Specific protein-protein interactions are likely involved in the modulation of photosynthetic apparatus activity.
Abstract
Light is the primary energy source for photosynthetic organisms, but in excess, it can generate reactive oxygen species and lead to cell damage. Plants evolved multiple mechanisms to modulate light use efficiency depending on illumination intensity to thrive in a highly dynamic natural environment. One of the main mechanisms for protection from intense illumination is the dissipation of excess excitation energy as heat, a process called nonphotochemical quenching. In plants, nonphotochemical quenching induction depends on the generation of a pH gradient across thylakoid membranes and on the presence of a protein called PHOTOSYSTEM II SUBUNIT S (PSBS). Here, we generated Physcomitrella patens lines expressing histidine-tagged PSBS that were exploited to purify the native protein by affinity chromatography. The mild conditions used in the purification allowed copurifying PSBS with its interactors, which were identified by mass spectrometry analysis to be mainly photosystem II antenna proteins, such as LIGHT-HARVESTING COMPLEX B (LHCB). PSBS interaction with other proteins appears to be promiscuous and not exclusive, although the major proteins copurified with PSBS were components of the LHCII trimers (LHCB3 and LHCBM). These results provide evidence of a physical interaction between specific photosystem II light-harvesting complexes and PSBS in the thylakoids, suggesting that these subunits are major players in heat dissipation of excess energy.
Photosynthetic organisms exploit sunlight energy to support their metabolism. However, if absorbed in excess, light can produce harmful reactive oxygen species (Li et al., 2009; Murchie and Niyogi, 2011). In a natural environment, light intensity is highly variable and can rapidly change from being limited to being in excess. To survive and thrive in such a variable habitat, plants evolved multiple strategies to modulate their light use efficiency to limit reactive oxygen species formation when exposed to excess illumination while maintaining the ability to harvest light efficiently when required (Li et al., 2009; Murchie and Niyogi, 2011; Ruban, 2015). Among these different protection processes, the fastest, called nonphotochemical quenching (NPQ), is activated in a few seconds after a change in illumination, and it leads to the thermal dissipation of excess absorbed energy. NPQ is a complex phenomenon with different components that are distinguished according to their activation/relaxation time scale (Demmig-Adams et al., 1996; Szabó et al., 2005; Niyogi and Truong, 2013). The primary and fastest NPQ component, called qE (for energy-quenching component) or feedback deexcitation, depends on the generation of a pH gradient across the thylakoid membranes (Niyogi and Truong, 2013). In land plants, qE activation requires the presence of a thylakoid protein called PHOTOSYSTEM II SUBUNIT S (PSBS; Li et al., 2000, 2004). The Arabidopsis (Arabidopsis thaliana) PSBS-depleted mutant psbs KO (Li et al., 2000) is unable to activate qE and also showed reduced fitness when exposed to natural light variations in the field, supporting a major role for this protein in responding to illumination intensity fluctuations (Li et al., 2000; Külheim et al., 2002). Mutational analyses showed that the PSBS role in qE strictly depends on the presence of two protonable Glu residues, which are most likely involved in sensing the pH decrease in the lumen (Li et al., 2004). Despite several studies, however, the precise molecular mechanism by which PSBS controls NPQ induction remains debatable, and contrasting hypotheses have been presented (for review, see Ruban et al., 2012). PSBS has been hypothesized to bind pigments and to be directly responsible for energy dissipation based on its sequence similarity with LIGHT HARVESTING COMPLEX (LHC) proteins (Li et al., 2000; Aspinall-O’Dea et al., 2002). An alternative hypothesis instead suggested that PSBS is unable to bind pigments (Funk et al., 1995; Crouchman et al., 2006; Bonente et al., 2008a) and that it plays an indirect role in NPQ by modulating the PSII antenna protein transition from light harvesting to an energy dissipative state (Betterle et al., 2009; Johnson et al., 2011). This transition has been suggested to depend on the control of the macroorganization of the PSII-LHCII supercomplexes that are present in the grana membranes (Kiss et al., 2008; Betterle et al., 2009; Kereïche et al., 2010; Johnson et al., 2011). Consistent with this hypothesis, it was recently demonstrated that PSBS is able to induce a dissipative state in isolated LHCII proteins in liposomes (Wilk et al., 2013), suggesting that its interactions with antenna proteins play a key role in its biological activity. However, the precise identity of PSBS interactors (Teardo et al., 2007; Betterle et al., 2009), the PSBS oligomerization state (Bergantino et al., 2003), and its localization within PSII supercomplexes (Nield et al., 2000; Haniewicz et al., 2013) remain unclear or at least controversial, limiting the current understanding of PSBS molecular mechanisms.
The moss Physcomitrella patens has recently emerged as a valuable model organism in which to study NPQ. As in the model angiosperm Arabidopsis, PSBS accumulation modulates NPQ amplitude and protects plants from photoinhibition under strong light in P. patens (Li et al., 2000; Alboresi et al., 2010; Zia et al., 2011; Gerotto et al., 2012). PSBS-mediated NPQ in P. patens also showed zeaxanthin dependence as in other plants (Niyogi et al., 1998; Pinnola et al., 2013). The moss P. patens has another protein involved in NPQ, LHCSR, which is typically found in algae and is different from proteins found in vascular plants (Peers et al., 2009; Bailleul et al., 2010; Gerotto and Morosinotto, 2013). Even if LHCSR is present in P. patens, LHCSR- and PSBS-dependent NPQ mechanisms were shown to be independent and to have an additive effect without any significant functional synergy (Gerotto et al., 2012).
Previous data also demonstrated the possibility of achieving strong overexpression of PSBS in P. patens (Gerotto et al., 2012), which, however, was never observed in Arabidopsis (Li et al., 2002). This property was exploited in this work to overexpress a His-tagged PSBS isoform, which was afterward purified in its native state from dark-adapted thylakoid membranes. Several PSII antenna proteins were copurified with PSBS and identified by mass spectrometry analyses, demonstrating that they interact physically in dark-adapted thylakoid membranes. Components of LHCII trimers (LHCB3 and LHCBM) appear to be major, but not exclusive, components of PSBS interactors.
RESULTS
Generation of P. patens Lines Expressing Functional His-Tagged PSBS
In this work, the moss P. patens was exploited to overexpress His-tagged PSBS (Gerotto et al., 2012). PSBS N and C termini have some conserved positively/negatively charged residues, which may be affected by the tag addition (Supplemental Fig. S1). Thus, the functional role of these regions was first assessed by complementing the P. patens PSBS knockout mutant psbs KO with the N- or C-terminally truncated form of the protein (called ΔN- or ΔC-PSBS, respectively). In the case of ΔN-PSBS, the transit peptide required for the protein import was retained to drive protein localization in the thylakoids, thus removing only the N terminus of the mature protein (amino acids 3–27; Supplemental Fig. S1). After transformation, several independent lines complemented with either ΔN-PSBS or ΔC-PSBS were generated (Supplemental Fig. S2). The truncated ΔN-PSBS or ΔC-PSBS proteins were accumulated and successfully imported in the chloroplast, as shown by their smaller size with respect to the wild type (Supplemental Fig. S2, A and B). All complemented lines also showed stronger NPQ with respect to the background psbs KO (Supplemental Fig. S2, C and D), indicating that truncated proteins were indeed active. The ΔC-PSBS line 1, characterized by the highest protein accumulation levels (Supplemental Fig. S2B), also showed the strongest NPQ (Supplemental Fig. S2D), confirming the correlation between protein accumulation and NPQ intensity observed for the wild-type isoform (Gerotto et al., 2012).
Because the deletion of PSBS N and C termini did not impair protein accumulation or its ability to activate NPQ, we generated moss lines expressing PSBS with a His-tag sequence (6×His) either at its N or C terminus. In the case of the C terminus (called C-His-PSBS), the His tag was added at the end of the protein sequence, whereas in the case of the N terminus (N-His-PSBS), the His-tag sequence replaced four residues shown above to be dispensable for PSBS activity (amino acids 17–20 of the mature protein; Supplemental Fig. S1). In this case, the tag was added at the N terminus of the mature protein sequence, retaining the wild-type transit peptide to correctly drive protein insertion into the thylakoid membranes (for details, see “Materials and Methods”).
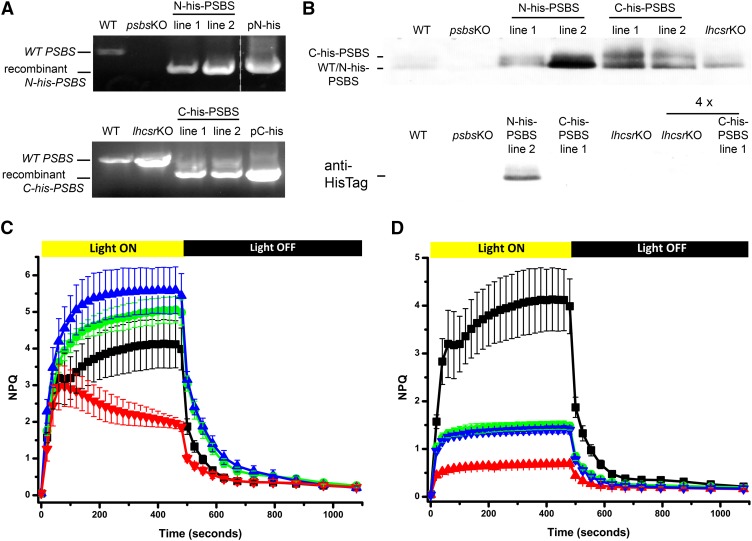
N-His-PSBS was used to complement the psbs KO background, thus generating lines in which only His-tagged PSBS was expressed (N-His-PSBS in psbs KO lines). By contrast, C-His-PSBS was expressed in the lhcsr KO mosses, obtaining lines in which both wild-type and His-tagged PSBS isoforms were simultaneously accumulated (C-His-PSBS in lhcsr KO lines). These different transformations allowed testing the possible influence of tag localization together with the verification of a possible influence of LHCSR on PSBS activity. Invariably, multiple stable resistant lines were obtained, and the characterization of two lines for each of the His-tagged (N- and C-terminal) proteins is shown in Figure 1. Additional lines were nevertheless analyzed, yielding indistinguishable results. The insertion of exogenous DNA was first verified by PCR (Fig. 1A). The presence of a recombinant PSBS-coding sequence in N-His-PSBS and C-His-PSBS lines was recognizable by its smaller size with respect to the wild-type gene, because N/C-His-PSBS sequences were obtained from complementary DNA (cDNA) and thus lacked introns (Fig. 1A). In the case of C-His-PSBS clones, in addition to the strong band corresponding to the recombinant C-His-PSBS coding sequence, a fainter one corresponding to the wild-type gene was also visible, consistent with its presence in the lhcsr KO background.
