Distinct transcriptional programs and hormonal pathways underlie the ovary-to-fruit transition.
Abstract
Fruit formation and early development involve a range of physiological and morphological transformations of the various constituent tissues of the ovary. These developmental changes vary considerably according to tissue type, but molecular analyses at an organ-wide level inevitably obscure many tissue-specific phenomena. We used laser-capture microdissection coupled to high-throughput RNA sequencing to analyze the transcriptome of ovaries and fruit tissues of the wild tomato species Solanum pimpinellifolium. This laser-capture microdissection-high-throughput RNA sequencing approach allowed quantitative global profiling of gene expression at previously unobtainable levels of spatial resolution, revealing numerous contrasting transcriptome profiles and uncovering rare and cell type-specific transcripts. Coexpressed gene clusters linked specific tissues and stages to major transcriptional changes underlying the ovary-to-fruit transition and provided evidence of regulatory modules related to cell division, photosynthesis, and auxin transport in internal fruit tissues, together with parallel specialization of the pericarp transcriptome in stress responses and secondary metabolism. Analysis of transcription factor expression and regulatory motifs indicated putative gene regulatory modules that may regulate the development of different tissues and hormonal processes. Major alterations in the expression of hormone metabolic and signaling components illustrate the complex hormonal control underpinning fruit formation, with intricate spatiotemporal variations suggesting separate regulatory programs.
The formation of fruit requires the coordinated growth and development of floral tissues following pollination. The molecular regulation of this process relies upon a complex interplay of gene expression changes, signaling events, and hormonal activity and is of critical importance for seed dispersal, plant fitness, and agricultural yield (Ruan et al., 2012).
In tomato (Solanum lycopersicum), a model system for fleshy fruit development (Kimura and Sinha, 2008), the fruit develops from the ovary, which is divided into two or more locules enclosed by the carpel wall, or pericarp, and separated by a septum that fuses with each of the carpels (Gasser and Robinson-Beers, 1993). Following pollination, the ovules, which are attached to the central septum via placental tissue, form seeds, with double fertilization of the egg and central cells producing the diploid embryo and triploid endosperm, respectively. Each seed is surrounded by a seed coat that is derived from the integuments that enclose the embryo sac (Nowack et al., 2010). Coinciding with seed initiation, the placenta, septum, and pericarp undergo rapid cell division, and the placenta proliferates to fill the locular cavity (Gillaspy et al., 1993). After a relatively short period, the rate of cell division in the pericarp declines and cell expansion contributes to the majority of the remaining increase in fruit size (Xiao et al., 2009; Pabón-Mora and Litt, 2011).
Previous studies have revealed complex patterns of gene expression during tomato fruit set and early growth, with prominent modulation of gene expression related to cell division, photosynthesis, sugar metabolism, and hormone biology, and hinted at the existence of distinct regulatory programs associated with specialized tissue development (Lemaire-Chamley et al., 2005; Vriezen et al., 2008; Mounet et al., 2009; Wang et al., 2009).
The concerted action of multiple hormones is also involved in the control of fruit development. In particular, auxin plays a major key role in the regulation of fruit set and growth (Wang et al., 2005; Goetz et al., 2007; de Jong et al., 2009), and there is increasing evidence of strict spatiotemporal control of auxin distribution and signaling during reproductive development in Arabidopsis (Arabidopsis thaliana; Fuentes and Vivian-Smith, 2009; Sundberg and Østergaard, 2009) and in tomato (Pattison et al., 2014). However, a comprehensive picture of tissue-specific, auxin-related gene expression to inform models of auxin action in fruit is currently lacking. Moreover, other hormones, including GAs (Serrani et al., 2008), cytokinins (Matsuo et al., 2012), ethylene (Llop-Tous et al., 2000), and brassinosteroids (Montoya et al., 2005), have been implicated in early tomato fruit development, but details of their spatiotemporal activity and related patterns of gene expression are limited.
Despite recent advances in the regulation of fruit formation and early development (Ruan et al., 2012), the mechanisms by which transcriptional control and hormonal activity interact to coordinate the growth and differentiation of diverse fruit tissues remain poorly understood. While previous transcriptomic studies have suggested distinct expression profiles in different parts of the fruit, they have typically lacked tissue specificity, being performed on whole ovaries and fruit (Wang et al., 2009) or using relatively coarse manual dissection (Lemaire-Chamley et al., 2005; Vriezen et al., 2008).
Tissue-specific expression profiling avoids the potential dilution of spatially restricted regulatory events and has proved effective in uncovering biological pathways and regulatory networks that would otherwise have been undetectable (Taylor-Teeples et al., 2011; Belmonte et al., 2013). Highly precise dissection can be achieved using laser-capture microdissection (LCM), which enables the separation of tissues or cell types in histological sections (Nelson et al., 2006). The benefits of LCM were demonstrated by a study that profiled the transcriptome of individual cell types within the pericarp of tomato fruit, revealing distinct patterns of cell type- and tissue-dependent expression (Matas et al., 2011). Furthermore, more complete transcriptome coverage and better quantitative assessment of gene expression than were previously possible can now be achieved using next-generation RNA sequencing (RNA-seq; Martin et al., 2013). RNA-seq has allowed in-depth, quantitative transcriptome profiling during ripening (Tomato Genome Consortium, 2012; Zhong et al., 2013) and other tomato developmental processes (Park et al., 2012; Gupta et al., 2013) but has yet to be applied to the early stage of fruit development.
In this study, we used a coupled LCM-RNA-seq approach to obtain a comprehensive view of gene expression during the early stage of fruit development in Solanum pimpinellifolium, a wild relative and direct ancestor (Blanca et al., 2012) of cultivated tomato. S. pimpinellifolium has highly consistent and predictable rates of flower and fruit development and so is particularly suitable for developmental studies (Xiao et al., 2009). We report extremely high spatial resolution and deep coverage of fruit and seed tissue-specific transcriptomes and present new insights into the regulatory framework underlying early fleshy fruit development.
RESULTS
RNA-seq Analysis of Microdissected Ovary and Fruit Tissues
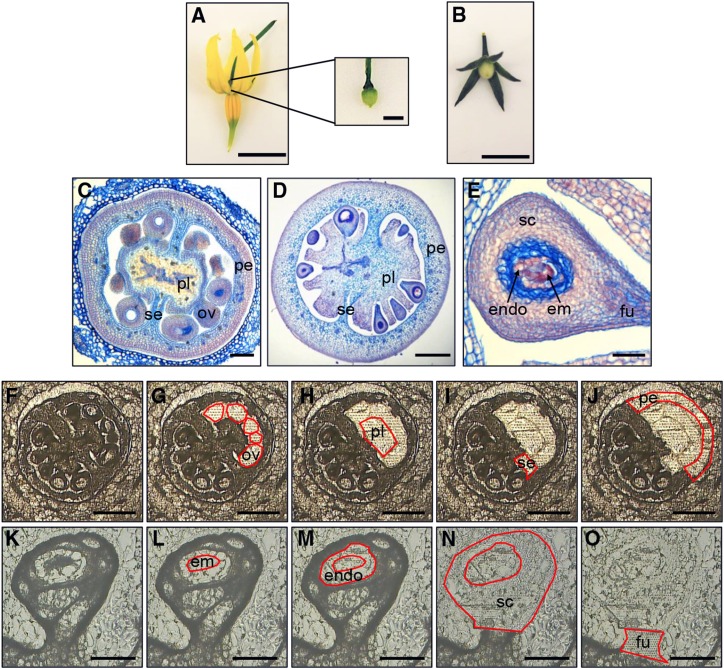
LCM was used to dissect S. pimpinellifolium (Fig. 1) ovaries at anthesis (0 DPA), a time point immediately before fruit initiation (Fig. 1A), and fruit at 4 DPA (Fig. 1B). The 4-DPA fruit is a key developmental time point at the beginning of exponential fruit growth, where active cell division is still taking place and the embryo is at the four- to 16-cell stage (Xiao et al., 2009). A total of 11 samples were dissected from cryosections, which allow efficient preservation and extraction of RNA from tomato fruit tissues following LCM (Matas et al., 2011). Samples from the 0-DPA ovary comprised the ovules, placenta, septum, and pericarp (Fig. 1, C and F–J). The equivalent tissues of placenta, septum, and pericarp were collected from 4-DPA fruits, together with the embryo, endosperm, seed coat, and funiculus from the developing seed (Fig. 1, D, E, and K–O).
Figure 1.
LCM of S. pimpinellifolium ovaries and fruit. A and B, Photographs of flower and ovary (A) at 0 DPA and fruit (B) at 4 DPA. Bars = 5 mm and 500 μm (inset). C to E, Transverse paraffin sections of 0-DPA ovaries (C), 4-DPA fruit (D), and 4-DPA seeds (E). Labels indicate ovules (ov), placenta (pl), septum (se), pericarp (pe), endosperm (endo), embryo (em), seed coat (sc), and funiculus (fu). Bars = 50 μm (C and E) and 300 μm (D). F to J, LCM of ovaries at 0 DPA. Cryosections show a whole ovary before (F) and after collection of ovules (G), placenta (H), septum (I), and pericarp (J). Bars = 300 μm. K to O, Cryosections of seed from a 4-DPA fruit before (K) and after removal of embryo (L), endosperm (M), seed coat (N), and funiculus (O). Bars = 100 μm.
Three biological replicates of each tissue type were collected for the production of RNA-seq libraries and, given the low nucleotide divergence between S. pimpinellifolium and S. lycopersicum (estimated at 0.6%; Tomato Genome Consortium, 2012), the sequencing reads were readily aligned to the S. lycopersicum genome (Supplemental Table S1). Read counts per gene were expressed as reads per million mapped reads (RPM; Supplemental Table S2). A total of 22,046 genes, approximately two-thirds of the annotated genes in the tomato genome (Tomato Genome Consortium, 2012), were expressed in at least one sample (RPM ≥ 2; Supplemental Table S3), and more than half of the expressed genes (13,855) were present in all samples. There was little variation in the total number of genes represented in each tissue type, similar to other fruit transcriptomic studies (Kang et al., 2013).
