Two different variants of polycomb group proteins, epigenetic regulators, coordinate the acquisition of flowering competence during the juvenile-to-adult phase transition in Arabidopsis through the regulation of specific miRNA levels.
Abstract
Polycomb group (PcG) proteins play important roles in regulating developmental phase transitions in plants; however, little is known about the role of the PcG machinery in regulating the transition from juvenile to adult phase. Here, we show that Arabidopsis (Arabidopsis thaliana) B lymphoma Moloney murine leukemia virus insertion region1 homolog (BMI1) POLYCOMB REPRESSIVE COMPLEX1 (PRC1) components participate in the repression of microRNA156 (miR156). Loss of AtBMI1 function leads to the up-regulation of the primary transcript of MIR156A and MIR156C at the time the levels of miR156 should decline, resulting in an extended juvenile phase and delayed flowering. Conversely, the PRC1 component EMBRYONIC FLOWER (EMF1) participates in the regulation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE and MIR172 genes. Accordingly, plants impaired in EMF1 function displayed misexpression of these genes early in development, which contributes to a CONSTANS-independent up-regulation of FLOWERING LOCUS T (FT) leading to the earliest flowering phenotype described in Arabidopsis. Our findings show how the different regulatory roles of two functional PRC1 variants coordinate the acquisition of flowering competence and help to reach the threshold of FT necessary to flower. Furthermore, we show how two central regulatory mechanisms, such as PcG and microRNA, assemble to achieve a developmental outcome.
Polycomb group (PcG) proteins are conserved epigenetic regulators that mediate gene repression through the incorporation of histone-modifying marks (Calonje, 2014). As far as it is known, PcG proteins associate in two multiprotein complexes in Arabidopsis (Arabidopsis thaliana): POLYCOMB REPRESSIVE COMPLEX1 (PRC1) and PRC2. The combined activity of the two complexes is required for stable repression of the target genes.
The major function of PRC2 is to perform histone H3 lysine-27 trimethylation (H3K27me3) through the methyltransferase activity of CURLY LEAF (CLF) and SWINGER (SWN) during sporophyte development or of MEDEA in the endosperm (Chanvivattana et al., 2004). Other PRC2 components are the Drosophila melanogaster suppressor of zeste12 homologs VERNALIZATION2 (VRN2), EMBRYONIC FLOWER2 (EMF2), and FERTILIZATION-INDEPENDENT SEED2, which confer specificity to the resulting PRC2s even though they have some overlapping functions (Chanvivattana et al., 2004), and finally MULTICOPY SUPPRESSOR OF INHIBITORY REGULATOR OF THE RAS-CYCLIC AMP PATHWAY and FERTILIZATION-INDEPENDENT ENDOSPERM, which are common subunits for the different PRC2s (Derkacheva and Hennig, 2014). On the other hand, the identity of Arabidopsis PRC1 is not defined yet. PRC1-mediated function can be histone 2A monoubiquitination (H2Aub) dependent, through the E3 ubiquitin ligase activity of the PRC1 RING finger proteins Arabidopsis B lymphoma Moloney murine leukemia virus insertion region1 homolog 1A (AtBMI1A)/B/C and AtRING1A/B, or H2Aub independent, which requires the activity of the PRC1 component EMF1 (Bratzel et al., 2010, 2012; Yang et al., 2013a; Calonje, 2014). These different PRC1 activities suggest the existence of PRC1 functional variants that may target different subsets of genes (Merini and Calonje, 2015). Another protein used to be considered as a putative PRC1 component is LIKE-HETEROCHROMATIN PROTEIN1 (LHP1), which has the ability to bind H3K27me3 marks (Turck et al., 2007); however, it was recently shown that LHP1 copurifies with PRC2, changing the notion of LHP1 as a PRC1 component (Derkacheva et al., 2013).
From a mechanistic point of view, recent data indicated that the binding and activity of PRC1 are required for H3K27me3 marking at some target genes, which challenges the classical hierarchical model for the recruitment of PcG complexes (Yang et al., 2013a; Calonje, 2014; Merini and Calonje, 2015). Whether this happens at all PcG targets is not yet known. In any case, both PRC1 and PRC2 play important roles in regulating developmental phase transitions in Arabidopsis. For instance, the combined activity of AtBMI1 and PRC2 is crucial for the transition from embryonic to vegetative development (Bratzel et al., 2010; Bouyer et al., 2011; Yang et al., 2013a); EMF1 and PRC2 regulate the transition from vegetative to reproductive development (Sung et al., 1992; Kinoshita et al., 2001; Schubert et al., 2006); and AtRING1A was recently shown to be involved in the regulation of several flowering repressors, suggesting its participation in the transition to flowering (Shen et al., 2014). However, thus far, little is known about the implication of PcG proteins in another important developmental change, the transition from juvenile to adult phase that marks the acquisition of reproductive competence.
Following germination, plants pass through a phase of vegetative growth that can be further divided into a juvenile and an adult vegetative phase. During the juvenile-to-adult phase transition, plants acquire competence to flower as well as undergo changes in multiple traits, such as leaf size and shape, internode length, and trichome distribution (Huijser and Schmid, 2011; Poethig, 2013). Although PcG proteins may have a role in regulating this developmental transition, the severity of the phenotype in some PcG mutants or the lack of phenotype in others has concealed their possible implication. Conversely, two microRNAs (miRNAs), miR156 and miR172, and their targets have been identified as key components of the mechanisms that underlie juvenile-to-adult phase changes. The miR156 targets transcripts of a subset of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors that have been shown to promote the transition from juvenile to adult and to flowering (Wu and Poethig, 2006; Schwarz et al., 2008). By contrast, miR172 targets APETALA2 (AP2)-like factors that have been shown to repress both the transition to flowering and flower development (Aukerman and Sakai, 2003; Schmid et al., 2003; Jung et al., 2007; Mathieu et al., 2009). The expression of these miRNAs is temporally regulated by age; thus, as the plant ages, miR156 levels decrease, resulting in an increase in SPL expression. In the shoot apical meristem (SAM), the SPL proteins activate the floral pathway integrators SUPPRESSOR OF CONSTANS1 (SOC1) and AGAMOUS-LIKE24 (AGL24) and the floral meristem identity genes FRUITFULL, LEAFY (LFY), and AP1; and in leaves, the SPLs activate miR172 expression that in turn down-regulates the AP2-like floral repressors, which inhibit the floral integrator FLOWERING LOCUS T (FT; Wang, 2014). The so-called age pathway is proposed to prevent flowering during the juvenile phase and ensure plant flowering even in the absence of exogenous inductive cues.