Figure 1.
Characterization of P. patens lines expressing His-tagged PSBS. A, Genotypic characterization of lines expressing N-His-PSBS (background psbs KO) and C-His-PSBS (background lhcsr KO) in top and bottom gels, respectively. PCR products of wild-type (WT) and recombinant genes are distinguishable by their different size. The dashed line indicates that other lanes were present on the same gel. pN-His and pC-His indicate amplification from plasmids containing N-His-PSBS (top) and C-His-PSBS (bottom) coding sequences, used as positive controls. B, Immunoblot with antibody against P. patens PSBS (top) and anti-His tag (bottom). One and 2.5 µg of Chl thylakoid extracts were loaded, respectively. In the latter, 4× indicates lanes loaded with 10 µg of Chl. C, NPQ kinetics of N-His-PSBS lines. The wild type is shown in black squares, psbs KO in red triangles, and N-His-PSBS lines 1 and 2 in green circles and blue triangles, respectively. D, NPQ kinetics of C-His-PSBS-expressing lines. The wild type is shown in black squares, the lhcsr KO in red triangles, and C-His-PSBS lines 1 and 2 in green circles and blue triangles, respectively. In C and D, data reported are the result of at least three independent replicates expressed as averages ± sd.
After genotype characterization, PSBS polypeptide accumulation in thylakoid extracts was verified using specific antibodies. Figure 1B shows that PSBS is present in the thylakoids of all N-His-PSBS lines, with line 2 showing a particularly strong accumulation. This result clearly demonstrates that protein sequence modifications did not affect PSBS accumulation. The presence of the His tag was also confirmed by detection with an anti-His-tag antibody, which showed a distinct band in N-His-PSBS line 2 and no cross-reactivity in any background line (the wild type, psbs KO, and lhcsr KO; Fig. 1B). In C-His-PSBS line thylakoids, two distinct bands were identifiable with anti-PSBS antibody (Fig. 1B), one the same size as wild-type PSBS and a larger one corresponding to the tagged isoform. However, no distinct band was visible with the anti-His-tag antibody (Fig. 1B), most likely because protein accumulation is below the sensitivity limit. It is notable that immunoblotting was performed on purified thylakoid extracts; thus, these results also imply that the N-/C-His-tagged proteins were correctly imported in the chloroplast and inserted in the membrane. In vivo chlorophyll (Chl) fluorescence kinetics were also measured for all lines to assess their ability to activate an NPQ response (Fig. 1, C and D). All plants showing recombinant PSBS accumulation also featured stronger NPQ with respect to their background, demonstrating that N-His-PSBS and C-His-PSBS proteins were indeed functionally active. The comparison of different lines with variable PSBS accumulation levels showed that the latter correlated with NPQ ability, as observed previously for wild-type PSBS (Gerotto et al., 2012; Fig. 1, C and D).
Purification of His-Tagged PSBS Suggests the Presence of Pigment-Binding Interactors
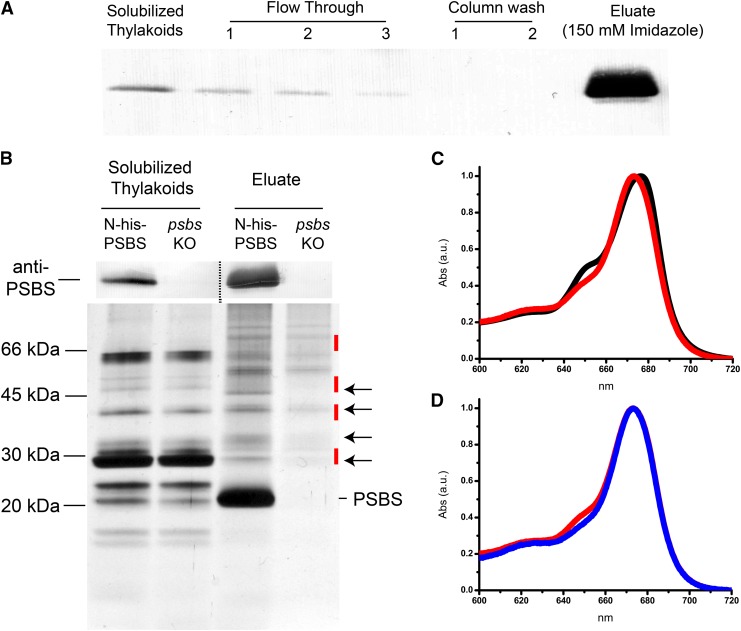
The results reported above showed that both N- and C-His-PSBS are expressed, correctly imported in the thylakoids, and active in NPQ, making these plants a suitable starting material from which to purify PSBS protein in its native state. Therefore, a purification protocol was optimized using the line with the highest expression levels of His-tagged PSBS (N-His-PSBS line 2 in psbs KO, hereafter named N-His-PSBS). Different detergent concentrations were tested for thylakoid solubilization, with 0.6% (w/v) dodecyl-α-d-maltoside (α-DM) selected because it was the mildest condition yielding a complete membrane solubilization (Alboresi et al., 2008). Solubilized thylakoids were then loaded onto a nickel affinity column, and after several washes, PSBS was eluted with 150 mm imidazole, as shown in Figure 2A. Silver-stained SDS-PAGE of the purified fraction showed that PSBS was the major protein present in the eluate, but several other polypeptides were also present (Fig. 2B). To distinguish contaminants from potential PSBS interactors, identical purification was performed with the background line lacking PSBS (psbs KO). The comparison of SDS-PAGE profiles showed the presence of multiple bands also in psbs KO (Fig. 2B), thus representing the contaminants of the purification. Some bands were nevertheless only detectable in the N-His-PSBS line (Fig. 2B, arrows), suggesting their identification as PSBS interactors.
Figure 2.
N-His-PSBS purification by affinity chromatography. A, Immunoblotting with anti-His-tag antibody on different purification fractions obtained from affinity chromatography of N-His-PSBS line 2 thylakoids. Fractions corresponding to starting material (solubilized thylakoids), flow through, column washes, and eluate with 150 mm imidazole are shown. B, PSBS immunodetection (top) and SDS-PAGE profile (bottom) of solubilized thylakoids and eluate with 150 mm imidazole from N-His-PSBS and psbs KO samples. Arrows indicate the main profile differences in N-His-PSBS and psbs KO eluates. Red segments indicate the gel regions where the proteins identified in Table II should be localized. One microgram of Chl was loaded for solubilized thylakoids and flow through in both A and B. For chromatography eluates, a volume corresponding to 15 (immunoblotting) and 130 (SDS-PAGE) µg of Chl in starting thylakoids was loaded. C, Absorption spectra of solubilized thylakoids (black) and N-His-PSBS eluate (red). D, Absorption spectra of eluates from N-His-PSBS (red) and psbs KO (blue). Spectra were normalized to the maximum of the red absorption band. a.u., Arbitrary units.
Pigment analysis of the different purification fractions showed that most of the Chls and carotenoids of the solubilized thylakoids were found in the flow through and column washes (Supplemental Fig. S3A), but a few pigments were also recovered in the eluate containing PSBS (Table I; Fig. 2C; Supplemental Fig. S3B). The solubilized thylakoids and this eluate also showed qualitative differences in native absorption spectra (Fig. 2C) and the Chl a/b ratio (Table I). Additionally, Chls were 3 times more abundant in the eluate from N-His-PSBS than in that from the psbs KO control (Table I; Fig. 2D; Supplemental Fig. S3B). Eluate from N-His-PSBS also showed increased Chl b content, as revealed by absorption signals at 470 and 650 nm (Supplemental Fig. S3B) and by the Chl a/b ratio determination (Table I). Notably, the difference in Chl b content between N-His-PSBS and psbs KO was absent in the starting thylakoids but was the result of the purification, suggesting that this was caused by the presence of PSBS (Table I).
Table I. Chl content of affinity chromatography eluates from different genotypes.
Chl content is shown for fractions eluted from affinity chromatography with 150 mm imidazole for different genotypes. Values are expressed as fractions of starting material (solubilized thylakoids). Chl a/b ratios of chromatography eluates and starting thylakoids are also shown. Values are reported as averages ± sd (n = 9 for Chl content, n = 3 for Chl a/b ratios). Asterisks indicate values significantly different from psbs KO (Student’s t test: *, P = 0.05; and **, P = 0.01).
| Genotype |
Chl Content in Chromatography Eluate | Chl a/b Ratio | |
|---|---|---|---|
| Chromatography Eluate | Thylakoids | ||
| % of solubilized thylakoids | |||
| N-His-PSBS | 0.081 ± 0.020** | 2.95 ± 0.17* | 2.41 ± 0.03 |
| psbs KO | 0.029 ± 0.005 | 4.41 ± 0.59 | 2.38 ± 0.09 |
| C-His-PSBS | 0.049 ± 0.010** | 3.15 ± 0.15* | 2.38 ± 0.03 |
| lhcsr KO | 0.031 ± 0.003 | 4.38 ± 0.18 | 2.34 ± 0.03 |
| Wild type | 0.030 ± 0.003 | 4.60 ± 0.59 | 2.39 ± 0.03 |
Several other protocols were conducted to further purify PSBS from contaminants. Unfortunately, any further purification step attempted was not successful, because it did not completely eliminate contaminants and also led to the loss of all PSBS interactors, likely because of the instability of interactions between PSBS and its partners. This conclusion is supported by results from affinity chromatography performed after different thylakoid solubilization, such as 1% (w/v) dodecyl-β-d-maltoside (β-DM; Alboresi et al., 2008; Pagliano et al., 2012). In this case, the qualitative and quantitative differences between N-His-PSBS and background lines remained, but they were smaller (Supplemental Fig. S4), suggesting that the harsher detergent solubilization also caused at least partial dissociation of pigments/pigment-binding proteins copurified with PSBS.