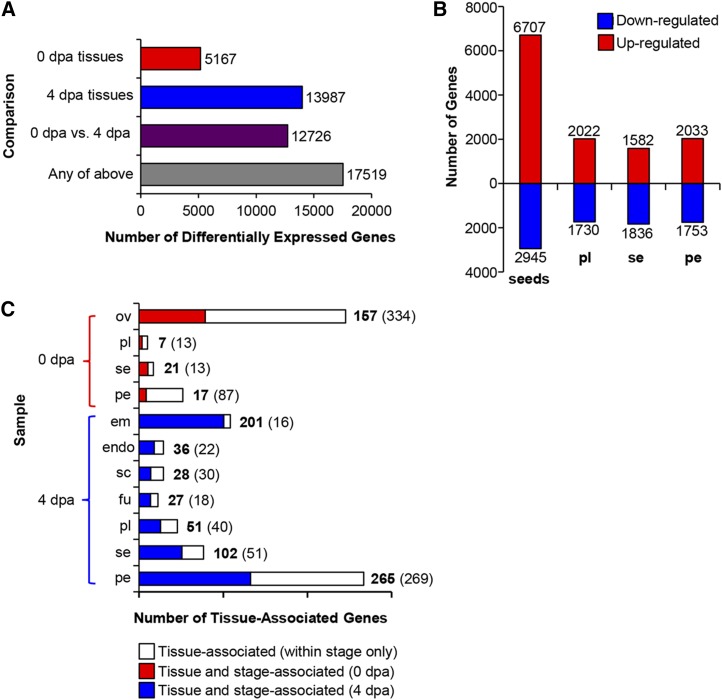
Differentially expressed genes (Fig. 2) were identified by pairwise sample comparisons, and the majority of genes (17,519) were identified as differentially expressed in at least one comparison (Fig. 2A; Supplemental Table S2). We calculated the number of up- and down-regulated genes according to tissue type (Fig. 2B). A similar number of genes were up- or down-regulated in placenta, septum, and pericarp. A higher number were up-regulated in any one of the seed tissues (embryo, endosperm, seed coat, and funiculus) relative to the ovules, although this is likely due to the greater number of seed tissues collected. Comparatively fewer genes were down-regulated in seed tissues relative to the ovule (Fig. 2B).
Figure 2.
Differential gene expression in ovary and fruit tissues. A, Total number of differentially expressed genes identified by pairwise comparisons between all tissues at 0 DPA (dpa), at 4 DPA, between both stages, or in at least one pairwise comparison. B, Number of genes up- and down-regulated at 4 DPA relative to 0 DPA. For seeds, the up-regulated genes include all those higher in at least one of the four seed tissues (embryo, endosperm, seed coat, and funiculus) relative to the ovules. Down-regulated genes include those showing down-regulation in all four tissues relative to the ovules. C, Number of tissue- and stage-associated genes. Tissue-associated genes are significantly higher expressed in one tissue relative to all others within the stage (white bars, numbers in parentheses). Tissue- and stage-associated genes (numbers in boldface, colored bars) show significantly higher expression in a single tissue relative to all other samples. Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; and se, septum.
The data set was also queried to identify genes strongly associated with an individual tissue or stage (Fig. 2C; Supplemental Table S4). Genes were defined as being strongly tissue associated (i.e. significantly higher expression in a single tissue relative to all tissues within a developmental stage) or tissue and stage associated (significantly higher expression in a single tissue relative to all other samples). The number of tissue-associated genes ranged from as low as 20 for the placenta from ovaries at anthesis to as high as 534 for the pericarp of 4-DPA fruit. In some cases, the majority of genes that were tissue associated were also stage associated. For example, 93% of genes associated with the embryo at 4 DPA were also stage associated, meaning that they showed significantly higher expression than in any ovary tissue, suggesting particular specialization of the embryo transcriptome. In contrast, only 16% of genes associated with the pericarp in ovaries were also stage associated, indicating a higher degree of tissue dependency and a reduced level of stage dependency of the pericarp transcriptome.
To assess transcriptome similarity between samples, principal component analysis and hierarchical clustering were performed (Fig. 3). Both analyses revealed three discrete groupings: one group consisting of the four tissues from the ovary and another formed by tissues from the 4-DPA fruit except embryo and endosperm, which constituted a distinct set highlighting the specialized nature of their transcriptomes. Interestingly, samples clustered according to developmental stage rather than tissue type; thus, placenta, septum, and pericarp pairs did not cluster together. However, principal component analysis using two components (Fig. 3A) revealed a shorter distance between the two pericarp samples than between any other cross-stage pairing, again suggesting a relatively high degree of tissue dependency of the pericarp transcriptome compared with other tissues.
Figure 3.
Transcriptional relationships between tissues in the developing fruit. A, Principal component analysis based on all expressed genes showing three distinct groups of tissues: 0-DPA (dpa) ovaries (orange), embryo and endosperm (light blue), and the rest of tissues from 4-DPA fruit (blue). B, Hierarchical clustering of all differentially expressed genes across the different tissues. Each row of the heat map represents an individual gene, and coloring represents relative expression level. Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; and se, septum.
Spatiotemporal Gene Expression Dynamics during Early Fruit Development
To identify the major transcriptional dynamics associated with the transition from ovary to fruit, a K-means clustering approach was used to group genes with similar expression profiles. A total of 30 coexpression clusters were defined (Supplemental Table S5; Supplemental Fig. S1), and approximately three-quarters (11,952 out of 16,249) of the differentially expressed genes were strongly correlated with the average profile of these clusters (Pearson correlation > 0.8). Each cluster was tested for overrepresentation of genes belonging to 31 functional categories (Supplemental Table S6), and 17 clusters were chosen for further analysis on the basis of these functional annotations and average expression profiles (Figs. 4–6).
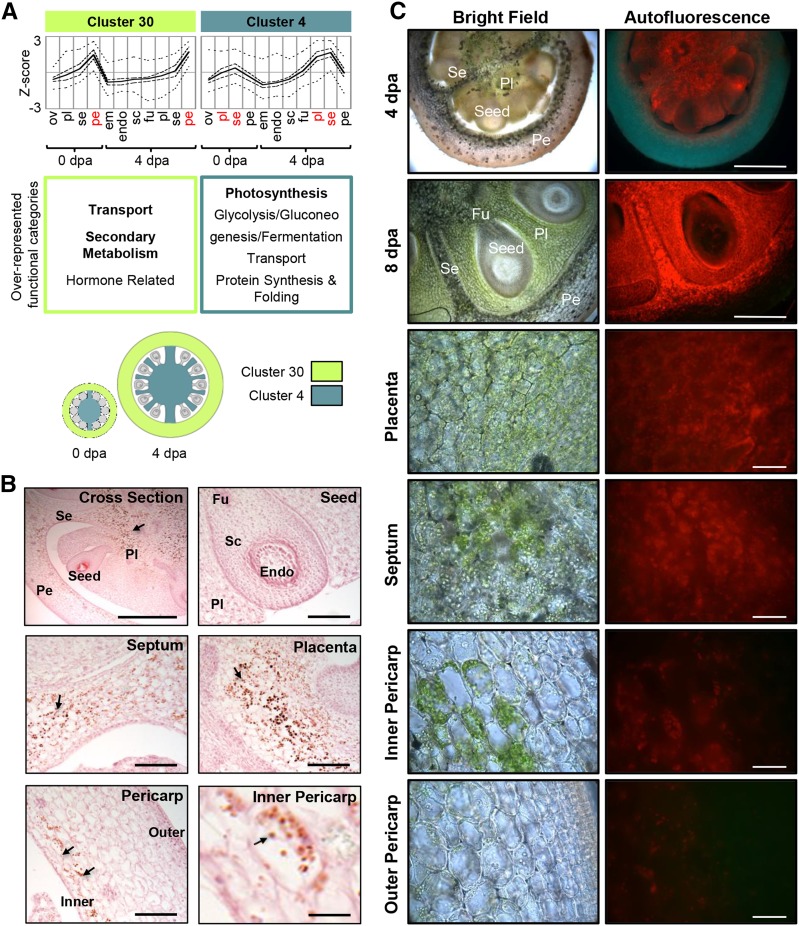
Figure 4.
Stage-independent gene clusters. A, Average profiles of cluster 30 and cluster 4. Cluster-wide average expression is plotted with solid lines, first and third quartiles by dashed lines, and maximum and minimum by dotted lines. Overrepresented functional category annotations are indicated below each cluster (P < 0.05, regular type; P < 0.01, boldface type). Clusters are represented graphically with coloring indicating the tissues where each is predominantly expressed. B, Starch accumulation in 4-DPA (dpa) fruit. Sections were stained with Lugol’s solution and counterstained with Safranin O (pink) to show anatomy. Starch granules (arrows, dark brown) are visible in the placenta, septum, and inner pericarp. Bars = 500 μm (cross section), 100 μm (seed, placenta, septum, and pericarp), and 20 μm (inner pericarp). C, Chlorophyll distribution in 4-DPA (top row) and 8-DPA (other rows) fruit. Chlorophyll is green in bright-field images and red in fluorescence images. Bars = 500 μm (top two rows) and 50 μm (other rows). Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; and se, septum.