FT, in addition to being regulated by the age pathway, is strongly controlled by photoperiod; in fact, the level of FT expression at the end of long days plays a primary role in determining when Arabidopsis flowers (Turck et al., 2008; Wigge, 2011). The circadian clock sets a high CONSTANS (CO) mRNA expression in the late afternoon in long days, which coincides with light exposure, resulting in CO protein accumulation as light stabilizes the CO protein. The vasculature-expressed CO protein promotes FT expression activation in the phloem companion cells, specifically at the end of long days (Imaizumi and Kay, 2006; Turck et al., 2008). During the night, CO is rapidly degraded by the proteasome and FT expression is repressed (Valverde et al., 2004). Upon its production at dusk, the FT protein moves from phloem to the SAM, where it interacts with the locally transcribed FLOWERING LOCUS D (FD) transcription factor to activate floral integrators like SOC1 and AGL24 to induce flowering (Amasino, 2010; Matsoukas et al., 2012). Accordingly, genetic studies have placed the age pathway parallel with the photoperiodic pathway (Wang, 2014), both being required to determine the threshold of FT necessary for flowering competence.
Several direct regulators of miR172-encoding genes have been identified, including the MADS box factor SHORT VEGETATIVE PHASE, which downregulates the levels of miR172 (Cho et al., 2012), GIGANTEA, which mediates the photoperiod activation of miR172 (Jung et al., 2007), and SPL9, which leads to an accumulation of miR172 (Wu et al., 2009). On the other hand, recent evidence indicates that the seed maturation gene FUSCA3 (FUS3) contributes to the direct expression of the primary transcripts of MIR156A and MIR156C (pri-MIR156A and pri-MIR156C) in the developing seed and that this expression is important after germination to delay the juvenile-to-adult vegetative phase transition (Wang and Perry, 2013). However, upstream effectors mediating the age-dependent decline in miR156 levels are largely unknown. Interestingly, several recent studies showed a correlation between plant nutritional status and miR156 levels. The accumulation of metabolically active sugars, such as Suc and Glc, acts as a signal to selectively repress the expression of the miR156A and miR156C genes (Wahl et al., 2013; Yang et al., 2013b; Yu et al., 2013), but the molecular mechanism by which this repression take place and is maintained is not yet understood.
In this work, we show that loss of function of the PRC1 component AtBMI1 leads to the up-regulation of pri-MIR156A/C at the time the levels of miR156 should decline, resulting in an extended juvenile phase and delayed flowering. We found that atbmi1a/b mutants display reduced levels of H3K27me3 marks at the transcriptional start site (TSS) of these genes, suggesting the participation of the PcG machinery in regulating miR156 expression. According to our results, AtBMI1-mediated repression of pri-MIR156A/C allows the age-dependent expression of FT and the development of adult traits. Interestingly, the PRC1 component EMF1 does not regulate pri-MIR156A/C expression; instead, EMF1 participates in the regulation of miR172. Our findings show how the combined regulatory roles of two functional PRC1 variants are crucial to coordinate the acquisition of flowering competence.
RESULTS
Loss of EMF1 Function Leads to CO-Independent FT Up-Regulation, But Not the Loss of AtBMI1 Function
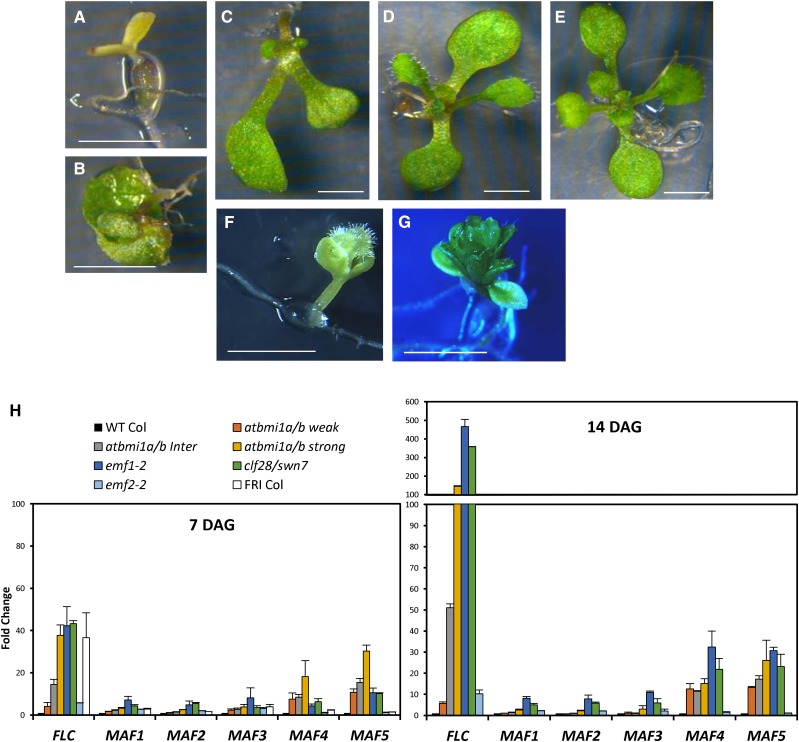
Mutant plants severely compromised in AtBMI1 activity do not undergo the transition from embryonic to vegetative development, remaining in an embryonic stage similar to that of mutants impaired in PRC2 function, like clf/swn (Chanvivattana et al., 2004). Unfortunately, the severity of atbmi1 strong mutant phenotypes or the lack of phenotype in atbmi1 single mutants has masked the possible implication of the AtBMI1 proteins in regulating other developmental transitions. To explore other possible roles of AtBMI1 proteins, we took advantage of the different penetrance of the atbmi1b allele (Bratzel et al., 2010) that causes a gradient of phenotypes in atbmi1a/b mutants. Early in development, atbmi1a/b phenotypes ranged from seedlings arrested in an embryo-like stage (strong mutants; Fig. 1A) and seedlings with twisted or embraced green cotyledons (intermediate mutants; Fig. 1, B and C) to seedlings with a wild-type-like phenotype (weak mutants; Fig. 1D). Later on, strong and intermediate atbmi1a/b mutants remained in an embryonic stage, in which they generated embryo-like structures, while atbmi1a/b weak mutants were able to flower and generate viable seeds (Bratzel et al., 2010), allowing us to analyze other developmental processes.
Figure 1.
FLC, MAF4, and MAF5 expression is significantly altered in atbmi1 mutants. A to G, Phenotypes of strong (A), intermediate (B and C), and weak (D) atbmi1a/b, wild-type (WT) Columbia (Col; E), emf1-2 (F), and emf2-2 (G) at 10 d after germination (DAG). Bars = 2 mm. H, Expression levels of FLC, MAF1, MAF2, MAF3, MAF4, and MAF5 in 7- and 14-d-old plants at ZT1 under LD conditions. The expression levels of these genes were also analyzed in 7-d-old FRI-Col seedlings. Quantifications were normalized to ACTIN2 (ACT2). The y axis indicates fold change compared with wild-type Col.
Interestingly, atbmi1a/b weak mutants did not show an early-flowering phenotype as other PcG mutants like emf1 or emf2 (Sung et al., 1992; Kinoshita et al., 2001). It is noteworthy that emf1 and emf2 display the earliest flowering phenotypes described in Arabidopsis. emf1-2 strong mutants produce a carpel right after germination without developing any leaf (Fig. 1F), and the emf1-1 mutant produced a small inflorescence after developing a few sesile leaves, which is the same phenotype displayed by emf2-2 (Fig. 1G).