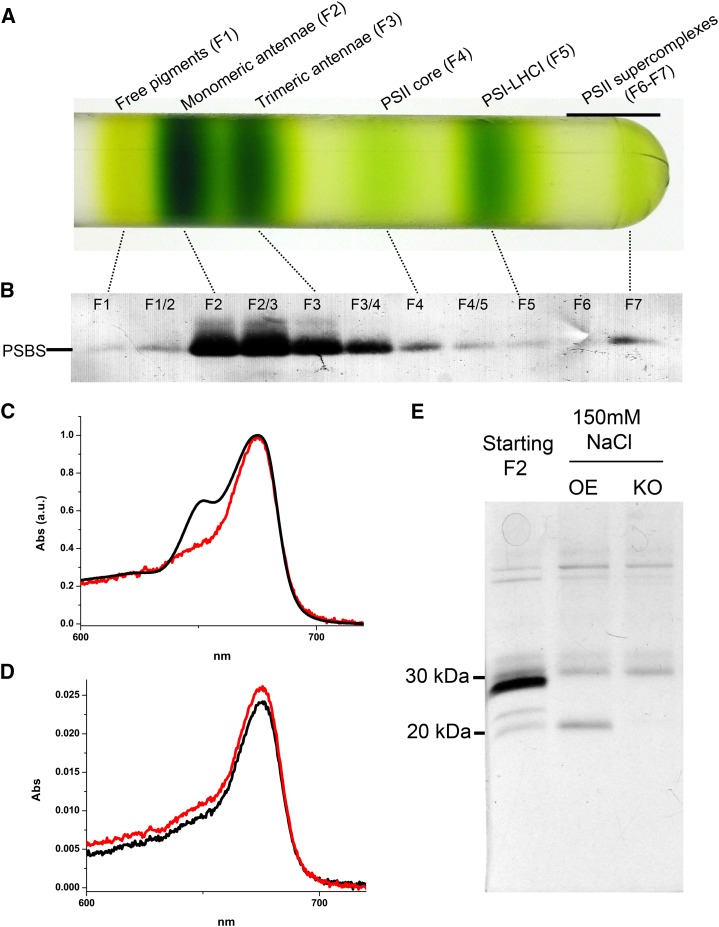
Additional support of this hypothesis came from purification from thylakoids with cation-exchange chromatography (Dominici et al., 2002). In this case, the PSBS polypeptide was copurified with PSI, thus requiring a preliminary purification step with Suc gradient centrifugation (Fig. 3A). Bands showing the strongest PSBS accumulation (the ones corresponding to antennae subunits) were then used for chromatography (Fig. 3B). The majority of pigments were washed out in the column flow through, but a few were recovered in PSBS-enriched fractions (Fig. 3C). In contrast to the affinity chromatography results, however, absorption spectra (Fig. 3D) and SDS-PAGE (Fig. 3E) showed that fractions purified from PSBS-expressing lines were indistinguishable from those obtained from the control psbs KO plants, except for the presence of PSBS in the former. This result was found independently of differences in the starting material used (either monomeric or trimeric antennae bands) or from the pH employed during purification (pH 6 or 7.5). Such an independence of pigment content from the presence/absence of PSBS supports some recent suggestions that this protein does not directly bind pigments (Crouchman et al., 2006; Bonente et al., 2008a). Additionally, this suggested that pigments found in PSBS fractions after affinity chromatography from α-DM solubilized thylakoids were instead most likely bound to some pigment-binding interactors that were copurified with PSBS, and the interaction did not survive harsher detergent treatments or cation-exchange chromatography.
Figure 3.
PSBS purification by cation-exchange chromatography. A, Suc gradient of 0.6% α-DM solubilized thylakoids purified from a PSBS-overexpressing line. B, PSBS detection with specific antibody in the different Suc gradient fractions. An equal volume for each fraction was loaded on the gel. F1 to F7 indicate the different pigmented bands, while F1/2, F2/3, and F3/4 indicate fractions harvested between two Chl-containing bands. C, Absorption spectrum of a PSBS-containing fraction resulting from cation-exchange chromatography (eluted with 150 mm NaCl; red) compared with one of a Suc gradient fraction used as starting material (F2; black). All spectra are normalized to the maximum. a.u., Arbitrary units. D, Absorption spectra of the corresponding chromatography fractions (eluted with 150 mm NaCl) obtained from a PSBS-expressing line (red) and psbs KO (black). E, Cation-exchange chromatography SDS-PAGE profile. Starting material (F2 fraction of the Suc gradient) is compared with samples purified from a PSBS-overexpressing line (OE) and psbs KO (KO).
C-His-PSBS Confirms the Presence of Pigment-Binding Proteins Interacting with PSBS
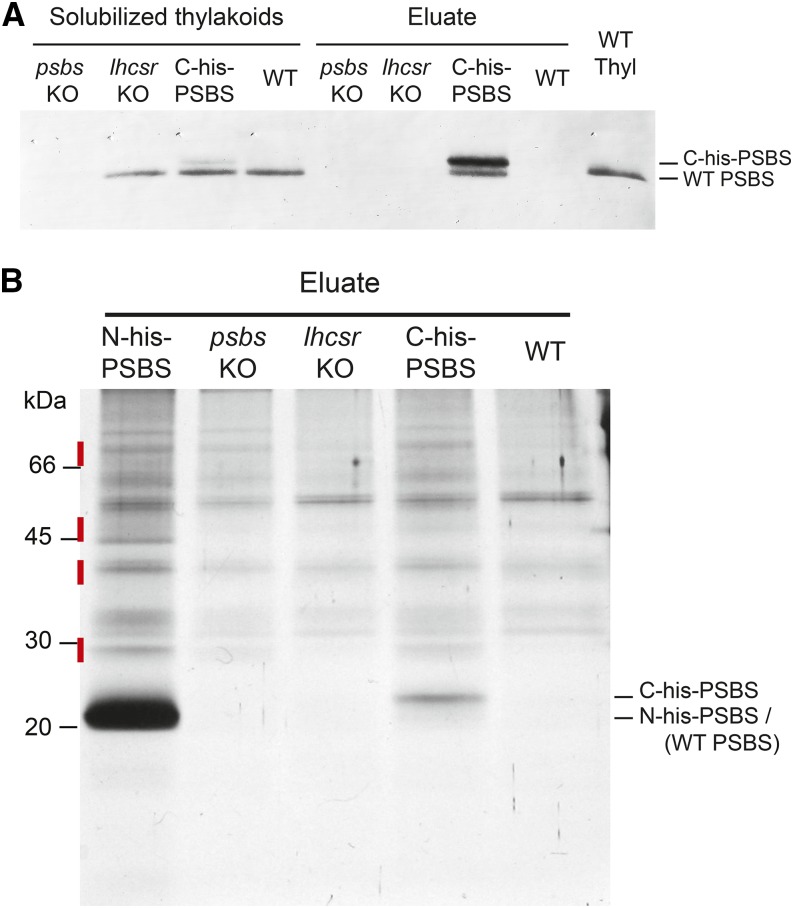
Affinity chromatography purification after mild thylakoid solubilization (0.6% [w/v] α-DM) was repeated with C-His-PSBS line 1 (hereafter C-His-PSBS) to confirm previous results and assess possible effects because of His-tag localization and possible artifacts due to the high PSBS accumulation in N-His-PSBS. Additionally, this line was obtained from an lhcsr KO background; thus, it still expressed the PSBS wild-type isoform while lacking LHCSR. Hence, lhcsr KO and wild-type mosses were also analyzed as controls. Pigment content (Table I) and protein profile (Fig. 4) in eluates from the wild type and lhcsr KO controls were very similar to those obtained with psbs KO, suggesting that, in all these genotypes, eluted fractions contain identical contaminant proteins unspecifically interacting with the stationary phase during chromatography. By contrast, PSBS was successfully purified from C-His-PSBS, as confirmed by PSBS detection in purification eluate (Fig. 4A). Eluate from the C-His-PSBS line also showed a higher total Chl content and lower Chl a/b ratio with respect to all backgrounds (Table I), confirming a correlation between PSBS presence and a Chl enrichment of Chl b in particular. Qualitative and quantitative differences with respect to control lines were smaller for the C-His-PSBS sample compared with the N-His-PSBS eluate, consistent with the smaller PSBS accumulation in the former plants (Fig. 1; Table I).
Figure 4.
C-His-PSBS purification by affinity chromatography. A, PSBS immunodetection in solubilized thylakoids (1 µg of Chl) and eluate from affinity chromatography from C-His PSBS and different controls (the wild type [WT], lhcsr KO, and psbs KO). In all chromatography eluates, a volume corresponding to 75 µg of Chl starting thylakoids was loaded. Wild-type thylakoids (WT Thyl) were also loaded as a positive control. B, SDS-PAGE of affinity chromatography eluates. For all samples, the same volume, corresponding to purification from 130 µg of Chl, was loaded. The wild type, lhcsr KO, and psbs KO are shown as controls. Red segments indicate the gel regions where proteins identified in Table II should be localized.
Notably, immunoblotting with anti-PSBS also showed two bands in the eluate from C-His-PSBS (Fig. 4A). The upper band, more intense, represents the tagged isoform, which is consistently enriched after purification. The weakest band instead corresponds to wild-type PSBS, which is thus copurified with C-His-PSBS. Because PSBS was not detectable in the chromatographic eluate in the case of lhcsr KO and wild-type plants, its presence is not attributable to the unspecific interactions with the nickel column that occurred but rather to the association of wild-type and His-tagged protein. However, these data cannot completely exclude that wild-type and His-tagged PSBS are copurified as members of a multiprotein complex and not because of a direct PSBS-PSBS interaction.
To verify this point, we performed cross-linking experiments on thylakoids purified from different genotypes. In wild-type plants after cross-linking, the monomeric PSBS band (apparent mass of 22 kD) became fainter while a second band at around 45 kD appeared, suggesting the presence of PSBS dimers. A similar result was found also for N-His-PSBS and C-His-PSBS lines (Supplemental Fig. S5), confirming PSBS presence as dimers.