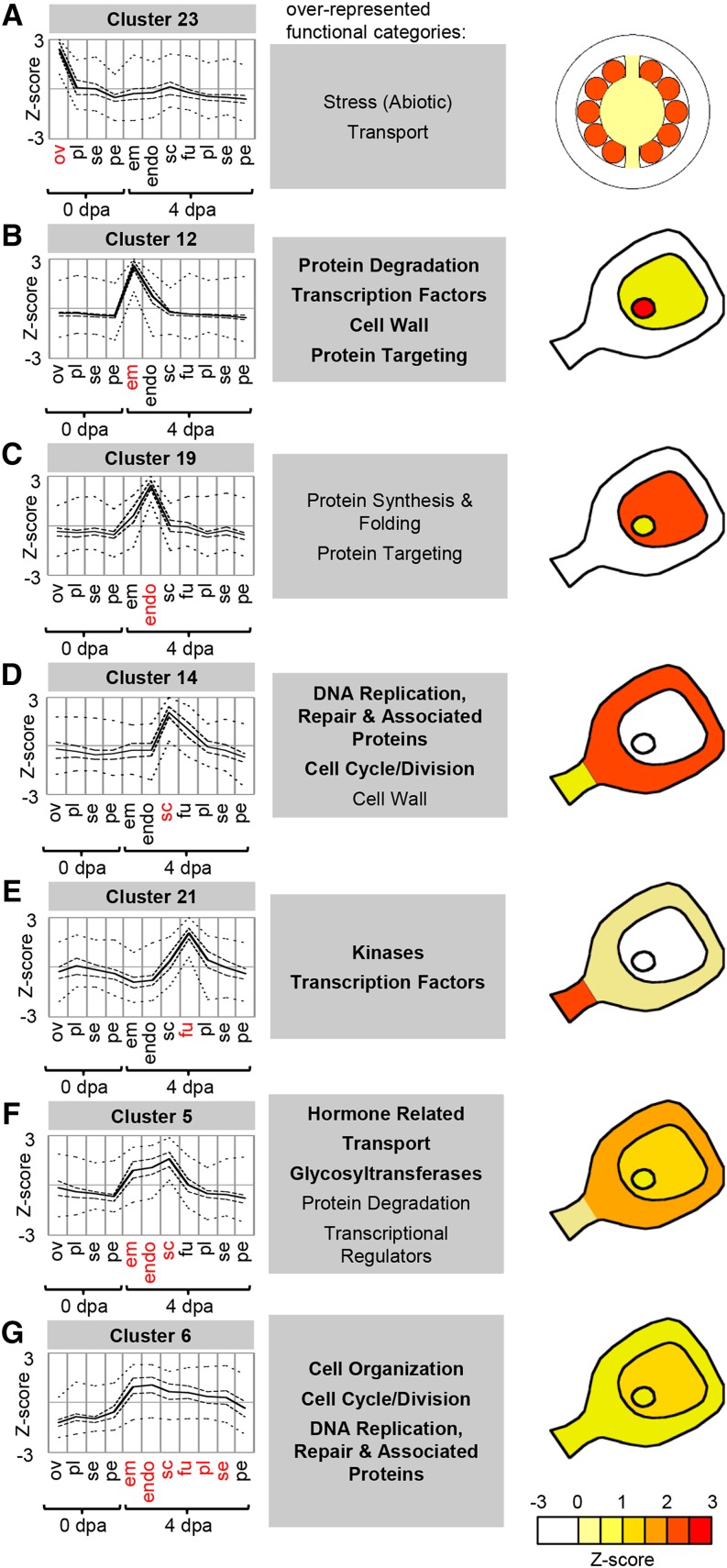
Figure 6.
Clusters associated with ovule/seed tissues. Cluster-wide average expression is plotted with solid lines, first and third quartiles by dashed lines, and maximum and minimum by dotted lines. Overrepresented functional category annotations are indicated below each cluster (P < 0.05, regular type; P < 0.01, boldface type). Clusters are represented graphically with coloring indicating the average level of relative gene expression. Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; se, septum; and dpa, DPA.
Stage-Independent Expression Clusters
Most clusters showed a distinct peak in expression in either ovaries or fruit, with relatively few spanning both developmental stages. However, clusters 30 and 4 were exceptions, having profiles that peaked in the same tissue(s) at both stages (Fig. 4).
Cluster 30, which comprised genes primarily expressed in the pericarp, had overrepresentation of genes from the transport, secondary metabolism, and hormone-related categories and contained many genes involved in isoprenoid biosynthesis and the production of steroid precursors in a variety of biosynthetic pathways (Vranová et al., 2012). Several of the genes in the hormone-related category were part of the connected brassinosteroid biosynthetic pathway, in which cycloartenol, an isoprenoid-derived steroid, is the starting point (Ohyama et al., 2009).
Cluster 4, which comprised genes expressed in the placenta and septum, was characterized by an abundance of genes associated with photosynthesis. Eighty-two out of the 376 genes in this cluster belonged to the photosynthesis category and included 28 encoding chlorophyll-binding proteins, 11 PSI subunits, and eight PSII subunits. An analysis of chlorophyll distribution in 4-DPA fruit confirmed a close correlation between photosynthetic tissues and the expression profile of this cluster (Fig. 4C, top row). Chlorophyll autofluorescence was strong in placenta and septum but was restricted to only the inner part of the pericarp and was mostly absent from seeds and the outer pericarp. Chlorophyll distribution is also shown at 8 DPA to have better spatial resolution (Fig. 4C, other rows). However, we cannot be certain that the spatial organization of gene expression is maintained at this time point. Cluster 4 also included several genes related to starch metabolism or catabolism (Supplemental Fig. S2), and staining for starch confirmed its accumulation in these same photosynthetic tissues. Starch granules were visible in the placenta, septum, and inner pericarp but were largely absent from the seed and outer pericarp (Fig. 4B). This is consistent with starch being a major storage form of fixed carbon (Schaffer and Petreikov, 1997) and with previous reports of continual starch synthesis and degradation in growing fruit (Nguyen-Quoc and Foyer, 2001).
Stage-Dependent Expression Clusters
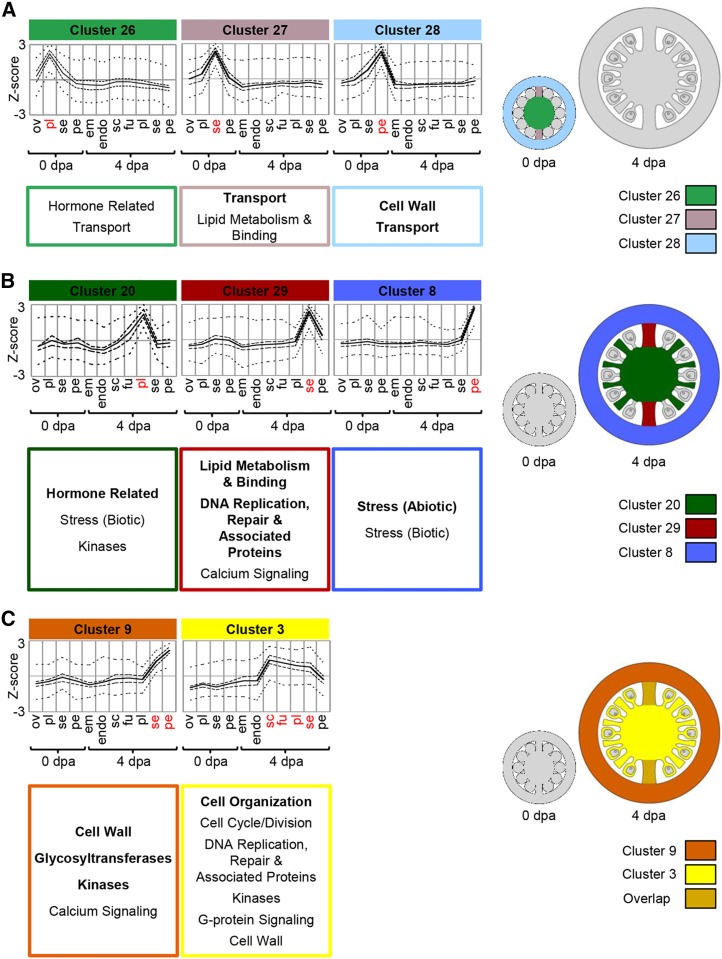
This category includes clusters primarily associated with specific tissues at a single developmental stage. Clusters 26, 27, and 28 peaked in individual tissues of the ovary and are associated with the placenta, septum, and pericarp, respectively (Fig. 5A). All three had overrepresentation of transporter genes encoding a diverse range of proteins with various substrate specificities. For example, cluster 27 (septum) contained genes encoding transporters of mineral nutrients, organic macromolecules, and the hormone auxin. There was a particular abundance of ATP-binding cassette transporters in cluster 28 (pericarp), a class of transporter associated with the transport of secondary metabolites (Yazaki, 2006). Their presence in the pericarp, therefore, may be related to secondary metabolite production in this tissue (see cluster 30 above). Cluster 28 also had overrepresentation of genes in the cell wall category, including those related to pectin modification.
Figure 5.
Stage-dependent gene clusters. Average profiles of clusters are primarily expressed at a single developmental stage. Cluster-wide average expression is plotted with solid lines, first and third quartiles by dashed lines, and maximum and minimum by dotted lines. Overrepresented functional category annotations are indicated below each cluster (P < 0.05, regular type; P < 0.01, boldface type). Clusters are represented graphically with coloring indicating the tissues where each is predominantly expressed. Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; se, septum; and dpa, DPA.
Clusters 20, 29, and 8 showed peaks of expression in the placenta, septum, and pericarp of young fruit, respectively, but relatively low expression in the ovary (Fig. 5B). Many genes in these clusters encode proteins with similar functions to their counterparts in clusters that peak in ovaries. For example, genes in the hormone-related category were overrepresented in both cluster 26 (ovary placenta) and cluster 20 (young fruit placenta). Likewise, genes from the lipid metabolism and binding category were overrepresented in both cluster 29 (young fruit septum) and cluster 27 (ovary septum). These clusters comprised several genes encoding lipases containing the GDSL motif and lipid-transfer proteins; in addition, some genes in cluster 29 encode proteins related to cutin metabolism, including the tomato cutin synthase CUTIN DEFICIENT1 (Yeats et al., 2014) and homologs of Arabidopsis ECERIFERUM8 and CYP86A2, a member of the cytochrome p450 CYP86A subfamily, which are involved in fatty acid modification (Xiao et al., 2004; Lü et al., 2009).
Functional categories uniquely associated with clusters expressed in young fruit were also present. For example, cluster 8, which peaked in 4-DPA pericarp, had an overrepresentation of genes in both the biotic and abiotic stress categories. These included multiple genes encoding heat-shock proteins, which may be induced by various stress responses or during development (Löw et al., 2000; Sun et al., 2002), and other proteins that are frequently induced under stress, such as MILDEW LOCUS O family proteins (Piffanelli et al., 2002) and PATHOGENESIS-RELATED proteins (Sels et al., 2008).
