A PHYL1 effector protein interferes with miR396-mediated transcriptional regulator mRNA decay, enhancing the transcription factor for abnormal flower development.
Abstract
Leafy flowers are the major symptoms of peanut witches’ broom (PnWB) phytoplasma infection in Catharanthus roseus. The orthologs of the phyllody symptoms1 (PHYL1) effector of PnWB from other species of phytoplasma can trigger the proteasomal degradation of several MADS box transcription factors, resulting in leafy flower formation. In contrast, the flowering negative regulator gene SHORT VEGETATIVE PHASE (SVP) was up-regulated in PnWB-infected C. roseus plants, but most microRNA (miRNA) genes had repressed expression. Coincidentally, transgenic Arabidopsis (Arabidopsis thaliana) plants expressing the PHYL1 gene of PnWB (PHYL1 plants), which show leafy flower phenotypes, up-regulate SVP of Arabidopsis (AtSVP) but repress a putative regulatory miRNA of AtSVP, miR396. However, the mechanism by which PHYL1 regulates AtSVP and miR396 is unknown, and the evidence of miR396-mediated AtSVP degradation is lacking. Here, we show that miR396 triggers AtSVP messenger RNA (mRNA) decay using genetic approaches, a reporter assay, and high-throughput degradome profiles. Genetic evidence indicates that PHYL1 plants and atmir396a-1 mutants have higher AtSVP accumulation, whereas the transgenic plants overexpressing MIR396 display lower AtSVP expression. The reporter assay indicated that target-site mutation results in decreasing the miR396-mediated repression efficiency. Moreover, degradome profiles revealed that miR396 triggers AtSVP mRNA decay rather than miRNA-mediated cleavage, implying that AtSVP caused miR396-mediated translation inhibition. We hypothesize that PHYL1 directly or indirectly interferes with miR396-mediated AtSVP mRNA decay and synergizes with other effects (e.g. MADS box transcription factor degradation), resulting in abnormal flower formation. We anticipate our findings to be a starting point for studying the posttranscriptional regulation of PHYL1 effectors in symptom development.
Phytoplasmas are obligate bacteria without cell walls that infect numerous plants, causing severe agricultural losses (Lee et al., 2000). The typical symptoms on phytoplasma-infected plants include witches’ broom, proliferative branches, leaf yellowing, and small leaves (Lee et al., 2000). The flowers of infected plants exhibit sterility, abnormal morphology, virescence, and phyllody (herein referred to as leafy flower; Himeno et al., 2011). Peanut witches’ broom (PnWB) phytoplasma-infected Catharanthus roseus plants exhibit leafy flower symptoms, which were classified into five stages depending on their severity as described previously (Yang, 1985; Liu et al., 2014). Stage 4 (S4) leafy flowers exhibited leaf vein patterning with phyllody petals and intumescent ovaries (Fig. 1A; Liu et al., 2014).
Figure 1.
The leafy flower phenotypes of PnWB phytoplasma-infected C. roseus plants. A, The healthy flower (HF) and S4 PnWB-infected leafy flower symptoms are presented in the left and right columns, respectively. Bars = 1 cm. B and C, The mRNA (B) and microRNA (miRNA; C) expression of the HF and S4 in C. roseus flowers are represented by heat maps. Red indicates up-regulated expression, and blue indicates down-regulated expression. The values at right indicate the results of the log2 (S4/HF) formula. CrAG, AGAMOUS of C. roseus; CrFYF, FOREVER YOUNG FLOWER of C. roseus; CrPI, PISTILLATA of C. roseus.
The draft genome sequence of the PnWB phytoplasma contains 13 chromosomal contigs with 562,473 bp and 421 protein-coding genes (Chung et al., 2013). Chung et al. (2013) suggested that the 48 putative PnWB effectors are similar to known pathogenicity effectors of phytoplasmas. In aster yellows witches’ broom (AYWB) phytoplasma, the secreted AYWB PHYTOPLASMA PROTEIN54 effector (SAP54) causes a leafy flower phenotype in transgenic Arabidopsis (Arabidopsis thaliana) plants (SAP54 plants; MacLean et al., 2011). Moreover, the phyllody symptoms1 (PHYL1) effector of onion yellows phytoplasma wild-type line, a SAP54 ortholog, also manifested an identical leafy flower phenotype in PHYL1-overexpressing plants (Maejima et al., 2014).
PHYL1 and SAP54 interact with MADS domain transcription factors (MTFs) in Arabidopsis, such as SEPALLATA3 (AtSEP3), APETALA1 (AtAP1), and CAULIFLOWER (AtCAL; MacLean et al., 2014; Maejima et al., 2014). These MTFs are key regulators in floral development and were triggered by SAP54/PHYL1-mediated ubiquitin/26S proteasomal degradation by interacting with the RADIATION SENSITIVE23 (RAD23) family, resulting in interference in floral homeotic gene expression levels (MacLean et al., 2014; Maejima et al., 2014). Consistently, the ap1 mutant, ap1/cal double mutant, and sep1/sep2/sep3/sep4 quadruple mutant manifest various types of leafy flower (e.g. leaf-like sepals, secondary flowers, cauliflower-like inflorescences, or loss of floral meristem determinacy; Bowman et al., 1993; Ditta et al., 2004; Gregis et al., 2006). These results suggest that phytoplasma SAP54/PHYL1 is a key effector for triggering positive regulators of MTF degradation to switch the phase transition of plants from the reproductive stage to the vegetative stage.
In PnWB-infected C. roseus plants, the expression levels of SHORT VEGETATIVE PHASE1 (CrSVP1) and CrSVP2 were up-regulated in S4 leafy flowers (Liu et al., 2014). The SVP of Arabidopsis (AtSVP) is an MTF and functions as a negative regulator of floral development as part of a complex with FLOWERING LOCUS C (AtFLC) to control flowering time by negatively regulating FLOWERING LOCUS T (AtFT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (AtSOC1) by directly binding their promoter sequences (Lee et al., 2007b; Li et al., 2008; Jang et al., 2009; Irish, 2010). Moreover, AtSVP also interacts with other components, such as AP1 and AGAMOUS-LIKE24 (AGL24), during stages 1 and 2 of flower development to repress floral homeotic genes that control petal, stamen, and carpel identity (Gregis et al., 2009). The svp mutant (svp32) of Arabidopsis exhibits an early-flowering phenotype, whereas constitutive expression of AtSVP in Arabidopsis delays flowering (Hartmann et al., 2000; Gregis et al., 2006; Lee et al., 2007a). Moreover, transgenic Arabidopsis expressing AtSVP orthologs from Actinidia spp. or rice (Oryza sativa) showed leaf-like sepals and secondary flower phenotypes (Trevaskis et al., 2007; Wu et al., 2012). In addition to MTF down-regulation, phytoplasma might induce leafy flower symptom development via up-regulated SVP expression.
MTF plays a role in the regulation of gene expression, whereas miRNA plays a role in posttranscriptional regulation. Whether phytoplasma effectors alter the miRNA expression levels for symptom development is an interesting question. MiRNAs regulate numerous transcription factors involved in plant development through RNA cleavage or translation inhibition (Kidner and Martienssen, 2005; Voinnet, 2009). In contrast to animal miRNAs, plant miRNAs display a high degree of sequence complementarity to their target mRNAs and guide cleavage of the target RNA via ARGONATUTE1 (AGO1; Kidner and Martienssen, 2005; Voinnet, 2009). In animals, the imperfect miRNA/target pairwise region results in translation inhibition and, consequently, decay of the target mRNA (Bazzini et al., 2012; Djuranovic et al., 2012). However, the mechanism of translation inhibition in plants is not well known.