To understand the differences in the flowering phenotypes among these PcG mutants, we examined the expression levels of several flowering time master regulators in atbmi1a/b, emf1-2, emf2-2, clf-28/swn-7, and wild-type Col plants. For this purpose, 7- and 14-d-old seedlings growing under long-day (LD) conditions were collected at Zeitgeber time 1 (ZT1; i.e. 1 h after light on; Fig. 1H). We included in the analysis 7-d-old FRIGIDA (FRI)-Col plants in which a functional FRI allele was introgressed into Col. FRI up-regulates the flowering repressor FLOWERING LOCUS C (FLC), which represses the expression of the flowering promoter gene FT, leading to late flowering (Searle et al., 2006).
We found that FLC was strongly up-regulated in the atbmi1a/b intermediate and strong phenotypes, emf1-2, clf-28/swn-7, and FRI-Col, compared with wild-type Col. The expression of FLC was also increased in atbmi1a/b weak and emf2-2 mutants, although to a lesser extent (Fig. 1H). When we measured the expression levels of the FLC-related flowering genes MADS AFFECTING FLOWERING1 (MAF1) to MAF5 (Scortecci et al., 2001; Ratcliffe et al., 2003), we found that the levels of MAF1, MAF2, and MAF3 were not or were slightly altered in the analyzed mutants with the exception of emf1-2 and clf-28/swn-7. On the other hand, MAF4 and MAF5 expression levels were dramatically increased in the different atbmi1a/b phenotypes, emf1-2 and clf-28/swn-7, whereas they were not significantly affected in emf2-2 and FRI-Col (Fig. 1H). The fact that emf2-2 did not show misregulation of MAF4 and MAF5 while clf-28/swn-7 did can indicate that these genes are regulated by a different paralog, such as VRN2 (Chen et al., 2009). Interestingly, atring1a/b mutants displayed similar expression levels of FLC, MAF4, and MAF5 to those of atbmi1a/b and emf1-2 mutants (Supplemental Fig. S1), suggesting that the PRC1 components AtBMI1, AtRING1, and EMF1 act together in the repression of these genes.
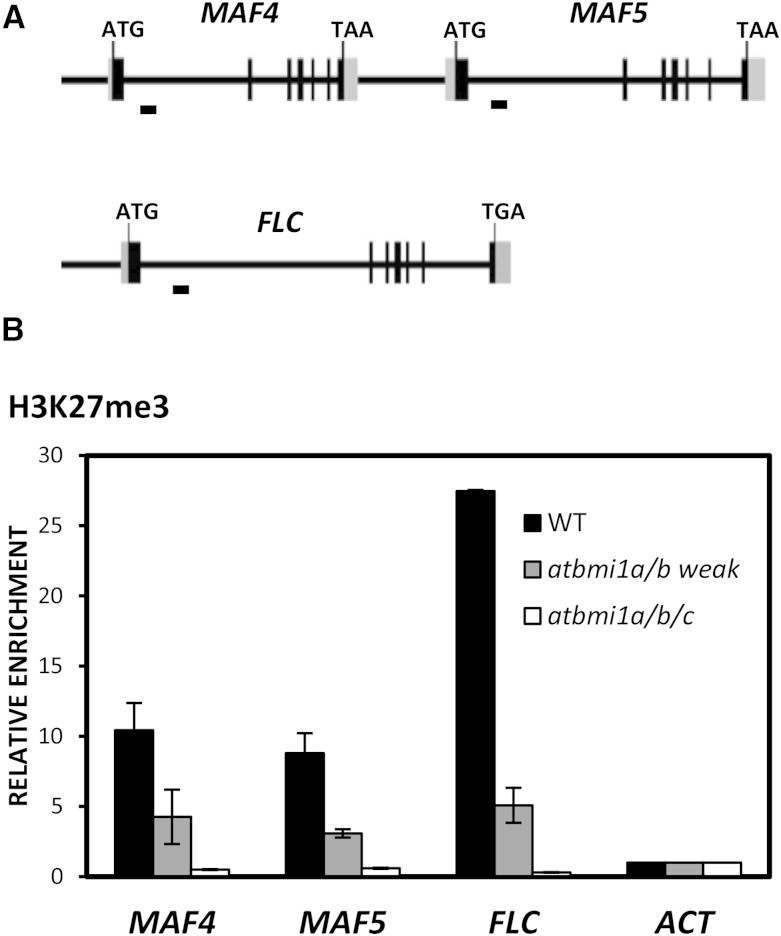
Consistent with the misexpression of FLC, MAF4, and MAF5 in the mutants, it has been shown previously that the levels of H3K27me3 marks at these genes were altered in PRC2 mutants (Jiang et al., 2008), emf1, and atring1a (Kim et al., 2012b; Shen et al., 2014). Therefore, to investigate whether AtBMI1 loss of function also affected the levels of H3K27me3 marks at FLC, MAF4, and MAF5, we examined the levels of this histone modification in atbmi1a/b mutants at the first intron of the genes, which has been shown to display an enrichment of H3K27me3 marks in wild-type seedlings at 9 to 10 DAG (Shen et al., 2014; Fig. 2A). Indeed, we found that the levels of H3K27me3 were decreased in atbmi1a/b weak mutants (Fig. 2B); furthermore, the H3K27me3 marks were eliminated in the very strong atbmi1a/b/c mutants (Fig. 2B), indicating that the loss of AtBMI1 function causes the loss of H3K27me3 marks at FLC, MAF4, and MAF5.
Figure 2.
H3K27me3 levels at MAF4, MAF5, and FLC are altered in atbmi1 mutants. A, Schematic diagram of MAF4, MAF5, and FLC genomic regions. Exons and untranslated regions are represented by black and gray boxes, respectively, while introns and other genomic regions are represented by black lines. The translation start site (ATG) and stop codon (TAA or TAG) are indicated. DNA fragments amplified in chromatin immunoprecipitation (ChIP) assays are indicated below the genomic regions. B, ChIP analysis of H3K27me3 levels at the FLC, MAF4, and MAF5 first intron region in wild-type (WT), atbmi1a/b weak, and atbmi1a/b/c seedlings at 10 DAG. ACT7 was used as a negative control. The immunoprecipitated DNAs were quantified and normalized to ACT7. Error bars indicate the sd of two biological replicates.