Clusters 9 and 3 also showed peak expression in young fruit, but their profiles spanned multiple tissues (Fig. 5C). Cluster 9, which comprised septum and pericarp, had an overrepresentation of genes from the cell wall and glycosyltransferase categories, including five XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE genes. On the other hand, cluster 3, which spanned all tissues except embryo, endosperm, and pericarp, was characterized by an overrepresentation of the cell organization and cell cycle/cell division categories. Many of the genes in the cell organization category were related to the cytoskeleton and included several α- and β-tubulin and kinesin genes whose expression is associated with dividing cells (Breyne et al., 2002).
Ovule/Seed-Associated Expression Clusters
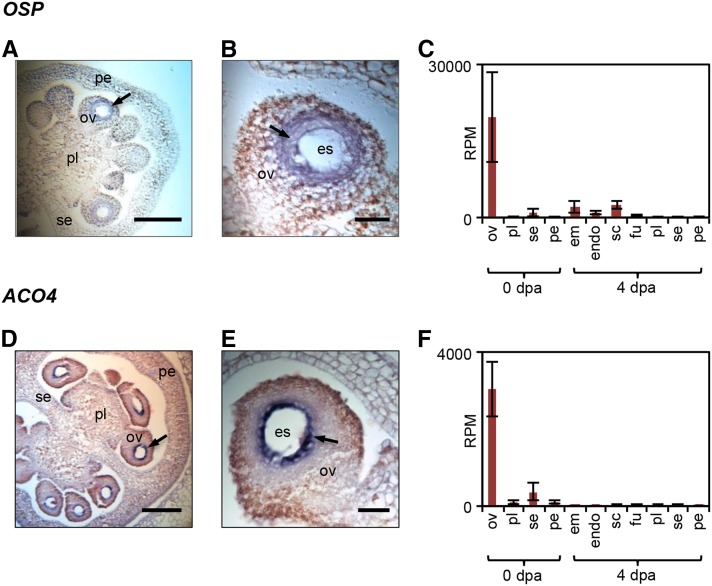
To evaluate the transcriptional changes associated with seed development after fertilization, we examined a number of gene clusters with peak expression in the ovules or seed tissues (Fig. 6). Genes in cluster 23 were predominantly expressed in the ovules and then strongly down-regulated in the fruit (Fig. 6A). Their expression profiles, therefore, negatively correlate with fruit initiation. The functional category transport was enriched in this cluster, which contained a set of six ABCG family transporters, ion channels, and transporters of amino acids, sugars, organic acids, and phosphate. Interestingly, a large proportion of genes in cluster 23 encoded small proteins (38% were less than 200 amino acids compared with only 25% among all expressed genes), many of them containing putative signal peptides. Genes in this category included all three tomato homologs of the Arabidopsis EC1 proteins, which are secreted by the egg cell upon interaction with the pollen sperm cell (Sprunck et al., 2012). However, a large number of genes had no known function, including a gene encoding a protein of 77 amino acids with extremely high levels of expression in ovules (an average of 19,682 RPM; Fig. 7), but relatively little expression in other fruit or vegetative tissues, except fruit entering the ripening stage (Fig. 7C; Supplemental Fig. S3C; Supplemental Methods and References S1). We have named this gene OVULE SECRETED PROTEIN (OSP; Solyc03g007780), as it has a signal peptide but no other known domains and has no known homologs other than an uncharacterized transcript from tobacco (Nicotiana tabacum; Koltunow et al., 1990). The preferential expression of this gene in ovules was confirmed by quantitative PCR (Supplemental Fig. S3B; Supplemental Methods and References S1) and by in situ hybridization, which showed expression of OSP in the tissue surrounding the embryo sac (Fig. 7, A and B; Supplemental Fig. S4). This cluster also contained genes encoding proteins associated with the secretory pathway, including dehydrodolichyl diphosphate synthase, an enzyme that catalyzes the formation of lipids used in the production of N-glycans (Cunillera et al., 2000), and two annexins, potentially involved in exocytosis (Carroll et al., 1998).
Figure 7.
Expression of OSP and ACO4 in ovules. RNA in situ hybridization of OSP (A and B) and ACO4 (D and E) is shown in ovaries at anthesis. Images show whole ovaries (A and D) and individual ovules (B and E). Signal (arrows) is detected as dark purple color. LCM-RNA-seq expression data are shown for OSP (C) and ACO4 (F). Sample labels are as follows: em, embryo; endo, endosperm; es, embryo sac; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; se, septum; and dpa, DPA. Bars = 100 μm (A and D) or 20 μm (B and E).
Cluster 23 also contained several genes encoding ethylene biosynthetic enzymes, including a 1-aminocyclopropane-1-carboxylic acid oxidase, ACO4, which was primarily expressed in the ovules. The predominant expression of ACO4 in the ovules was confirmed by in situ hybridization, which showed strong expression localized in the innermost layers around the embryo sac (Fig. 7, D–F; Supplemental Fig. S4).
Clusters 12 and 19 contained genes predominantly expressed in the embryo and endosperm, respectively (Fig. 6, B and C). The overrepresentation of functional categories such as protein degradation, protein targeting, and protein synthesis and folding suggests high rates of protein turnover in both of these tissues. Cluster 12 (embryo) also contained overrepresentation of transcription factors, possibly indicating a major shift in regulation of the embryo transcriptome compared with the other fruit tissues.
Cluster 14 corresponded to genes predominantly expressed in the seed coat and showed an abundance of genes associated with DNA replication, the cell cycle, and the cell wall (Fig. 6D), which may reflect rapid cell division and elongation during seed coat growth (Haughn and Chaudhury, 2005). Cluster 21, expressed in the funiculus, contained an overrepresentation of the transcription factor category (Fig. 6E). This cluster included AUXIN RESPONSE FACTOR3 (ARF3) and ARF19-1, which encode transcription factors involved in auxin signaling (Zouine et al., 2014), and homologs of the Arabidopsis transcription factors PLETHORA, SUPERMAN, and INDEHISCENT, which regulate auxin biosynthesis and activity (Nibau et al., 2011; Kay et al., 2013; Pinon et al., 2013), which is congruent with previous reports of high auxin activity in this tissue (Pattison and Catalá, 2012). This cluster also contained a large number of genes encoding kinases, in particular several members of Leu-rich repeat receptor-like kinase subfamily XI/XII (iTAK database family 1.12.4), such as the Arabidopsis HAESA proteins, which are expressed during abscission zone differentiation (Jinn et al., 2000). This suggests that the tomato HAESA homologs could be involved in specifying eventual seed abscission zones at the end of fruit development.
Clusters 5 and 6 had broader expression profiles that spanned several seed tissues (Fig. 6, F and G). Cluster 5 contained genes expressed in the seed coat, embryo, and endosperm and had an overrepresentation of genes in the hormonal function category, particularly genes related to auxin and GA activity. These included several members of the AUXIN (AUX)/INDOLE-3-ACETIC ACID (IAA) family of transcriptional regulators and several Gretchen Hagen (GH3) family genes, which encode enzymes putatively involved in auxin conjugation (Ludwig-Müller, 2011). Genes in cluster 6 were predominantly expressed in seed tissues but also showed relatively high expression in other fruit tissues, except the pericarp. Similar to cluster 3, this cluster had overrepresentation of the cell cycle/cell division and cell organization categories (Fig. 5C).
Predicted Regulatory Modules Link Transcription Factor Expression, Regulatory Motifs, and Coexpression Cluster Function
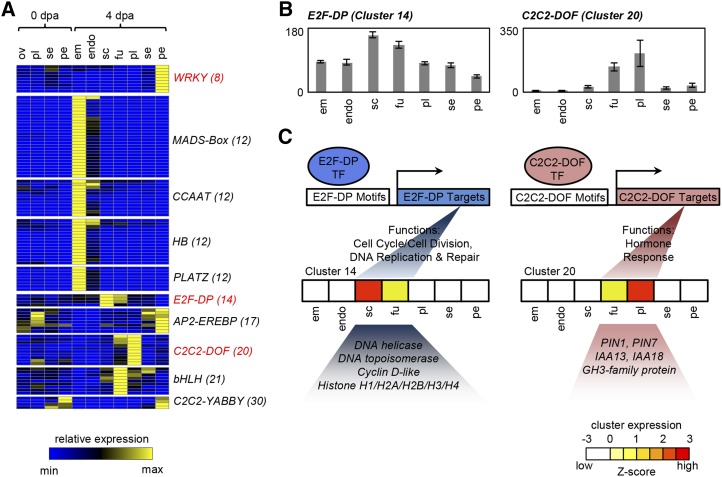
To investigate potential regulators of the transcriptome during early fruit development (Fig. 8), coexpressed gene clusters were examined for the overrepresentation of families of transcription factors or transcriptional regulators from the iTAK database (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi; Supplemental Table S7, A and B). The expression profiles of members of transcription factor families that were overrepresented in coexpressed gene clusters are shown in Figure 8A.
Figure 8.
Transcription factor profiling and examples of hypothetical regulatory modules. A, Heat map showing the expression of transcription factor families that are overrepresented among coexpression clusters. Cluster numbers are given in parentheses. B, Cumulative expression of E2F-DP (eight members) and C2C2-DOF (10 members) transcription factors in 4-DPA fruit. C, Hypothetical models of E2F- and C2C2-DOF-mediated transcriptional regulation of genes in cluster 14 (internal tissues) and cluster 20 (placenta). The expression of each cluster is represented by a heat map showing overlap with the respective transcription factor expression. Functional categories associated with each transcription factor are overrepresented in these clusters. E2F and C2C2-DOF proteins are suggested to interact with E2F- and C2C2-DOF-binding motifs, respectively, which are enriched in the promoters of genes in cluster 14 or cluster 20 (Supplemental Table S8). Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; se, septum; and dpa, DPA.
WRKY and C2C2-YABBY transcription factor families were overrepresented in clusters 8 and 30, respectively, both of which peak in the pericarp, either in fruit (cluster 8) or in both ovaries and fruit (cluster 30). WRKY transcription factors have been implicated in the regulation of the transcriptional response to both abiotic and biotic stresses (Pandey and Somssich, 2009; Chen et al., 2012), a functional category that is overrepresented among genes in cluster 8 (Fig. 5). The enrichment of C2C2-YABBY transcription factors in the pericarp is consistent with their reported role in Arabidopsis, where several family members, including CRABS CLAW, are involved in fruit development and show preferential expression in valve margins (Alvarez and Smyth, 2002).