Chorostecki et al. (2012) proposed a predicted miR396 target site on the MADS box region of the AtSVP mRNA. Nevertheless, the lack of direct evidence from 5′ RACE data left them unable to demonstrate miR396-mediated AtSVP cleavage (Chorostecki et al., 2012). In contrast, the miR396-mediated GROWTH-REGULATING FACTOR (AtGRF) family of Arabidopsis features a bulge between the seventh and eighth positions of miR396 that is highly conserved among different plant species. The 5′ RACE data indicated that the bulge structure did not affect miR396-mediated AtGRF cleavage. However, interaction along the 5′ portion of the miRNA is the feature most relevant to its activity and cleavage efficiency (Mallory et al., 2004; Schwab et al., 2005; Lin et al., 2009), and modifying the pairing status of the 5′ portion of miR396 through mutagenesis alters the efficiency of AtGRF2 cleavage (Debernardi et al., 2012). Moreover, Li et al. (2013) demonstrated that a 5′ miRNA mismatch triggers miRNA-mediated target RNA translation inhibition in Arabidopsis. These results suggest that mispairing of the 5′ portion of the miRNA plays a role in modulating cleavage efficiency and translation inhibition during target gene regulation.
We previously identified miR396 of C. roseus via whole-transcriptome analysis and small RNA profiling by next-generation sequencing (Chang, 2012; Liu et al., 2014). There are two mismatches in the 5′ portion of the miR396 (fourth and fifth positions) on the pairwise region of miR396/CrSVPs (Chang, 2012). Therefore, this study seeks to answer several questions: (1) whether CrSVP1/2 are targets of miR396; (2) whether the regulation approaches differ between miR396/CrSVPs and miR396/CrGRFs due to the two mismatches in the 5′ portion of the miRNA; (3) whether the PHYL1 effector of PnWB also determines leafy flower phenotype in Arabidopsis; and (4) whether PHYL1 alters MTF and miRNA expression levels.
In this study, transgenic Arabidopsis expressing the PHYL1 gene of PnWB (PHYL1 plant) was generated and showed an identical leafy flower phenotype. The leafy flowers of PHYL1 plants exhibited a lower miR396 expression level but higher AtSVP and AtGRF1 expression levels compared with normal flowers of Columbia-0 (Col-0) plants. Moreover, we demonstrated that miR396 triggers SVP down-regulation via mRNA decay rather than cleavage based on degradome analysis. The mechanisms of PHYL1-altered miR396/SVP expression levels and miR396-mediated SVP down-regulation were investigated.
RESULTS
Abnormal Gene and miRNA Expression in PnWB-Infected C. roseus
Deep sequencing was used to compare the whole transcriptome with the small RNA profiles of C. roseus HF and S4 PnWB-infected leafy flowers (Fig. 1A). The floral homeotic gene expression levels were compared between HF and S4 flowers. The expression levels of CrFT, PISTILLATA of C. roseus, FOREVER YOUNG FLOWER of C. roseus, CrAP1.1, CrAP3.1, and CrAP3.2, but not CrAP1.2, CrSOC1.1, and CrSOC1.2, which enhance flowering, were repressed in the S4 flowers (Fig. 1B). In contrast, the expression levels of CrSVP1 and CrSVP2 were increased in S4 flowers (Fig. 1B). AGAMOUS of C. roseus and CrAP2 were not differentially expressed between HF and S4 flowers (Fig. 1B). In addition, we observed that most C. roseus miRNAs were significantly down-regulated in the S4 flowers, with only the up-regulation of miR162, miR169, and miR398 observed (Fig. 1C). These results suggest that PnWB effectors or other bacterial proteins might directly or indirectly alter either the miRNA gene expression profile of C. roseus plants (CrMIR genes) or the floral homeotic gene expression levels in PnWB-infected plants.
The PHYL1 Effector of PnWB Causes Leafy and Abnormal Flower Phenotypes and Represses miR396 Expression in Arabidopsis
The SAP54 effector of AYWB causes leafy flower in transgenic Arabidopsis (MacLean et al., 2011). We identified a PHYL1 effector gene in the PnWB genome sequence (Chung et al., 2013) that shares 61.3% amino acid sequence similarity with AYWB and 56% similarity with PHYL1 of the onion yellows phytoplasma wild-type line (Fig. 2A) and determined whether PHYL1 of PnWB can cause leafy flower and alter miRNA expression. PHYL1 plants, which overexpress the PHYL1 of PnWB, exhibit two types of leafy flowers (Fig. 2B) and three types of abnormal flowers (Fig. 2C), depending on the severity of the abnormal phenotype in the flowers. The type I leafy flower phenotype exhibits sepal enlargement, petal shrinking, and stigma elongation (Fig. 2B). The type II leafy flower phenotype features a secondary flower growing out from the primary flower (Fig. 2B). In some cases, a cauliflower-like inflorescence with narrow interspaces and short pedicles results (Fig. 2B). Three types of abnormal flowers include asymmetric petals, abnormal petal, and stamen numbers (five to six petals and seven to eight stamens; AF-I), swollen stigma, and secondary flower growth (Fig. 2C). We assume that various phenotypes occurred on PHYL1 plants because of the various expression levels of PHYL1.
Figure 2.
The phytoplasma effectors trigger the leafy flower phenotype. A, Alignment of orthologous amino acids of PHYL1. OY-W, Onion yellows phytoplasma wild-type line. B, Leafy flower phenotypes of PHYL1 and SAP54 plants. The type I leafy flower is indicated as LF-I, and the type II leafy flower is indicated as LF-II. Bars = 0.5 cm. C, The abnormal flower phenotype of PHYL1 plants. The type I to III phenotypes are indicated as AF-I, AF-II, and AF-III, respectively. Bars = 0.5 cm.
Interestingly, in PHYL1 and SAP54 plants, most miRNAs were up-regulated compared with Col-0 plants, whereas miR156h, miR159, miR390, and miR396 were repressed, indicating that both effectors have similar functions in CrMIR gene regulation (Fig. 3A). However, PnWB infection inhibits most CrMIR genes (Fig. 1C). We hypothesize that the other effectors of PnWB might be involved in the repression of CrMIR genes. Furthermore, PHYL1 and SAP54 have specificity in the repression of particular miRNAs. Indeed, the northern-blot data also indicated that the leafy flowers of PHYL1 plants exhibited lower miR396 accumulation (0.57-fold) compared with Col-0 plants (Fig. 3B).
Figure 3.
Phytoplasma effectors interfere with miRNA expression. A, MiRNA expression in flowers of Col-0, PHYL1, and SAP54 plants as represented by a heat map. Red indicates up-regulated expression, and blue indicates down-regulated expression. The values at right indicate the results of the log2 (Col-0/PHYL1 or Col-0/SAP54) formula. B, MiR396 detection in Col-0, PHYL1, and svp32 plants by northern blot. U6 was used as a loading control. The numbers indicate the relative expression of miR396 compared with Col-0 plants.
Identification of the MIR396 Gene of C. roseus
The hairpin structures of the miR396 precursor of C. roseus (DDS44007) and its miRNA/miRNA* pairing positions are shown in Figure 4A. The miR396/miR396* duplex exhibits a two-nucleotide overhang at the 3′ end (Fig. 4A). Remarkably, 14,256 reads of the mature form of miR396 were obtained for the HF flowers, whereas only 1,482 reads of miR396* were obtained (Fig. 4A). These results satisfy the general rule that less miRNA* than the mature form of the miRNA is required in vivo because the miRNA* exhibits a rapid turnover rate due to cleavage by AGO1 (Guo and Lu, 2010). Importantly, the amount of miR396 was reduced (2,587 reds) in S4 flowers, which is consistent with previous observations (Fig. 4A). In addition, the complementary DNA (cDNA) of the miR396 precursor was cloned from the C. roseus flower by reverse transcription (RT)-PCR for further investigation (data not shown), suggesting that the miR396 precursor exists in C. roseus plants.
Figure 4.
