Aspartate family amino acid contents increase in loss-of-function mutants for specific isoforms of the early steps of the biosynthesis pathways.
Abstract
Biosynthesis of aspartate (Asp)-derived amino acids lysine (Lys), methionine (Met), threonine (Thr), and isoleucine involves monofunctional Asp kinases (AKs) and dual-functional Asp kinase-homoserine dehydrogenases (AK-HSDHs). Four-week-old loss-of-function Arabidopsis (Arabidopsis thaliana) mutants in the AK-HSDH2 gene had increased amounts of Asp and Asp-derived amino acids, especially Thr, in leaves. To explore mechanisms behind this phenotype, we obtained single mutants for other AK and AK-HSDH genes, generated double mutants from ak-hsdh2 and ak mutants, and performed free and protein-bound amino acid profiling, transcript abundance, and activity assays. The increases of Asp, Lys, and Met in ak-hsdh2 were also observed in ak1-1, ak2-1, ak3-1, and ak-hsdh1-1. However, the Thr increase in ak-hsdh2 was observed in ak-hsdh1-1 but not in ak1-1, ak2-1, or ak3-1. Activity assays showed that AK2 and AK-HSDH1 are the major contributors to overall AK and HSDH activities, respectively. Pairwise correlation analysis revealed positive correlations between the amount of AK transcripts and Lys-sensitive AK activity and between the amount of AK-HSDH transcripts and both Thr-sensitive AK activity and total HSDH activity. In addition, the ratio of total AK activity to total HSDH activity negatively correlates with the ratio of Lys to the total amount of Met, Thr, and isoleucine. These data led to the hypothesis that the balance between Lys-sensitive AKs and Thr-sensitive AK-HSDHs is important for maintaining the amounts and ratios of Asp-derived amino acids.
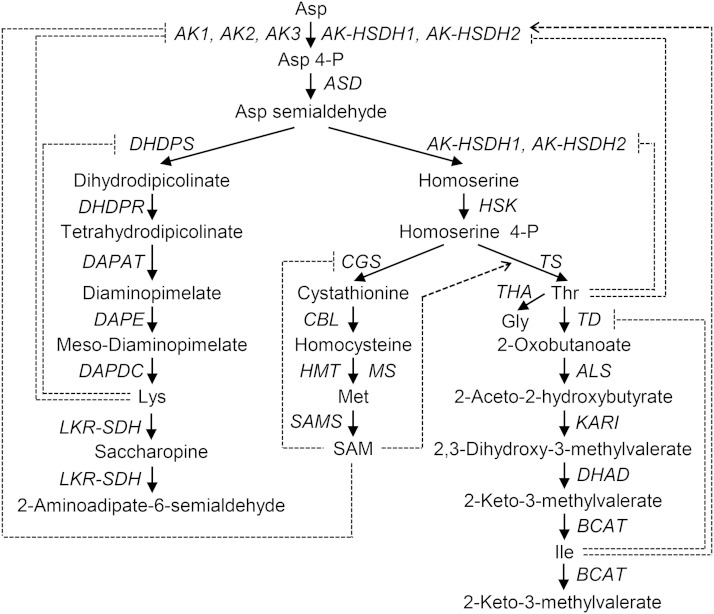
Humans and other animals rely on plants as a dietary source of essential amino acids (Coruzzi and Last, 2000; Galili et al., 2008). In major field crops, such as corn (Zea mays), rice (Oryza sativa), soybean (Glycine max), and potato (Solanum tuberosum), Asp-derived amino acids Lys, Met, Thr, and Ile are present at levels that limit the nutritional values (Müntz et al., 1998; Debabov, 2003; Pfefferle et al., 2003; Stiller et al., 2007). Increasing the contents of these amino acids in crop plants has long been a major goal of traditional breeding and genetic engineering (Galili et al., 2008; Ufaz and Galili, 2008; Jander and Joshi, 2009). Genetic improvement requires a better understanding of the pathways and regulation of Asp-derived amino acids. The first step of the biosynthetic pathway for Asp-derived amino acids is the activation of Asp to Asp-4-P by monofunctional Asp kinases (AKs) and dual-functional Asp kinase-homo-Ser dehydrogenases (AK-HSDHs; Fig. 1; Jander and Joshi, 2009). Asp-4-P is then converted to Asp semialdehyde, the branch-point intermediate for Lys or Met, Thr, and Ile biosynthesis. The committing step leading to Met, Thr, and Ile biosynthesis is the formation of homo-Ser from Asp semialdehyde, which is catalyzed by dual-functional AK-HSDHs.
Figure 1.
Biosynthesis and degradation of Asp-derived amino acids. Feedback inhibition is shown as a dotted line with a bar. Activation is shown as a dotted line with an arrowhead. ALS, Acetolactate synthase; ASD, Asp semialdehyde dehydrogenase; CBL, cystathionine β-lyase; DAPAT, diaminopimelate aminotransferase; DAPDC, diaminopimelate decarboxylase; DAPE, diaminopimelate epimerase; DHAD, dihydroxyacid dehydratase; DHDPR, dihydrodipicolinate reductase; HMT, homo-Cys S-methyltransferase; HSK, homo-Ser kinase; KARI, ketol-acid reductoisomerase; MS, Met synthase.
Higher plants have multiple genes that encode AKs and AK-HSDHs. For example, the Arabidopsis (Arabidopsis thaliana) nuclear genome contains three AK-encoding genes (AK1 [At5g13280], AK2 [A5g14060], and AK3 [At3g02020]) and two AK-HSDH-encoding genes (AK-HSDH1 [At1g31230] and AK-HSDH2 [At4g19710]; Jander and Joshi, 2009). Public microarray data indicated that the five enzymes are expressed in all plant tissues (Schmid et al., 2005; Winter et al., 2007). Transcription of these enzymes is repressed by basic Leu zipper transcription factors in dark and other low-sugar conditions (Ufaz et al., 2011). In addition, both AKs and AK-HSDHs are subject to feedback inhibition by their downstream products. For example, the activity of monofunctional AKs is feedback inhibited by Lys (Relton et al., 1988; Dotson et al., 1989; Frankard et al., 1997; Tang et al., 1997; Curien et al., 2007; Wang et al., 2007). The inhibition is mediated through the two Lys-binding ACT domains (for small regulatory domains initially found in AK, chorismate mutase and Tyr A [prephenate dehydrogenase]) at the C terminus of the proteins (Supplemental Fig. S1; Chipman and Shaanan, 2001; Jander and Joshi, 2009). The activity of AK-HSDHs is feedback inhibited by Thr (Muehlbauer et al., 1994; Paris et al., 2003; Rognes et al., 2003; Curien et al., 2005). The inhibition is mediated through the two Thr-binding ACT domains in the middle of the proteins (Supplemental Fig. S1; Chipman and Shaanan, 2001; Jander and Joshi, 2009).
Genetic improvement of Asp-derived amino acid contents in crop plants is complicated and will benefit from the knowledge of the relative importance of different isozymes when overlapping functions exist (Galili et al., 2008). However, most of the previous studies on AKs and AK-HSDHs focused on kinetic properties and allosteric regulation of purified enzymes in the absence of other isozymes (Dotson et al., 1989, 1990; Rognes et al., 2003; Curien et al., 2005, 2007). In addition to allosteric regulation, enzyme abundance is key for determining the relative importance of individual enzymes. Therefore, it is difficult to predict the relative significance of individual isozymes solely based on the kinetic properties of the enzymes. On the contrary, characterization of a series of loss-of-function single and double ak and ak-hsdh mutants will help to elucidate the contributions of individual AK and AK-HSDH enzymes to the biosynthesis of Asp-derived amino acids and how plants respond to the loss of individual enzymes. Understanding how plants adjust resources among competing branches within Asp-derived amino acid biosynthesis in response to the loss of an AK enzyme and/or an AK-HSDH enzyme will provide additional insights into metabolic regulation of branched pathways in general. Because monofunctional AKs catalyze the committing step leading to Asp-derived amino acid biosynthesis (Fig. 1), one would expect that loss-of-function mutations in the AK genes result in reduced amounts of Asp-derived amino acids. Similarly, because dual-functional AK-HSDHs catalyze the committing step in Asp-derived amino acid biosynthesis as well as the branching step leading to Met, Thr, and Ile biosynthesis (Fig. 1), one would anticipate that loss-of-function mutations in the AK-HSDH genes cause reductions in the contents of Asp-derived amino acids, especially Met, Thr, and Ile.
In this work, we described the identification and analysis of two previously uncharacterized Arabidopsis mutants with high levels of Asp and Asp-derived amino acids, especially Thr, in leaves. This observation was further investigated with free and protein-bound amino acid profiling, transcript abundance, and activity assays on a series of single and double loss-of-function ak and ak-hsdh mutants. Pairwise Pearson’s correlation analysis revealed that the ratio of total AK activity to total HSDH activity negatively correlates with the ratio of Lys to the total amount of Met, Thr, and Ile. These results suggested that the balance between Lys-sensitive monofunctional AKs and Thr-sensitive dual-functional AK-HSDHs is important for maintaining the levels and ratios of Asp-derived amino acids.
RESULTS
A Transfer DNA Mutant Line with Increased Leaf Thr Content
In an attempt to identify functions for chloroplast proteins and mechanisms for processes occurring in the chloroplast, thousands of Arabidopsis transfer DNA (T-DNA) lines of nuclear-encoded, chloroplast-targeted genes were analyzed for a variety of phenotypes, including leaf and seed free amino acid contents, using an HPLC-tandem mass spectrometry method (Lu et al., 2008, 2011a, 2011b, 2011c; Ajjawi et al., 2010; Bell et al., 2012). This screen identified a homozygous mutant (SALK_082155) in the Columbia-0 (Col-0; CS60000) background with high-leaf Thr content (Supplemental Fig. S2A). Analysis of the public T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress; O’Malley and Ecker, 2010) revealed that this mutant contains a T-DNA insertion in the single exon of the At1g55805 gene (Supplemental Fig. S2B), which encodes a BolA-like family protein. A second mutant (SALK_059678) in the At1g55805 gene did not have an increased level of Thr in leaves (Supplemental Fig. S2A). These results indicated that the phenotype in SALK_082155 is caused by a second-site mutation.