Notably, cluster 12, which peaked in embryo, had overrepresentation of four separate families, MADS (for MINICHROMOSOME MAINTENANCE1, AGAMOUS, DEFICIENS, SERUM RESPONSE FACTOR) box, CCAAT-binding, homeobox (HB), and plant AT-rich sequence- and zinc-binding protein (PLATZ), consistent with previous findings that gene regulation during embryo development is controlled by a complex transcriptional network (Santos-Mendoza et al., 2008). In particular, several CCAAT transcription factors are expressed specifically in seeds (Laloum et al., 2013); accordingly, homologs of LEAFY COTYLEDON1, a key regulator of embryo development in Arabidopsis (Kwong et al., 2003), were found in this cluster.
E2F-DP, Basic Helix-Loop-Helix (bHLH), and C2C2-DOF are other transcription factor families overrepresented in gene clusters with the highest expression in seed or internal fruit tissues. The bHLH family, which comprises transcription factors that are known to act during gynoecium and fruit development in Arabidopsis (Gremski et al., 2007; Girin et al., 2011; Kay et al., 2013), was enriched in cluster 21, which peaked in the funiculus.
Coexpressed gene clusters were also analyzed for the enrichment of regulatory motifs in promoter regions (Supplemental Table S7, C and D). In several cases, transcription factors were overrepresented in the same coexpression clusters as their target motifs, and a correlation could be established with functional categories overrepresented in the clusters, allowing transcriptional modules to be predicted (Fig. 8, B and C; Supplemental Fig. S5). One such potential regulatory module linked E2F-DP transcription factors to cell division in internal fruit tissues (Fig. 8C). E2F-DP transcription factors are key regulators of the cell cycle associated with the G1-to-S phase transition and S phase progression, and their targets include various genes directly related to cell division and DNA replication (De Veylder et al., 2007). Several related motifs known to participate in E2F-DP binding, including E2F-binding site motif, E2FCONSENSUS, and E2FAT motifs (Supplemental Fig. S5), were overrepresented in clusters 3, 6, and 14, which span internal tissues but show lower expression in the pericarp. Likewise, E2F-DP transcription factor expression was highest in these tissues, and they were overrepresented in cluster 14, which peaked in the seed coat (Fig. 8B). The presence of E2F-DP transcription factors in these clusters is consistent with the overrepresentation of the cell cycle/cell division and DNA replication functional categories. Potential E2F-DP targets include genes encoding cyclins, kinetochore proteins, histones, and DNA topoisomerases, all of which contain E2F-binding site motifs in their promoters (Supplemental Table S8A).
In addition to the predicted E2F-DP-mediated transcriptional module, where the association between transcription factor, target genes, and biological function has been described in other systems, the motif analysis also uncovered novel potential associations. C2C2-DOF transcription factors were overrepresented in cluster 20, which peaked in the placenta, and two known C2C2-DOF-interacting motifs (-300CORE and NTBBF1ARROLB; Mena et al., 1998; Baumann et al., 1999) were overrepresented in the same cluster (Fig. 8C; Supplemental Fig. S5). The NTBBF1ARROLB motif is necessary for auxin induction of the Agrobacterium rhizogenes rolB oncogene in tobacco, likely mediated by the C2C2-DOF transcription factor NtBBF1 (Baumann et al., 1999), and is overrepresented between auxin-induced genes in Arabidopsis (Nemhauser et al., 2004). Cluster 20 contained a tomato homolog of NtBBF1 (Solyc08g082910) and several auxin-related genes with the NTBBF1ARROLB motif in their promoter regions. They included PIN-FORMED1 (PIN1) and PIN7, which encode auxin-efflux carriers and have three copies of the NTBBF1ARROLB motif in their promoters, two AUX/IAA family genes (IAA13 and IAA17) involved in auxin signal transduction, and a GH3 family gene involved in auxin conjugation (Supplemental Table S8C). Transcription of these genes, or their homologs, in other species is often auxin inducible (Hagen and Guilfoyle, 2002; Vieten et al., 2005; Audran-Delalande et al., 2012). We note that these predicted regulatory modules await further experimental validation.
Transcriptional Framework of Auxin Synthesis, Transport, and Signaling Components
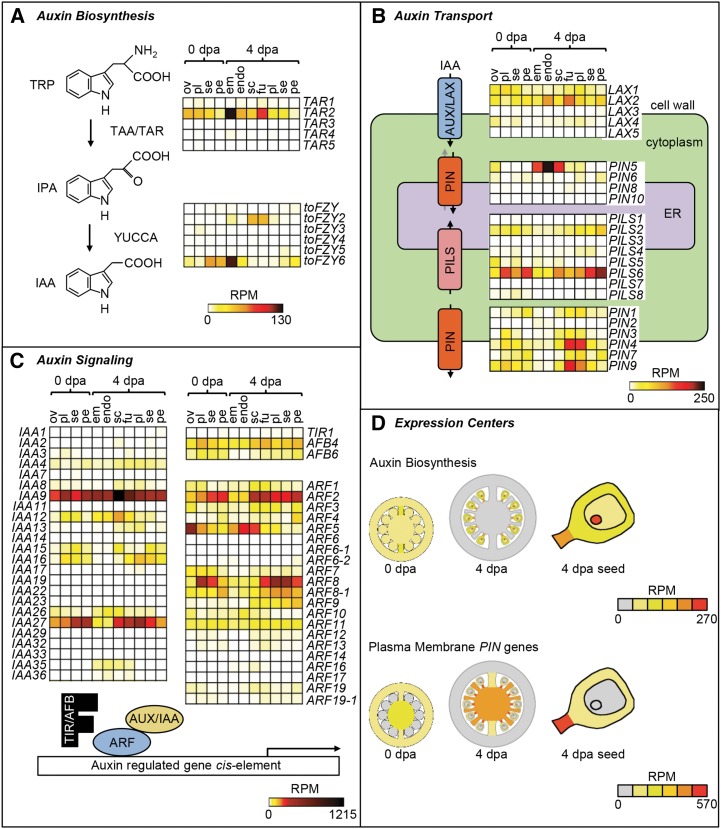
Despite auxin being a central regulator of tomato fruit development, current models of auxin distribution and activity lack detailed spatial resolution (Pattison et al., 2014). The LCM data set, therefore, was used to investigate auxin biosynthesis, transport, and signaling (Fig. 9; Supplemental Table S9). Only three genes, TRYPTOPHAN AMINOTRANSFERASE RELATED2 (TAR2) and the YUCCA genes tomato FLOOZY2 (toFZY2) and toFZY6 (Expósito-Rodríguez et al., 2011), encoding TAR and YUCCA enzymes in the major auxin biosynthetic pathway (Brumos et al., 2014), were expressed at levels greater than 10 RPM. They increased in expression in 4-DPA fruit relative to ovaries and accumulated primarily in seed tissues and the funiculus, with TAR2 and toFZY6 showing peak expression in the embryo (Fig. 9A).
Figure 9.
Profile of auxin-related gene expression. A to C, Heat maps showing absolute expression values (RPM) for genes related to auxin biosynthesis (A), transport (B), and signaling (C). D, Graphical illustration of the center of expression for genes encoding auxin biosynthetic enzymes and plasma membrane-localized PIN genes. Coloring of tissues represents the cumulative expression of each gene category in 0-DPA (dpa) ovaries, 4-DPA fruit, and 4-DPA seed. Sample labels are as follows: em, embryo; endo, endosperm; fu, funiculus; ov, ovules; pe, pericarp; pl, placenta; sc, seed coat; and se, septum.
Auxin transporters also generally showed increased expression in 4-DPA fruit compared with ovaries (Fig. 9B). The influx carrier LIKE-AUX2 and the plasma membrane-targeted efflux carriers PIN1, PIN3, PIN4, PIN7, and PIN9 displayed the highest expression, which peaked in either the funiculus or placenta at 4 DPA. In contrast, PIN transporters thought to localize to the endomembrane system (Mravec et al., 2009; Dal Bosco et al., 2012) showed a divergent expression profile. Specifically, PIN5 was highly expressed in endosperm, whereas PILS2 and PILS5, PIN-LIKE proteins found in the endoplasmic reticulum in Arabidopsis (Barbez et al., 2012), showed highest expression in the pericarp of 4-DPA fruit (Fig. 9B).
Key components of auxin signaling include the TRANSPORT INHIBITOR RESPONSE (TIR)/AFB family of auxin-receptor F-box proteins, AUX/IAA coreceptors, and ARF transcription factors (Sauer et al., 2013), which had broad patterns of expression (Fig. 9C). Within the TIR/AFB genes, AFB4 and AFB6 were expressed widely, with no evidence of tissue specificity. The most highly expressed AUX/IAA and ARF genes were IAA9 (Wang et al., 2005) and IAA27 (Bassa et al., 2012), which have previously been implicated in fruit set and development, and three ARF genes with peak expression in ovule and seed (ARF5) or in other fruit tissues (ARF2 and ARF8).
In contrast to the relatively diverse expression patterns of auxin signaling genes, common centers of expression could be defined related to auxin biosynthesis and transport (Fig. 9D). The cumulative expression of auxin biosynthesis genes was higher in 4-DPA fruit relative to anthesis and restricted to seed tissues. Similarly, the cumulative expression of genes encoding plasma membrane-localized PIN proteins increased in 4-DPA fruit and concentrated in the funiculus and placenta.
Spatiotemporal Distribution of Hormone-Related Gene Expression
The data set was also used to profile gene expression related to the metabolism and signaling of other major hormones (Supplemental Fig. S6; Supplemental Table S9; Supplemental Methods and References S1). In each case, the expression of individual genes showed evidence of strong tissue and developmental stage dependency. However, common trends and coexpression were evident among genes involved in the metabolism and signaling of certain hormones. For example, several genes with a role in brassinosteroid biosynthesis were preferentially expressed in the ovary and fruit pericarp (Supplemental Fig. S6C). These include genes encoding squalene epoxidase, cycloartenol synthase, and homologs of STEROL METHYLTRANSFERASE, FACKEL, DWARF1 (DWF1), DWF5, and DWF7, which are all implicated in the biosynthesis of the steroid precursors of brassinosteroids (Clouse, 2011). However, these steroid precursors are not necessarily specific for brassinosteroids and, as discussed above (see cluster 30), may reflect the biosynthesis of other isoprenoid-derived metabolites.