Characterization of MIR396, CrSVP1, and CrSVP2 in C. roseus. A, Illustration of the hairpin structures of the MIR396 precursors of C. roseus and their paired sequences. The mature miR396 (solid line) and miR396* (dashed line) are indicated on the precursor structure (top). The pairing of the miR396 (black) and miR396* (gray) sequences are indicated at bottom. The numbers indicate the deep-sequence reads of miR396 or miR396*. B, Predicted targets of miR396 in the MADS box domain of SVP genes. The nucleotide sequences of the CrSVP1 (DDS_51524), CrSVP2 (DDS_57797), and AtSVP (AT2G22540) targets were aligned, and the paired regions with miR396 are indicated. In the SVP alignment, dots indicate identical nucleotides, whereas boldface letters indicate the nucleotides that are perfectly paired between SVP and miR396. The translated SVP amino acids are presented below the alignment. C, Analysis of miR396 expression between HF and S4 PnWB-infected leafy flower by small RNA northern blot. The small RNA loading quantity was adjusted to 200 ng using the 2100 Bioanalyzer (Agilent). The numbers below indicate the relative fold change in the expression of miR396 regarding the S4 flowers relative to the HFs. CrU6 small nuclear RNA was used as a loading control. D, Semi-RT-PCR analyses of the CrSVP1 and CrSVP2 expression levels in an HF, S4 flower, and healthy leaf (HL). UBIQUITIN (UBQ) of C. roseus was used as an internal control. The numbers indicate the fold change in CrSVP1 and CrSVP2 expression in the S4 flower and HL samples relative to the HF. E, Phylogenetic tree of SVP proteins. The genes sharing greater than 80% amino acid similarity are classified and shaded in gray. AtSOC1 is referred to as an outgroup. The numbers indicate the percentage of 1,000 bootstrap replicates at the appropriate nodes. The scale bar indicates a 0.1 divergence of amino acid substitutions per site.
The MADS Box Domain of the SVP Gene Contains a Potential miR396 Target Site
Based on sequence pairing, a putative miR396 target site was identified on the MADS box domain of the SVPs (Fig. 4B); however, the miR396-mediated cleavage of SVP was not confirmed by 5′ RACE (Chorostecki et al., 2012). The target sequences in CrSVP1 and CrSVP2 were highly conserved with that of AtSVP (AT2G22540) of Arabidopsis, and four mismatches (the fourth, fifth, 19th, and 20th positions of miR396) were identified in the pairwise region of miR396/CrSVP1 or miR396/AtSVP (Fig. 4B). However, the pairwise region of miR396/CrSVP2 had an extra G-to-A mismatch at the seventh position of miR396 (Fig. 4B).
MiR396 was down-regulated 0.06-fold in the S4 leafy flower compared with the HF (Fig. 4C). Moreover, semi-RT-PCR results indicated that the levels of CrSVP1 were up-regulated 3.2- and 3.5-fold in the S4 leafy flower and healthy leaf samples, respectively, compared with the HF (Fig. 4D). In addition, CrSVP2 was up-regulated 3.4- and 4.6-fold in S4 and healthy leaf, respectively (Fig. 4D). These data suggest that PnWB infection alters the expression of CrSVP1/2 and miR396.
SVPs Contain Mismatch Pairings at the Target Site in the 5′ Portion of miR396
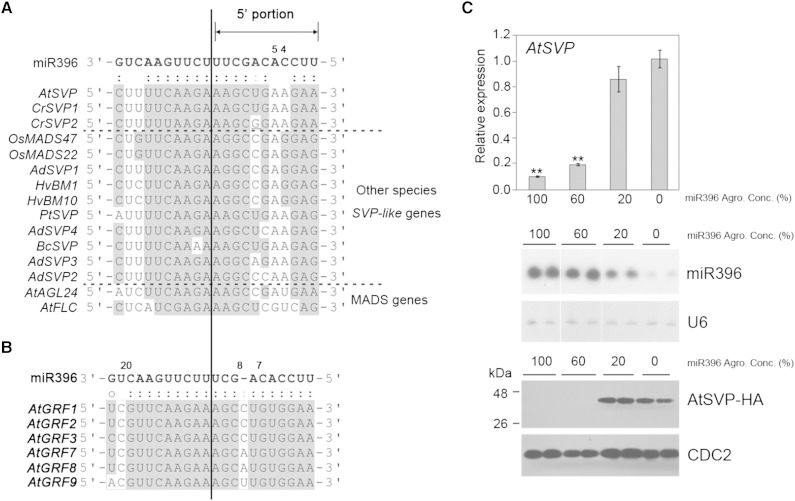
Alignment of the SVP genes revealed highly conserved miR396 target sites in various species. Fewer than five mismatches were observed on these SVP-miR396 pairwise regions, whereas 94% of MADS box genes had more than six mismatches spread throughout parallel regions of AtSVP, such as AtAGL24 and AtFLC (Fig. 5A; Supplemental Fig. S1). The AtGRF gene family is also the target of miR396 (Jones-Rhoades and Bartel, 2004; Wang et al., 2004; Debernardi et al., 2012). However, the AtGRF-miR396 pairwise region exhibits mostly complementary pairing that differs from the SVP-miR396 pairing (Fig. 5B). Thus, we assume that the miR396-mediated degradation efficiency or approaches between miR396/SVPs and miR396/AtGRFs might differ (Fig. 5, A and B).
Figure 5.
SVP contains variation pairing of the 5′ portion of miR396 with its target genes. A, Highly conserved miR396 target sites on the SVP genes of various species. The alignment of the miR396 target sites in SVP genes of various species is shown. The GenBank accession numbers of the SVP genes are as follows: AtSVP (NM127820), AtAGL24 (NM118587), AdSVP1 (JF838212), AdSVP2 (JF838213), AdSVP3 (JF838214), AdSVP4 (JF838215), HvBM1 (AJ249142), HvBM10 (EF043040), OsMADS22 (AB107957), OsMADS47 (AY345221), BcSVP (DQ922945), and PtSVP (FJ373210). AtAGL24 and AtFLC (AT5G10140) were chosen as representative MADS genes for target-site comparison. The SVP nucleotides that pair with the miR396 sequences are indicated in gray. B, The miR396 target-site alignment for GRFs (AtGRF1, AT2G22840; AtGRF2, AT4G37740; AtGRF3, AT2G36400; AtGRF7, AT5G53660; AtGRF8, At4G24150; and AtGRF9, AT2G45480). Gray shading indicates the nucleotides of AtGRFs paired with the miR396 sequences. C, In vivo analysis of miR396-mediated AtSVP-HEMAGGLUTININ (HA) efficiency by transient expression. The relative expression levels of AtSVP were normalized to that of NbACTIN via real-time RT-PCR. Error bars represent se (n = 6). The relative expression levels were significantly different from those in Col-0 plants for each RNA sample, based on Student’s t test: **, P < 0.01. The various percentages of miR396 were determined by a small RNA northern blot. U6 small nuclear RNAs were used as loading controls. The various percentages of miR396 are indicated below the real-time RT-PCR and northern-blot data. The AtSVP-HA protein level was detected by western blotting with a 1:10,000 dilution of HA antibody. CELL DIVISION CONTROL2 (CDC2) was used as a protein loading control.
MiR396-Mediated Down-Regulation of SVPs by Transient Expression
Different concentrations of agrobacteria carrying the MIR396a gene of Arabidopsis (AtMIR396a) were transiently coexpressed with AtSVP in Nicotiana benthamiana plants and were then analyzed via agroinfiltration to evaluate whether miR396 can down-regulate AtSVP. The AtSVP gene was expressed using a Cauliflower mosaic virus 35S promoter to avoid the side effects of endogenous transcription factors that are regulated by miR396. The AtSVP RNA levels were decreased by increasing miR396 expression levels (Fig. 5C). Similar results were observed for the miR396-mediated down-regulation of CrSVP1, CrSVP2, and AtGRF1 (Supplemental Fig. S2). No CrSVP1 down-regulation occurred when amiRGFP4 was present (an artificial miRNA that targets the GFP gene used as a negative control; Supplemental Fig. S3). These data indicate that miR396 directly triggers SVP posttranscriptional down-regulation.