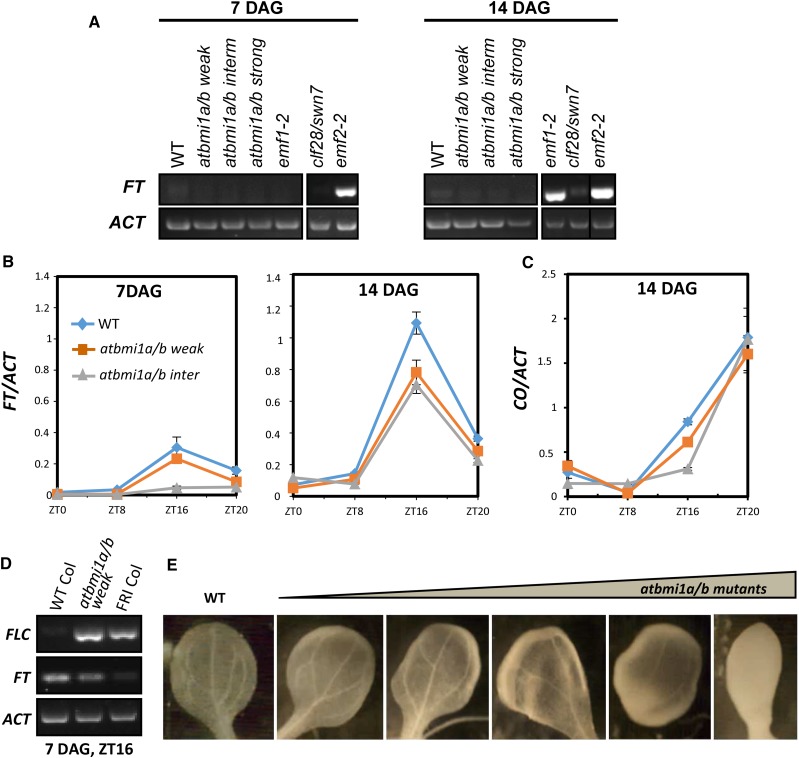
Then, we assessed the levels of FT in the different seedlings. In agreement with their early-flowering phenotype (Sung et al., 1992), emf1-2 and emf2-2 displayed a strong up-regulation of FT, despite the high levels of FLC expression (Fig. 3A). A recent report proposed that FLC recruits a PRC1-containing EMF1 (EMF1-PRC1) to FT chromatin for PcG repression and that CO activity antagonizes this repression by reducing the levels of EMF1-PRC1 at FT in the evening (Wang et al., 2014). This would explain why FLC up-regulation did not lead to FT repression in emf1, as FLC could not mediate FT repression in the absence of EMF1, and also in emf2 mutants, as EMF1 activity may be required for PRC2 recruitment. Since the Arabidopsis Col accession contains a nonfunctional FRI allele, and therefore the levels of FLC expression are very low (Kim and Sung, 2014; Fig. 1H), other FLC-related genes might recruit the EMF1-PRC1 for FT repression in this background, which could explain why emf1 mutants are also unresponsive to MAF4 and MAF5 overexpression.
Figure 3.
FT expression in atbmi1 mutants is CO dependent. A, Expression levels of FT in 7- and 14-d-old plants at ZT1 under LD conditions. ACT2 was used as an internal control (samples are as in Fig. 1H). B, FT mRNA levels in the indicated seedlings over an LD cycle at 7 and 14 DAG. C, CO mRNA levels over an LD cycle at 14 DAG. FT and CO transcript levels were normalized to ACT2; error bars indicate the sd of two biological repeats. D, FLC and FT transcript levels in 7-d-old wild-type (WT) Col, atbmi1a/b weak, and FRI-Col seedlings under LD conditions at Zeitgeber time 16 (ZT16). E, Vasculature organization of 10-d-old cotyledons from wild-type Col and different atbmi1a/b phenotypes.
As CO transcription is low at ZT1 and its expression is not altered in emf1 and emf2 mutants (Kim et al., 2010), the FT misexpression in these mutants may be CO independent. In support of this, it has been shown that emf1-1/co and emf2/co double mutant phenotypes were indistinguishable from their respective emf1 and emf2 single mutant parents, while emf1-1/ft double mutants usually did not flower and emf2/ft double mutants bolted after producing a higher number of sessile leaves than emf2 single mutants (Haung and Yang, 1998).
Surprisingly, we did not find a significant FT expression in any of the atbmi1a/b phenotypes at ZT1 (Fig. 3A); hence, we wondered whether FT levels were altered at other times of the day. When we measured the levels of FT transcripts over a 24-h LD cycle in atbmi1a/b weak, intermediate, and wild-type Col seedlings (Fig. 3B), we found that the expression of FT was photoperiod dependent in both the wild-type and atbmi1a/b mutants, but the levels of FT in atbmi1a/b were lower than in wild-type plants, despite the fact that CO levels were not affected in these mutants (Fig. 3C). Also, we found that FT expression seemed to decrease along with the severity of the atbmi1a/b phenotype. It might be argued that the decrease in FT levels was a consequence of FLC up-regulation; however, the expression levels of FLC in atbmi1a/b mutants were as high as in FRI-Col plants, but FT was not down-regulated to FRI-Col levels (Fig. 3D). Therefore, it seems that FLC is not able to mediate FT repression in atbmi1a/b, emf1, or PRC2 mutants in spite of the differences in FT expression among mutants.
Interestingly, like atbmi1a/b mutants, clf-28/swn-7 did not show misexpression of FT. Low levels of FT in clf/swn compared with clf single mutants have been reported before (Farrona et al., 2011). Alterations in vascular development and differentiation were proposed to be the basis for FT down-regulation in clf/swn double mutants (Farrona et al., 2011). Similarly, atbmi1a/b mutant phenotypes displayed different degrees of altered vascular development (Fig. 3E), which might explain the gradual decrease of FT expression correlated with the strength of the phenotype.