We also observed common patterns of expression among genes related to ethylene metabolism and signaling (Supplemental Fig. S6E). Several ethylene biosynthetic genes, such as ACO4 and ACO5 and a 1-aminocyclopropane-1-carboxylic acid synthase family gene, together with the ethylene receptor gene ETR6, were highly expressed in the ovule at anthesis but were then strongly down-regulated in the fruit. Similarly, other sets of ethylene biosynthetic and signaling genes preferentially expressed in the ovary placenta or septum were strongly down-regulated in 4-DPA fruit.
In contrast to the down-regulation of genes related to ethylene, several key genes in GA biosynthesis were expressed at higher levels in developing fruit than ovaries (Supplemental Fig. S6F; Supplemental Table S9). The expression of GA20ox1, GA20ox2, and GA20ox3, which encode GA 20-oxidase, a rate-limiting enzyme, was highest in the funiculus, and two uncharacterized GA20ox homologs showed highest expression in the embryo and endosperm. In contrast, GA2ox4, encoding a GA 2-oxidase that converts bioactive GAs to inactive forms (Serrani et al., 2008), was highly expressed in ovules and placenta in anthesis-stage ovaries but was strongly down-regulated in fruit tissues (Supplemental Fig. S6F; Supplemental Table S9).
DISCUSSION
A comprehensive transcriptome profile of the major tissue types comprising the tomato ovary and young fruit was obtained using LCM and RNA-seq, revealing enormous diversity in gene expression associated with tissue type and developmental stage. The tissue-specific profiles indicated a dramatic shift in gene expression during the transition from ovary to fruit, of greater magnitude than the differences between individual tissues, highlighting the central role of fruit set in shaping the transcriptome (Fig. 3). Gene expression was highly dynamic, and while many genes were expressed constitutively, very few were expressed evenly among all the tissues. This emphasizes the importance of considering tissues individually and highlighting potential pitfalls inherent in analyzing the fruit as a whole. Differential gene expression analysis uncovered distinct tissue- and stage-associated gene sets (Fig. 2), including rare transcripts expressed in typically understudied tissues, such as the funiculus. Several of the tissue-associated genes have no known function and represent potential novel regulators of fruit development (Supplemental Table S4).
Tissue-Specific Analysis of Transcriptional Dynamics Outlines a Map of Core Molecular Processes Underlying Fruit Formation
Coexpressed gene cluster analysis illustrated the spatial distribution of core molecular events in early fruit development and suggested that transcriptome specialization is already established at the ovary stage for some biological processes. At anthesis, genes related to secondary metabolism and photosynthesis were already overrepresented among pericarp- and placenta/septum-associated clusters, respectively, and this profile was maintained in young fruit (Fig. 4). This suggests an early metabolic specialization of the pericarp for the production of isoprenoid-derived metabolites, such as carotenoids, neoxanthin, and the phytol tail of chlorophylls, all of which accumulate in young fruit (Carrari et al., 2006). Our results also indicate that photosynthesis-related genes are tightly coexpressed and show strong tissue dependency, being expressed at higher levels in internal tissues. The elevated expression of these genes in internal tissues may seem counterintuitive but is consistent with previous studies suggesting that parenchymatous tissues surrounding the seeds may play an active role in CO2 scavenging and the provision of carbon assimilates to the seed (Smillie et al., 1999; Lytovchenko et al., 2011). However, the significance of fruit photosynthesis in the support of fruit and/or seed growth is still controversial (Cocaliadis et al., 2014).
The identification of genes associated with single tissues and stages highlighted a number of specialized features of the ovary and fruit transcriptomes (Fig. 5). Cluster 28 revealed the importance of cell wall remodeling by active pectin metabolism in the growing pericarp of the ovary at anthesis, which correlates with reports of rapid pectin synthesis and demethylesterification in the pericarp during the period spanning fruit set and early growth (Terao et al., 2013). In addition, the enrichment of genes within the transport category in clusters 27 and 28 associated with the anthesis-stage septum and pericarp, respectively, is indicative of the active transport of mineral nutrients in the septum and of small molecules likely to be involved in secondary metabolism in the pericarp. A distinct gene set associated with the pericarp of 4-DPA fruit is related to stress responses, perhaps reflecting the fact that the pericarp is at the boundary of the fruit and the external environment. In other cases, a functional relationship was apparent between gene clusters in equivalent tissues but at different stages, even though the genes themselves were different. For example, different genes formed clusters preferentially expressed in the septum of ovaries and young fruit, but both clusters had overrepresentation of genes related to lipid metabolism. Likewise, hormone-related gene expression appeared to be significant in the placenta both at anthesis and in growing fruit (Fig. 5, A and B). The presence of separate gene sets at anthesis and in young fruit, but with seemingly overlapping roles, suggests that the actions of specific members of a gene family are required at different stages of development as a fine-tuning mechanism.
In contrast to specialized transcriptional activity associated with single tissues, there is also evidence of shared gene expression programs among fruit tissues at 4 DPA. This is exemplified by clusters 3 and 6, which indicate a functional partition between internal tissues and pericarp. These clusters were absent in the pericarp but either spanned all internal tissues (cluster 6) or all internal tissues except embryo and endosperm (cluster 3) and were characterized by genes related to cell division. This indicates an activation of cytokinesis-related gene expression in the rapidly proliferating inner tissues of young fruit. In contrast, the pericarp and septum shared a common expression of genes (cluster 9) involved in cell wall modification processes commonly associated with cell expansion (Cosgrove, 2005).
Gene Sets Associated with Seed Development
We also examined the ovule and early seed transcriptomes, which are often omitted from tomato fruit transcriptome studies (Vriezen et al., 2008; Osorio et al., 2011). Analysis of a gene cluster predominantly expressed in the ovules revealed the presence of a set of genes encoding small proteins that are predicted to be secreted and also of secretion-associated factors, which were strongly down-regulated post anthesis. The functional relevance of this gene set may be related to a role of small secreted proteins in pollen tube guidance (Chae and Lord, 2011) or in gamete interaction (Sprunck et al., 2012). Many genes encoding small secreted proteins are expressed in Arabidopsis embryo sacs, where they may function in extracellular signaling during embryo sac development and fertilization (Jones-Rhoades et al., 2007). Although mostly annotated as of unknown function, the corresponding tomato proteins may play a similar role. Of particular interest is OSP, which had extremely high expression in ovules (Fig. 7, A–C) and for which database searches revealed no apparent homologs outside non-Solanaceae species. This underscores the value of the high spatial resolution of this study in uncovering genes likely to play a specialized role in the regulation of reproductive development.
Several coexpressed gene sets were associated with various regions of the developing seed (Fig. 6, B–G). Cluster 6 contained many genes related to cell division and spanned multiple seed tissues, possibly reflecting the need to coordinate nuclear division and cell proliferation across the embryo, endosperm, seed coat, and tissues surrounding the seed upon fertilization (Nowack et al., 2010). Another gene set, cluster 5, indicates a spatial focus of hormone-related gene expression and suggests the coregulation of genes involved in controlling the levels of auxin in seed tissues. Up-regulation of GH3 family genes, putatively involved in auxin conjugation, in seeds relative to ovules has also been described in strawberry (Fragaria vesca; Kang et al., 2013) and may represent a mechanism for reducing active auxin concentration following fruit set.
In addition, we found clusters that revealed regulatory programs associated with specific seed tissues. Of particular note was cluster 21, associated with the funiculus, a tissue that is seldom studied but one that our data indicate has importance in regulating hormone activity (Fig. 6E; Supplemental Table S5).
Integration of Transcription Factor Expression and Regulatory Motif Analysis Uncovers Potential Gene Expression Networks
The LCM-RNA-seq data set can serve as a resource to develop and test hypotheses regarding the regulatory networks that govern early fruit development, in part by delineating the spatial limits of transcription factor expression. For example, MADS box, CCAAT, HB, and PLATZ transcription factors were found to accumulate preferentially in the embryo and are likely part of the transcriptional regulatory complex controlling the gene set in cluster 12. Two other transcription factor families, C2C2-YABBY and WKRY, are likely involved in regulating pericarp development and stress responses in this tissue, respectively (Fig. 8A).
It is significant that a tomato homolog of INDEHISCENT is found in cluster 21 expressed in the funiculus. INDEHISCENT is an important regulator or auxin activity involved in specifying the valve margins of Arabidopsis siliques and controlling auxin distribution via the regulation of genes involved in auxin transport (Sorefan et al., 2009). As several genes mediating auxin transport are also predominantly expressed in the funiculus (Fig. 9), we propose that INDEHISCENT may control auxin distribution in the funiculus.
Interestingly a potential E2F-mediated network controlling cell division was found to be predominantly restricted to the internal tissues, further highlighting the separation between the rapidly dividing internal tissues and the elongating pericarp and suggesting molecular regulators of division-based processes in the fruit. In addition, a novel transcriptional association between C2C2-DOF transcription factors and their potential target genes involved in auxin transport and signaling was suggested. C2C2-DOF transcription factors regulate diverse processes, including seed development, light responses, and hormone signal transduction (Yanagisawa, 2004). However, the potential role of NtBBF1 and other C2C2-DOF transcription factors in the auxin induction of endogenous plant genes is still unknown, and our data suggest that they may act as upstream regulators of auxin homeostasis in the fruit (Fig. 8, B and C). These networks present hypotheses that may be tested by future experimental work.