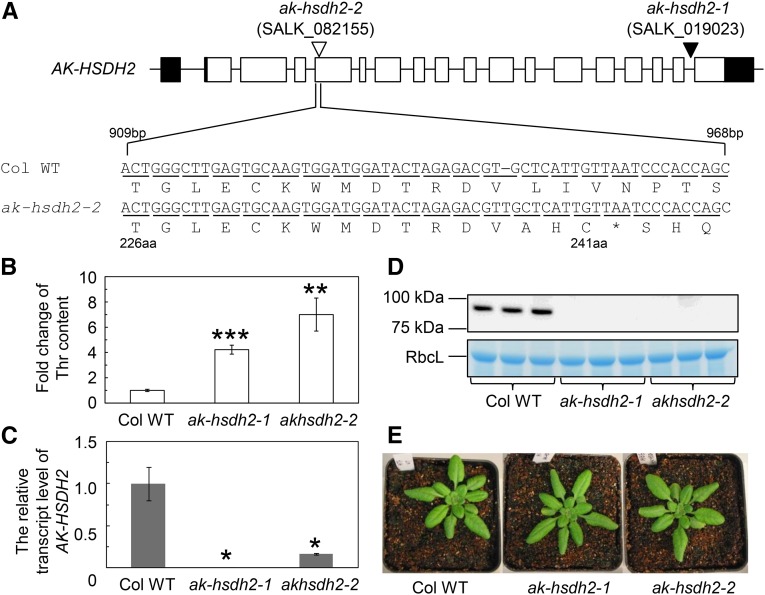
To identify the mutation in SALK_082155 that caused the increases in Asp-derived amino acids, a mapping population was made by crossing a SALK_082155 plant to the Landsberg erecta (Ler) wild type. In total, 181 F2 plants were analyzed for leaf free amino acid contents (Supplemental Fig. S2, C and D). DNA from 11 F2 plants with the highest Thr contents and 13 F2 plants with the lowest Thr contents was extracted and used for array mapping. The output from array mapping showed that the mutation is located in a 1.75-Mb region of chromosome 4 (At4g17440–At4g21580; Supplemental Fig. S2, C and D). Based on gene descriptions of 492 genes in this region, one candidate gene (AK-HSDH2, At4g19710) was identified. This gene encodes a dual-functional AK-HSDH. Targeted genomic DNA sequencing showed that there is a thymine insertion in the fourth exon of the coding strand for the AK-HSDH2 gene. This single-base insertion resulted in a premature stop codon and a predicted truncated protein of 241 amino acids (Fig. 2) instead of the wild-type 961-amino acid protein. The truncated protein is predicted to be missing part of the AK domain (amino acids 88–371), the two Thr-binding ACT domains (amino acids 411–471 and 491–552), and the HSDH domain (amino acids 564–906).
Figure 2.
Mutations in the AK-HSDH2 gene cause the Thr increase. A, Schematic representation of the AK-HSDH2 gene (At4g19710) and two mutations: SALK_019023 and SALK_082155. Black rectangles represent 5′- and 3′-untranslated regions, white rectangles represent exons, black lines represent introns and intergenic regions, the white triangle represents a single-base insertion, and the black triangle represents a T-DNA insertion. SALK_019023 (ak-hsdh2-1) contains a T-DNA insertion in the last intron of the AK-HSDH2 gene. SALK_082155 (ak-hsdh2-2) contains an extra base of thymine (T) in the fourth exon of the same gene. The location of this extra thymine base was determined by array mapping and targeted genomic DNA sequencing. This single-base insertion results in a premature stop codon and a truncated protein of 241 amino acids in size. aa, Amino acid; Col WT, Col-0 wild type. The asterisk indicates the placement of a stop codon. B, Fold change of Thr content in the Col-0 wild type and two mutants. The values (mean ± se; n = 5) are presented as ratios to the Thr content in the Col-0 wild type. Asterisks indicate significant differences between the mutant and the Col-0 wild type (Student’s t test; **, P < 0.01; and ***, P < 0.001). C, Relative amounts of the AK-HSDH2 transcript. Total RNA was extracted from mature leaves and analyzed with quantitative RT-PCR. The level of the AK-HSDH2 transcript was normalized to that of the ACTIN2 transcript (AT3g18780). The values (mean ± se; n = 3–6) are presented as ratios to the transcript level in the Col-0 wild type. *, Significant differences between the mutant and the Col-0 wild type (Student’s t test; P < 0.05). D, Representative immunoblot of the AK-HSDH2 protein and Coomassie Brilliant Blue G250-stained gel showing the Rubisco large subunit (RbcL; loading control). The lanes were loaded on an equal protein basis. E, Images of 4-week-old Col-0 wild type and ak-hsdh2 plants under a 12-h-light/12-h-dark photoperiod.
The Phenotype Is Confirmed in a Second ak-hsdh2 Mutant
To confirm that the phenotype in SALK_082155 is caused by the single-base insertion in the AK-HSDH2 gene, we analyzed leaf amino acid contents in a second homozygous mutant (SALK_019023) of the same gene. SALK_019023 has a T-DNA insertion in the last intron of the AK-HSDH2 gene (Fig. 2). SALK_019023 was renamed ak-hsdh2-1. The mutant with a single-base insertion in the AK-HSDH2 gene after genetic background cleanup (SALK_082155) was renamed ak-hsdh2-2. As shown in Figure 2B, the amount of Thr in 4-week-old ak-hsdh2 mutants was 4 to 7 times higher than that in the wild type at the same age. Quantitative reverse transcription (RT)-PCR showed that the AK-HSDH2 transcript was completely abolished in ak-hsdh2-1 and substantially reduced in ak-hsdh2-2 (Fig. 2C). The reduction of the AK-HSDH2 transcript in the ak-hsdh2-2 mutant could be caused by a nonsense-mediated decay pathway (Kalyna et al., 2012). Although there was still a small amount of the AK-HSDH2 transcript in the ak-hsdh2-2 mutant, the transcript is frame shifted, and the corresponding protein is truncated and probably not functional (Fig. 2A). Consistent with the quantitative RT-PCR data, mature AK-HSDH2 protein was detected in wild-type Arabidopsis plants but undetectable in either ak-hsdh2 mutant (Fig. 2, D and E). These results revealed that loss-of-function mutations in the AK-HSDH2 gene cause an increased level of Thr.
Isolation and Generation of Other ak and ak-hsdh Single and Double Mutants
To further understand the regulation of monofunctional AKs and dual-functional AK-HSDHs, we obtained ak1-1 (WiscDsLox461-464J6), ak2-1 (SALK_003685), ak3-1 (SALK_043533), and ak-hsdh1-1 (SALK_125957) single mutants (Fig. 3) and generated three double-homozygous mutants: ak-hsdh2-1 ak1-1, ak-hsdh2-1 ak2-1, and ak-hsdh2-1 ak3-1. Attempts to make the double-homozygous ak-hsdh2-1 ak-hsdh1-1 mutant were not successful; thus, this combination was not analyzed. About one-fifth to one-fourth of seeds from ak-hsdh2-1/ak-hsdh2-1 AK-HSDH1-1/ak-hsdh1-1 plants did not develop into normal seeds, suggesting that the double-homozygous ak-hsdh2-1 ak-hsdh1-1 mutant is not viable (Supplemental Fig. S3). The six single mutants and three double mutants appeared to be healthy and normal (Figs. 2E and 3B).
Figure 3.
T-DNA locations and plant images of other single or double ak and ak-hsdh mutants. A, Schematic representation of the AK1, AK2, AK3, and AK-HSDH1 genes and four T-DNA insertion alleles. Black rectangles represent 5′- and 3′-untranslated regions, white rectangles represent exons, black lines represent introns and intergenic regions, and black triangles represent T-DNA insertions. ak1-1, WiscDsLox461-464J6; ak2-1, SALK_003685; ak3-1, SALK_043533; ak-hsdh1-1, SALK_125957. B, Images of 4-week-old Col-0 wild-type (Col WT) and single- or double-mutant plants under a 12-h-light/12-h-dark photoperiod.
Free and Protein-Bound Amino Acids in 4-Week-Old ak and/or ak-hsdh Mutants
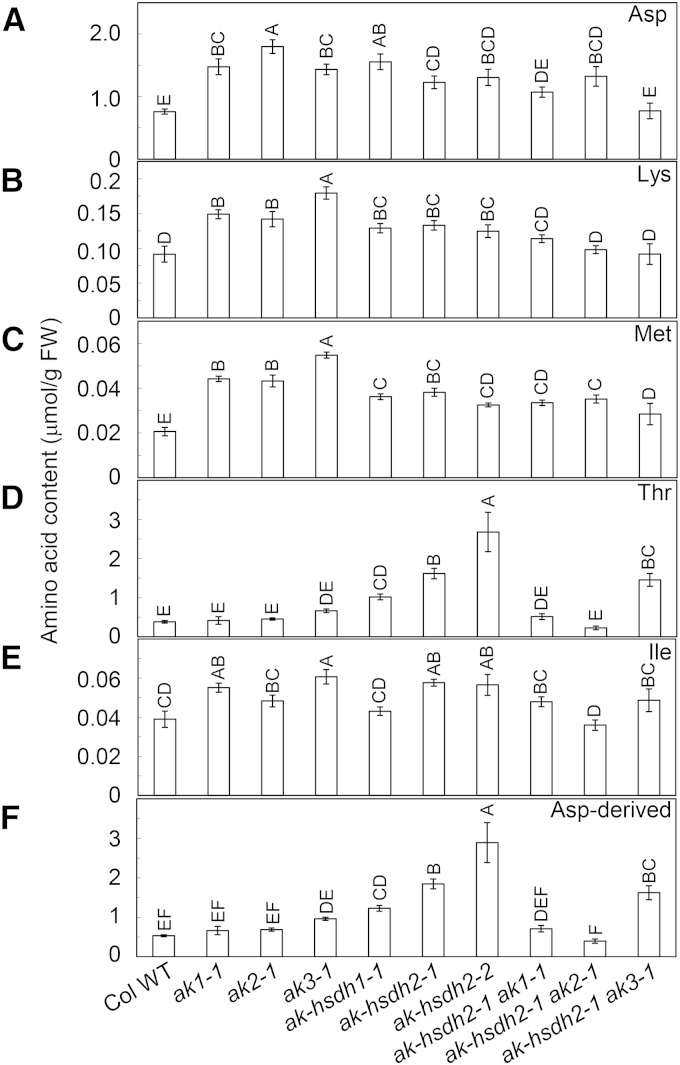
We determined free amino acid contents in mature rosette leaves from the 4-week-old wild types and single and double mutants. The contents of Asp, Lys, and Met in ak1-1, ak2-1, ak3-1, ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 were significantly higher than those in the wild type (Fig. 4). On average, these amino acids increased 93%, 56%, and 101%, respectively, in six single mutants compared with the wild type (Fig. 4). The Ile levels in ak1-1, ak3-1, ak-hsdh2-1, and ak-hsdh2-2 were also significantly higher than those in the wild type (Fig. 4). Similar to the two ak-hsdh2 mutants, the ak-hsdh1-1 mutant had an increased amount of Thr (Fig. 4). The total amount of Asp-derived amino acids behaved similarly to Thr, which is not surprising, because Thr is the most abundant Asp-derived amino acid in Arabidopsis leaves (Fig. 4).