To further demonstrate the target gene down-regulation by miR396, a reporter assay was used in this study. A DNA fragment (129 nucleotides in length) that contains a target site for AtSVP and AtGRF1 was fused at the 5′ end of the YELLOW FLUORESCENT PROTEIN (YFP) gene to generate SVP129-YFP and GRF1129-YFP constructs, respectively (Fig. 6A). Moreover, SVP129m-YFP is a mutant construct that has several mutations at the target site, resulting in eight mismatches in the pairwise region of miR396 (Fig. 6A). The SVP129-YFP and GRF1129-YFP constructs resulted in an approximately 85% reduction of YFP fluorescence when these constructs were coexpressed with miR396, whereas no significant differences were observed regarding YFP fluorescence in the YFP negative control with/without miR396 (Fig. 6, B and C). In contrast, the SVP129m-YFP construct resulted in an approximately 40% reduction of YFP fluorescence when coexpressed with miR396 (Fig. 6, B and C). Notably, the fluorescent spots are present in nuclei (Fig. 6B). The reporter assay demonstrated that miR396 specifically down-regulated AtSVP and AtGRF1.
Figure 6.
The reporter assay for miR396-mediated targeted down-regulation. A, Schematic of miR396 and its target-site pairing on the YFP gene. All of the constructs were constructed on a binary vector under the control of a 35S promoter. A YFP-only construct (i) was used as a negative control. The SVP129-YFP construct (ii) represents 129 nucleotides of the 5′ end of the SVP gene containing the miR396 target site fused with the YFP gene. The SVP129m-YFP construct (iii) represents 129 nucleotides of the 5′ end of the SVP gene containing a mutated miR396 target site fused with the YFP gene. The GRF1129-YFP construct (iv) represents 129 nucleotides of the 5′ end of the GRF1 gene containing the miR396 target site fused with the YFP gene. B, The YFP reporter assay was analyzed using confocal microscopy. Bar = 100 µm. C, Relative average YFP expression levels (log2) of the reporter assay as determined by confocal microscopy (n = 3). Relative YFP expression levels were significantly different from that of the negative control construct without miR396 present in each sample, based on Student’s t test: **, P < 0.01; and *, P < 0.05. MiR396 was detected by northern blot for each sample (two replicates). U6 small nuclear RNA was used as a loading control.
Flowering Phenotypes of miR396 in Loss and Gain of Function
We used genetic approaches to examine gain of function and loss of function in miR396-mediated SVP regulation. A transfer DNA (T-DNA) insertion mutant line, SALK064047, was demonstrated to be an AtMIR396a mutant (atmir396a-1) in Arabidopsis (Bao et al., 2014). T-DNA was inserted at the −450-bp promoter region of AtMIR396a (AT2G10606; Fig. 7A). The homozygous atmir396a-1 plants exhibited only 0.6-fold miR396 expression levels compared with Col-0 plants (Fig. 7B). We assumed that this expression represents a background signal of miR396b in atmir396a-1 plants (Fig. 7B). The atmir396a-1 plants exhibit normal flower and flowering time phenotypes, whereas the svp32 plants display early flowering (Fig. 7, C and D). In addition, atmir396a-1 plants exhibit 0.5- and 0.3-fold increased AtSVP and AtGRF1 expression, respectively, compared with Col-0 plants (Fig. 7E), indicating that lower miR396 levels induce up-regulation of AtSVP and AtGRF1.
Figure 7.
Characterization of miR396 and SVPs in Arabidopsis plants. A, Diagram of the atmir396a-1 mutant (SALK064047). The black arrows indicate the gene regions for AT2G10605, AtMIR396a (AT2G10606), and AT2G10608 and their translation directions. The T-DNA insertion site is located at the promoter region (450 bp upstream) of AtMIR396a. The left border (LB) and right border (RB) are indicated at the two borders of the T-DNA. The primers LP, RP, and LBb1.3 were used for genotyping via PCR. Bar = 150 bp. B, Analysis of miR396 by small RNA northern blot in Col-0, svp32 mutant, atmir396a-1 mutant, two independent CrMIR396 lines (lines 8 and 27), and AtMIR396a-overexpressing plants. U6 small RNA was used as a loading control. The numbers below indicate fold changes in miR396 expression relative to the Col-0 plant. C, CrSVP1, CrSVP2, atmir396a-1, and svp32 (SALK072930C) mutant plants exhibited normal flower phenotypes. CrMIR396 and AtMIR396a plants exhibited abnormal flower phenotypes. Plants with AdSVP2 or AtSVP overexpression exhibited leafy or abnormal flower phenotypes, respectively. Bars = 0.1 cm. D, Evaluation of the flowering time of various Arabidopsis mutant or transgenic plants. Leaf number represents flowering time. Error bars represent se (n = 20). E, MiR396-mediated AtSVP and AtGRF1 regulation. AtSVP and AtGRF1 were detected by real-time RT-PCR. Relative expression levels were normalized to the level of AtUBQ. Error bars represent se (n = 3). Relative expression levels were significantly different from those of the Col-0 plants in each RNA sample, based on Student’s t test: *, P < 0.05; and **, P < 0.01.
In contrast, transgenic Arabidopsis expressing CrMIR396 (CrMIR396 plant) exhibits abnormal flowers with curved sepals and petals and enlarged stigmas, consistent with transgenic Arabidopsis expressing AtMIR396a (AtMIR396a plant; Fig. 7C; Liang et al., 2014). Two individual CrMIR396 plants (lines 8 and 27) exhibited 4.6- and 3.5-fold increased miR396 expression compared with Col-0 plants. Furthermore, AtMIR396a plants exhibited a 1.9-fold increase in miR396 expression (Fig. 7B). CrMIR396 and AtMIR396a plants exhibited late-flowering phenotypes that were correlated with the expression levels of miR396 (Fig. 7D). Moreover, AtSVP and AtGRF1 were down-regulated in CrMIR396 and AtMIR396a plants (Fig. 7E). AtGRF1 was down-regulated by more than 80% by the overexpression of miR396, while AtSVP was down-regulated by only 20% to 40% in MIR396 plants (Fig. 7E). These results indicated that CrMIR396 can generate miR396 and has a genetic function similar to that of AtMIR396a in Arabidopsis plants. Moreover, miR396 regulates AtGRF1 more efficiently than AtSVP.
SVP Orthologs from Different Species Cause Various Flower Phenotypes
Two orthologous SVP genes (CrSVP1 and CrSVP2) from C. roseus were overexpressed in Arabidopsis (CrSVP1 and CrSVP2 plants), and transgene overexpression was confirmed by RT-PCR (Supplemental Fig. S4). However, both plants exhibited normal flower phenotype and flowering time (Fig. 7, C and D). In addition, transgenic Arabidopsis expressing the SVP2 gene of Actinidia spp. (AdSVP2 plants) exhibited enlarged sepals, small green petals, and a curving pistil mimicking the leafy flower phenotype of PHYL1 plants (Fig. 7C); however, the flowering time of the AdSVP2 plant was similar to that of Col-0 plants (Fig. 7D). In contrast, the AtSVP plants showed asymmetric petals of an abnormal flower phenotype identical to PHYL1 plants (Figs. 2C and 7C). In some cases, the severely abnormal flowers of AtSVP plants had five petals with eight stamens (Fig. 7C). These data suggested that the up-regulation of AtSVP causes an abnormal flower phenotype. The abnormal flower phenotype in AtSVP plants is consistent with the AF-I of PHYL1 plants, which have five to six asymmetric petals and seven to eight stamens (Figs. 2C and 7C).