A Map of Hormone-Related Gene Expression during Early Fruit Formation
An analysis of gene families involved in the biosynthesis and signaling of different plant hormones suggested spatiotemporal specialization of distinct family members and demonstrates that the data set can be used to identify biologically relevant genes based on expression profile. For example, within the multigene TAR and YUCCA/toFZY families, only TAR2, toFZY2, and toFZY6 were expressed in ovaries and young fruit (Fig. 9A). Interestingly, the expression profiles of these genes in the seed and funiculus overlap with maximal auxin activity, as indicated by DR5 reporter expression (Pattison and Catalá 2012), suggesting that they are the main genes involved in auxin synthesis in developing seeds. Common centers of gene expression for auxin biosynthesis and transport genes (Fig. 9D) are consistent with evidence that the seeds are the predominant site of auxin biosynthesis (Mapelli et al., 1978), and the funiculus and placenta represent important transport routes (Pattison and Catalá, 2012). The diverse expression profiles of genes related to auxin signaling (Fig. 9B) may constitute the basis for tissue-specific responses, as has been reported in Arabidopsis (Weijers et al., 2005; Hayashi, 2012) and suggests diversification of signaling pathways between the seed and surrounding fruit tissues.
The data related to GA metabolism (Supplemental Fig. S6F) are consistent with previous reports of increased bioactive GA content and GA20ox transcription after pollination (Serrani et al., 2007, 2008; de Jong et al., 2011). However, it was not previously known that a GA20ox transcript increases particularly in the funiculus, which is also a site of high auxin activity (Pattison and Catalá, 2012; Fig. 9F). Auxin and GA signaling are known to interact during fruit set and development, and it is thought that auxin activity after fertilization induces GA biosynthesis by increasing GA20ox transcription (Serrani et al., 2008). We suggest that up-regulation of GA20ox transcription in the funiculus is controlled by the high auxin activity in this tissue.
We also saw evidence for the involvement of different members of gene families associated with ethylene biosynthesis and signaling according to tissue type (Supplemental Fig. S6E), consistent with a general down-regulation of ethylene-related expression after anthesis (Vriezen et al., 2008; Wang et al., 2009). Pollination is known to induce a burst of ethylene in tomato ovaries accompanied by a transient increase in ACO gene expression (Llop-Tous et al., 2000), and it has been suggested that ethylene is involved in controlling ovule senescence and the fruit-set response (Carbonell-Bejerano et al., 2010). By spatially resolving gene expression, our data implicate the ovule as a likely site of ethylene biosynthesis in ovaries, with specific ACO isoforms, such as ACO4 (Fig. 7, D–F), playing a key role during fruit set.
In conclusion, the LCM-RNA-seq data set represents a valuable new resource that can be queried to identify patterns of gene expression associated with individual tissue type and developmental stage. We have illustrated this utility with several examples of how tissue-specific profiling can provide biological information that may be masked in composite samples, adding insights into the cellular processes that occur in the diverse fruit tissues and the hormonal control underpinning these processes. This includes evidence of separate transcriptional programs related to cell division, photosynthesis, and auxin transport in internal tissues and of secondary metabolism, cell wall modification, and stress responses in the pericarp. The existence of tightly coexpressed gene groups and highly ordered gene expression networks further underlines the value of the LCM-RNA-seq approach as a platform to investigate regulatory processes during early fruit development.
MATERIALS AND METHODS
Plant Material and Sample Collection
Plant material was harvested from greenhouse-grown Solanum pimpinellifolium (LA1589) plants (26°C/20°C day/night conditions, 16-h photoperiod with supplemental 400-W sodium lights). Ovaries at 0 DPA were identified based on the definition of Xiao et al. (2009) and harvested immediately before the point when they would otherwise be self-fertilized. For 4-DPA fruit collection, flowers were manually pollinated at anthesis, and fruit was harvested 4 d later. The extreme abaxial and adaxial ends of the fruit were removed before further processing of the medial segment.
Histology
Tissue was fixed with 3.7% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol and infiltrated with paraffin using standard protocols (Ruzin, 1999). Ten-micrometer sections were prepared using a Leica RM2255 microtome and stained with Safranin O and Astra Blue as described (Xiao et al., 2009).
For LCM, tissue was fixed overnight in ice-cold Farmer’s fixative (3:1 ethanol:acetic acid) and processed essentially as described by Matas et al. (2011). Briefly, fixed tissue was vacuum infiltrated sequentially with 1× phosphate-buffered saline medium containing 10% and 20% (w/v) Suc before embedding in Tissue Tek O.C.T. medium (Sakura). Fourteen-micrometer cryosections were prepared using an HM 550 cryostat (Thermo Fisher) and transferred to CryoJane 4× CFSA slides (Leica). Slides were dehydrated using a graded ethanol-HistoClear II (National Diagnostics) series consisting of 70%, 95%, and 100% (v/v) ethanol, and two washes of 100% (v/v) HistoClear II.
Laser Microdissection and RNA Isolation
Tissues were isolated using a P.A.L.M microbeam LCM system (Carl Zeiss) and a minimum 2.5- × 105-μm2 area, pooled from sections of at least five ovaries/fruits, collected for each sample. Embryo- and endosperm-enriched areas were identified on the basis of tissue density, which was higher for the tightly packed embryo cells than the less compacted endosperm. Total RNA was isolated using the RNeasy Micro kit (Qiagen), and RNA amplification was performed using the TargetAmp two-round aRNA amplification kit (Epicentre). Three biological replicates were collected from independent plant material for each sample.
RNA-seq
Strand-specific RNA-seq libraries were prepared as described (Zhong et al., 2011) omitting the poly(A) isolation step and using 6-bp barcode adapters to allow multiplexed sequencing. Equal amounts of up to six barcoded libraries were pooled prior to sequencing. Libraries were sequenced from a single end for 50 cycles using the Illumina HiSeq 2000 platform at the Genomics Resources Core Facility Cornell at Weill Cornell Medical College.
Read Mapping and Transcript Quantification
Raw RNA-seq reads were first aligned to a ribosomal RNA sequence database (Quast et al., 2013) using Bowtie, allowing three mismatches (Langmead et al., 2009), and the mapped reads were discarded. The resulting cleaned reads were aligned to the tomato (Solanum lycopersicum) genome (version 2.40; Tomato Genome Consortium, 2012) using Tophat, allowing one segment mismatch (Trapnell et al., 2009). As a result of RNA amplification using poly(A)-based priming techniques (Van Gelder et al., 1990), the majority of reads aligned toward the 3′ end of the gene models. Thus, sense-strand reads were counted after extending the tomato gene models 1 kb upstream of the translational start site and 1 kb downstream of the translational stop site, to include reads from untranslated region sequences that may not have been annotated. As in previous LCM studies (Takacs et al., 2012), read counts were normalized to the total number of reads mapped (RPM).
Expression Data Analysis
Differentially expressed genes were identified using EdgeR (Robinson et al., 2010). Genes were considered differentially expressed if the adjusted P value was less than 0.05.
Principal component analysis based on all expressed genes was performed on the replicate averaged data using cluster 3 (de Hoon et al., 2004). Only genes that had an average RPM value of 2 or more in at least one sample were included in the analysis. RPM values were centered around the mean and normalized using the sum-of-squares method. Hierarchical clustering of samples and K-means clustering were performed using Gene-E (www.broadinstitute.org/cancer/software/GENE-E/) and 1 − Pearson correlation as the distance metric. All differentially expressed genes from all comparisons were used for hierarchical clustering with the complete linkage method. Genes that were differentially expressed between tissues within a developmental stage, between equivalent tissues across stages and between 0-DPA ovule and 4-DPA embryo, endosperm, or seed coat, were used for K-means clustering. The Z-score was calculated for each gene per sample using the formula (X − Xav)/sd, where X is the average RPM value in a particular sample, and Xav and sd are the mean and sd of the average RPM values across all 11 samples, respectively. The expression profile of each cluster is represented by the maximum, minimum, first quartile, third quartile, and average Z scores for all genes in each cluster. Only genes that were correlated with the average profile of each cluster (Pearson correlation > 0.8) were retained for further analysis. This resulted in some genes not fitting closely to any of the defined clusters being excluded from this analysis.
Genes were functionally categorized based on previously defined Mapman bins (Thimm et al., 2004), with manual reassignment according to tomato genome annotations (Tomato Genome Consortium, 2012; Supplemental Table S6). Overrepresentation was determined using a hypergeometric test, and P values were adjusted for multiple testing using Benjamini-Hochberg correction (using the phyper function and multtest package in R; www.bioconductor.org/packages/2.14/bioc/html/multtest.html).
Promoter sequences corresponding to 1 kb upstream of the transcription start site for genes with defined transcription start sites (Zhong et al., 2013) or 1 kb upstream of the translational start site for other genes were analyzed using PatMatch (Yan et al., 2005) for the presence of motifs collected from the PLACE (Higo et al., 1999), PlantCARE (Lescot et al., 2002), and AtcisDB (Yilmaz et al., 2011) databases. A hypergeometric test was used to generate P values adjusted using the Benjamini-Hochberg correction.
Chlorophyll Autofluorescence and Starch Staining
Chlorophyll was detected in hand sections from 4- or 8-DPA fruit (to allow better resolution of fruit anatomy) using bright-field or fluorescence microscopy. Chlorophyll fluorescence was viewed using a Leica DM5500 epifluorescence microscope with excitation filter BP 450 to 490 nm and emission filter LP 515 nm.
For starch staining, paraffin sections were prepared as described above. Slides were deparaffinized in HistoClear II, passed through a graded ethanol series (100%, 95%, 70%, and 50% [v/v] ethanol), stained in Safranin O (1% [w/v] in 50% [v/v] ethanol) for 10 min, washed in 50% (v/v) ethanol and distilled water, and then stained in Lugol’s iodine solution (0.5% [w/v] iodine and 1% [w/v] potassium iodide) for 10 min. Sections were then dehydrated via an ethanol/HistoClear II series (50%, 70%, and 100% [v/v] ethanol followed by 100% [w/v] HistoClear II) and then mounted in Permount (Fisher Scientific).