Figure 4.
The contents of free Asp and Asp-derived amino acids in leaves of 4-week-old plants. Asp (A), Lys (B), Met (C), Thr (D), Ile (E), and the total amount of Asp-derived amino acids Lys, Met, Thr, and Ile (F). Values are presented as means ± se (n = 5). Values not connected by the same letter are significantly different (Student’s t test, P < 0.05). Values connected by the same letter are not significantly different. FW, Fresh weight; Col WT, Col-0 wild type.
The near-unanimous increases of Asp, Lys, and Ile in the single mutants were not observed in the double mutants at the same age; only ak-hsdh2-1 ak2-1 was found to have increased Asp content (Fig. 4). The Thr increase observed in ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 single mutants was absent in ak-hsdh2-1 ak1-1 and ak-hsdh2-1 ak2-1 double mutants (Fig. 4). However, the ak-hsdh2-1 ak3-1 double mutant still had a high amount of Thr (280% higher than the wild type), and all three double mutants had increased Met content (Fig. 4).
In addition to Asp and Asp-derived amino acids, the contents of Asn, Gln, Glu, His, and Leu showed significant increases in the six 4-week-old single mutants (Supplemental Fig. S4). These amino acids did not show corresponding changes in the three double mutants at the same age (Supplemental Fig. S4). Furthermore, the contents of Ala, Gly, Phe, Pro, Trp, and Tyr showed significant increases in 4-week-old ak1-1, ak2-1, and ak3-1 mutants (Supplemental Fig. S4). These changes were less evident or not observed in ak-hsdh1-1 and ak-hsdh2 mutants and the three double mutants (Supplemental Fig. S4). Consequently, the total amount of free amino acids was substantially increased in the six single mutants, but it remained unchanged in the three double mutants (Supplemental Fig. S4). These observations showed that perturbation in Asp-derived amino acid biosynthesis may cause changes in the contents of other amino acids.
Because free amino acids represent a minor proportion of the total amino acids in plants, we determined the amounts of protein-bound amino acids in the leaves of 4-week-old plants after acid hydrolysis. During acid hydrolysis, Asn and Gln are converted to Asp and Glu, respectively, and Met is partially oxidized (Anders, 2002). Therefore, Asn and Asp were reported collectively, Glu and Gln were shown together, and Met was not reported. The only statistically significant differences in these amino acids between the mutants and the wild type were decreases in Asn + Asp and Gln + Glu contents in ak3-1 and ak-hsdh1-1 (Supplemental Table S1).
Free Amino Acid Profiling in 1-, 2-, and 3-Week-Old Plants
To further investigate the dynamics of Asp-derived amino acid metabolism, we measured leaf free amino acid contents in 1-, 2-, and 3-week-old wild-type and mutant plants. Because of the relative small plant size, the entire aboveground portion of 1-week-old plants was used for free amino acid assay, and the ak1-1, ak3-1, ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 mutants showed significant increases in the Lys content (Supplemental Table S2). For 2-week-old plants, the first two true leaves were used, and the six single mutants showed significant reductions in Lys, Met, and Ile contents (Supplemental Table S3). For 3-week-old plants, the second, third, and fourth true leaves were used, and the six single mutants exhibited significant increases in the contents of Lys, Met, and Ile (Supplemental Table S4), similar to the same mutants at 4 weeks old (Fig. 4). Unlike other Asp-derived amino acids, the Thr increase was only observed in the ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 mutants among 3-week-old plants.
Many of these changes in Asp-derived amino acids were not observed in the three double mutants. For example, the Lys increase in the six 1-week-old single mutants was not observed in the three double mutants at the same age (Supplemental Table S2). Also, the unanimous decreases of Lys, Met, and Ile contents in six 2-week-old single mutants were only observed in some double mutants at the same age (Supplemental Table S3). None of the 3-week-old mutants showed increases of Lys, Met, or Ile content, unlike the six single mutants at the same age (Supplemental Table S4). Taken together, the amino acid data from plants at multiple growth stages consistently showed that changes of Asp-derived amino acids in the ak and ak-hsdh single mutants were often not observed in the double mutants.
Quantitative RT-PCR Analysis of Select Genes Involved in the Biosynthesis and Catabolism of Asp-Derived Amino Acids
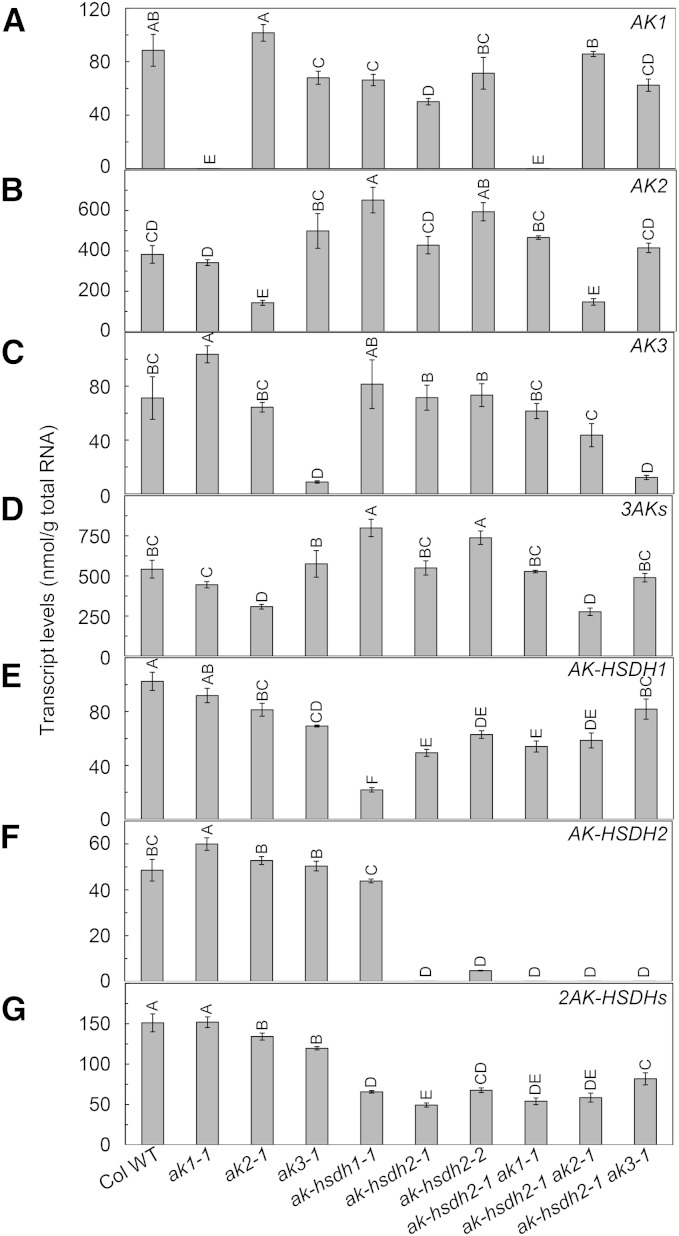
To investigate the transcript levels of the five AK or AK-HSDH genes in the wild type and nine ak and/or ak-hsdh mutants, we performed quantitative RT-PCR in mature rosette leaves from 4-week-old plants. In the wild type, the AK2 transcript was more abundant than AK1 and AK3, and the AK-HSDH1 transcript was more abundant than AK-HSDH2 (Fig. 5). In the ak1-1 mutant, the AK1 transcript was completely abolished, which was accompanied by a 46% increase in AK3 and a 22% increase in AK-HSDH2 (Fig. 5). Overall, there was no significant change in total AK or AK-HSDH transcripts in ak1-1. In the ak2-1 mutant, there was a 63% decrease in AK2, a 43% decrease in total AK transcripts, and an 11% decrease in total AK-HSDH transcripts (Fig. 5). In the ak3-1 mutant, there was an 87% reduction in AK3, but because AK3 is the least abundant AK transcript, there was no significant change in total AK transcripts (Fig. 5). In the ak-hsdh1-1 mutant, there was a 78% decrease in AK-HSDH1, a 47% increase in total AK transcripts, and a 56% decrease in total AK-HSDH transcripts (Fig. 5). In the ak-hsdh2-1 mutant, the AK-HSDH2 transcript was completely abolished, and there was a 67% decrease in total AK-HSDH transcripts, with no net change in total AK transcripts (Fig. 5). In the ak-hsdh2-2 mutant, there was a 90% decrease in AK-HSDH2, a 36% increase in total AK transcripts, and a 55% decrease in total AK-HSDH transcripts (Fig. 5). Overall, some significant changes in the transcript levels of other AK or AK-HSDH gene(s) were observed in the six single mutants. However, loss-of-function mutations in one AK or AK-HSDH gene did not cause systematic up- or down-regulation of other AK or AK-HSDH gene(s).
Figure 5.
Transcript levels of AK and AK-HSDH genes in leaves of 4-week-old plants. AK1 (A), AK2 (B), AK3 (C), 3AKs (D), AK-HSDH1 (E), AK-HSDH2 (F), and 2AK-HSDHs (G). Values are presented as means ± se (n = 3–4). Values not connected by the same letter are significantly different (Student’s t test, P < 0.05). Values connected by the same letter are not significantly different. Col WT, Col-0 wild type.
In the ak-hsdh2-1 ak1-1 double mutant, both AK1 and AK-HSDH2 transcripts were completely abolished, and there was a 64% decrease in total AK-HSDH transcripts (Fig. 5). However, there was no net change in total AK transcripts in this double mutant, possibly because the AK2 transcript is much more abundant than AK1 and AK3. In the ak-hsdh2-1 ak2-1 double mutant, the AK-HSDH2 transcript was completely abolished, the AK2 transcript was reduced by 62%, total AK transcripts were reduced by 49%, and total AK-HSDH transcripts were reduced by 61% (Fig. 5). In the ak-hsdh2-1 ak3-1 double mutant, the AK-HSDH2 transcript was completely abolished, the AK3 transcript was reduced by 83%, total AK-HSDH transcripts were reduced by 46%, and there was no significant change in total AK transcripts (Fig. 5). To summarize, among the three double mutants, ak-hsdh2-1 ak2-1 showed the most pronounced reductions in the amounts of AK and AK-HSDH transcripts.