Mass Spectrometry Identification of Proteins Interacting with PSBS
Liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analyses were performed on fractions purified from the N/C-His-PSBS and psbs KO genotypes to identify those proteins copurifying with PSBS. Almost 350 proteins were identified in each replicate, and nearly 300 were common to all three independent biological replicates. Proteins were quantified across the different samples using a label-free quantification approach to estimate relative protein abundances from the chromatographic areas of the identified peptides. For all proteins identified, the areas under the peaks in the N- and C-His-PSBS samples were compared with those obtained from proteins in psbs KO, and this was used as an internal reference (for details on peptide identification and quantification, see “Materials and Methods”; Supplemental Tables S1–S3). In addition to PSBS, only eight other proteins showed an abundance at least 5 times higher in N-His-PSBS eluate compared with the psbs KO control (Table II). Among them, three were PSII antenna subunits (LHCBM, LHCB3, and LHCB4), three were calcium sensor (CaS) protein isoforms, one was a plastid terminal oxidase (PTOX), and the last one (A9SC57) was a protein similar to several uncharacterized proteins (Table II). All the above-mentioned proteins were also enriched in purifications from C-His-PSBS, but to a lower extent compared with the N-His-PSBS eluate, consistent with PSBS abundance in the two lines, lower in the case of the C-His line (Fig. 1B). Moreover, with the possible exception of A9SC57, all retrieved proteins are involved in regulating photosynthesis (Klimmek et al., 2006; Peltier et al., 2010; Petroutsos et al., 2011; Ruban et al., 2012; Caffarri et al., 2014). Indeed, PTOX was recently recognized as an alternative electron sink in the thylakoids, being involved in the so-called chlororespiration process (Peltier et al., 2010; Houille-Vernes et al., 2011), and CaS proteins were recently found to be involved in photoprotection in the model alga Chlamydomonas reinhardtii as well (Petroutsos et al., 2011; Terashima et al., 2012). Although their role in plant photoprotection is far from being fully understood, the identification of CaS and PTOX polypeptides in this study supports the emerging view of a network of mechanisms responsible for fine-tuning photosynthesis efficiency (Allahverdiyeva et al., 2015). According to the number of peptide spectral matches (Supplemental Table S4), which provides a rough estimate of the relative abundance of a protein compared with the others retrieved in the eluates, LHCB isoforms were certainly the most common proteins in PSBS eluate among the ones listed in Table II, suggesting that they are the major PSBS interactors, as hypothesized in recent literature (Betterle et al., 2009; Kereïche et al., 2010; Ruban et al., 2012).
Table II. Mass spectrometry quantification of proteins enriched in affinity chromatography eluates.
Proteins showing at least a 5 times enrichment in N-His-PSBS eluate compared with psbs KO according to mass spectrometry-based label-free quantification are shown. Each protein was quantified from the average of its three most intense peptides. The content of each protein was normalized to the one in psbs KO. Relative enrichment in N-His-PSBS compared with C-His-PSBS is also shown. LHCBM and LHCB4 include multiple polypeptides that were not individually distinguished, as detailed in “Materials and Methods.” n.d., Protein not detected in at least one replicate. All data are expressed as averages ± se of three independent replicates. Expected mass of the mature protein for each identified protein is also reported. Signal peptide length was estimated with the ChloroP tool and mature protein mass with the Compute pI/Mw tool (Expasy).
| Uniprot Accession | Mass | Annotation | N-His-PSBS | C-His-PSBS | psbs KO | N-His-PSBS/C-His-PSBS |
|---|---|---|---|---|---|---|
| kD | ||||||
| A9TJ15 | 23 | PSBS | n.d. | 3.60 ± 0.60 | ||
| A9T914, A9SGM1, A9S6S7, A9TL35, A9RT62, Q765P5, A9RJS3 | 24.8 | LHCBM | 5.69 ± 1.35 | 1.85 ± 0.07 | 1 | 3.07 ± 0.43 |
| A9U3M1, A9T2F8 | 28 | LHCB4 | 8.48 ± 3.90 | 1.66 ± 0.30 | 1 | 5.12 ± 1.46 |
| A9TL15 | 36.9 | PpCAS1 | 7.12 ± 1.16 | 3.17 ± 0.63 | 1 | 2.25 ± 0.33 |
| A9TKU6 | 25 | LHCB3 | 6.85 ± 1.55 | 1.90 ± 0.15 | 1 | 3.60 ± 0.50 |
| A9SC57 | 74 | Uncharacterized protein | 7.14 ± 0.92 | 2.70 ± 0.47 | 1 | 2.65 ± 0.33 |
| A9TW76 | 39.3 | CAS3 | 8.94 ± 2.19 | 4.28 ± 0.52 | 1 | 2.09 ± 0.33 |
| A9SZ30 | 40.2 | PpCAS2 | n.d. | 2.87 ± 0.57 | ||
| A9TEL5 | 48 | PTOX1 | n.d. | 2.29 ± 0.30 |
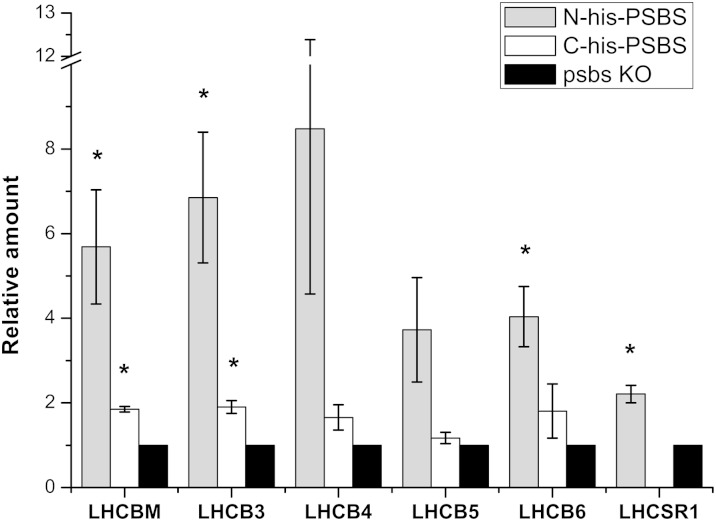
Considering these results, the content of all PSII antennae subunits (LHCB) was evaluated in more detail. The results of the mass spectrometry-based relative antennae quantification, shown in Figure 5, indicate that all LHCB subunits actually display an increased accumulation after purification in the presence of PSBS, although to varying extents. LHCB3 and LHCBM, components of LHCII trimers, were significantly enriched in both PSBS-expressing lines, approximately 6- and 2-fold in N-His-PSBS and C-His-PSBS eluates, respectively. Additionally, LHCB4, despite high variability across N-His-PSBS replicates, showed a strong average enrichment in samples containing PSBS compared with the psbs KO control (Fig. 5). In the case of LHCB6, the increase is less obvious but remains significant in the N-His-PSBS line. Instead, LHCB5 appears to be the isoform with the lowest enrichment, particularly in C-His-PSBS purification, where its accumulation was near that of the negative control. It is notable that, in all cases, enrichment is higher in N-His-PSBS compared with C-His-PSBS, supporting the conclusion that LHCB presence indeed depends on PSBS. The accumulation of LHCSR, the polypeptide involved in NPQ induction in algae (Peers et al., 2009), which is present in P. patens (Alboresi et al., 2010), was also evaluated. As expected, the protein was not found in C-His-PSBS, which has an lhcsr KO background, whereas the eluates obtained from the N-His-PSBS-expressing line showed a low but significant increase in this protein.
Figure 5.
Mass spectrometry quantification of LHCB subunits in PSBS eluates. Quantification (data from Table III) is shown for all PSII antennae subunits (LHCB), including LHCSR, in affinity chromatography eluates from N-His-PSBS (gray bars) and C-His-PSBS (white bars) genotypes, normalized to their amount in the psbs KO negative control (black bars). Data are expressed as averages ± se of three independent replicates. Asterisks indicate values significantly different from psbs KO (Student’s t test: P = 0.05; n = 3). For details about LHCB quantification, see “Materials and Methods.”
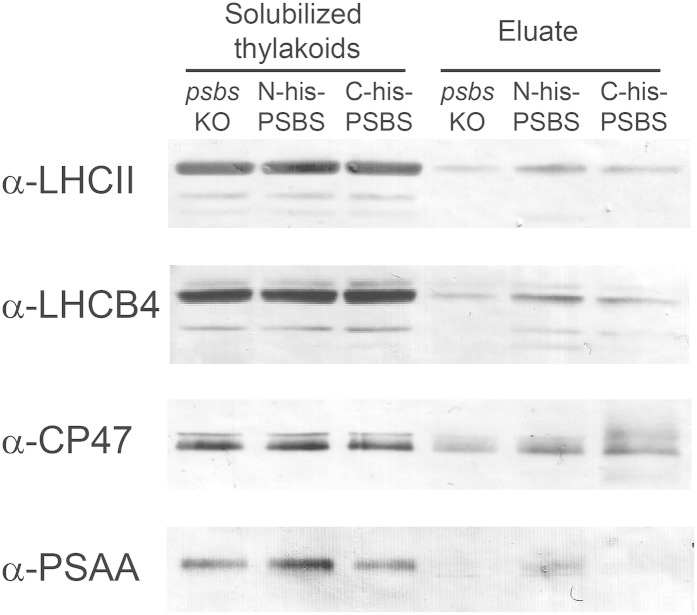
An immunoblot with different available antibodies recognizing P. patens antennae, anti-LHCII (LHCBM and LHCB3 trimers), and anti-LHCB4 were also employed to confirm the results obtained from the mass spectrometry analysis. All antennae had a very similar content in the thylakoids of all genotypes (Fig. 6). By contrast, after purification in both cases, bands were stronger in N-His-PSBS- and C-His-PSBS-expressing lines compared with the psbs KO negative control (Fig. 6), further supporting the conclusion that their presence was caused by copurification with PSBS. Band intensities also showed a dependence on the accumulation of PSBS when comparing N-His-PSBS and C-His-PSBS eluates, confirming the mass spectrometry results. Unfortunately, other LHCB isoforms, namely LHCB3, LHCB5, and LHCB6, were not testable with this approach, because available antibodies did not specifically recognize the moss isoforms (Alboresi et al., 2008).
Figure 6.