Two individual lines of AtSVP plants (lines 1 and 2) exhibited late flowering time with 16 to 19 rosette leaves. Indeed, phylogenetic analysis revealed that AtSVP, AdSVP2, CrSVP1, and CrSVP2 are in different groups based on 80% amino acid similarity (Fig. 4E). Moreover, the C-terminal regions of these SVPs differ (Supplemental Fig. S5). These results imply that the SVP genes in other species have functions regarding flower phenotypes and flowering times that differ from those in Arabidopsis. In addition, the abnormal flower phenotypes in AdSVP2 and AtSVP plants suggest that overexpression of SVP may alter flower development. To confirm this hypothesis, AtSVP expression was detected in PHYL1 plants.
SVP Is Up-Regulated in the Sepal and Petals of PHYL1 Plants
Because miR396 expression was reduced in PHYL1 plants, we evaluated AtSVP and AtGRF1 expression in transgenic plants. Real-time RT-PCR analysis revealed that AtSVP and AtGRF1 are up-regulated in the PHYL1 leafy flower (Fig. 8A). Moreover, in situ hybridization data indicate strong accumulation of AtSVP signals in the leafy flower tissue of PHYL1 plants, particularly in the basal part of the petals (Fig. 8B, image ii), whereas few AtSVP signals were observed in the sepals of Col-0 plants (Fig. 8B, image i). The sense probe for AtSVP was used as a negative control for in situ hybridization, and no signal was observed in flower buds of PHYL1 or Col-0 plants (Fig. 8B, images iii and iv). These results support the hypothesis of miR396-mediated AtSVP expression in flower tissue.
Figure 8.
PHYL1 alters the expression of miR396 and its targets. A, AtSVP and AtGRF1 expression levels in Col-0, PHYL1, and svp32 plants were detected by real-time RT-PCR. Error bars represent se (n = 9). Relative expression levels were normalized to the level of AtUBQ. Relative expression levels were significantly different from those of the Col-0 plants in each RNA sample, based on Student’s t test: *, P < 0.05; and **, P < 0.01. B, Detection of SVP gene expression in floral bud tissue by in situ hybridization. The buds of Col-0 (i) and PHYL1 (ii) plants were sectioned and hybridized with an antisense probe for SVP detection. The sense probe for AtSVP was used as a negative control (iii and iv). The black arrows indicate the AtSVP signal at the sepal, and the red arrows indicate the AtSVP signals at the basal part of the petal. an, Anther; pe, petal; se, sepal; st, stigma. Bar = 50 µm.
MiR396 Triggers SVP Decay
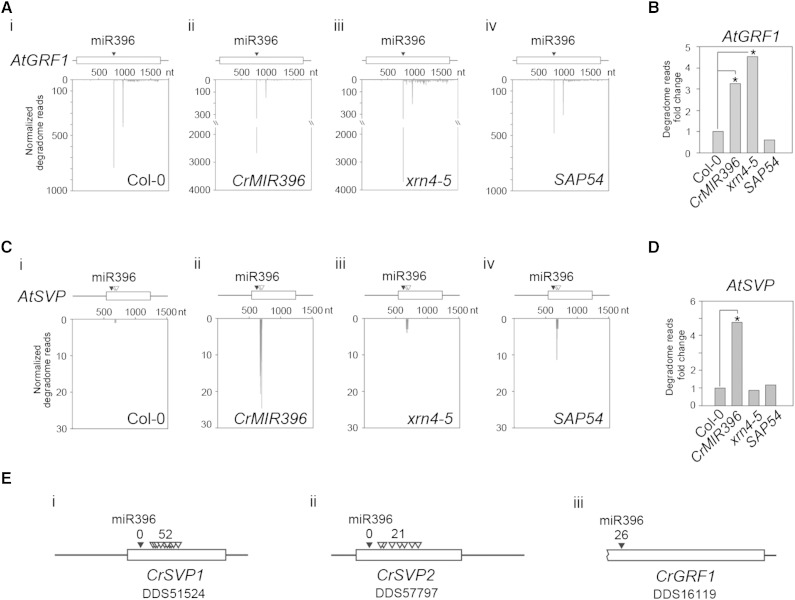
The pairwise fourth and fifth positions are reportedly critical for the efficiency of low miRNA-guided cleavage of the target RNA (Lin et al., 2009). We assumed that the two mismatches at the fourth and fifth positions of the pairwise region will yield differences in cleavage between miR396/AtSVP and miR396/AtGRF1. To test this hypothesis, a high-throughput degradome approach was used to analyze the mRNA degradation products. Overexpression of miR396 yielded 22,705 degradome reads of degraded 3′ AtGRF1 fragments in the CrMIR396 plant, whereas only 800 degradome reads were obtained from Col-0 plants (Fig. 9A). In addition, 3,744 degradome reads of AtGRF1 were obtained from EXORIBONUCLEASE4 mutant (xrn4-5), a 5′-to-3′ exoribonuclease mutant, whereas approximately 500 degradome reads were detected in SAP54 plants (Fig. 9A). These data indicate that miR396-mediated AtGRF1 cleavage is dependent on miR396 dosage and that the cleaved 3′ AtGRF1 fragment is highly accumulated in xrn4-5 mutants. A 3- to 5-fold increase in AtGRF1 degradome reads was detected in CrMIR396 and xrn4-5 plants compared with Col-0 plants (Fig. 9B). In addition, the other AtGRF targets, Arabidopsis BASIC HELIX-LOOP-HELIX74 (AT1G10120) and AtMMG4.7 (AT5G43060), which are also targets of miR396, also produced significant degradome reads on the corrected target site (Supplemental Fig. S6). These results indicate that miR396 mediated AtGRF1 cleavage at the target site.
Figure 9.
Degradome analysis of C. roseus and Arabidopsis. A and C, Degradome patterns of AtGRF1 (A) and AtSVP (C) in flower buds of Col-0, CrMIR396, xrn4-5 mutant, and PHYL1 plants. nt, Nucleotides. B and D, Fold changes in the degradome of AtGRF1 (B) and AtSVP (D) in flower buds of Col-0, CrMIR396, xrn4-5 mutant, and PHYL1 plants. The fold changes were significantly different from those of the Col-0 plants in each RNA sample based on Student’s t test: *, P < 0.01. E, Degradome patterns of CrSVP1, CrSVP2, and CrGRF1 in flowers of C. roseus. The black arrowheads indicate the miR396 target site. The white arrowheads indicate the nonspecific RNA degradation position.
While no degradome reads were detected for the target site of AtSVP in these plants, approximately 280 various degradome reads were identified at the rear position of the miR396 target site in CrMIR396 plants, suggesting the presence of miR396-mediated AtSVP decay (Fig. 9C, white arrowheads). Total degradome reads of AtSVP were increased 5-fold in CrMIR396 plants compared with Col-0 plants, but no significant reads were detected in either xrn4-5 or SAP54 plants (Fig. 9C). Identical results were obtained for the degradome profile of C. roseus flowers (Fig. 9E, images i and ii). In total, 52 and 21 degradome reads were detected for the rear positions of the target sites on CrSVP1 and CrSVP2, respectively, suggesting the presence of miR396-mediated CrSVP1/2 decay. In contrast, 26 degradome reads of CrGRF1 were detected in the target position, indicating miR396-mediated cleavage (Fig. 9E, image iii, black arrowhead). In addition, one predicted miR396 target gene (AT1G71350) exhibits mismatch pairing at the 5′ portion (Supplemental Fig. S7). No significant miR396-cleaved degradome reads at the target site were observed for the gene. However, AT2G31880 has approximately 5-fold more degradation reads in CrMIR396 plants (Supplemental Fig. S7, B and C). In contrast, under the miR396-decreasing condition, AT2G31880 expression was increased 50% in SAP54 plants, which suggests the presence of miR396-mediated AT2G31880 down-regulation (Supplemental Fig. S7D). In summary, based on the 5′ portion pairing differences between miR396 and its targets, miR396-mediated SVP or AT2G31880 down-regulation might differ from the GRF1 control. In addition, this phenomenon is conserved in various plant species.