We investigated the transcript levels of other biosynthetic genes in the mutants. The dihydrodipicolinate synthase1 (DHDPS1) and DHDPS2 genes encode DHDPSs, the branch-point enzymes leading to Lys biosynthesis. There were significant increases in total DHDPS transcripts in ak1-1 and ak3-1, but no significant change was observed in other single or double mutants (Supplemental Fig. S5). The cystathionine γ-synthase1 (CGS1) and Thr synthase1 (TS1) transcripts encode CGS and TS, which are the branch-point enzymes leading toward Met and Thr biosynthesis, respectively (Fig. 1). The CGS1 and TS1 transcripts did not show significant changes in any of the mutants (Supplemental Fig. S5). The Thr deaminase1 (TD1) gene encodes TD, the branch-point enzyme toward Ile biosynthesis (Fig. 1). There were significant increases in the TD1 transcript level in all six single mutants, but there was no significant change in the three double mutants (Supplemental Fig. S5).
We also analyzed the transcript levels of select catabolic genes in the nine mutants. The Lys-ketoglutarate reductase-saccharopine dehydrogenase1 (LKR-SDH1) gene encodes a Lys catabolic enzyme: LKR-SDH (Fig. 1). The LKR-SDH1 transcript level was significantly increased in the ak-hsdh2-1 and ak-hsdh2-2 single mutants and the ak-hsdh2-1 ak1-1 and ak-hsdh2-1 ak2-1 double mutants (Supplemental Fig. S6). The S-adenosyl-Met Synthase1 (SAMS1), SAMS2, SAMS3, and SAMS4 genes encode S-adenosyl-Met (SAM) synthases, which convert Met to SAM (Fig. 1). The total amount of SAMS transcripts was increased in ak1-1 and ak2-1 but decreased in ak-hsdh2-1, ak-hsdh2-2, ak-hsdh2-1 ak1-1, and ak-hsdh2-1 ak2-1 (Supplemental Fig. S6). The Thr aldolase1 (THA1) and THA2 genes encode THA, which convert Thr to Gly (Fig. 1). The total amount of THA transcripts was only significantly increased in ak-hsdh2-1 (Supplemental Fig. S6). Although branched-chain aminotransferases (BCATs) are involved in both biosynthesis and catabolism of branched-chain amino acids Ile, Leu, and Val, BCAT1 has been shown to initiate catabolism (Schuster and Binder, 2005). The BCAT1 transcript level showed universal decreases in the six single mutants and the ak-hsdh2-1 ak1-1 and ak-hsdh2-1 ak2-1 double mutants (Supplemental Fig. S6). Taken together, loss-of-function mutations in the AK and/or AK-HSDH gene(s) can result in changes to related anabolic and catabolic genes, such as a near-unanimous decrease of the BCAT1 transcript.
AK2 Is the Major Contributor to Overall AK Activity
To dissect the relative importance of different AK and AK-HSDH isozymes, we measured the overall AK activity in leaf protein extracts from the wild type and six single mutants. In the absence of allosteric effectors, the ak1-1, ak2-1, ak3-1, ak-hsdh1-1, and ak-hsdh2 mutants show 17%, 56%, 22%, 34%, and 15% to 22% reductions in the overall AK activity, respectively (Fig. 6). If we use the average reduction of the two ak-hsdh2 mutants and scale the total amount of reductions caused by different mutations in the five genes to 100%, the individual contributions from AK1, AK2, AK3, AK-HSDH1, and AK-HSDH2 can be calculated as 12%, 38%, 15%, 23%, and 12%, respectively. Thus, the group contributions of AKs and AK-HSDHs were 65% and 35%, respectively. Taken together, AK2 is the major contributor to the overall AK activity, and monofunctional AKs collectively contribute more to the overall AK activity than dual-functional AK-HSDHs, which is consistent with their relative transcript abundance in leaves (Fig. 5).
Figure 6.
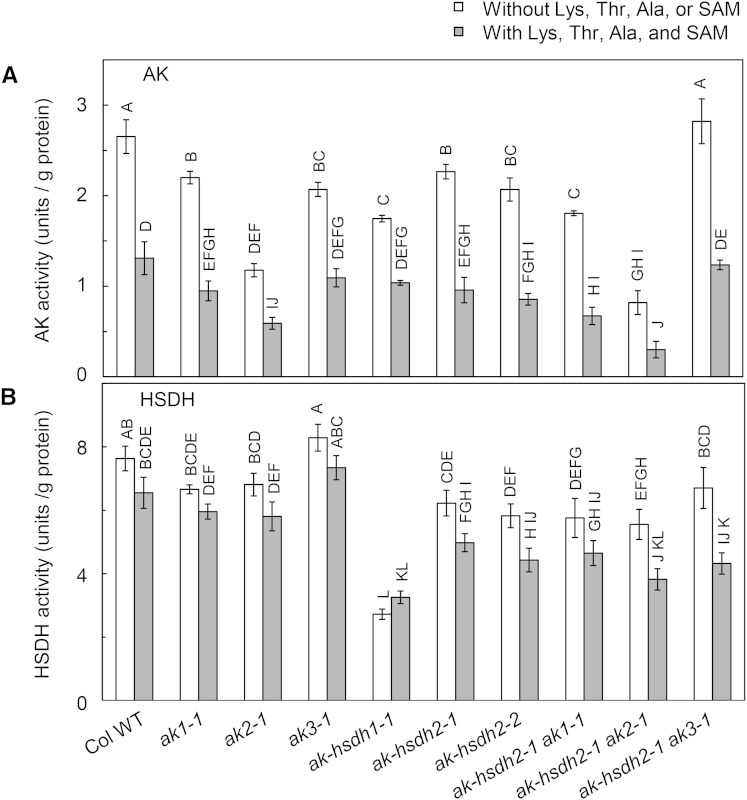
AK and HSDH activities in leaf protein extracts. A, The AK activity measured with the hydroxamate-ferric chloride method. B, The HSDH activity measured with the NADP+ reduction method. Values are represented as means ± se (n = 3). The AK and HSDH activities in physiological concentrations of effectors were measured in the presence of 70 μm Lys, 300 μm Thr, 400 μm Ala, and 20 μm SAM. Values not connected by the same letter are significantly different (Student’s t test, P < 0.05). Values connected by the same letter are not significantly different. Col WT, Col-0 wild type.
AKs are feedback inhibited by Lys, and AK-HSDHs are feedback inhibited by Thr. In addition, Curien et al. (2005) reported that the AK activity of AK-HSDH1 and AK-HSDH2 is activated by Ala, Cys, Ile, Ser, and Val and inhibited by Leu (Curien et al., 2005). Among the six additional effectors, Ala is the most abundant in chloroplast stroma (Curien et al., 2005). Moreover, Curien et al. (2007) reported that AK1 is synergistically inhibited by Lys and SAM (Curien et al., 2007). Therefore, we measured the AK activity in near-physiological concentrations of Lys, Thr, Ala, and SAM, which are 70, 300, 400, and 20 μm, respectively (Curien et al., 2009). Under this condition, the mutation in AK1, AK2, AK3, AK-HSDH1, or AK-HSDH2 gene reduced the overall AK activity by 28%, 55%, 17%, 21%, or 27% to 35%, respectively (Fig. 6A). In the same way as we calculated the AK activity measured in the absence of allosteric effectors, we can calculate the individual contributions from AK1, AK2, AK3, AK-HSDH1, and AK-HSDH2 to be 18%, 36%, 11%, 14%, and 20%, respectively, and the group contributions of AKs and AK-HSDHs to be 66% and 34%, respectively. This is in close agreement with the AK activity data measured in the absence of allosteric effectors.
Simultaneous Loss of AK-HSDH2 and Monofunctional AK1 or AK2 Has an Additive Effect on the Reduction of Overall AK Activity
To test the impact of simultaneous loss of AK-HSDH2 and a monofunctional AK on the overall AK activity, we measured the AK activity in ak-hsdh2-1 ak1-1, ak-hsdh2-1 ak2-1, and ak-hsdh2-1 ak3-1 double mutants. Because the overall AK activity is contributed by both monofunctional AKs and dual-functional AK-HSDHs, one would expect that simultaneous loss of the AK-HSDH2 activity and a monofunctional AK activity would have an additive effect on the reduction of the overall AK activity. Indeed, in the absence of allosteric effectors, the overall AK activity in the ak-hsdh2-1 ak1-1 double mutant was 18% and 20% lower than those in the ak1-1 and ak-hsdh2-1 single mutants, respectively (Fig. 6A). Under the same condition, the overall AK activity in the ak-hsdh2-1 ak2-1 double mutant was 30% and 64% lower than those in the ak2-1 and ak-hsdh2-1 single mutants, respectively (Fig. 6A). In near-physiological concentrations of Lys, Thr, Ala, and SAM, the overall AK activity in the ak-hsdh2-1 ak1-1 double mutant was approximately 30% lower than that in the ak1-1 and ak-hsdh2-1 single mutants, and the overall AK activity in the ak-hsdh2-1 ak2-1 double mutant is 49% and 69% lower than those in the ak2-1 and ak-hsdh2-1 single mutants (Fig. 6A). To summarize, these data showed that simultaneous loss of AK-HSDH2 and monofunctional AK1 or AK2 has an additive effect on the reduction of the overall AK activity.
The AK activity in the ak-hsdh2-1 ak3-1 double mutant was quite different from that in the other two double mutants. In the absence of allosteric effectors, the overall AK activity in the ak-hsdh2-1 ak3-1 double mutant was 28% and 25% higher than those in the ak3-1 and ak-hsdh2-1 single mutants, respectively (Fig. 6A). In near-physiological concentrations of Lys, Thr, Ala, and SAM, the overall AK activity in the ak-hsdh2-1 ak3-1 double mutant was approximately 30% higher than those in the ak3-1 and ak-hsdh2-1 single mutants (Fig. 6A). It is intriguing that the ak-hsdh2-1 ak3-1 double mutant showed an unexpectedly higher AK activity than the corresponding single mutants.