Identification of photosynthetic subunits copurified with His-tagged PSBS using immunoblotting. Solubilized thylakoids and eluates after affinity chromatography from psbs KO, N-His-PSBS, and C-His-PSBS were compared using antibodies recognizing different subunits of the photosynthetic apparatus, LHCII, LHCB4, CP47, and PSAA. A total of 0.35 µg of Chl starting solubilized thylakoids was loaded for α-LHCII, α-CP47, and α-PSAA, and 0.12 µg of Chl was loaded for α-LHCB4. For chromatography eluates, a volume equivalent to purification from 100 µg of Chl starting thylakoids was loaded.
For a more comprehensive view of the PSBS interaction with the photosystems, we also evaluated the accumulation of other subunits of both PSI and PSII, also including the reaction centers. We observed a small enrichment in PSII core subunits (PSBA, PSBB, PSBC, and PSBD) in purification eluates from N-His-PSBS (Table III), which was also visible in immunoblotting with anti-CP47 (Fig. 6), whereas in the case of C-His-PSBS, the accumulation was comparable to the psbs KO control. It is also notable that, within the same eluate, all detected PSII core subunits showed similar relative amounts, approximately 2 in N-His-PSBS and approximately 1 in C-His-PSBS samples (Table III). This result suggests that these polypeptides were purified as intact PSII complexes, possibly partly as multiprotein complexes of the PSII core with its antennae (PSII-LHCII supercomplexes), which are partially maintained under the employed solubilization conditions (Alboresi et al., 2008; Fig. 3A).
Table III. Mass spectrometry quantification of PSI and PSII subunits in chromatography eluates from different genotypes.
Relative quantification by mass spectrometry-based label-free quantification of PSI and PSII subunits in affinity chromatography eluates is shown. All quantifications are normalized to the content in psbs KO. Relative enrichment in PSBS-expressing genotypes (N-His-PSBS/C-His-PSBS) is also shown. LHCBM, LHCB4, and LHCB5 include multiple polypeptides that were not individually distinguished, as detailed in “Materials and Methods.” n.d., Protein not detected. All data are expressed as averages ± se of three independent replicates.
| Uniprot Accession | Annotation | N-His-PSBS | C-His-PSBS | psbs KO | N-His-PSBS/C-His-PSBS |
|---|---|---|---|---|---|
| Q6YXN7 | PSBA | 1.94 ± 0.43 | 0.90 ± 0.18 | 1 | 2.16 ± 0.37 |
| Q6YXM8 | PSBB | 2.56 ± 0.55 | 0.92 ± 0.19 | 1 | 2.77 ± 0.47 |
| Q6YXN9 | PSBC | 2.07 ± 0.32 | 0.88 ± 0.19 | 1 | 2.34 ± 0.35 |
| Q6YXN8 | PSBD | 2.43 ± 0.50 | 0.78 ± 0.02 | 1 | 3.11 ± 0.37 |
| A9T914, A9SGM1, A9S6S7, A9TL35, A9RT62, Q765P5, A9RJS3 | LHCBM | 5.69 ± 1.35 | 1.85 ± 0.07 | 1 | 3.07 ± 0.43 |
| A9TKU6 | LHCB3 | 6.85 ± 1.55 | 1.90 ± 0.15 | 1 | 3.60 ± 0.50 |
| A9U3M1, A9T2F8 | LHCB4 | 8.48 ± 3.90 | 1.66 ± 0.30 | 1 | 5.12 ± 1.46 |
| A9U5B1, A9RFL7 | LHCB5 | 3.73 ± 1.23 | 1.17 ± 0.13 | 1 | 3.18 ± 0.64 |
| A9RU32 | LHCB6 | 4.04 ± 0.71 | 1.81 ± 0.64 | 1 | 2.23 ± 0.51 |
| A9TED6 | LHCSR1 | 2.21 ± 0.21 | n.d. | 1 | |
| A9T399 | LHCA1.1 | 4.51 ± 0.87 | 1.48 ± 0.08 | 1 | 3.05 ± 0.35 |
| Q8MFA3 | PSAA | 1.95 ± 0.22 | 1.02 ± 0.13 | 1 | 1.92 ± 0.19 |
| Q8MFA2 | PSAB | 2.89 ± 0.53 | 1.08 ± 0.21 | 1 | 2.68 ± 0.41 |
| Q6YXQ2 | PSAC | 4.03 ± 0.74 | 1.90 ± 0.50 | 1 | 2.11 ± 0.39 |
Some PSI-LHCI subunits also were found enriched in N-His-PSBS but not in the C-His-PSBS samples (Table III), as confirmed by the immunoblot against PSI subunit AA (PSA; Fig. 6). However, in this case, quantifications of different PSI subunits showed more variability in contrast to PSI, known to be a supercomplex, including the core and the antenna proteins (LHCA; Ben-Shem et al., 2003; Caffarri et al., 2014). This variability, together with the missed detection of some primary subunits (e.g. LHCA2-3), suggests the absence of interactions between PSBS and intact PSI-LHCI supercomplexes. By contrast, PSBS most likely interacts unspecifically with some polypeptides that were dissociated from PSI during purification, as observed with other approaches (Teardo et al., 2007).
PSBS Purification from High Light-Treated Plants
All putative interactions found in this study should be light modulated and reorganized upon lumen acidification and protonation of Glu-122/Glu-226 in PSBS (Li et al., 2004). Unfortunately, any attempt to identify light-induced alterations in PSBS interactions failed (Supplemental Fig. S6). Although we cannot exclude that PSBS is always near the same proteins regardless of light intensity, this negative outcome is most likely because the entire protocol, from light treatment, tissue harvesting, thylakoid purification, and affinity chromatography, lasted several hours and therefore is unsuitable to study changes correlated with a fast mechanism, such as PSBS-dependent NPQ, which is largely inactivated after 1 min in the dark (Fig. 1; Supplemental Fig. S5A). Moreover, nickel affinity chromatography works efficiently only at neutral or basic pH, therefore preventing the use of an acidic pH, which could possibly keep PSBS in its active state, and hindering the possibility of analyzing a genuine NPQ active state.
DISCUSSION
PSBS Preferentially Interacts with One Side of PSII-LHCII Supercomplexes in a Dark-Adapted State
NPQ has a seminal role in plant survival in a highly dynamic natural environment (Külheim et al., 2002); thus, it has attracted the attention of many researchers in the past few years (Szabó et al., 2005; Niyogi and Truong, 2013). PSBS plays a key role in NPQ activation in plants, but despite many studies, its molecular mechanism remains unclear (Ruban et al., 2012). An emerging consensus, however, supports the idea that PSBS plays an indirect role in energy dissipation by inducing a structural rearrangement of pigment-binding complexes in the thylakoid membrane, thus stimulating the transition of PSII antenna proteins from a light-harvesting state to an energy dissipation state (Betterle et al., 2009; Kereïche et al., 2010; Ruban et al., 2012). This hypothesis suggests that PSBS must interact with other photosynthetic apparatus proteins to be able to facilitate these structural conformational changes, and the search for these interactors is seminal to clarifying its molecular mechanism. Previous attempts in this direction provided some evidence of interaction but failed to identify any preferred PSBS partner (Dominici et al., 2002; Teardo et al., 2007; Betterle et al., 2009; Ruban et al., 2012).
The moss P. patens shows the same dependence of NPQ on PSBS and zeaxanthin as the model plant Arabidopsis but also exhibits the ability to express PSBS at high levels, which has not been found in other model organisms (Li et al., 2002; Gerotto et al., 2012). Thus, we exploited P. patens lines expressing His-tagged PSBS to purify the protein with its potential interactors. This approach was complicated by the requirement that purification conditions be finely calibrated to ensure sufficient purification levels without disrupting the protein-protein interactions. As shown above, to maintain these interactions, we had to renounce achieving a complete PSBS purification and eliminate all contaminants after affinity chromatography. Indeed, unfortunately, any additional purification step attempted also led to the disruption of all interactions, as shown by the results from alternative purification methods, such as cation-exchange chromatography (Fig. 3). Another factor to be considered is that potential interactors, such as LHC proteins, are highly abundant in thylakoid membranes and also have several His residues in their sequences, factors that can increase the probability of having a few weak unspecific interactions during nickel affinity chromatography (Passarini et al., 2014). To overcome these issues, it was fundamental to analyze several control lines, namely the wild type, lhcsr KO, and psbs KO, and two independent overexpressing lines with different PSBS accumulation levels, genetic backgrounds, and His-tag positions to distinguish the contaminants from legitimate PSBS interactors. The first clear observation was that samples purified from N-His-PSBS and C-His-PSBS lines showed higher Chl content with respect to all controls, which were instead indistinguishable. Additionally, eluates from PSBS-expressing lines showed an altered pigment composition, with a relative increase in Chl b with respect to all controls (the wild type, psbs KO, and lhcsr KO; Table I; Fig. 2; Supplemental Fig. S3B), despite the fact that the Chl a/b ratio of the starting thylakoids was indistinguishable (Table I; Fig. 6). The increase in Chl content and the Chl a/b ratio alteration also correlate with PSBS expression level in the two lines, supporting the conclusion that the change in pigment content indeed depended on the PSBS presence during purification.
Purification under harsher conditions (cation-exchange chromatography) showed no pigment differences in PSBS-containing fractions compared with the psbs KO control (Fig. 3), suggesting that Chls copurified in milder conditions are unlikely to bind to the protein, unless such an interaction is very labile (Bonente et al., 2008a; Ruban et al., 2012).
Independently from the ability of PSBS to bind pigments, several pigment-binding proteins were copurified with PSBS as interactors. Immunoblotting and mass spectrometry analysis identified some LHCB isoforms as the major polypeptides enriched in purification eluates (Table II; Figs. 5 and 6). It is notable that all LHCB isoforms, with the exception of LHCB5, showed an increased accumulation in the presence of PSBS (Fig. 5), which correlated with their PSBS expression level (Table II). These observations suggest that PSBS shows a significant promiscuity and interacts with several LHCB proteins and not with a single specific antenna. The list of different partners should also include PSBS, since the results clearly show that wild-type PSBS is copurified with the His-tagged form (Fig. 4). The presence of PSBS dimers in dark-adapted native thylakoids was confirmed by cross-linking (Supplemental Fig. S5), providing an independent verification of results from purification with a nickel affinity column and confirming previous suggestions that PSBS could form dimers in the dark (Bergantino et al., 2003).