In Situ Hybridization
Tissue was fixed in 4% (v/v) formaldehyde, 0.1% (v/v) Tween 20, and 0.1% (v/v) Triton X-100, dehydrated, and embedded in paraffin. In situ hybridization was performed using digoxigenin-labeled probes as described by Wu et al. (2011) with minor modifications. Gene-specific fragments for probe synthesis were amplified by PCR using primers 5′-ATGGCTTCCATGAAGTTGC-3′ and 5′-TCAGATACGAAGAAAATTATTAGGG-3′ for OSP and 5′-GAACAGATACCAAACACAAGCAA-3′ and 5′-AAACCCCAGTTTTCACATGC-3′ for ACO4. Sections were treated with 1 μg mL−1 proteinase K for 30 min at 37°C. After hybridization, slides were washed with 2× SSC and 0.2× SSC in 50% (v/v) formamide.
RNA-seq data have been deposited in the Tomato Functional Genomics Database and are available at http://ted.bti.cornell.edu. Illumina reads have been submitted to the Sequencing Read Archive at the National Center for Biotechnology Information (accession no. SRP048699).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. K-means clustering of the ovary and fruit transcriptome.
Supplemental Figure S2. Expression of genes related to starch metabolism.
Supplemental Figure S3. Quantitative reverse transcription-PCR validation of RNA-seq expression profiling and expression of OSP in various tissues of S. pimpinellifolium.
Supplemental Figure S4. RNA in situ negative controls.
Supplemental Figure S5. Putative regulatory modules in early fruit development.
Supplemental Figure S6.> Expression profiling of hormone-related genes.
Supplemental Table S1. Mapping statistics.
Supplemental Table S2. Gene expression in ovaries and developing fruits.
Supplemental Table S3. Transcript representation in the LCM-RNA-seq data set.
Supplemental Table S4. Tissue- and stage-associated genes at 0 DPA and 4 DPA.
Supplemental Table S5. K-means clustering of the ovary and fruit transcriptome.
Supplemental Table S6. Overrepresentation of functional category annotations among coexpressed gene clusters.
Supplemental Table S7. Overrepresentation of transcription factors and regulatory motifs among coexpressed gene clusters.
Supplemental Table S8. Target genes of putative regulatory modules in early fruit development.
Supplemental Table S9. Expression of genes related to hormone metabolism, transport, and signaling.
Supplemental Methods and References S1. Methods and references for the “Supplemental Data.”
Supplementary Material
Acknowledgments
We thank Michael J. Scanlon for the use of the laser microdissection instrumentation, Antonio Matas Arroyo for advice on performing laser microdissection and RNA amplification and assistance with data analysis, and Jocelyn Rose for reading the article and providing helpful comments.
Glossary
- LCM
laser-capture microdissection
- RNA-seq
RNA sequencing
- RPM
reads per million mapped reads
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program (grant nos. DBI–0922661 and IOS–1339287).
Articles can be viewed without a subscription.
References
- Alvarez J, Smyth DR (2002) CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int J Plant Sci 163: 17–41 [Google Scholar]
- Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53: 659–672 [DOI] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122 [DOI] [PubMed] [Google Scholar]
- Bassa C, Mila I, Bouzayen M, Audran-Delalande C (2012) Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol 53: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca J, Cañizares J, Cordero L, Pascual L, Diez MJ, Nuez F (2012) Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS ONE 7: e48198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al. (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99: 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Alonso JM, Stepanova AN (2014) Genetic aspects of auxin biosynthesis and its regulation. Physiol Plant 151: 3–12 [DOI] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA (2010) A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol 154: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AD, Moyen C, Van Kesteren P, Tooke F, Battey NH, Brownlee C (1998) Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. Plant Cell 10: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Lord EM (2011) Pollen tube growth and guidance: roles of small, secreted proteins. Ann Bot (Lond) 108: 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocaliadis MF, Fernández-Muñoz R, Pons C, Orzaez D, Granell A (2014) Increasing tomato fruit quality by enhancing fruit chloroplast function: a double-edged sword? J Exp Bot 65: 4589–4598 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Arró M, Forés O, Manzano D, Ferrer A (2000) Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett 477: 170–174 [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, Eimer S, Hegermann J, Paponov IA, Ruperti B, et al. (2012) The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J 71: 860–870 [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, García-Martínez JL, Mariani C, Vriezen WH (2011) The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J Exp Bot 62: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2011) Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol Biochem 49: 782–791 [DOI] [PubMed] [Google Scholar]
- Fuentes S, Vivian-Smith A (2009) Fertilisation and fruit initiation. In Østergaard L, ed, Annual Plant Reviews, Vol 38: Fruit Development and Seed Dispersal. Wiley-Blackwell, Oxford, UK, pp 107–171 [Google Scholar]
- Gasser CS, Robinson-Beers K (1993) Pistil development. Plant Cell 5: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, Paicu T, Stephenson P, Fuentes S, Körner E, O’Brien M, Sorefan K, Wood TA, Balanzá V, Ferrándiz C, et al. (2011) INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23: 3641–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Vivian-Smith A, Koltunow AM (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF (2007) The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601 [DOI] [PubMed] [Google Scholar]
- Gupta S, Shi X, Lindquist IE, Devitt N, Mudge J, Rashotte AM (2013) Transcriptome profiling of cytokinin and auxin regulation in tomato root. J Exp Bot 64: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Hayashi K. (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay P, Groszmann M, Ross JJ, Parish RW, Swain SM (2013) Modifications of a conserved regulatory network involving INDEHISCENT controls multiple aspects of reproductive tissue development in Arabidopsis. New Phytol 197: 73–87 [DOI] [PubMed] [Google Scholar]
- Kimura S, Sinha N (2008) Tomato (Solanum lycopersicum): a model fruit-bearing crop. Cold Spring Harb Protoc 2008: pdb.emo105. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2: 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18: 157–166 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139: 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llop-Tous I, Barry CS, Grierson D (2000) Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol 123: 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löw D, Brändle K, Nover L, Forreiter C (2000) Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo. Planta 211: 575–582 [DOI] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59: 553–564 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A, Eickmeier I, Pons C, Osorio S, Szecowka M, Lehmberg K, Arrivault S, Tohge T, Pineda B, Anton MT, et al. (2011) Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol 157: 1650–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli S, Frova C, Torti G, Soressi GP (1978) Relationship between set, development and activities of growth regulators in tomato fruits. Plant Cell Physiol 19: 1281–1288 [Google Scholar]
- Martin LBB, Fei Z, Giovannoni JJ, Rose JKC (2013) Catalyzing plant science research with RNA-seq. Front Plant Sci 4: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo S, Kikuchi K, Fukuda M, Honda I, Imanishi S (2012) Roles and regulation of cytokinins in tomato fruit development. J Exp Bot 63: 5569–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 16: 53–62 [DOI] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Yokota T, Farrar K, Harrison K, Jones JD, Kaneta T, Kamiya Y, Szekeres M, Bishop GJ (2005) Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J 42: 262–269 [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T (2006) Laser microdissection of plant tissue: what you see is what you get. Annu Rev Plant Biol 57: 181–201 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH (2001) A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52: 881–889 [DOI] [PubMed] [Google Scholar]
- Nibau C, Di Stilio VS, Wu HM, Cheung AY (2011) Arabidopsis and tobacco superman regulate hormone signalling and mediate cell proliferation and differentiation. J Exp Bot 62: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Ungru A, Bjerkan KN, Grini PE, Schnittger A (2010) Reproductive cross-talk: seed development in flowering plants. Biochem Soc Trans 38: 604–612 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Suzuki M, Kikuchi J, Saito K, Muranaka T (2009) Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc Natl Acad Sci USA 106: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N, Litt A (2011) Comparative anatomical and developmental analysis of dry and fleshy fruits of Solanaceae. Am J Bot 98: 1415–1436 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Schatz MC, Lippman ZB (2012) Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci USA 109: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Catalá C (2012) Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J 70: 585–598 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Csukasi F, Catalá C (2014) Mechanisms regulating auxin action during fruit development. Physiol Plant 151: 62–72 [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B (2013) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA 110: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Bouzayen M, Osorio S, Fernie AR (2012) Molecular regulation of seed and fruit set. Trends Plant Sci 17: 656–665 [DOI] [PubMed] [Google Scholar]
- Ruzin SE. (1999) Plant Microtechnique and Microscopy. Oxford University Press, Oxford [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Sauer M, Robert S, Kleine-Vehn J (2013) Auxin: simply complicated. J Exp Bot 64: 2565–2577 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Petreikov M (1997) Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol 113: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J, Mathys J, De Coninck BMA, Cammue BPA, De Bolle MFC (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46: 941–950 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, García-Martínez JL (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J 56: 922–934 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuán R, Ruiz-Rivero O, Fos M, García-Martínez JL (2007) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol 145: 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie RM, Hetherington SE, Davies WJ (1999) Photosynthetic activity of the calyx, green shoulder, pericarp, and locular parenchyma of tomato fruit. J Exp Bot 50: 707–718 [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L (2009) A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Østergaard L (2009) Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol 1: a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs EM, Li J, Du C, Ponnala L, Janick-Buckner D, Yu J, Muehlbauer GJ, Schnable PS, Timmermans MC, Sun Q, et al. (2012) Ontogeny of the maize shoot apical meristem. Plant Cell 24: 3219–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M, Ron M, Brady SM (2011) Novel biological insights revealed from cell type-specific expression profiling. Curr Opin Plant Biol 14: 601–607 [DOI] [PubMed] [Google Scholar]
- Terao A, Hyodo H, Satoh S, Iwai H (2013) Changes in the distribution of cell wall polysaccharides in early fruit pericarp and ovule, from fruit set to early fruit development, in tomato (Solanum lycopersicum). J Plant Res 126: 719–728 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 87: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wiśniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W (2012) Structure and dynamics of the isoprenoid pathway network. Mol Plant 5: 318–333 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177: 60–76 [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Xiao H, Cabrera A, Meulia T, van der Knaap E (2011) SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Radovich C, Welty N, Hsu J, Li D, Meulia T, van der Knaap E (2009) Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY (2005) PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 33: W262–W266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45: 386–391 [DOI] [PubMed] [Google Scholar]
- Yazaki K. (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Huang W, Chatterjee S, Viart HMF, Clausen MH, Stark RE, Rose JKC (2014) Tomato Cutin Deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. Plant J 77: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen Y, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, Audran C, Roustan JP, Bouzayen M (2014) Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 9: e84203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.