AK-HSDH1 Is the Major Contributor to Overall HSDH Activity
The relative contributions of AK-HSDH1 and AK-HSDH2 to the overall HSDH reaction were estimated by comparing the HSDH activities in protein extracts from wild-type, ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 leaves. In the absence of Lys, Thr, Ala, or SAM, the mutation in the AK-HSDH1 gene decreased the activity by 64%, whereas the mutation in the AK-HSDH2 gene decreased the activity by 18% in the ak-hsdh2-1 mutant and 24% in the ak-hsdh2-2 mutant (Fig. 6B). In near-physiological concentrations of Lys, Thr, Ala, and SAM, the ak-hsdh1-1, ak-hsdh2-1, and ak-hsdh2-2 mutants showed 50%, 24%, and 32% reductions in the overall HSDH activity, respectively. These results suggested that AK-HSDH1 is the primary contributor to the overall HSDH activity. We also measured the overall HSDH activity in the three ak single mutants and three double mutants. In the absence or presence of Lys, Thr, Ala, and SAM, the overall HSDH activity in the ak1, ak2, and ak3 mutants was not statistically different from that in the corresponding wild type (Fig. 6B). The overall HSDH activity in the ak-hsdh2-1 ak1-1, ak-hsdh2-1 ak2-1, and ak-hsdh2-1 ak3-1 double mutants was also not significantly different from that in the single ak-hsdh2-1 mutant (Fig. 6B). These observations are consistent with the fact that monofunctional AKs do not have an HSDH domain (Supplemental Fig. S1).
Correlations Exist among Transcript Levels, Enzyme Activities, Amino Acid Contents, and Their Ratios
To look for systematic patterns among transcript levels, enzyme activities, and amino acid contents, we performed pairwise Pearson’s correlation analysis among these parameters and their ratios. In total, 7,381 correlations were analyzed (Supplemental Table S5). For simplicity reasons, only select correlations are described below.
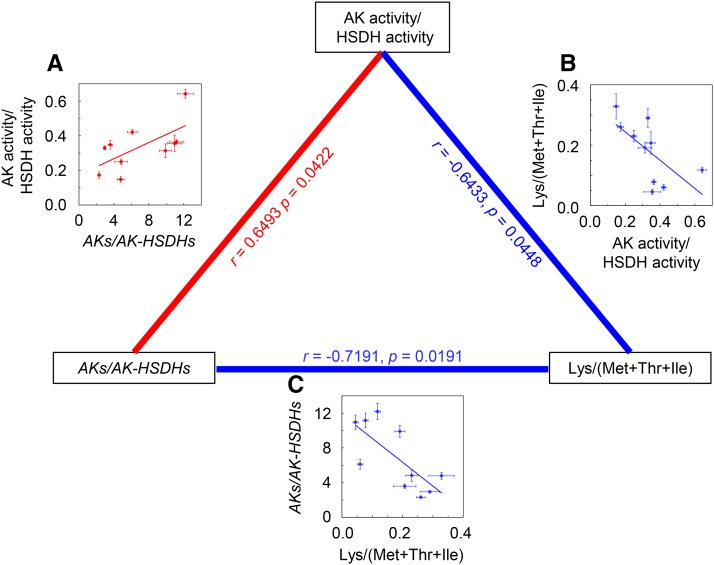
First, we focused on the relationships between transcript levels and enzyme activities (Supplemental Table S5). The total amount of AK transcripts had a moderate positive correlation with the Lys-sensitive AK activity (r = 0.6449, P = 0.0441). The total amount of AK-HSDH transcripts had a moderate positive correlation with the Thr-sensitive AK activity (r = 0.537, P = 0.1095); it also had a moderate positive correlation with the HSDH activity measured in the absence of allosteric effectors (r = 0.5897, P = 0.0728).
Second, we studied the relationships between enzyme activities and free amino acid contents (Supplemental Table S5). The AK activity measured in the absence of allosteric effectors had a moderate positive correlation (r = 0.5918, P = 0.0715) with the ratio of free Asp-derived amino acids to free Asp (i.e. free [Lys + Met + Thr + Ile] to Asp). This is in agreement with the AK activity being responsible for the committing enzymatic step leading to the biosynthesis of Asp-derived amino acids (Fig. 1; Jander and Joshi, 2009). Free Lys had a weak negative correlation (r = −0.3984, P = 0.2541) with Lys-sensitive AK activity, consistent with Lys being a major inhibitor of monofunctional AKs (Jander and Joshi, 2009). Free Thr had a moderate negative correlation (r = −0.5430, P = 0.1048) with the Thr-sensitive AK activity. These correlations are in agreement with Thr being a major inhibitor of dual-functional AK-HSDHs. The ratio of the AK activity to the HSDH activity measured in the absence of allosteric effectors had a moderate negative correlation with the free Lys to (Met + Thr + Ile) ratio (r = −0.6433, P = 0.0448). This negative correlation remained when the AK and HSDH activities were measured in the presence of allosteric effectors (r = −0.7320, P = 0.0161). These data suggested that the balance between Lys-sensitive AKs and Thr-sensitive AK-HSDHs is important to maintain the ratio between Lys and the total amount of Met, Thr, and Ile.
Third, we analyzed whether the transcript levels of genes encoding the committing/branch-point enzymes have any relationships with the amino acid products (Supplemental Table S5). We found that the total amount of AK transcripts had a moderate positive correlation (r = 0.6144, P = 0.0588) with the total amount of free Asp-derived amino acids. This is consistent with the AK gene products being the committing enzymes in the biosynthetic pathway of Asp-derived amino acids. The total amount of DHDPS transcripts had a strong positive correlation (r = 0.7704, P = 0.0091) with free Lys, which is in agreement with the DHDPS gene products being the branch-point enzymes leading to Lys biosynthesis. The total amount of AK-HSDH transcripts had a weak negative correlation (r = −0.4547, P = 0.1868) with the total amount of free Met, Thr, and Ile. This could be the result of two antagonistic factors: the HSDH activity of AK-HSDHs catalyzes the branch-point step, leading to the biosynthesis of Met, Thr, and Ile, but it is feedback inhibited by Thr. The CGS1 transcript had a weak positive correlation (r = 0.3576, P = 0.3103) with free Met, which is consistent with the CGS1 gene product being the committing enzyme of Met biosynthesis. The TD1 transcript had a moderate positive correlation (r = 0.5921, P = 0.0713) with free Ile, which is in agreement with the TD1 gene product being the branch-point enzyme leading to Ile biosynthesis.
Fourth, we investigated whether the transcript levels of genes encoding the catabolic enzymes have any relationships with the amino acid substrates. We found that the SAMS1 and SAMS2 transcripts had moderate positive correlations with free Met (r = 0.6725, P = 0.0331 and r = 0.4836, P = 0.1568, respectively). The THA2 transcript had a moderate positive correlation with free Thr (r = 0.5148, P = 0.1279). The BCAT1 transcript had a moderate negative correlation with free Ile (r = −0.4659, P = 0.1748). This is surprising, because BCAT1 has been shown to initiate catabolism of branched-chain amino acids, including Ile (Schuster and Binder, 2005).
DISCUSSION
Asp-derived amino acids Lys, Met, Thr, and Ile are essential to humans and other animals, but they are present in major food crops at suboptimal levels (Müntz et al., 1998; Debabov, 2003; Pfefferle et al., 2003; Stiller et al., 2007). Genetic engineering of crop species with improved contents of Asp-derived amino acids requires a deep and comprehensive understanding of the metabolic pathways of these essential amino acids (Galili et al., 2008). Although the biosynthesis of Asp-derived amino acids has been previously studied, there is still much to learn about the regulation and components of this pathway (Jander and Joshi, 2009). In this work, we reported that loss-of-function mutations in the AK-HSDH2 gene cause increased contents of Asp and Asp-derived amino acids, especially Thr, in leaves. Additional analyses revealed that increases in Lys and Met are also observed in other ak and ak-hsdh single mutants. These findings underscore the complexity and previously unknown features of this pathway.
Accumulation of Downstream Amino Acid Products
It is common for loss-of-function mutants of a downstream enzyme to accumulate upstream products. For example, Zhu et al. (2001) reported that deficiency in a Lys catabolic enzyme (Lys ketoglutarate reductase-saccharopine dehydrogenase) results in Lys overaccumulation in seeds. Recently, it was reported that loss-of-function mutations in genes encoding branched-chain amino acid catabolic enzymes (isovaleryl-CoA dehydrogenase, methylcrotonyl-CoA carboxylase α- and β-subunits, and hydroxymethylglutaryl-CoA lyase) cause overaccumulation of branched-chain amino acids in seeds (Gu et al., 2010; Lu et al., 2011b). Moreover, it is not uncommon for feedback-insensitive mutants of an upstream enzyme to accumulate downstream products. For instance, Heremans and Jacobs (1995, 1997) reported that a single mutant with decreased AK sensitivity to Lys overaccumulates Lys and that a single mutant with decreased AK sensitivity to Thr overaccumulates Thr. Mourad and King (1995) reported that feedback-insensitive mutants of an Ile biosynthetic enzyme (TD) overaccumulate Ile in leaves. Furthermore, it is not unusual for loss-of-function mutants of a branch-point enzyme to shunt substrates into other branches. For example, multiple studies have shown that loss-of-function mutants of the DHDPS2 protein, a branch-point enzyme leading to Lys biosynthesis, overaccumulate Thr (Craciun et al., 2000; Sarrobert et al., 2000; Jones-Held et al., 2012). Another example is the Thr increase in transgenic plants with reduced expression of CGS1, the branch-point enzyme toward Met biosynthesis (Kim and Leustek, 2000).
However, it is rare for loss-of-function mutants of an upstream enzyme to accumulate downstream products. In this study, we showed that loss-of-function mutations in the AK-HSDH2 gene cause increased contents of free Asp-derived amino acids, especially Thr, in the leaves of 4-week-old Arabidopsis plants (Fig. 4). The increases in free Asp, Lys, and Met were also observed in the ak1-1, ak2-1, ak3-1, and ak-hsdh1-1 single mutants (Fig. 4). However, the increase in free Thr was not seen in the ak1-1, ak2-1, and ak3-1 mutants. These results indicated that loss-of-function mutations in the AK and AK-HSDH genes have different effects on different Asp-derived amino acids. The contents of many other amino acids, such as Asn, Gln, Glu, His, and Leu, showed coordinated increases in 4-week-old single mutants (Supplemental Fig. S4), which show that perturbation in Asp-derived amino acid biosynthesis in single mutants may cause changes in the contents of other amino acids. These coordinated changes were not observed in double mutants at the same age.