atbmi1a/b Mutants Have an Extended Juvenile Phase
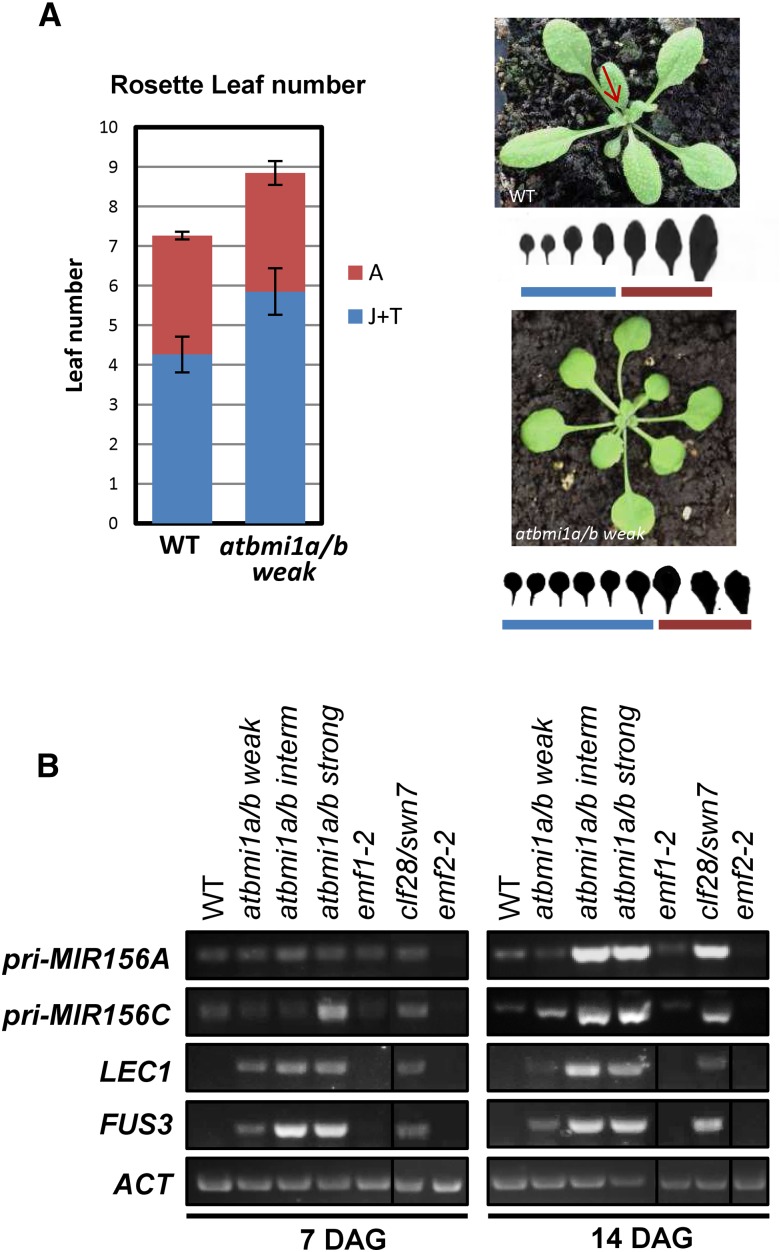
As we mentioned before, in contrast to emf1 or PRC2 mutants like emf2, atbmi1a/b weak mutants did not show an early-flowering phenotype; moreover, the most affected mutants never flowered. To investigate if flowering time was altered in atbmi1a/b weak mutants, we compared the flowering time in days and number of rosette leaves before bolting between atbmi1a/b weak mutant and wild-type Col plants under LD conditions (Fig. 4A). We found that flowering was delayed for 3 d in atbmi1a/b weak mutants compared with wild-type plants (22 ± 1 and 19 ± 1 d, respectively) and that the mutants generated two extra leaves before bolting (Fig. 4A, left), which was consistent with FT levels in the mutants but not with FLC, MAF4, or MAF5 levels. Surprisingly, these two extra leaves displayed round shape and a long petiole (Fig. 4A, right), which are considered juvenile traits (Wu et al., 2009), suggesting a prolonged juvenile phase in the mutants.
Figure 4.
atbmi1a/b mutants misexpress MIR156A and MIR156C. A, Flowering time of wild-type (WT) Col and atbmi1a/b weak plants (left). The time was measured by the number of rosette leaves produced from the SAM prior to flowering; 16 to 20 plants for each line were scored. Error bars indicate sd. Juvenile (J) and transition (T) leaves were differentiated from adult leaves (A) by shape (right). B, Expression levels of pri-MIR156A, pri-MIR156C, and the seed maturation genes LEAFY COTYLEDON1 (LEC1) and FUS3 in the different mutants at 7 and 14 DAG growing under LD conditions at ZT1.
Overexpression of miR156 prolongs the expression of juvenile vegetative traits and delays flowering. miR156 is encoded by eight genes in Arabidopsis (MIR156A to MIR156H; Reinhart et al., 2002). Among these genes, MIR156A and MIR156C were recently shown to be direct targets of the seed maturation gene FUS3. FUS3 activates MIR156A/C expression during seed development, and this expression is important after germination to delay the juvenile-to-adult vegetative phase transition (Wang and Perry, 2013). MIR156A and MIR156C contain RY elements at their 5′ end and into/through the gene, which are DNA elements specifically recognized by the B3 DNA-binding domain of FUS3 (Wang and Perry, 2013).
Since FUS3 is misexpressed in atbmi1 mutants and clf-28/swn-7 but not in emf1 or emf2 (Yang et al., 2013a; Fig. 4B), we investigated levels of the pri-MIR156A/C transcripts in these mutants (Fig. 4B). Strikingly, we found that the levels of pri-MIR156A/C displayed a drastic increase at 14 DAG in the three atbmi1a/b mutants, especially in intermediate and strong phenotypes, and in clf-28/swn-7 (Fig. 3B), but they were not altered in emf1-2 and emf2-2 (Fig. 4B). In addition, we found that the pri-MIR156s displayed similar levels in atring1a/b mutants than in atbmi1a/b weak mutants (Supplemental Fig. S2), indicating that both AtBMI1 and AtRING1 proteins are required to regulate miR156 levels. According to these results, the prolonged juvenile phase in atbmi1a/b weak mutants may be a consequence of miR156 misexpression; however, since FUS3 is ectopically expressed in these mutants, the high levels of pri-MIR156A/C might be an indirect effect of AtBMI1 loss of function.
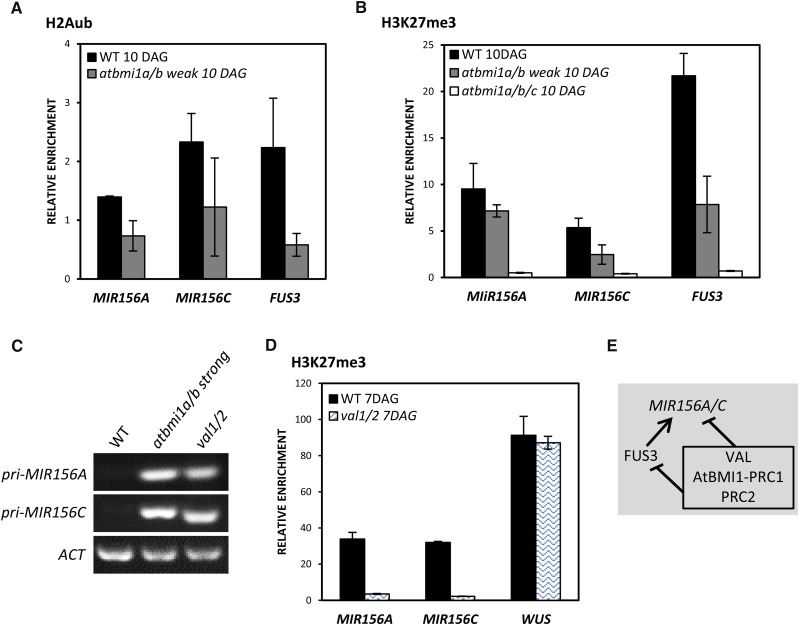
The Levels of H2Aub and H3K27me3 Marks in atbmi1 Mutants Are Decreased at MIR156A/C
To determine whether the AtBMI1 proteins play a role in regulating pri-miR156A/C expression, we investigated the levels of H2Aub marks at the TSS region of MIR156A and MIR156C in wild-type and atbmi1a/b weak seedlings at 10 DAG. We found that the levels of these marks at MIR156A were decreased in atbmi1a/b mutants and that the levels at MIR156C seemed to be reduced, although the experimental variation was large (Fig. 5A). Since AtBMI1 activity is required for PRC2-mediated H3K27me3 marking at several target genes (Yang et al., 2013a), we examined the levels of H3K27me3 marks at the TSS of these genes (Fig. 5B). We found that the levels of H3K27me3 were decreased at the TSS of all these genes in atbmi1a/b weak mutants (Fig. 5B); furthermore, the H3K27me3 marks were eliminated in the very strong atbmi1a/b/c mutants (Fig. 5B), indicating that MIR156A and MIR156C are regulated by the PcG machinery.