DISCUSSION
Multifunctional effectors are common in many pathogens. In addition to MTF degradation, we demonstrated that PHYL1 effector can up-regulate SVP MTF expression and alter miRNA expression to switch the phase transition back to the vegetative stage. In addition, we also provided evidence to show that miR396 mediated SVP mRNA decay.
The Effectors of PnWB Alter Gene and miRNA Expression in Host Plants
The virescence and phyllody symptoms on phytoplasma-infected plants are remarkable phenotypes. Phytoplasma infection alters the physiological and morphological characteristics of the host cells to enhance pathogenic multiplication and infection in the C. roseus plants (Sugio et al., 2011). To revert the reproductive stage to the vegetative stage, PHYL1/SAP54 effectors suppress positive regulators of flowering but enhance the expression levels of negative regulators, such as CrSVP1 and CrSVP2. However, CrSOC1.1, CrSOC1.2, and CrAP1.2, the orthologs of which function as positive regulators in flowering in Arabidopsis, were not repressed by PnWB. This phenomenon can be explained by two forces that originate from the pathogen and host to counteract each other; they are abnormally coexpressed in the floral meristem, resulting in leafy flower formation (Liu et al., 2014).
Rather than altering gene expression, the miRNA expression levels were also altered during PnWB infection. Interestingly, the miRNA expression patterns in PnWB-infected S4 leafy flowers differ in PHYL1 and SAP54 plants. We hypothesize that the other effectors or bacterial proteins in PnWB directly or indirectly control CrMIR gene expression, resulting in different miRNA expression patterns between PnWB-infected plants and transgenic plants. Moreover, the PHYL1/SAP54 that triggers MTF degradation might alter abnormal miRNA expression to cause a synergistic effect, thereby resulting in the leafy flower phenotype.
Like SAP54 or PHYL1 of onion yellows phytoplasma mutant plants, PHYL1 of PnWB effector causes leafy flowers in transgenic Arabidopsis plants. Moreover, the miRNA expression patterns of PHYL1 plants (PHYL1 of PnWB) were also consistent with those of SAP54 plants. SAP54 and PHYL1 of onion yellows phytoplasma plants mediate AtAP1, AtSEP3, and AtCAL proteasomal degradation (MacLean et al., 2014; Maejima et al., 2014); therefore, MTF degradation might result in abnormal miRNA expression. These results indicate that these effectors contain common motif(s) for triggering leafy flower formation and altering miRNA expression. Furthermore, PHYL1 mediates the down-regulation of miR396 expression; however, it did not interfere with miR396 biogenesis in vivo. PHYL1 might trigger MTF degradation or alter the expression levels of other factors that regulate miR396 expression.
In addition, the atmir396a-1 mutant exhibited AtSVP accumulation without manifesting any abnormal flower phenotype. Our explanation for this observation is that the functional redundancy of miR396b might play a role in regulating AtSVP. Moreover, the 0.5-fold accumulation of AtSVP relative levels in atmir396a-1 plants might not be sufficient to trigger the abnormal flower phenotype, whereas the 3- and 90-fold accumulation of AtSVP in PHYL1 and AtSVP plants, respectively, might trigger this phenotype (Fig. 8; Supplemental Fig. S4B). In contrast, AtMIR396a and CrMIR396 plants did not show the same early-flowering phenotype as the svp32 mutant, possibly because miR396 also regulates other genes; therefore, the synergistic effect resulted in a milder late flowering.
The 5′ Portion of miR396 Determines Target RNA Degradation by miRNA Cleavage or mRNA Decay
The degradome patterns indicated that miR396-mediated SVP and GRF1 degradation profiles differ because of the discrepant pairing on the 5′ portion of the miRNA. The 5′ portion (i.e. the fourth and fifth positions) of the miRNA is critical for miRNA-mediated cleavage, whereas the 19th and 20th positions are nonessential (Lin et al., 2009). The 5′ mismatch of miRNA reduces the efficiency of target RNA cleavage and may trigger mRNA decay (Lin et al., 2009; Bazzini et al., 2012; Djuranovic et al., 2012), which may explain the failure to confirm the miR396 target site of SVP by 5′ RACE (Chorostecki et al., 2012). Indeed, the degradome profiles showed that CrSVP1, CrSVP2, and AtSVP have nonspecific mRNA decay, whereas a clear degradome pick was shown on the target site of CrGRF1 and AtGRF1. In miRNA regulation of mRNA by translation repression in animal models, miRNA-mediated silencing complex bound of mRNA is typically degraded by another endogenous RNase system (Stefani and Slack, 2008; Bazzini et al., 2012; Djuranovic et al., 2012). In plants, the miR398-mediated cleavage of the COPPER/ZINC SUPEROXIDE DISMUTASE (CSD1 and CSD2) family, which exhibits a mismatch at the fourth position of miR398, demonstrated that miR398 regulates CSD2 expression through translation repression (Sunkar et al., 2006; Li et al., 2013). Therefore, we hypothesize that SVP mRNA decay might be triggered by miR396-mediated translation inhibition.
PHYL1-Mediated MTF Degradation Might Interfere with the Heterodimer Formation of the SVP Complex
PHYL1/SAP54 interacts with the RAD23 family to trigger MTF (AP1, SEP3, and CAL) degradation, resulting in the formation of leafy flower phenotypes (MacLean et al., 2014; Maejima et al., 2014). However, AtSVP interacts with AP1 or other components to form different complexes that regulate floral gene expression (Gregis et al., 2009). Therefore, the lack of AP1 in PHYL1/SAP54 plants might prevent AtSVP from forming a heterodimer complex, thereby resulting in misregulation during the phase transition and floral organ identity. In addition, the degrees of phyllody and virescence of SAP54 plants in the rad23 family mutant background were reduced, especially in the rad23BCD triple mutant that has no strong leafy flower formation in the presence of phytoplasma infection or SAP54 overexpression (MacLean et al., 2014). Interestingly, rad23BCD mutant plants have shown the abnormal flower (five to six petals) phenotype (MacLean et al., 2014) identical with the AF-I of PHYL1 and AtSVP plants. Therefore, these data implied that the gain of function of the AtSVP-AP1 complex might also affect flower formation, while AP1 shows no degradation in the rad23BCD mutant background (Fig. 10). In addition, the SVP complex has species specificity with regard to interference with flowering time and flower development in Arabidopsis, because only AtSVP plants show the late-flowering and AF-I phenotypes in Arabidopsis, whereas other SVP plants have normal flowering time and normal flowers (except AdSVP plants), similar to Col-0 plants.
Figure 10.
Model for PHYL1-regulated miR396-mediated SVP expression in the flowering pathway resulting in the abnormal flower phenotype.
A Working Model for PHYL1 Repression of miR396-Mediated SVP Regulation
We propose a model for PnWB repression of the miR396-mediated SVP silencing pathway (Fig. 10). Multiple functions of PHYL1 trigger positive regulators of MTF (AtAP1, AtSEP3, and AtCAL) proteasomal degradation, resulting in leafy flower formation. PHYL1-mediated MTF degradation also affects many floral homeotic gene expression levels and miRNA expression levels, including the up-regulation of AtSVP and the repression of miR396. MiR396 regulates SVP through an mRNA decay approach that might be triggered by translation inhibition. Therefore, the down-regulation of miR396 by PHYL1 results in the up-regulation of SVP to change the complex formation to repress flowering and cause an abnormal flower phenotype.
CONCLUSION
This study provides evidence to demonstrate that PHYL1/SAP54 effectors can change miRNA expression levels. In addition, we also demonstrate that miR396 can down-regulate SVP, and the overexpression of AtSVP causes the abnormal flower phenotype. In the future, we will explore how PHYL1 controls these miRNA gene expression levels and examine symptom development with regard to the pathogen effector-host factor interaction at the posttranscriptional level.