Relative Contributions of Different Isoforms to the Activities
Dissecting the individual contributions of different AK and AK-HSDH isoforms will help determine the target isoform for genetic engineering in crop species. We first attempted to use publically available transcriptome data to retrieve the relative transcript intensities for five AK and AK-HSDH genes. Because the probes for AK2 and AK3 genes on the Arabidopsis ATH1 GeneChip are identical (Schmid et al., 2005), the individual signal intensities for AK2 and AK3 cannot be extrapolated. Therefore, we used quantitative RT-PCR to determine transcript abundances. The transcript abundances of AK1, AK-HSDH1, and AK-HSDH2 in the wild type determined with quantitative RT-PCR were comparable with their relative signal intensities in wild-type Arabidopsis rosette leaves in the AtGenExpress developmental baseline experiment determined by microarray analysis (Schmid et al., 2005). The Pearson’s correlation coefficient between the two sets of data was 0.9105, which provides a strong and independent validation of our quantitative RT-PCR data. The RT-PCR data showed that AK2 is the most abundant transcript among the three AKs and that AK-HSDH1 is more abundant than AK-HSDH2.
Our AK activity data showed that monofunctional AKs collectively contribute more to the overall AK activity than the dual-functional AK-HSDHs, which is consistent with the estimated values reported by Curien et al. (2009). Curien et al. (2009) proposed that AK1 is the primary contributor to overall AK activity, because among the three AKs, AK1 is least sensitive to Lys inhibition (Curien et al., 2007). The estimation by Curien et al. (2009) was based on the relative protein abundances in chloroplast stroma and the sensitivities of different AK and AK-HSDH isoforms to physiological concentrations of allosteric effectors. However, because of the cross reactions of antibodies between isoforms, Curien et al. (2009) were only able to determine the total amount of AK isoforms and the total amount of AK-HSDH isoforms and unable to determine the relative abundances of each isoform. Thus, Curien et al. (2009) estimated the individual contributions of AK and AK-HSDH isoforms based on the hypothetic assumptions that AK1 and AK2 are equally abundant and that AK-HSDH1 and AK-HSDH2 are equally abundant. We had three lines of evidence suggesting that AK2 is the primary contributor to the overall AK activity and that AK-HSDH1 is the primary contributor to the overall HSDH activity: transcript abundances measured with quantitative RT-PCR, the activities measured in the absence of allosteric effectors, and activities measured in near-physiological concentrations of Lys, Thr, Ala, and SAM.
Calculating from the values in Figure 6, the ratio of the AK activity to the HSDH activity measured in wild-type Arabidopsis leaves in the absence of allosteric effectors was 0.35:1.00. This ratio was close to the ratio (0.4:1.00) of the total AK protein abundance to the total AK-HSDH protein abundance determined by ELISA with antibodies against purified recombinant proteins (Curien et al., 2009). In the presence of near-physiological concentrations of Lys, Thr, Ala, and SAM, the ratio of the AK activity to the HSDH activity in wild-type Arabidopsis leaves decreased to 0.20:1.00. This suggested that the total AK activity is more inhibited than the total HSDH activity in near-physiological concentrations of Lys, Thr, Ala, and SAM.
Our AK activity data showed that simultaneous loss of AK-HSDH2 and monofunctional AK1 or AK2 has an additive effect on the reduction of the overall AK activity. However, the ak-hsdh2-1 ak3-1 double mutant showed an unexpectedly higher AK activity than single mutants. This result could be the combined effect of several factors. First, among the three AK genes, the AK3 transcript is least abundant, and it is preferentially expressed in the xylem of leaves (Yoshioka et al., 2001). Second, among the three AK proteins, AK3 is most sensitive to Lys inhibition (Curien et al., 2007). Third, although AK-HSDH1 is not the primary contributor to the overall AK activity, the AK-HSDH1 transcript in the ak-hsdh2-1 ak3-1 double mutant was 67% higher than that in the ak-hsdh2-1 single mutant (Fig. 5).
Loss-of-function mutations in one AK or AK-HSDH gene did cause some changes in the expression of other AK or AK-HSDH gene(s), but none of these changes were systematic up- or down-regulations (Fig. 5). Loss-of-function mutations in an AK or AK-HSDH gene also resulted in changes in the transcript levels of other biosynthetic and catabolic genes, and some of the changes were shared among different mutants (Supplemental Figs. S5 and S6). For example, the TD1 transcript level was unanimously increased in the six single mutants. Other examples include the increase of the SAMS1 transcript and the decrease of the BCAT1 transcript in most of the single and double mutants.
Correlations among Transcript Abundance, Enzyme Activities, and Free Amino Acids
To identify systematic patterns among transcript abundance, enzyme activities, and free amino acids, we carried out pairwise Pearson’s correlation analysis (Fig. 7 shows representative correlations; Supplemental Table S5 has a complete list of correlations). First, the total amount of AK transcripts positively correlated with the Lys-sensitive AK activity, and the total amount of AK-HSDH transcripts positively correlated with both the Thr-sensitive AK activity and the total HSDH activity. These data suggested that transcriptional regulations play an important role in controlling the activities of the corresponding AK and AK-HSDH proteins. Second, there were correlations between enzyme activity and free amino acid content: the total AK activity positively correlated with the free (Lys + Met + Thr + Ile) to Asp ratio, the Lys- and Thr-sensitive AK activities negatively correlated with free Lys and free Thr, respectively, the total HSDH activity (i.e. the HSDH activity of AK-HSDHs) negatively correlated with free Thr, and the ratio of the AK activity to the HSDH activity negatively correlated with the free Lys to (Met + Thr + Ile) ratio. These data led to the hypothesis that the balance between Lys-sensitive AKs and Thr-sensitive AK-HSDHs is important for determining the amounts and ratios of Asp-derived amino acids and vice versa. This hypothesis is also supported by previous studies. Heremans and Jacobs (1997) reported that a mutant with decreased AK sensitivity to Lys had an increased amount of Thr, that a second mutant with decreased AK sensitivity to Thr had an increased amount of Lys, and that the double mutant had wild-type levels of Lys and Thr. Third, the total amount of Asp-derived amino acids positively correlated with the total AK transcripts, and free Lys, Met, and Ile positively correlated with the DHDPS, CGS1, and TD1 transcripts, respectively. These data are consistent with AKs being the committing enzymes for Asp-derived amino acid biosynthesis and DHDPSs, CGS1, and TD1 being the committing enzymes for Lys, Met, and Ile biosynthesis, respectively (Fig. 1). Fourth, free Met positively correlated with the transcripts of Met catabolic genes SAMS1 and SAMS2, and free Thr negatively correlated with the total AK-HSDH transcripts and positively correlated with the transcript of Thr catabolic gene THA2. These correlations suggested the possibilities that Thr may inhibit the expression of its biosynthetic genes and that Met and Thr may induce the expression of their catabolic genes. This is not uncommon: Zhu-Shimoni and Galili (1998) reported that the regulation of AK-HSDH could occur at the transcriptional level, and Rébeillé et al. (2006) found that Met strongly induced the expression of a gene encoding Met catabolic enzyme Met γ-lyase.
Figure 7.
Correlations among the ratio of total AK transcripts to total AK-HSDH transcripts, the ratio of total AK activity to total HSDH activity, and the ratio of free Lys to the total amount of free Met, Thr, and Ile. A, Correlation between the ratio of total AK transcripts to total AK-HSDH transcripts and the Lys to (Met + Thr + Ile) ratio in 4-week-old plants. B, Correlation between the ratio of total AK activity to total HSDH activity and the ratio of total AK transcripts to total AK-HSDH transcripts in 4-week-old plants. C, Correlation between the Lys to (Met + Thr + Ile) ratio and the ratio of total AK activity to total HSDH activity in 4-week-old plants. AKs/AK-HSDHs stands for the ratio of total AK1, AK2, and AK3 transcripts to total AK-HSDH1 and AK-HSDH2 transcripts. Lys/(Met + Thr + Ile) stands for the ratio of free Lys to the total amount of free Met, Thr, and Ile. The total AK and HSDH activities shown were measured in the absence of Lys, Thr, Ala, and SAM. Each point represents one genotype. Error bars represent sem (n = 3–5). The dotted lines are the linear regressions between free and protein-bound Asp-derived amino acids. The correlation coefficient (r) and the P value are shown for each plot on the edges of the triangle: Positive correlations are in red, and negative correlations are in blue.
The influence of changes in Asp-derived amino acid biosynthesis on the contents of other amino acids highlights the idea that metabolic regulation is far more complex than illustrated in the textbook. Some of the other amino acids, such as Asn, Glu, and Gln, are either involved in or related to Asp-derived amino acid biosynthesis. Glu is directly involved in the biosynthesis of Lys and Ile (Jander and Joshi, 2009). In the presence of ATP, Asp and Gln can be converted to Asn and Glu by Asn synthetases (Coruzzi and Last, 2000). Because Asp, Asn, Glu, and Gln are interrelated, it is somewhat expected that the four amino acids showed coordinated increases in the six 4-week-old single mutants (Fig. 4; Supplemental Fig. S4). In addition to Asn, Glu, and Gln, the contents of His and Leu, which are not related to Asp-derived amino acid biosynthesis, showed increased accumulation in the six single mutants and no change in the three double mutants (Supplemental Fig. S4). One possible reason is that the biosynthetic pathways of His and Leu involve Glu and Gln. His biosynthesis uses Gln and Glu as substrates (Stepansky and Leustek, 2006); free His had a very strong positive correlation with free Gln (r = 0.9028, P = 0.0003) and a moderate positive correlation with free Glu (r = 0.6327, P = 0.0497). Leu can be synthesized from the transamination between Glu and 4-methyl-2-oxopentanoate (Binder, 2010); free Leu had a moderate positive correlation with free Glu (r = 0.5385, P = 0.1083). It is not uncommon that perturbation in the biosynthesis of Asp-derived amino acids results in changes in other amino acids. For instance, van Bochaute et al. (2013) reported that long-term inhibition of DHDPS activity caused increased levels of His, Leu, Phe, and Val. Additional studies are needed to understand why changes in Asp-derived amino acids have such wide-ranging impact on leaf amino acid accumulation in general.