No evidence was found for a strong interaction between PSBS and LHCSR, the protein responsible for NPQ in algae, which is active in P. patens as well (Alboresi et al., 2010). This is consistent with previous results (Gerotto et al., 2012) showing that PSBS and LHCSR are present simultaneously in this moss but are autonomous in their activity.
The idea that PSBS is capable of interacting with different PSII antennae is consistent with all data obtained from mutants depleted in different individual LHCB isoforms. Indeed, whereas the Arabidopsis lhcb5 KO mutant showed indistinguishable qE with respect to the wild type (Dall’Osto et al., 2005), depletion of the LHCB6, LHCB4, and LHCII subunits all showed alterations of qE to some extent (Andersson et al., 2003; Betterle et al., 2009; Damkjaer et al., 2009; de Bianchi et al., 2011). By contrast, however, no antenna mutant showed a complete depletion of NPQ and its fast PSBS component qE (Andersson et al., 2003; Betterle et al., 2009; Damkjaer et al., 2009; de Bianchi et al., 2011), supporting the idea that PSBS does not have an exclusive partner and that it can be active by interacting with different antenna complexes. Thus, in their involvement in energy dissipation, antenna complexes appear to have an overlapping function, with the possible exception of LHCB5, which does not influence the qE component but is involved in the slower, zeaxanthin-dependent, qZ component (Dall’Osto et al., 2005).
Although the data presented here support a functional overlap between different antenna isoforms, mass spectrometry data also revealed that LHCB3 and LHCBM, components of LHCII trimers, showed the strongest enrichment in both N-His-PSBS and C-His-PSBS eluates (Fig. 5), suggesting that PSBS preferentially interacts with LHCII trimers. This is consistent with reports proposing LHCII trimers as the primary sites for energy quenching (Ruban et al., 2007; Caffarri et al., 2011; Valkunas et al., 2011). The previously mentioned PSBS promiscuity, however, also explains why the controversy on the prominent role of minor or major antennae in NPQ activation was never satisfactorily settled, with evidence supporting both hypotheses (Ruban et al., 2012).
The highest accumulation of LHCB3 and LHCM isoforms, together with the low enrichment of LHCB5, also suggest that PSBS might preferentially interact with one side of the PSII-LHCII supercomplexes, in the region of LHCII trimer M (LHCII-M; Fig. 7). According to the most recent data on PSII-LHCII supercomplex structure, LHCB3 is a specific component of LHCII-M trimers in Arabidopsis (Caffarri et al., 2009), which is connected to minor antennae LHCB4 and LHCB6 and another LHCII trimer, LHCII-S. Conversely, LHCB5 is at the opposite side of the PSII-LHCII supercomplexes, consistent with its limited PSBS interaction shown here.
Figure 7.
Hypothesis of PSBS interaction with PSII-LHCII supercomplexes. A schematic view of the C2S2M2 PSII-LHCII supercomplex, derived from recent literature (Dekker and Boekema, 2005; Caffarri et al., 2009), is shown. The PSII dimeric core is shown in blue, LHCB1/2 (LHCBM in P. patens) in dark green, LHCB3 in light green, LHCB4 (CP29) in yellow, LHCB5 (CP26) in orange, and LHCB6 (CP24) in red. The gray circle indicates the putative region of PSBS interaction according to purification results, thus suggesting that PSBS interacts with the PSII-LHCII supercomplex in proximity to the LHCII-M trimer.
A direct experimental validation for the PSBS localization model in Figure 7 is clearly still lacking, but the hypothesized preferential localization at one side of the PSII-LHCII supercomplexes is also consistent with the distribution of different LHC subunits in photosynthetic eukaryotes. LHCB3 and LHCB6 subunits emerged more recently during the evolution of Viridiplantae (Koziol et al., 2007; Engelken et al., 2010), and they are both found in all plants but absent in most green algae, where NPQ relies on a different protein, LHCSR. Notably, a recent characterization of PSII-LHCII from the model chlorophyte C. reinhardtii (Tokutsu et al., 2012; Drop et al., 2014) showed important differences with respect to Arabidopsis in the association of the LHCII-M trimer, mediated by another LHCII trimer and not by LHCB6, as in plants. If, as suggested previously, this is the region of the PSII-LHCII supercomplex responsible for interaction with PSBS, the different structural organization could also explain why PSBS in C. reinhardtii appears to be unable to activate NPQ, even if overexpressed (Bonente et al., 2008b).
It is notable that, despite limited available genomic resources, LHCB6 isoforms can be identified in several species of streptophyte algae (Supplemental Fig. S7), the group of green algae that later diverged from plant ancestors. LHCB6 distribution is highly similar to that of PSBS, which was found to accumulate and play a role in NPQ in the same later-diverging streptophyte algae (Gerotto and Morosinotto, 2013). Thus, it is tempting to propose a scenario in which the appearance of LHCB6 (and possibly LHCB3) and a modification of the supramolecular organization of the PSII-LHCII supercomplexes were correlated with PSBS assuming its role in NPQ activation by interacting with LHCB.
The results presented in this study thus point to a model in which PSBS in a dark, relaxed, state interacts with one side of the PSII-LHCII supercomplex, preferring LHCII trimer subunits, but not exclusively. These interactions likely play a role in PSBS’s ability to induce, upon illumination, a reorganization of the photosystems and a quenched conformation in antenna proteins (Betterle et al., 2009; Johnson et al., 2011).
MATERIALS AND METHODS
Plant Material
Protonemal tissue of Physcomitrella patens wild-type strain as well as psbs KO and/or lhcsr KO lines (Alboresi et al., 2010) and lines (over)expressing mutated/His-tagged PSBS isoforms (obtained in this work) were grown in rich PpNH4 or minimum PpNO3 medium (Ashton et al., 1979) and solidified with 0.8% (w/v) plant agar (Duchefa Biochemie) for transformation or physiological characterization of phenotypes, respectively. In the case of biomass production for PSBS purification, growth in a liquid-rich medium was also used. All cultures were placed in a growth chamber under controlled conditions: 24°C, a 16-h-light/8-h-dark photoperiod, and a light intensity of 40 µmol m−2 s−1.
Sequence Retrieval and Analysis
Sequences were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments were generated using ClustalW and manually corrected using BioEdit (Hall, 1999). Sequence logos were constructed using the WebLogo server (http://weblogo.berkeley.edu/logo.cgi).
Generation of Mutated PSBS Coding Sequences
To generate mosses expressing truncated/His-tagged PSBS proteins, cDNA obtained from P. patens protonema grown in control conditions was used as starting material. A coding sequence for PSBS (XM_001778511.1) was amplified by PCR with different strategies according to the required mutation and then cloned into a pMAK1 vector (kindly provided by Mitsuyasu Hasebe, National Institute for Basic Biology).
To obtain truncated PSBS isoforms, amplifications with PSBS_FOR and PSBS_ΔCtermREV (Supplemental Table S5) primers led to the generation of a PSBS protein with deleted C-terminal amino acids (Supplemental Fig. S1). To obtain the N-terminal deletion, we first separately obtained the sequences of the transit peptide, which is necessary to correctly import the protein to the thylakoids (primers PSBS_FOR and PSBS_pepREV) and of the mature protein depleted in the N-terminal amino acids (primers PSBS_ΔNterFOR and PSBS_REV). The primers PSBS_pepREV and PSBS_ΔNterFOR were designed to anneal to one another; this annealing allowed us to fuse the two amplicons in an additional PCR using the two fragments as templates and PSBS_FOR and PSBS_REV as primers; thus, the resulting amplificate presented the transit peptide fused with the mature PSBS coding sequence shortened in the N terminus of the polypeptide.
Similarly, to generate coding sequences for PSBS isoforms with His-tag peptide (6×His), we employed two different strategies for the N- and C-terminal tags. To obtain PSBS with the C-terminal 6×His peptide, the His-tagged coding sequence was obtained by PCR from wild-type cDNA (primers PSBS_for/PSBS_Chis_rev), with the 6×His sequence added because of its presence in the reverse primer. To generate PSBS with an N-terminal His tag, the entire PSBS wild-type cDNA amplicon was first cloned into the pTOPOblunt vector (Invitrogen). N-terminal His-tag insertion in the coding sequence was obtained by site-directed mutagenesis on the resulting PSBS-pTOPOblunt vector employing PSBS_Nhis_for/rev primers with the QuikChange Site-Directed Mutagenesis Kit (Stratagene), designed according to the manufacturer’s instructions. The primers led to the replacement of four amino acids with 6×His peptide in the N terminus of the mature protein. We then digested all coding sequences with XhoI/HpaI restriction enzymes to clone them into the pMAK1 vector.
Moss Transformation
To obtain all the genotypes presented here, P. patens transformation was performed as described (Schaefer and Zrÿd, 1997) with minor modifications, as reported previously (Alboresi et al., 2010; Gerotto et al., 2012). Five-day-old protonemal tissue was collected for protoplast generation and polyethylene glycol-mediated transformation. Resistant colony selection was started 6 d after transformation by transferring regenerated plants on culture medium supplemented with the antibiotic zeocin (50 µg mL−1; Duchefa Biochemie). After 7 to 10 d of growth in selective medium, resistant colonies were transferred for 10 d to nonselective plates. Stable mutant lines were then obtained with a second round of selection in culture medium supplemented with zeocin. Genomic DNA from controls and resistant lines was obtained with EuroGOLD Plant DNA mini kits (EuroClone) according to the manufacturer’s instructions and used as templates to confirm DNA insertion by PCR.
In Vivo Chl Fluorescence Measurement
In vivo Chl fluorescence of P. patens wild-type and mutant lines grown for 10 d in minimum medium was measured at room temperature on a Dual PAM-100 fluorometer (Walz) with saturating light at 6,000 µmol m−2 s−1 and actinic light at 830 µmol m−2 s−1. Before measurements were made, plates were dark adapted for 40 min at room temperature. Parameters Fv/Fm (variable PSII fluorescence in the dark-adapted state/maximum PSII fluorescence in the dark-adapted state) and NPQ were calculated as (Fm − F0)/Fm and (Fm − Fm′)/Fm′, where F0 = initial (minimum) PSII fluorescence in the dark-adapted state and Fm′ = maximum PSII fluorescence in the light-adapted state (Demmig-Adams et al., 1996). Data are presented as averages ± sd of at least three independent experiments.