MATERIALS AND METHODS
Plant Materials and Phytoplasma Infection
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized and chilled at 4°C for 2 d prior to sowing on Murashige and Skoog (MS) medium with or without suitable antibiotics for selection. One-week-old seedlings were germinated on MS plates and transferred to soil. Catharanthus roseus and Nicotiana benthamiana seeds were sown in soil. All seedlings and plants were maintained in a growth chamber or greenhouse (16 h of light/8 h of dark, 20°C–25°C).
Two-month-old C. roseus plants were inoculated with PnWB phytoplasma via grafting. The PnWB-infected plants developed virescence symptoms 3 to 4 weeks post grafting, whereas phyllody symptoms developed at 5 to 6 weeks post grafting.
Screening of atmir396a Mutants of Arabidopsis
Mutant lines of atmiR396a-1 (SALK064047) for T-DNA insertion were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/). Genomic DNA from 1-week-old seedlings was used for genotyping. The primers LP and RP were used to detect the wild-type genome region of the AtMIR396a gene, whereas the primers LBb1.3 and RP were used to detect the T-DNA insertion region. The primers used for genotyping are listed in Supplemental Table S1.
Gene Construction and Transgenic Plants
The genes AtSVP, CrSVP1, CrSVP2, AtMIR396a, CrMIR396, and PnWB PHYL1 were identified based on the Arabidopsis (The Arabidopsis Information Resource) and C. roseus transcriptome databases (Chang, 2012; Liu et al., 2014). AtSVP was cloned via RT-PCR amplification using the primers F-AtSVP and R-AtSVP. For AtGRF1, the 5′ end of AtGRF1 (792 nucleotides) was cloned via RT-PCR amplification using the primers F-AtGRF1 and R-AtGRF1-792. CrSVP1 was cloned via RT-PCR amplification using the primers 51524_F and 51524_R. CrSVP2 was cloned via RT-PCR amplification using the primers 57797_F and 57797_R. AtMIR396a was cloned via RT-PCR amplification using the primers F-AtMIR396a and R-AtMIR396a. CrMIR396 was cloned via RT-PCR amplification using the primers 34004-29-F and 34004-235-R. The PHYL1 gene was identified from the PnWB genome sequence (Chung et al., 2013) and amplified from the cDNA of PnWB-infected C. roseus by RT-PCR with the primers FG-PHYL1-Pn and RG-PHYL1-Pn. PCR fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) according to the manufacturer’s instructions to generate pENTR-CrSVP1, pENTR-CrSVP2, pENTR-CrMIR396, and pENTR-PHYL1.
For the miR396 target-site mutation of AtSVP, the AtSVP cDNA was used as a template for PCR-based mutagenesis with MAtSVPm2-118 and PAtSVPm2-70. The PCR fragments were cloned into pENTR/D-TOPO vector to generate pENTR-AtSVPm.
For the reporter assay, the AtSVP129 and AtSVP129m DNA fragments (nucleotides 1–129 of the 5′ end of either AtSVP or AtSVPm) were amplified by PCR from pENTR-AtSVP as well as pENTR-AtSVPm with F-AtSVP and MAtSVP-129 primers. In addition, the GRF1129 DNA fragment was amplified by PCR from pENTR-AtGRF1 with PAtGRF1-731 and MAtGRF1-817 primers. These PCR fragments were cloned into the pENTR/D-TOPO vector to generate pENTR-AtSVP129, pENTR-AtSVP129m, and pENTR-AtGRF1129. Gateway system procedures were used to transfer these genes to a pBA-DC-YFP binary vector to generate pBA-AtSVP129-YFP, pBA-AtSVP129m-YFP, and pBA-AtGRF1129-YFP.
These binary plasmids were transformed into an Agrobacterium tumefaciens ABI strain for agroinfiltration or used for transgenic Arabidopsis transformation using a floral dip procedure (Zhang et al., 2006). The transformant lines were screened on MS medium containing 10 μg mL−1 Basta (Sigma).
The artificial miRNA precursor that targets the GFP gene (preamiR-GFP4) was constructed based on the miR159 precursor of Arabidopsis via PCR mutagenesis using the primers PamiR-FG and MamiR. The PCR product was cloned into a pENTR/D-TOPO vector to generate pENTR-amiR-GFP4. Gateway system procedures were used to transfer preamiR-GFP4 to a pBA-DC-HA binary vector (Zhang et al., 2005) and generate pBA-amiR-GFP4. The primers used for cloning are listed in Supplemental Table S1.
Transient Expression via Agroinfiltration
An A. tumefaciens ABI strain-containing binary vector was incubated in Luria-Bertani medium containing 10 mm MES, pH 5.6, 40 μm acetosyringone, 100 μg mL−1 spectinomycin, and 50 μg mL−1 kanamycin at 28°C for 16 h. The A. tumefaciens was suspended, the absorbance was adjusted to optical density at 600 nm = 0.5 using an appropriate buffer (10 mm MgCl2 and 150 μm acetosyringone), and the A. tumefaciens was incubated at room temperature for 3 h (Lin et al., 2013). Four days after agroinfiltration, the infiltrated leaves of the N. benthamiana plants were collected and analyzed by semi-RT-PCR and northern blot.
For the observation of reporter assay results, YFP fluorescence in the infiltrated leaf was monitored using the Leica TCS SP5 II confocal laser-scanning microscope (Joint Center for Instruments and Research, College of Bioresources and Agriculture, National Taiwan University) equipped with a multiline argon laser with a filter set for YFP fluorescence (excitation filter, Acousto-optic Tunable filter 488; emission bandwidth, 496 to 574 nm; PMT2 offset [−1]/gain [895]). All images were graphically arranged used Adobe Photoshop CS3 software.
Total RNA Extraction and Small RNA Detection
Total RNA was extracted from the plant tissues using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For small RNA northern blotting, 10 μg of total RNA was separated on a 15% polyacrylamide/1× Tris-borate/EDTA/8 m urea gel and transferred to a Hybond-N+ membrane. The miR396-AS DNA probe (5′-CAGTTCAAGAAAGCTGTGGAA-3′) and the amiRGFP4-AS DNA probe (5′-TACTCCAATTGGCGATGGCCC-3′) were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Free radioisotopes were filtered out using a mini Quick Spin Oligo Column (Roche). The membrane was hybridized with ULTRAhyb-Oligo hybridization buffer (Ambion) at 42°C for 16 h, and the signal was detected using X-film (GE Healthcare) at −80°C for 16 h.
Real-Time RT-PCR, Semi-RT-PCR, and Northern Blot
For real-time RT-PCR, cDNA was first reverse transcribed from total RNA using oligo(dT)20 and the SuperScript III First Strand Synthesis System (Invitrogen). Real-time PCR was performed using a LightCycler 480 instrument (Roche) and three sets of primers for AtSVP, AtGRF1, and AtUBQ expression detection. The primers used for real-time RT-PCR are listed in Supplemental Table S2. For semi-RT-PCR detection, the primers F-CrSVP1_A and R-CrSVP1_C were used to detect CrSVP1. The primers F-CrSVP2_A and R-CrSVP2_C were used to detect CrSVP2. The primers F-AtSVP and R-AtSVP were used to detect AtSVP. The primers F-AdSVP2 and R-AdSVP2 were used to detect AdSVP2. The primers CrUBQc-F and CrUBQc-R were used to detect CrUBQ. The primers NbActin-F1 and NbActin-R1 were used to detect NbACTIN. The primers AtUBQ-F1 and AtUBQ-R1 were used to detect AtUBQ. The primers used for semi-RT-PCR are listed in Supplemental Table S3.