Overall, this study showed that loss-of-function mutations in the AK and/or AK-HSDH genes result in reduced transcript levels and gene product activities of the corresponding genes, alterations in the transcript levels of other biosynthetic and catabolic genes, and increases in the contents of Asp-derived (and some other) amino acids. Pairwise Pearson’s correlation analysis revealed that the ratio of total AK activity to total HSDH activity negatively correlates with the free Lys to (Met + Thr + Ile) ratio. Although the amounts and ratios of Asp-derived amino acids were also found to correlate with the transcript levels of a few other biosynthetic and catabolic genes, these patterns were identified from analyzing the responses of amino acid contents and transcript levels to loss-of-function mutations in the AK and/or AK-HSDH genes. Therefore, we hypothesize that the balance between Lys-sensitive AKs and Thr-sensitive AK-HSDHs is important in controlling the amounts and ratios of Asp-derived amino acids. The results from this research revealed some complexity in the regulation of Asp-derived amino acid contents (for example, the impact of changes in Asp-derived amino acid biosynthesis on amino acids that are not directly related to this pathway).
MATERIALS AND METHODS
Plant Materials, Array Mapping, Sequencing, Genotyping, and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA lines (SALK_003685, SALK_019023, SALK_043533, SALK_059678, SALK_082155, SALK_125957, and WiscDsLox461-464J6) used in this study were obtained from the Arabidopsis Biological Resource Center. All are in the Col-0 ecotype (Alonso et al., 2003; Woody et al., 2007). Homozygosity was confirmed by PCR as described by Ajjawi et al. (2010).
To locate the mutation in SALK_082155 that causes the Thr increase, a SALK_082155 plant was crossed to an Ler wild-type plant. The F2 plants were tested for their leaf free amino acid contents as described below. For array mapping, leaf tissues were harvested (one leaf per plant) from 13 F2 plants with the highest Thr concentrations and 27 F2 plants with the lowest Thr concentrations. Leaf tissues from high- and low-Thr plants were pooled and ground separately, and approximately 100 mg of powder was used for DNA extraction using the DNeasy Plant Mini Kit (Qiagen; http://www.qiagen.com). The two resulting DNA samples were labeled with biotin using the BioPrime DNA Labeling System (Life Technologies; http://www.lifetechnologies.com) according to the manufacturer’s instructions. Labeled DNAs were hybridized to two Arabidopsis ATH1 gene chips (Affymetrix; http://www.affymetrix.com). As described by Hazen et al. (2005), the raw data were analyzed with R scripts, which is available at http://naturalvariation.org/methods/. Hybridization and targeted gene sequencing (see below) were done at the Michigan State University (MSU) Research Technology Support Facility.
After the mapping internal was narrowed to a 1.75-Mb region on chromosome 4, the candidate gene AK-HSDH2 (At4g19710) of SALK_082155 was sequenced. Primers AK-HSDH2_5′L and AK-HSDH2_5′R were used to amplify with PCR the 5′ one-half of the gene, and primers AK-HSDH2_3′L and AK-HSDH2_3′R were used to amplify the 3′ one-half of the gene (Supplemental Table S6).
SALK_082155 was backcrossed to the Col-0 wild type to segregate the single-base insertion in the AK-HSDH2 gene and the T-DNA insertion in the At1g55805 locus. The segregating F2 plants were genotyped for both loci. Primers SALK_082155LP and SALK_082155RP were used to test for the presence of the T-DNA insertion in the At1g55805 locus (Supplemental Table S6). Primers Exon2_L and Exon4_R were used to amply a 716-bp region flanking the single-base thymine insertion in the fourth exon (Supplemental Table S6). In the absence of the single-base thymine insertion, the PCR product can be digested into two fragments (197 and 519 bp) by restriction enzyme Bsp1286I. In the presence of the single-base thymine insertion, the PCR product cannot be digested by Bsp1286I. Seeds from F2 plants with the single-base thymine insertion in both copies of the AK-HSDH2 gene and no T-DNA insertion in either copy of the At1g55805 gene were harvested and grown for downstream analyses, including quantitative RT-PCR, immunoblots, activity assays, and free amino acid measurements.
Mutants and wild-type (Col-0 and Ler) Arabidopsis plants were grown in a growth chamber on a 12-h-light/12-h-dark photoperiod. The light intensity was 150 μmol photons m−2 s−1, the temperature was 20°C, and the relative humidity was 50%. Unless otherwise stated, plants used for quantitative RT-PCR, immunoblots, activity assays, and free amino acid measurements were 4 weeks old. To minimize the impact of growth conditions on transcript levels, enzyme activities, and amino acid contents, the wild-type and nine mutant plants were grown in the same growth chamber and harvested 1 h after the lights came on.
Quantitative RT-PCR
Quantitative RT-PCR was performed as described by Lu et al. (2011a). For consistency purposes, the 8th to 10th oldest true leaves from 4-week-old plants were harvested (Supplemental Fig. S7), frozen, ground to a fine powder, and used for quantitative RT-PCR analysis. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), digested with RNase-Free DNase I (Qiagen), and reverse transcribed with oligo dT(15) primer and Moloney murine leukemia virus reverse transcriptase (Promega; http://www.promega.com). Quantitative PCR was performed on a StepOnePlus Real-Time PCR System (Life Technologies). Primers used for quantitative RT-PCR are listed in Supplemental Table S6. Quantification of transcript levels was achieved by comparing the threshold cycle numbers of unknown samples with a standard curve with known copy numbers.
Immunoblots
For consistency purposes, the 8th to 10th oldest true leaves from 4-week-old plants were harvested (Supplemental Fig. S7), frozen, ground to a fine powder, and used for soluble protein extraction and immunodetection of the AK-HSDH2 protein. Total soluble proteins were extracted in a buffer containing 100 mm HEPES (pH 7.5), 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 50 mg mL−1 polyvinylpolypyrrolidone as described by Lu et al. (2006). The protein concentration was determined using the Bradford method, with bovine serum albumin as the standard. In total, 43.8 μg of protein per lane was separated on a precast NuPAGE Novex 4% (w/v) to 12% (w/v) Bis-Tris Gel (Life Technologies) according to the manufacturer’s instructions. After electrophoresis, the proteins were electroblotted onto a polyvinylidene difluoride membrane as described by Lu et al. (2011a). Immunodetection of the AK-HSDH2 protein on the polyvinylidene difluoride membrane was performed using the SuperSignal West Pico Rabbit IgG Detecting Kit (Thermo Fisher Scientific; www.piercenet.com) and analyzed by the Gel Logic 1500 Imaging System (Kodak; www.kodak.com). The anti-AK-HSDH2 polyclonal antibody was made by Open Biosystems (now part of Thermo Scientific; http://www.thermoscientificbio.com/openbiosystems). A 14-amino acid peptide (corresponding to amino acids 820–833 of the full-length AK-HSDH2 protein) with an additional N-terminal Cys residue, C-DGDLAKERLDAENS, was synthesized, conjugated with keyhole limpet hemocyanin, and used to raise the AK-HSDH2-specific antibody.
Enzyme Extraction for AK and HSDH Activity Assays
Protein samples for AK and HSDH activity assays were prepared as described by Di Marco and Grego (1975) and Matthews et al. (1975). Because of the large amount of tissue required for enzyme extraction, the entire aboveground portion of 4-week-old plants was harvested (Supplemental Fig. S7), frozen in liquid nitrogen, and ground into a fine powder. Soluble proteins were homogenized in the extraction buffer (0.2 m Tris-HCl, pH 8.5, 0.1 m KCl, 1 mm EDTA, 1.4 mm 2-mercaptoethanol, 5 mm Thr, and 30% [v/v] glycerol) in a ratio of 2.5 mL g−1 fresh weight. Unground residues were removed by filtering through two layers of cheesecloth and centrifuging at 6°C for 30 min at 20,000g. To remove endogenous ions or metabolites that would potentially influence the downstream activity assays, leaf protein extracts were purified by adding ammonium sulfate to 50% saturation (0.27 g mL−1) and shaking on ice for 30 to 60 min. Proteins were collected by centrifuging at 6°C for 10 min at 3,000 rpm and redissolved in the resuspension buffer (50 mm potassium phosphate buffer, pH 7.5, 1.4 mm 2-mercaptoethanol, 1 mm EDTA, and 20% [v/v] glycerol). To prevent the loss of HSDH activity, the resuspension buffer for protein samples to be used in HSDH activity assays contains 0.5 mm Thr (Bryan, 1969). The protein concentration was determined using the Bradford method as described above.
AK Activity Assay
The AK activity was quantified using the hydroxamate-ferric chloride method as described by Dotson et al. (1989) and Paris et al. (2002). The product of the AK reaction, l-aspartyl-4-P, has an ester group that undergoes a substitution reaction with hydroxylamine (Pechère and Capony, 1968). This results in the formation of a hydroxamate acid. When ferric chloride is added, the iron binds to the hydroxamate acid and induces a color change (Pechère and Capony, 1968). The reaction mixtures contained 100 mm Tris-HCl (pH 8.0), 400 mm NH2OH-HCl, 515 mm KOH, 20 mm MgCl2, 150 mm KCl, 40 mm ATP, and 100 μL of enzyme extracts. The reactions were initiated by the addition of 50 mm Asp in a final reaction volume of 0.5 mL. After a 1-h incubation at 37°C, the assays were stopped by the addition of 0.25 mL of 20% (w/v) trichloroacetic acid, 370 mm FeCl3, and 360 mm HCl. To prepare the blank reactions, Asp was omitted during the enzyme incubation and added to the reaction mixture immediately before the reaction was stopped. Stopped reactions were mixed briefly and centrifuged for 15 min at 10,000g. The absorbance of the supernatant at 505 nm was measured immediately after centrifuging. An extinction coefficient of 750 m−1 cm−1 was used to convert absorbance to enzyme activity (1 unit = 1 μmol min−1; Paris et al., 2002).
HSDH Activity Assay
The HSDH activity was quantified by measuring the rate of NADP+ reduction (Matthews et al., 1975; Paris et al., 2002). The reaction mixtures contained 100 mm Tris-HCl (pH 8.0), 150 mm KCl, 50 mm homo-Ser, 1 mm NADP+, and 112.5 μL of enzyme preparation in a final volume of 0.75 mL. Reactions were initiated by the addition of enzyme preparation and allowed to proceed for 2 min before the A340 was measured every 15 s for 3 min. An extinction coefficient of 6,250 m−1 cm−1 was used to convert absorbance to enzyme activity (1 unit = 1 μmol min−1; Paris et al., 2002).