Thylakoid Extraction
Thylakoids from 10-d-old protonemal tissue were prepared using an Arabidopsis (Arabidopsis thaliana) protocol with minor modifications, as described (Gerotto et al., 2012), frozen in liquid nitrogen, and stored at −80°C until used.
SDS-PAGE, Gel Staining, and Immunoblotting
Thylakoid extracts and purification fractions were analyzed for their protein content after SDS-PAGE separation followed by silver staining or immunoblotting. For silver staining, gels were incubated overnight in fixing solution (50% [v/v] methanol, 12% [v/v] acetic acid, and 0.0185% [v/v] formaldehyde) and then in pretreatment solution (0.02% [w/v] Na2S2O3∙5H2O) for 1 min. Gels were then incubated for 20 min in silver staining solution (0.2% [w/v] AgNO3 and 0.028% [v/v] formaldehyde). Incubation with 6% (w/v) Na2CO3, 2% (v/v) pretreatment solution, and 0.0185% (v/v) formaldehyde allowed us to visualize the bands. The reaction was blocked by incubating with 50% (v/v) methanol and 12% (v/v) acetic acid. After each step, gels were rinsed with water.
For the immunoblot analysis after SDS-PAGE separation, proteins were transferred to nitrocellulose membranes (Pall) and detected with specific homemade polyclonal antibodies for PSBS, LHC, and PSII core subunits. The anti-His-tag peptide antibody was obtained from Sigma-Aldrich, and anti-PSAA antibody was obtained from Agrisera.
His-Tagged PSBS Purification by Affinity Chromatography
Thylakoids from dark-adapted plants (24 h) or light-treated plants (30 min at 1,000 µmol m−2 s−1) were used as starting material. They were washed with 5 mm EDTA and then washed three times with 20 mm HEPES, pH 7.5, to completely remove EDTA traces from the sample. After centrifugation at 12,000g at 4°C for 10 min, the membranes were suspended in 20 mm HEPES, pH 7.5, at a Chl concentration of 1 µg µL−1. Then, they were solubilized by adding 1.2% (w/v) α-DM in 20 mm HEPES, pH 7.5, to a final Chl concentration of 0.5 µg µL−1 (final detergent concentration of 0.6%). After vortexing for 1 min, the solubilized samples were centrifuged at 12,000g for 10 min at 4°C to pellet insolubilized material. The supernatant (solubilized thylakoids) was then diluted 10 times in the chromatographic equilibration buffer (EB; 20 mm HEPES, pH 7.5, 100 mm NaCl, 0.06% [w/v] α-DM, and 10% [w/v] glycerol) and loaded onto a nickel-nitrilotriacetic acid agarose column (HIS-Select Nickel Affinity Gel; Sigma-Aldrich). Chromatography was performed by gravity flow. The flow through was recovered and loaded onto the column two additional times to increase the purification yield. After sample loading, the column was washed with 10 column volumes of EB and 10 column volumes of 5 mm imidazole in EB. His-tagged PSBS was then eluted with 10 column volumes of 150 mm imidazole in EB. Purification fractions were analyzed for pigment content both with native absorption spectra in EB solution and after pigment extraction with 80% (v/v) acetone. The Chl a/b ratio was obtained by fitting the spectrum of 80% acetone pigment extracts with spectra of the individual purified pigments, as described (Croce et al., 2002). Protein content was analyzed by SDS-PAGE and silver staining or immunoblotting.
Wild-Type PSBS Purification: Suc Gradients and Cation-Exchange Chromatography
Thylakoid membranes corresponding to 500 μg of Chl were washed with 50 mm EDTA. After centrifugation at 12,000g at 4°C for 10 min, the membranes were suspended in 500 μL of 10 mm HEPES, pH 7.5, solubilized by adding 500 μL of 1.2% (w/v) α-DM and 10 mm HEPES, pH 7.5, and vortexed for 1 min. The solubilized samples were centrifuged at 12,000g for 10 min at 4°C to eliminate insolubilized material and then fractionated by ultracentrifugation in a 0.1 to 1 m Suc gradient, 10 mm HEPES, pH 7.5, and 0.06% (w/v) α-DM for 19 h in an SW41 rotor at 40,000 rpm (254,000g) at 4°C (Beckman OPTIMA LE-8-K).
The resulting monomeric/trimeric antenna bands were harvested with a syringe and used as starting material for cation-exchange chromatography, using the Vivapure Ion-Exchange S Mini M column (Sartorius Stedim). After column equilibration with 1 mL of column equilibration buffer (CEB; 20 mm MES/NaOH, pH 6, 400 mm Suc, 0.15% [w/v] β-DM, and 0.2 mm CaCl2 or 0.15% [w/v] β-DM, 20 mm HEPES, pH 7.5, 0.2 mm CaCl2, and 400 mm Suc in the case of purification at neutral pH), the sample was diluted six times in CEB and loaded onto the column. All centrifugation steps were performed at 500g for 5 min at 4°C. Proteins were eluted using increasing NaCl concentrations (25, 75, 100, 125, 150, 200, and 250 mm or 1 m) in CEB.
Cross-Linking
Thylakoids (final concentration of 300 µg Chl mL−1 in 20 mm HEPES, pH 7.5) were incubated for 30 min with 0.5 mm dithiobis-succinimidyl propionate or 0.5 mm disuccinimidyl suberate (in dimethyl sulfoxide) at room temperature in the dark. Reaction was blocked by adding 1 m Tris, pH 7.5. Samples were solubilized with nonreducing sample buffer before loading for SDS-PAGE (4 µg of Chl was loaded in each lane).
Mass Spectrometry Analysis
In-Gel Tryptic Digestion and LC-Mass Spectrometry
Three independent biological replicates of LC coupled to high-resolution MS/MS were performed to identify proteins interacting with PSBS. After affinity chromatography, which was performed starting with the same amount of thylakoid extracts (700 µg of Chl) and under identical conditions for the N-His-PSBS, C-His-PSBS, and psbs KO lines, all samples were loaded onto NuPAGE 12% (w/v) Bis-Tris gels (Life Technologies). Proteins were allowed to enter for approximately 1 cm on the resolving gel, which was then stained with SimplyBlue Coomassie (Invitrogen) and destained with water. For each gel lane, a single band was excised, cut into approximately 1-mm2 pieces, and incubated with 10 mm dithiothreitol (Fluka) in 50 mm NH4HCO3 for 1 h at 56°C. Alkylation was successively performed with 55 mm iodoacetamide (Sigma-Aldrich) dissolved in 50 mm NH4HCO3 for 45 min at room temperature in the dark. In-gel protein digestion was performed overnight by treating the samples with 12.5 ng µL−1 sequencing-grade modified trypsin (Promega) in 50 mm NH4HCO3 at 37°C. Peptides were extracted from the gel with 50% (v/v) acetonitrile/0.1% (v/v) formic acid and analyzed with an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled with a nano-HPLC Ultimate 3000 device (Dionex-Thermo Fisher Scientific). Peptides were loaded onto a homemade pico-frit column packed with C18 material (Aeris Peptide 3.6 µm XB-C18; Phenomenex) and separated using a linear gradient of acetonitrile/0.1% formic acid from 3% to 50% in 90 min at a flow rate of 250 nL min−1. Spray voltage was set to 1.3 kV with an ion source capillary temperature of 200°C. The instrument was operating in data-dependent mode with a top-four acquisition method (a full scan at 60,000 resolution on the Orbitrap followed by MS/MS fragmentation in a linear trap of the four most intense ions).
Protein Identification
The acquired raw data files were processed using Proteome Discoverer 1.4 software (Thermo Fisher Scientific) coupled with a Mascot server (version 2.2.4; Matrix Science) as the search engine. Protein identification was performed against the P. patens protein database (release December 2013; 34,952 sequences). A list of the most common contaminants found in proteomics experiments was also added to the database as reported (Arrigoni et al., 2013). Enzyme specificity was set to trypsin, and a maximum of two missed cleavages were allowed. The precursor and fragment mass tolerances were set to 10 ppm and 0.6 D, respectively. Carbamidomethylation of Cys was set as a static modification, whereas oxidation of Met was set to variable modification. Percolator (Wright et al., 2012) was used to assess peptide and protein identification confidence: only proteins identified with at least two independent unique peptides with high confidence (q < 0.05) were considered positive hits. Proteins were grouped into protein families according to the principle of maximum parsimony.
Protein Quantification
To perform a label-free quantification, a precursor ion area detector node was also used in the processing method of Proteome Discoverer, which extracted and integrated the area under the peak for each identified peptide. Protein quantification was then computed by averaging the area under the peak of the three most represented peptides for each protein. Therefore, quantitative data were associated only with proteins identified with at least three unique independent peptides. Protein abundance was compared between samples by normalizing their amount (area) to the values from the psbs KO line. Finally, the relative abundances of the three biological replicates were mediated, and statistical analysis was performed.
For LHCB subunits, which exist in several different isoforms that are characterized by a high degree of similarity, peptide and protein quantification was manually edited. Because it was not possible to individually quantify all the different LHCB isoforms found in the P. patens genome (Alboresi et al., 2008), we present the data by grouping the different types of LHCB: LHCBM (including all LHCBM1–LHCBM13 isoforms), LHCB3, LHCB4 (including LHCB4.1 and LHCB4.2), LHCB5 (LHCB5.1 and LHCB5.2), and LHCB6. To increase the accuracy of the LHCB relative quantification across the samples, all the identified peptides (not only the three most abundant ones) were considered for these proteins.
Sequence data from this article can be found in the GenBank data libraries under accession number XM_001778511 (P. patens PSBS; Phytozome Pp1s241_86V6); additional PSBS and LHC sequence accession numbers are reported in Supplemental Figures S1 and S7.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PSBS sequence conservation in different plant species.
Supplemental Figure S2. PSBS accumulation in ΔN- and ΔC-PSBS complemented lines.
Supplemental Figure S3. Additional data on purification from the N-His-PSBS complemented line and psbs KO.
Supplemental Figure S4. Comparison of PSBS purification from N-His-PSBS mosses after solubilization with α-DM and β-DM.