Statistical Analyses and Deep Sequencing
The AtSVP, AtGRF1, and AtUBQ transcripts were quantified via a relative cycle threshold (Ct) method. All experiments were performed with three independent replicates to compensate for possible loading errors. The relative expression levels were calculated based on the ∆∆Ct value, and each sample was normalized according to the expression level of AtUBQ. The miR396 expression levels were normalized via U6 internal/loading controls, which were quantified using ImageQuant 5.2 (GE Healthcare). The ∆∆Ct values were used to explain the fold changes in the expression levels.
The small RNA profiles of the PHYL1 plant, SAP54 plant, and HF and S4 flowers were analyzed using Illumina HiSeq 2000. The degradome profiles of the Col-0 plant, CrMIR396 plant, xrn4-5 mutant, SAP54 plant, and HF of C. roseus plant were also analyzed using Illumina HiSeq 2000. The miRNA and degradome profiles were analyzed using CLC Genomics Workbench 5.5.2 (CLC Bio) and the ContigViews analysis platform (www.contigviews.bioagri.ntu.edu.tw). The read normalization for the miRNA and degradome assays was calculated by the ratio of total reads across various samples to Col-0 plants. The raw small RNA reads reported here have been submitted to the National Center for Biotechnology Information Short Read Archive under accession numbers SRR1818286 (normal flower of Col-0 plant), SRR1818303 (leafy flower of PHYL1 plant), and SRR1818289 (leafy flower of SAP54 plant). The raw degradome reads reported are available in the Short Read Archive under accession numbers SRR1821048 (flower of Col-0 plant), SRR1821008 (flower of CrMIR396 plant), SRR1821172 (flower of xrn4-5 mutant), SRR1821156 (leafy flower of SAP54 plant), and SRR1821217 (HF of C. roseus plant).
In Situ Hybridization
A digoxigenin (DIG)-labeled SVP RNA probe (1,117–1,416 bp) was produced using an in vitro transcription kit (Roche) following the manufacturer’s protocol. The embedded Col-0 and PHYL1 inflorescence tissues were sectioned to 8-µm thickness and hybridized with DIG-labeled RNA probes. The hybridization signal was detected using an anti-DIG antibody conjugated to alkaline phosphatase (Roche) with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as the substrate. The results were examined with a bright-field microscope as purple dotted signals. The protocol followed that of a previous study (Wang et al., 2008).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment and comparison of the parallel positions of the miR396 target sites from MADS box-containing Arabidopsis genes.
Supplemental Figure S2. In vivo analysis of the miR396-mediated CrSVP1, CrSVP2, and AtGRF1 cleavage efficiency by transient expression.
Supplemental Figure S3. In vivo analysis of the miR396-mediated CrSVP1 expression efficiency by transient expression.
Supplemental Figure S4. Detection of different SVP genes in various transgenic plants.
Supplemental Figure S5. Amino acid sequence alignment of AtSVP, CrSVP1, CrSVP2, and AdSVP2.
Supplemental Figure S6. Degradome analysis of Arabidopsis.
Supplemental Figure S7. Degradome data regarding the mismatches in the 5′ portion of the miR396 pairwise region.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Primers for real-time RT-PCR.
Supplemental Table S3. Primers for semi-RT-PCR.
Supplementary Material
Acknowledgments
We thank Dr. Erika Varkonyi-Gasic for providing AdSVP2 transgenic Arabidopsis seeds, Dr. Han Ning for AtSVP transgenic Arabidopsis seeds, and Dr. Saskia Hogenhout for SAP54 transgenic Arabidopsis seeds.
Glossary
- PnWB
peanut witches’ broom
- S4
stage 4
- AYWB
Aster yellow witches’ broom
- MTF
MADS domain transcription factor
- miRNA
microRNA
- Col-0
Columbia-0
- HF
healthy flower
- AF-I
abnormal flower type I
- RT
reverse transcription
- T-DNA
transfer DNA
- MS
Murashige and Skoog
- cDNA
complementary DNA
- DIG
digoxigenin
Footnotes
This work was supported by the Ministry of Science and Technology (grant nos. NSC 97–2321–B–002–046, NSC–98–2313–B–002–047–MY3, NSC 100–2923–B–002–001–MY3, and NSC 102–2313–B–002–068–MY3) and the National Taiwan University (grant no. 102R7602B2).
S.-S.L. and C.-P.L. conceived the original screening and research plans; S.-S.L. and C.-P.L. supervised the experiments; C.-Y.Y. and Y.-H.H. performed most of the experiments; Y.-Y.L., H.-C.H., C.-N.W., and B.-N.S. provided technical assistance; L.-Y.D.L. performed the statistical assays; S.-S.L. designed the experiments and analyzed the data; S.-S.L. conceived the project and wrote the article with contributions of all authors.
References
- Bao M, Bian H, Zha Y, Li F, Sun Y, Bai B, Chen Z, Wang J, Zhu M, Han N (2014) miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol 55: 1343–1353 [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Chang TC (2012) Identification of conserved microRNAs and their targets in PnWB phytoplasma induced leafy flower of Catharanthus roseus. PhD thesis. National Taiwan University, Taipei, Taiwan [Google Scholar]
- Chorostecki U, Crosa VA, Lodeyro AF, Bologna NG, Martin AP, Carrillo N, Schommer C, Palatnik JF (2012) Identification of new microRNA-regulated genes by conserved targeting in plant species. Nucleic Acids Res 40: 8893–8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WC, Chen LL, Lo WS, Lin CP, Kuo CH (2013) Comparative analysis of the peanut witches’-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS ONE 8: e62770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF (2012) Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM (2009) The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J 60: 626–637 [DOI] [PubMed] [Google Scholar]
- Guo L, Lu Z (2010) The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE 5: e11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Himeno M, Neriya Y, Minato N, Miura C, Sugawara K, Ishii Y, Yamaji Y, Kakizawa S, Oshima K, Namba S (2011) Unique morphological changes in plant pathogenic phytoplasma-infected petunia flowers are related to transcriptional regulation of floral homeotic genes in an organ-specific manner. Plant J 67: 971–979 [DOI] [PubMed] [Google Scholar]
- Irish VF. (2010) The flowering of Arabidopsis flower development. Plant J 61: 1014–1028 [DOI] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8: 38–44 [DOI] [PubMed] [Google Scholar]
- Lee IM, Davis RE, Gundersen-Rindal DE (2000) Phytoplasma: phytopathogenic mollicutes. Annu Rev Microbiol 54: 221–255 [DOI] [PubMed] [Google Scholar]
- Lee JH, Park SH, Lee JS, Ahn JH (2007a) A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochim Biophys Acta 1769: 455–461 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007b) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D (2014) Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol 164: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Wu HW, Elena SF, Chen KC, Niu QW, Yeh SD, Chen CC, Chua NH (2009) Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog 5: e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Fang MM, Lin PC, Chiu MT, Liu LY, Lin CP, Lin SS (2013) Improving initial infectivity of the Turnip mosaic virus (TuMV) infectious clone by an mini binary vector via agro-infiltration. Bot Stud 54: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LY, Tseng HI, Lin CP, Lin YY, Huang YH, Huang CK, Chang TH, Lin SS (2014) High-throughput transcriptome analysis of the leafy flower transition of Catharanthus roseus induced by peanut witches’-broom phytoplasma infection. Plant Cell Physiol 55: 942–957 [DOI] [PubMed] [Google Scholar]
- MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RG, Hogenhout SA (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol 12: e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, Hogenhout SA (2011) Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 157: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, et al. (2014) Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J 78: 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Sugio A, MacLean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA (2011) Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu Rev Phytopathol 49: 175–195 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C (2007) Short Vegetative Phase-Like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol 143: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang CN, Chen YJ, Chang YC, Wu CH (2008) A step-by-step optimization guide for applying tissue specific RNA in-situ hybridization to non-model plant species. Taiwania 53: 383–393 [Google Scholar]
- Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RM, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E (2012) Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J Exp Bot 63: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IL. (1985) Host responses of peanut witches’ broom disease. J Agric Res China 34: 464–468 [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.