Figure 5.
MIR156A and MIR156C are direct targets of AtBMI1. A, ChIP analysis of H2Aub levels at MIR156A and MIR156C TSS in wild-type (WT) and atbmi1a/b weak seedlings at 10 DAG. FUS3 was used as a positive control. B, ChIP analysis of H3K27me3 levels at MIR156A and MIR156C TSS in wild-type, atbmi1a/b weak, and atbmi1a/b/c seedlings at 10 DAG. FUS3 was used as a positive control. The immunoprecipitated DNAs were quantified and normalized to ACT7. Error bars indicate the sd of at least two biological replicates. C, Expression levels of pri-MIR156A and pri-MIR156C in the wild type, atbmi1a/b strong, and val1/2 mutants at 10 DAG. ACT2 was used as an internal control. D, ChIP analysis of H3K27me3 levels at the TSS of MIR156A and MIR156C in wild-type and val1/2 seedlings at 7 DAG. WUSCHEL (WUS) was included as a negative target of VAL and a positive control of H3K27me3 (Yang et al., 2013a). The immunoprecipitated DNAs were quantified and normalized to ACT7. Error bars indicate the sd of two biological replicates. E, Schematic representation of MIR156A/C regulation by VAL-AtBMI1-PRC1/PRC2 and FUS3. Lines with bars indicate the repression of gene expression, and the line with the arrow indicates activation.
Then, we wondered whether the VIVIPAROUS1/ABSCISIC ACID INSENSITIVE3-LIKE1/2/3 (VAL1/2/3) proteins were involved in the recruitment of AtBMI1 and subsequently PRC2 to MIR156A/C, as is the case for the regulation of FUS3 (Yang et al., 2013a). The VAL proteins have a B3 DNA-binding domain that is proposed to recognize RY elements (Suzuki et al., 2007). Since MIR156A and MIR156C contain RY motifs (Wang and Perry, 2013), we reasoned that they might be targets of the VAL proteins. To investigate this, we first analyzed the expression levels of the pri-MIR156s in val1/2 mutants and compared them with the levels in wild-type and strong atbmi1a/b seedlings at 10 DAG (Fig. 5C). Indeed, we found that both pri-MIR156s were up-regulated in val1/2 to the same levels as in atbmi1a/b strong mutants. We further compared the levels of H3K27me3 at the TSS of MIR156A and MIR156C between the wild type and val1/2 mutants (Fig. 5D), and we found that the levels were dramatically reduced in the mutants. Together, these data suggest that the expression of pri-MIR156A/C is regulated by VAL and the AtBMI1 proteins. Therefore, the strong up-regulation of pri-MIR156 genes in atbmi1a/b mutants may be caused by both the loss of AtBMI1 function and the ectopic expression of FUS3 (Fig. 5E). It is possible that the activation of MIR156A/C by FUS3 only takes place in the absence of VAL-PcG-mediated repression, as must be the case during seed development.
emf1-2 Displays Up-Regulation of pri-MIR172b, SPL3, and SPL9
During the juvenile-to-adult phase transition, plants acquire competence to flower. In wild-type conditions, miR156 levels decrease as plants age, resulting in an increase in SPL expression. SPL9 has been shown to activate pri-MIR172b expression that, in turn, down-regulates the AP2-like floral repressors, which inhibit FT (Wang, 2014). Also, SPL3 directly regulates FT expression (Kim et al., 2012a). Consistent with this, it has been shown that the vasculature-specific expression of FT was notably increased in the cotyledons and distal regions of true leaves of plants overexpressing an miR156-resistant SPL3 and that FT::GUS expression was greatly reduced in the cotyledons and leaves of 35S::MIR156 plants (Kim et al., 2012a). In addition, it has been proposed that high miR156 levels reduce the ability of FT/FD to induce flowering by repressing SPL activity in the SAM (Wang et al., 2009). Therefore, SPLs and miR172 action contribute to set the threshold of FT necessary for flowering and to prepare the SAM to respond to the flowering signal.
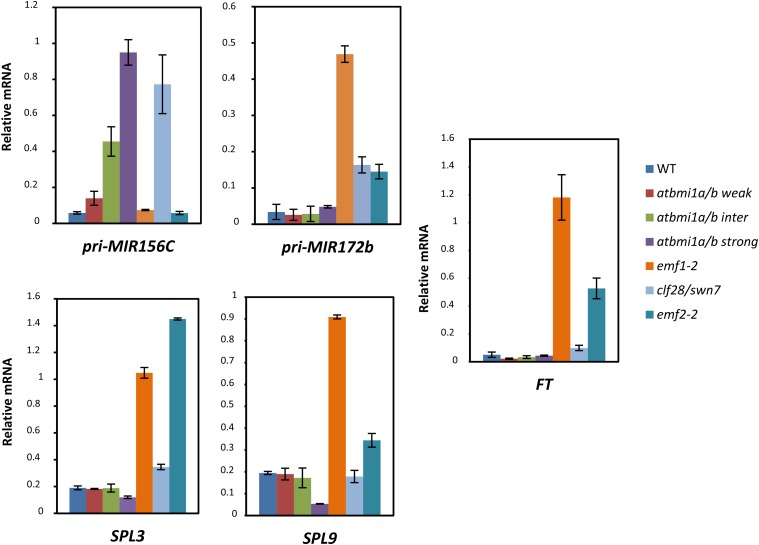
To determine whether the levels of pri-MIR156A/C expression in the different mutants correlate with the levels of SPL3, SPL9, pri-MIR172b, and FT, and if the expression pattern of the gene in each mutant explains the different flowering times, we analyzed the expression of all these genes in 10-d-old mutants and wild-type seedlings (Fig. 6). Consistent with the pri-MIR156A/C levels in atbmi1a/b mutants, we found low expression levels of SPL3, SPL9, and pri-MIR172b, confirming their juvenile stage. Accordingly, we found low levels of FT in these mutants, which are maintained later in development, leading to a delay in flowering time in atbmi1a/b weak mutants. In atbmi1a/b intermediate and strong mutants, misexpression of these genes along with the lack of a correctly differentiated phloem may be the cause of their never-flowering phenotype.
Figure 6.
AtBMI1-PRC1- and EMF1-PRC1-mediated regulation of miR156 and miR172. Expression levels of pri-MIR156C, pri-MIR172b, SPL3, SPL9, and FT are shown for wild-type (WT) and mutant seedlings at 10 DAG. Quantifications were normalized to ACT2. Error bars represent the sd of two biological replicates.