Supplemental Figure S5. Cross-linking with dithiobis-succinimidyl propionate or disuccinimidyl suberate in thylakoids from different genotypes.
Supplemental Figure S6. Purification from dark-adapted and high light-treated plants.
Supplemental Figure S7. Identification of putative LHCB6 sequences in late streptophyte algae.
Supplemental Table S1. Mass spectrometry raw data from biological replicate I.
Supplemental Table S2. Mass spectrometry raw data from biological replicate II.
Supplemental Table S3. Mass spectrometry raw data from biological replicate III.
Supplemental Table S4. Relative enrichment and peptide spectral matches for proteins copurified with PSBS from biological replicates I to III.
Supplemental Table S5. Primers employed for the amplification and cloning of truncated/His-tagged P. patens PSBS coding sequences from cDNA.
Supplementary Material
Acknowledgments
We thank Mitsuyasu Hasebe for providing plasmid for moss transformation and Cristina Sudiro for preliminary work.
Glossary
- NPQ
nonphotochemical quenching
- qE
energy-quenching component
- cDNA
complementary DNA
- β-DM
dodecyl-β-d-maltoside
- α-DM
dodecyl-α-d-maltoside
- Chl
chlorophyll
- MS/MS
tandem mass spectrometry
- EB
equilibration buffer
- LC
liquid chromatography
Footnotes
This work was supported by the University of Padova (grant no. CPDR104834 to T.M. and grant no. GRIC13V6YZ to C.G.), by the European Research Council (BIOLEAP grant no. 309485 to T.M.), by the Ministero dell'Istruzione dell'Università e della Ricerca (grant no. RIN 2012XSAWYM to T.M.), and by Cassa di Risparmio di Padova e Rovigo Holding (funding for the acquisition of the LTQ-Orbitrap XL mass spectrometer).
Articles can be viewed without a subscription.
References
- Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T (2008) In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Suorsa M, Tikkanen M, Aro EM (2015) Photoprotection of photosystems in fluctuating light intensities. J Exp Bot 66: 2427–2436 [DOI] [PubMed] [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S (2003) Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II: effects on photosynthesis, grana stacking and fitness. Plant J 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Arrigoni G, Tolin S, Moscatiello R, Masi A, Navazio L, Squartini A (2013) Calcium-dependent regulation of genes for plant nodulation in Rhizobium leguminosarum detected by iTRAQ quantitative proteomic analysis. J Proteome Res 12: 5323–5330 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427–435 [DOI] [PubMed] [Google Scholar]
- Aspinall-O’Dea M, Wentworth M, Pascal A, Robert B, Ruban A, Horton P (2002) In vitro reconstitution of the activated zeaxanthin state associated with energy dissipation in plants. Proc Natl Acad Sci USA 99: 16331–16335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G (2010) An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci USA 107: 18214–18219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bergantino E, Segalla A, Brunetta A, Teardo E, Rigoni F, Giacometti GM, Szabò I (2003) Light- and pH-dependent structural changes in the PsbS subunit of photosystem II. Proc Natl Acad Sci USA 100: 15265–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betterle N, Ballottari M, Zorzan S, de Bianchi S, Cazzaniga S, Dall’Osto L, Morosinotto T, Bassi R (2009) Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J Biol Chem 284: 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Howes BD, Caffarri S, Smulevich G, Bassi R (2008a) Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J Biol Chem 283: 8434–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Passarini F, Cazzaniga S, Mancone C, Buia MC, Tripodi M, Bassi R, Caffarri S (2008b) The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem Photobiol 84: 1359–1370 [DOI] [PubMed] [Google Scholar]
- Caffarri S, Broess K, Croce R, van Amerongen H (2011) Excitation energy transfer and trapping in higher plant photosystem II complexes with different antenna sizes. Biophys J 100: 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S, Kouril R, Kereïche S, Boekema EJ, Croce R (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S, Tibiletti T, Jennings RC, Santabarbara S (2014) A comparison between plant photosystem I and photosystem II architecture and functioning. Curr Protein Pept Sci 15: 296–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce R, Canino G, Ros F, Bassi R (2002) Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41: 7334–7343 [DOI] [PubMed] [Google Scholar]
- Crouchman S, Ruban A, Horton P (2006) PsbS enhances nonphotochemical fluorescence quenching in the absence of zeaxanthin. FEBS Lett 580: 2053–2058 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damkjaer JT, Kereïche S, Johnson MP, Kovacs L, Kiss AZ, Boekema EJ, Ruban AV, Horton P, Jansson S (2009) The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21: 3245–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bianchi S, Betterle N, Kouril R, Cazzaniga S, Boekema E, Bassi R, Dall’Osto L (2011) Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW III, Barker DH, Logan BA, Bowling DR, Verhoeven AS (1996) Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiol Plant 98: 253–264 [Google Scholar]
- Dominici P, Caffarri S, Armenante F, Ceoldo S, Crimi M, Bassi R (2002) Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J Biol Chem 277: 22750–22758 [DOI] [PubMed] [Google Scholar]
- Drop B, Webber-Birungi M, Yadav SK, Filipowicz-Szymanska A, Fusetti F, Boekema EJ, Croce R (2014) Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim Biophys Acta 1837: 63–72 [DOI] [PubMed] [Google Scholar]
- Engelken J, Brinkmann H, Adamska I (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Adamska I, Green BR, Andersson B, Renger G (1995) The nuclear-encoded chlorophyll-binding photosystem II-S protein is stable in the absence of pigments. J Biol Chem 270: 30141–30147 [DOI] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Giacometti GM, Bassi R, Morosinotto T (2012) Coexistence of plant and algal energy dissipation mechanisms in the moss Physcomitrella patens. New Phytol 196: 763–773 [DOI] [PubMed] [Google Scholar]
- Gerotto C, Morosinotto T (2013) Evolution of photoprotection mechanisms upon land colonization: evidence of PSBS-dependent NPQ in late Streptophyte algae. Physiol Plant 149: 583–598 [DOI] [PubMed] [Google Scholar]
- Hall T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Haniewicz P, De Sanctis D, Büchel C, Schröder WP, Loi MC, Kieselbach T, Bochtler M, Piano D (2013) Isolation of monomeric photosystem II that retains the subunit PsbS. Photosynth Res 118: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci USA 108: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Goral TK, Duffy CDP, Brain AP, Mullineaux CW, Ruban AV (2011) Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereïche S, Kiss AZ, Kouril R, Boekema EJ, Horton P (2010) The PsbS protein controls the macro-organisation of photosystem II complexes in the grana membranes of higher plant chloroplasts. FEBS Lett 584: 759–764 [DOI] [PubMed] [Google Scholar]
- Kiss AZ, Ruban AV, Horton P (2008) The PsbS protein controls the organization of the photosystem II antenna in higher plant thylakoid membranes. J Biol Chem 283: 3972–3978 [DOI] [PubMed] [Google Scholar]
- Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol 143: 1802–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külheim C, Agren J, Jansson S (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93 [DOI] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem 279: 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Niyogi KK (2002) Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J Biol Chem 277: 33590–33597 [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Murchie EH, Niyogi KK (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nield J, Funk C, Barber J (2000) Supermolecular structure of photosystem II and location of the PsbS protein. Philos Trans R Soc Lond B Biol Sci 355: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Truong TB (2013) Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr Opin Plant Biol 16: 307–314 [DOI] [PubMed] [Google Scholar]
- Pagliano C, Barera S, Chimirri F, Saracco G, Barber J (2012) Comparison of the α and β isomeric forms of the detergent n-dodecyl-D-maltoside for solubilizing photosynthetic complexes from pea thylakoid membranes. Biochim Biophys Acta 1817: 1506–1515 [DOI] [PubMed] [Google Scholar]
- Passarini F, Xu P, Caffarri S, Hille J, Croce R (2014) Towards in vivo mutation analysis: knock-out of specific chlorophylls bound to the light-harvesting complexes of Arabidopsis thaliana—the case of CP24 (Lhcb6). Biochim Biophys Acta 1837: 1500–1506 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier G, Tolleter D, Billon E, Cournac L (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res 106: 19–31 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Dall’Osto L, Gerotto C, Morosinotto T, Bassi R, Alboresi A (2013) Zeaxanthin binds to light-harvesting complex stress-related protein to enhance nonphotochemical quenching in Physcomitrella patens. Plant Cell 25: 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV. (2015) Evolution under the sun: optimizing light harvesting in photosynthesis. J Exp Bot 66: 7–23 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Berera R, Ilioaia C, van Stokkum IH, Kennis JT, Pascal AA, van Amerongen H, Robert B, Horton P, van Grondelle R (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Johnson MP, Duffy CD (2012) The photoprotective molecular switch in the photosystem II antenna. Biochim Biophys Acta 1817: 167–181 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Szabó I, Bergantino E, Giacometti GM (2005) Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep 6: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teardo E, de Laureto PP, Bergantino E, Dalla Vecchia F, Rigoni F, Szabò I, Giacometti GM (2007) Evidences for interaction of PsbS with photosynthetic complexes in maize thylakoids. Biochim Biophys Acta 1767: 703–711 [DOI] [PubMed] [Google Scholar]
- Terashima M, Petroutsos D, Hüdig M, Tolstygina I, Trompelt K, Gäbelein P, Fufezan C, Kudla J, Weinl S, Finazzi G, et al. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA 109: 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkunas L, Chmeliov J, Trinkunas G, Duffy CD, van Grondelle R, Ruban AV (2011) Excitation migration, quenching, and regulation of photosynthetic light harvesting in photosystem II. J Phys Chem B 115: 9252–9260 [DOI] [PubMed] [Google Scholar]
- Wilk L, Grunwald M, Liao PN, Walla PJ, Kühlbrandt W (2013) Direct interaction of the major light-harvesting complex II and PsbS in nonphotochemical quenching. Proc Natl Acad Sci USA 110: 5452–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JC, Collins MO, Yu L, Käll L, Brosch M, Choudhary JS (2012) Enhanced peptide identification by electron transfer dissociation using an improved Mascot Percolator. Mol Cell Proteomics 11: 478–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zia A, Johnson MP, Ruban AV (2011) Acclimation- and mutation-induced enhancement of PsbS levels affects the kinetics of non-photochemical quenching in Arabidopsis thaliana. Planta 233: 1253–1264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.