinese Academy of Sciences) for helping in microscope observation, and Wim J.J. Soppe (Max-Planck Institute for Plant Breeding Research) for discussion and critical reading of the article.
Glossary
- T-DNA
transfer DNA
- DAPI
4,6-diamino-phenylindole
- qRT
quantitative reverse transcription
- RNA-seq
mRNA-sequencing
- FDR
false discovery rate
- GO
Gene Ontology
- DEG
differentially expressed gene
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31301052 and 31171164) and the National Basic Research Program of China (973 Program; grant no. 2014CB943400).
Articles can be viewed without a subscription.
References
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Ménard R, Meyer D, Fuchs J, Dong A, Shen WH (2010) Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22: 3232–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Shafiq S, Shen WH (2011) Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta 1809: 567–576 [DOI] [PubMed] [Google Scholar]
- Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F (2012) Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet 8: e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Braun S, Madhani HD (2012) Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep 13: 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Musisi H, Bienz M (2005) Bre1 is required for Notch signaling and histone modification. Dev Cell 8: 279–286 [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron JB, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Feng J, Lu Y, Liu X, Xu X (2001) Pollen development and its stages in rice (Oryza sativa L.). Chinese J. Rice Sci 15: 21–28 [Google Scholar]
- Feng J, Shen WH (2014) Dynamic regulation and function of histone monoubiquitination in plants. Front Plant Sci 5: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al. (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jørstad TS, Wilson ZA, Aalen RB (2009) The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One 4: e7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Jiang D, Wang Y, Bachmair A, He Y (2009) Repression of the floral transition via histone H2B monoubiquitination. Plant J 57: 522–533 [DOI] [PubMed] [Google Scholar]
- He JH, Shahid MQ, Li YJ, Guo HB, Cheng XA, Liu XD, Lu YG (2011) Allelic interaction of F1 pollen sterility loci and abnormal chromosome behaviour caused pollen sterility in intersubspecific autotetraploid rice hybrids. J Exp Bot 62: 4433–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M (2012) Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J 72: 249–260 [DOI] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Pei BL, Zhang LF, Li YZ (2014) Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in Arabidopsis. Plant Physiol 164: 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant 6: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG (2009) RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20: 759–770 [DOI] [PubMed] [Google Scholar]
- Lee S, Jung KH, An G, Chung YY (2004) Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol Biol 54: 755–765 [DOI] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJ (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD (2010) The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X (2010) RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 18: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Luo MZ, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A (2008) UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R, Verdier G, Ors M, Erhardt M, Beisson F, Shen WH (2014) Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol 55: 455–466 [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N (2004) The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Ferrier P, Spicuglia S (2010) A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res 20: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder JB, Attwood KM, Dellaire G (2013) Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front Genet 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, Hintermair C, Eick D, Kremmer E, Simons M, et al. (2011) Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res 71: 5739–5753 [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, et al. (1996) Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86: 799–810 [DOI] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T (2008) Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK (2007) Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105: 13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tan H, Liang W, Hu J, Zhang D (2012) MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev Cell 22: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21: 2213–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen T, Grini PE, Aalen RB (2011) SET domain proteins in plant development. Biochim Biophys Acta 1809: 407–420 [DOI] [PubMed] [Google Scholar]
- Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 98: 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ (2005) Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Ejiri S, Hinata K (1994) Molecular characterization of rice genes specifically expressed in the anther tapetum. Plant Mol Biol 26: 1737–1746 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc Natl Acad Sci USA 100: 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, et al. (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, Li Y, Ding M, Zhang C, Deng X, et al. (2013) Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH (2009) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 57: 279–288 [DOI] [PubMed] [Google Scholar]
- Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56: 979–994 [DOI] [PubMed] [Google Scholar]
- Yuan W, Li X, Chang Y, Wen R, Chen G, Zhang Q, Wu C (2009) Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J 59: 303–315 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK (2007) Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One 2: e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tao J, Wang S, Chong K, Wang T (2006) The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60: 533–554 [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2003) Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev 17: 2733–2740 [DOI] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Wang Y, Li W, Zhao Z, Ren Y, Wang Y, Gu S, Lin Q, Wang D, Jiang L, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23: 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 20: 601–611 [DOI] [PubMed] [Google Scholar]
- Zou B, Yang DL, Shi Z, Dong H, Hua J (2014) Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol 165: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.