On the other hand, SPL3, SPL9, and pri-MIR172b expression was high in emf1-2 mutants. Interestingly, a recent report showed that SPL9 is a target of EMF1 (Kim et al., 2012b); thus, derepression of SPL9 may cause the activation of pri-MIR172b in emf1-2 mutants. Also, SPL3 is up-regulated in transgenic plants expressing an EMF1 antisense complementary DNA under the control of the floral meristem identity gene LFY promoter (LFY:asEMF1; Pu et al., 2013). Moreover, it has been shown that several MIR172 genes are direct targets of EMF1 (Kim et al., 2012b). emf2-2 also displayed increased expression levels of SPL3, SPL9, and pri-MIR172b, although the levels of the transcripts were not as high as in emf1-2, probably due to a redundant role of VRN2 in regulating these genes, as EMF2 and VRN2 regulate a common subset of targets (Lafos et al., 2011). Therefore, EMF1 and EMF2 directly and indirectly regulate miR172 levels. Remarkably, the levels of pri-MIR156, SPLs, and pri-MIR172b in emf1-2 and emf2-2 may explain the CO-independent expression of FT and the extremely early acquisition of flowering competence of these mutants.
Surprisingly, in the complete loss-of-PRC2-function clf-28/swn-7 mutants, the levels of SPL3 and SPL9 were only slightly higher than in the wild type (Fig. 6), and pri-MIR172b expression was not as high as in emf1-2. However, the high levels of pri-MIR156A/C in these mutants most likely affect pri-MIR172b expression by reducing SPL levels, thus explaining the expression pattern in these mutants. Consistent with this, clf-28/swn-7 did not display high levels of FT expression, which must be accentuated by alterations in vascular development.
DISCUSSION
PcG proteins have been shown to play important roles in regulating developmental phase transitions in plants; however, given that PcG components are present in the nuclei of most cells, whether they are targeted to distinct subsets of targets in specific cell types or developmental stages has been a major research problem. Recent findings regarding the PcG mechanism have shown that PRC1 is required for H3K27me3 marking at some target genes in both Arabidopsis (Yang et al., 2013a; Calonje, 2014) and animals (Comet and Helin, 2014; Schwartz and Pirrotta, 2014), placing PRC1 in a decisive position for the repression of some genes. In addition, several lines of evidence have suggested the existence of different mechanisms for PRC1-mediated repression in Arabidopsis (Kim et al., 2012b; Yang et al., 2013a; Calonje, 2014); however, it is not known whether a combination of different PRC1 subunits is required to exert the different mechanisms.
According to previous results in Arabidopsis, the PRC1 RING finger proteins AtBMI1 and AtRING1 are required for the repression of the seed maturation program after germination, whereas EMF1 is required for the repression of the floral program during vegetative development (Moon et al., 2003; Calonje et al., 2008; Bratzel et al., 2010; Chen et al., 2010), indicating that different PRC1 components are crucial for the regulation of different subsets of targets. On the other hand, other results suggest that all these components are required for the regulation of a different subset of target genes. For instance, AtRING1A has been shown to participate in the repression of FLC, MAF4, and MAF5 (Shen et al., 2014) and EMF1 in the repression of FLC (Kim et al., 2010). We show here that both EMF1 and AtBMI1 are required for FLC, MAF4, and MAF5 repression, suggesting a PRC1 in which AtRING1, AtBMI1, and EMF1 are required for repression. Whether these PRC1 proteins are always associated in the same complex or not remains to be investigated. In any case, the current data on PRC1-mediated gene regulation in Arabidopsis point to the existence of at least different PRC1 functional variants. Interestingly, despite the fact that AtBMI1 and EMF1 may participate in the regulation of FT through the repression of FLC, MAF4, and MAF5, loss of function in AtBMI1 and EMF1 does not have the same effect on FT expression, suggesting that the coordinated activity of different PRC1 functional variants may be required to give a specific developmental outcome. Therefore, to understand the role of PcG regulation in plant development, it will be necessary to determine the particular combination of PRC1s that regulates a specific process.
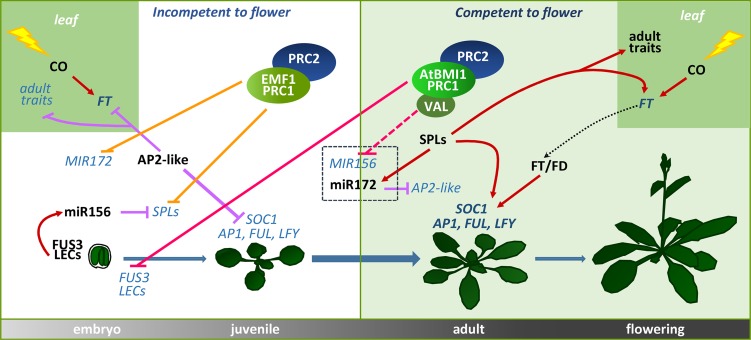
By exploring other possible roles of AtBMI1 proteins during plant development besides the repression of seed maturation genes after germination, we found that these proteins play a crucial role in the regulation of the transition from juvenile to adult phase. More importantly, our results point to a model in which two different functional PRC1 variants, an AtBMI1-PRC1 and an EMF1-PRC1 variant, coordinate the acquisition of flowering competence and contribute to reach the threshold of FT necessary to flower through the regulation of miR156 and miR172 levels, respectively (Fig. 7).
Figure 7.
Model of the roles of AtBMI1-PRC1 and EMF1-PRC1 variants in regulating juvenile-to-adult phase transition through miR156 and miR172 repression. EMF1-PRC1 represses MIR172 and SPLs to maintain the juvenile phase. As the plant ages, the levels of miR156 decrease by AtBMI1-PRC1-mediated repression, which allows the development of adult traits and the acquisition of flowering competence. Solid purple lines with bars indicate negative regulation; solid red lines with arrows indicate positive regulation; orange lines with bars indicate EMF1-PRC1/PRC2 repression; pink lines with bars indicate AtBMI1-PRC1/PRC2 repression (the dashed pink line indicates a possible negative regulation); and the dotted black line with arrow indicates the movement of FT from leaves to the SAM. Repressed genes are indicated in light blue italic type and activated genes in dark blue italic type; proteins and miRNAs are indicated in black type. FUL, FRUITFULL.
miR156 and miR172 have been identified as key components of the mechanisms that underlie the transition from juvenile to adult phase (Huijser and Schmid, 2011); however, although the roles of these miRNAs have been studied extensively, the mechanisms involved in their regulation are still largely unknown, especially those related to the age-dependent decline of miR156. We found that plants impaired in AtBMI1 function showed increased levels of MIR156A/C at the time the levels of miR156 should decline, which indicates that AtBMI1 proteins are required for miR156 repression. We propose that the high miR156 levels in atbmi1a/b contribute to reduce the levels of FT in leaves and to reduce the ability of FT/FD to induce flowering in the SAM by repressing SPL activity, leading to an extended juvenile phase. Conversely, we found that EMF1-PRC1 is required to maintain the repression of several SPL and MIR172 genes during the juvenile phase, thereby delaying the acquisition of flowering competence (Fig. 7). Accordingly, plants impaired in EMF1 function displayed up-regulation of SPL3, SPL9, and pri-MIR172 early in development, which may trigger a CO-independent up-regulation of FT and a precocious acquisition of flowering competence. In addition, AtBMI1-PRC1 and EMF1-PRC1 seem to be required for H3K27me3 marking at miR156 and miR172, respectively, supporting the idea that PRC1 triggers H3K27me3 at some target genes.
In summary, these results show how the coordinated roles of two functional PRC1 variants are required to regulate the transition from juvenile to adult phase; furthermore, we show how two central regulatory mechanisms, such as PcG and miRNA, assemble to control the acquisition of flowering competence, providing new insights into the paths actually used by the cell in order to achieve a developmental outcome.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) emf1-2, emf2-2, val1/2, atbmi1a/b, clf-28/swn-7, and atring1a/b mutants were described previously (Yang et al., 1995; Suzuki et al., 2007; Bratzel et al., 2010; Chen et al., 2010; Lafos et al., 2011). Plants were grown under LD conditions (16 h of light/8 h of dark) at 21°C on Murashige and Skoog agar plates containing 1.5% (w/v) Suc and 0.8% (w/v) agar. After germination, plants were transferred to soil and grown under the same conditions.
Seedlings at 10 DAG were fixed in ethanol:acetic acid (9:1, v/v) to analyze vasculature development in cotyledons.
Gene Expression Analysis
Total RNA was extracted using the ISOLATE II RNA Plant Kit (Bioline). Complementary DNAs were reverse transcribed from total RNAs with the QuantiTect reverse transcription kit (Qiagen). Quantitative reverse transcription-PCR was performed using the SensiFAST SYBR & Fluorescein Kit (Bioline) and the Bio-Rad iQ5 system. Primers used are specified in Supplemental Table S1.
ChIP
ChIP assays were carried out on fixed chromatin extracted from seedlings at 10 DAG using anti-H2Aub monoclonal (Cell Signaling; 8240) and anti-H3K27me3 polyclonal (Diagenode; pAb-069-050) antibodies. Buffers and procedures were as described previously (Yang et al., 2013a). Quantitative measurements of the immunoprecipitated DNA were performed using the SensiFAST SYBR & Fluorescein Kit (Bioline) and the Bio-Rad iQ5 system. Each of the immunoprecipitations was repeated independently at least once, and each sample was quantified in triplicate. Primers used are specified in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of flowering repressors in atring1a/b mutants.
Supplemental Figure S2. Expression of pri-MIR156A/C in atring1a/b mutants.
Supplemental Table S1. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Z. Renee Sung (University of California, Berkeley), Federico Valverde, José María Romero, and Teresa Ruiz (Instituto de Bioquímica Vegetal y Fotosíntesis) for helpful suggestions.
Glossary
- PcG
Polycomb group
- H2Aub
histone 2A monoubiquitination
- H3K27me3
histone H3 lysine-27 trimethylation
- miRNA
microRNA
- SAM
shoot apical meristem
- TSS
transcriptional start site
- Col
Columbia
- LD
long-day
- ZT1
Zeitgeber time 1
- DAG
days after germination
- ChIP
chromatin immunoprecipitation
Footnotes
This work was supported by the FP7–PEOPLE–2012 Marie Curie Career Integration Grant program (grant no. 333748) and by the Spanish Ministry of Economy and Competitiveness (grant no. BIO2013–44078–P).
References
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Bratzel F, Yang C, Angelova A, López-Torrejón G, Koch M, del Pozo JC, Calonje M (2012) Regulation of the new Arabidopsis imprinted gene AtBMI1C requires the interplay of different epigenetic mechanisms. Mol Plant 5: 260–269 [DOI] [PubMed] [Google Scholar]
- Calonje M. (2014) PRC1 marks the difference in plant PcG repression. Mol Plant 7: 459–471 [DOI] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen L, Sung ZR (2008) EMBRYONIC FLOWER1 participates in Polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen D, Molitor A, Liu C, Shen WH (2010) The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res 20: 1332–1344 [DOI] [PubMed] [Google Scholar]
- Chen LJ, Diao ZY, Specht C, Sung ZR (2009) Molecular evolution of VEF-domain-containing PcG genes in plants. Mol Plant 2: 738–754 [DOI] [PubMed] [Google Scholar]
- Cho HJ, Kim JJ, Lee JH, Kim W, Jung JH, Park CM, Ahn JH (2012) SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett 586: 2332–2337 [DOI] [PubMed] [Google Scholar]
- Comet I, Helin K (2014) Revolution in the Polycomb hierarchy. Nat Struct Mol Biol 21: 573–575 [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Hennig L (2014) Variations on a theme: Polycomb group proteins in plants. J Exp Bot 65: 2769–2784 [DOI] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of Polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haung MD, Yang CH (1998) EMF genes interact with late-flowering genes to regulate Arabidopsis shoot development. Plant Cell Physiol 39: 382–393 [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2014) Genetic and epigenetic mechanisms underlying vernalization. The Arabidopsis Book 12: e0171, doi/10.1199/tab.0171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH (2012a) The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol 159: 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR (2012b) EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 8: e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR (2010) Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol 152: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL (2001) Polycomb repression of flowering during early plant development. Proc Natl Acad Sci USA 98: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsoukas IG, Massiah AJ, Thomas B (2012) Florigenic and antiflorigenic signaling in plants. Plant Cell Physiol 53: 1827–1842 [DOI] [PubMed] [Google Scholar]
- Merini W, Calonje M (2015) PRC1 is taking the lead in PcG repression. Plant J 83: 110–120 [DOI] [PubMed] [Google Scholar]
- Moon YH, Chen L, Pan RL, Chang HS, Zhu T, Maffeo DM, Sung ZR (2003) EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. (2013) Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol 105: 125–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Liu MS, Kim SY, Chen LFO, Fletcher JC, Sung ZR (2013) EMBRYONIC FLOWER1 and ULTRAPETALA1 act antagonistically on Arabidopsis development and stress response. Plant Physiol 162: 812–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2014) Ruled by ubiquitylation: a new order for Polycomb recruitment. Cell Rep 8: 321–325 [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26: 229–236 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Thong Z, Gong X, Shen Q, Gan Y, Yu H (2014) The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis. Development 141: 1303–1312 [DOI] [PubMed] [Google Scholar]
- Sung ZR, Belachew A, Shunong B, Bertrand-Garcia R (1992) EMF, an Arabidopsis gene required for vegetative shoot development. Science 258: 1645–1647 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu X, Yuan W, Schmitz RJ, He Y (2014) Photoperiodic control of the floral transition through a distinct Polycomb repressive complex. Dev Cell 28: 727–736 [DOI] [PubMed] [Google Scholar]
- Wigge PA. (2011) FT, a mobile developmental signal in plants. Curr Biol 21: R374–R378 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M (2013a) VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol 23: 1324–1329 [DOI] [PubMed] [Google Scholar]
- Yang CH, Chen LJ, Sung ZR (1995) Genetic regulation of shoot development in Arabidopsis: role of the EMF genes. Dev Biol 169: 421–435 [DOI] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS (2013b) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.