Leaf Free Amino Acid Assay by HPLC-Tandem Mass Spectrometry
For 1-week-old plants, the entire aboveground portion of plants was harvested, and approximately 20 plants per replicate were used. For 2-week-old plants, the first two true leaves were harvested, and approximately 10 plants per replicate were used. For 3-week-old plants, the youngest three mature rosette leaves (e.g. the second to fourth oldest true leaves) were harvested, and one plant per replicate was used. For 4-week-old plants, the ninth oldest true leaf was harvested, and one plant per replicate was used. Five replicates per genotype were used for all ages tested. Leaf numbers of 1-, 2-, 3-, and 4-week-old plants are shown in Supplemental Figure S7. Free amino acids were extracted and analyzed with the HPLC-tandem mass spectrometry method (Jander et al., 2004; Lu et al., 2008; Gu et al., 2012). For quantification, mixtures of 20 protein amino acids plus l-Phe-α,β,β,2,3,4,5,6-d8 and l-Val-2,3,4,4,4,5,5,5-d8 of 0.9 μm were analyzed along with the leaf samples.
Leaf Protein-Bound Amino Acid Assay by Acid Hydrolysis and HPLC-Tandem Mass Spectrometry
For consistency purposes, the ninth oldest true leaves from 4-week-old plants were harvested (Supplemental Fig. S7), frozen, ground to a fine powder, and used for leaf protein-bound amino acid assay. Ground tissues were hydrolyzed at 120°C for 24 h in 6 m HCl according to Nguyen et al. (2012). After the evaporation of 6 m HCl, free and protein-bound amino acids were extracted and analyzed with the HPLC-tandem mass spectrometry method (Jander et al., 2004; Lu et al., 2008; Gu et al., 2012). For quantification of protein-bound amino acids, free amino acids were subtracted. During acid hydrolysis, Cys and Trp are destroyed, Met is partially oxidized, and Asn and Gln are converted to Asp and Glu, respectively (Anders, 2002). Therefore, Asn and Asp are reported together, and Glu and Gln are reported together.
Data Analysis
All quantitative analyses (amino acid contents, transcript levels, protein abundances, and enzyme activities) were performed on three to five biological independent replicates as described. All statistical analyses were performed with JMP 11 Statistical Software (SAS Institute). Student’s t test was used to compare means among each pair of plant groups. Pearson’s correlation was used to assess the relationships between transcript levels, enzyme activities, and amino acid contents and their ratios. The strength of correlations was categorized according to the work by Fowler et al., 1998: 0 ≤ |r| < 0.20, very weak or negligible; 0.20 ≤ |r| < 0.40, weak; 0.40 ≤ |r| < 0.70, moderate; 0.70 ≤ |r| < 0.90, strong; and 0.90 ≤ |r| ≤ 1.00, very strong.
Sequences data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK1, At5g13280; AK2, A5g14060; AK3, At3g02020; AK-HSDH1, At1g31230; AK-HSDH2, At4g19710; DHDPS1, At3g60880; DHDPS2, At2g45440; CGS1, At3g01120; TS1, At4g29840; TD1, At3g10050; LKR-SDH1, At4g33150; SAMS1, At1g02500; SAMS2, At4g01850; SAMS3, At3g17390; SAMS4, At2g36880; THA1, At1g08630; THA2, AT3G04520; and BCAT1, At1g10060.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Domains in full-length monofunctional AKs and dual-functional AK-HSDHs.
Supplemental Figure S2. The high-Thr phenotype in SALK_082155 is caused by a second-site mutation.
Supplemental Figure S3. Representative siliques from ak-hsdh2-1/ak-hsdh2-1 AK-HSDH1-1/ak-hsdh1-1 and AK-HSDH2-1/AK-HSDH2-1 AK-HSDH1-1/AK-HSDH1-1 plants.
Supplemental Figure S4. The contents of other free amino acids in leaves of 4-week-old plants.
Supplemental Figure S5. Transcript levels of other genes involved in Asp-derived amino acid biosynthesis in leaves of 4-week-old plants.
Supplemental Figure S6. Transcript levels of select genes involved in Asp-derived amino acid catabolism in leaves of 4-week-old plants.
Supplemental Figure S7. Leaf numbers of 1-, 2-, 3-, and 4-week-old Col-0 wild-type Arabidopsis plants grown under a 12-h-light/12-h-dark photoperiod.
Supplemental Table S1. The contents of protein-bound amino acids in leaves of 4-week-old plants.
Supplemental Table S2. Leaf free amino acid contents in 1-week-old plants.
Supplemental Table S3. Leaf free amino acid contents in 2-week-old plants.
Supplemental Table S4. Leaf free amino acid contents in 3-week-old plants.
Supplemental Table S5. Correlation coefficients and P values among transcript levels, AK activity, HSDH activity, free amino acid contents, and protein-bound amino acid contents and their ratios.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank James P. O’Donnell, Justin B. Hackett, Ryan L. Wessendorf, Krishna Nath, Meriah K. Lucas, Nguyen Nguyen, Colleen E. O’Brien, Qing Shu, Emma R. Trabue, and Scott Warner (Western Michigan University [WMU]), David A. Hall and Kathleen M. Imre (MSU), and Helen Hasegawa (Andrews University) for technical assistance; Thomas Leustek (Rutgers University) for comments on antibody production; and Robert L. Last (MSU) and Todd J. Barkman (WMU) for critical review of the article.
Glossary
- Col-0
Columbia-0
- Ler
Landsberg erecta
- RT
reverse transcription
- SAM
S-adenosyl-Met
- T-DNA
transfer DNA
Footnotes
This work was supported by the U.S. National Science Foundation (grant nos. MCB–0519740 and MCB–1244008) and Western Michigan University (Support for Faculty Scholars Award and Graduate Student Research Fund).
Articles can be viewed without a subscription.
References
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL (2010) Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anders JC. (2002) Advances in amino acid analysis. BioPharm Int 4: 32–67. [Google Scholar]
- Bell SM, Burgoon LD, Last RL (2012) MIPHENO: data normalization for high throughput metabolite analysis. BMC Bioinformatics 13: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8: e0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JK. (1969) Studies on the catalytic and regulatory properties of homoserine dehydrogenase of Zea mays roots. Biochim Biophys Acta 171: 205–216 [DOI] [PubMed] [Google Scholar]
- Chipman DM, Shaanan B (2001) The ACT domain family. Curr Opin Struct Biol 11: 694–700 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Last RL (2000) Amino acids. In Buchanan RB, Gruissem W, Jones R, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 358–410 [Google Scholar]
- Craciun A, Jacobs M, Vauterin M (2000) Arabidopsis loss-of-function mutant in the lysine pathway points out complex regulation mechanisms. FEBS Lett 487: 234–238 [DOI] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cárdenas ML, Dumas R (2009) Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol 5: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Laurencin M, Robert-Genthon M, Dumas R (2007) Allosteric monofunctional aspartate kinases from Arabidopsis. FEBS J 274: 164–176 [DOI] [PubMed] [Google Scholar]
- Curien G, Ravanel S, Robert M, Dumas R (2005) Identification of six novel allosteric effectors of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase isoforms. Physiological context sets the specificity. J Biol Chem 280: 41178–41183 [DOI] [PubMed] [Google Scholar]
- Debabov VG. (2003) The threonine story. In Faurie R, Thommel J, eds, Microbial Production of L-Amino Acids. Springer, Berlin, pp 113–136 [Google Scholar]
- Di Marco G, Grego S (1975) Homoserine dehydrogenase in Pisum sativum and Ricinus communis. Phytochemistry 14: 943–947 [Google Scholar]
- Dotson SB, Somers DA, Gengenbach BG (1990) Kinetic studies of lysine-sensitive aspartate kinase purified from maize suspension cultures. Plant Physiol 93: 98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson SB, Somers DA, Gengenbach BG (1989) Purification and characterization of lysine-sensitive aspartate kinase from maize cell cultures. Plant Physiol 91: 1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler J, Cohen L, Jarvis. P (1998) Practical Statistics for Field Biology. John Wiley and Sons, Chichester, UK [Google Scholar]
- Frankard V, Vauterin M, Jacobs M (1997) Molecular characterization of an Arabidopsis thaliana cDNA coding for a monofunctional aspartate kinase. Plant Mol Biol 34: 233–242 [DOI] [PubMed] [Google Scholar]
- Galili S, Amir R, Galili G (2008) Genetic engineering of amino acid metabolism in plants. In Bohnert HJ, Nguyen H, Lewis NG, eds, Advances in Plant Biochemistry and Molecular Biology. Elsevier, Amsterdam, The Netherlands, pp 49–80 [Google Scholar]
- Gu L, Jones AD, Last RL (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2012) Rapid LC-MS/MS profiling of protein amino acids and metabolically related compounds for large-scale assessment of metabolic phenotypes. In Alterman MAA, Hunziker P, eds, Amino Acid Analysis. Humana Press, New York, pp 1–11 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, Yanovsky MJ, Liljegren SJ, Ecker JR, Kay SA (2005) Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol 138: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans B, Jacobs M (1997) A mutant of Arabidopsis thaliana (L.) Heynh. with modified control of aspartate kinase by threonine. Biochem Genet 35: 139–153 [DOI] [PubMed] [Google Scholar]
- Heremans B, Jacobs M (1995) Threonine accumulation in a mutant of Arabidopsis thaliana (L.) Heynh. with an altered aspartate kinase. J Plant Physiol 146: 249–257 [Google Scholar]
- Jander G, Joshi V (2009) Aspartate-derived amino acid biosynthesis in Arabidopsis thaliana. Arabidopsis Book 7: e0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Jones-Held S, Ambrozevicius LP, Campbell M, Drumheller B, Harrington E, Leustek T (2012) Two Arabidopsis thaliana dihydrodipicolinate synthases, DHDPS1 and DHDPS2, are unequally redundant. Funct Plant Biol 39: 1058–1067 [DOI] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40: 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Leustek T (2000) Repression of cystathionine γ-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci 151: 9–18 [Google Scholar]
- Lu Y, Hall DA, Last RL (2011a) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber APM, et al. (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL (2011b) Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiol 155: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Last RL (2011c) Chloroplast phenomics: systematic phenotypic screening of chloroplast protein mutants in Arabidopsis. In Jarvis RP, ed, Chloroplast Research in Arabidopsis. Humana Press, New York, pp 161–185 [DOI] [PubMed] [Google Scholar]
- Lu Y, Steichen JM, Yao J, Sharkey TD (2006) The role of cytosolic α-glucan phosphorylase in maltose metabolism and the comparison of amylomaltase in Arabidopsis and Escherichia coli. Plant Physiol 142: 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BF, Gurman AW, Bryan JK (1975) Changes in enzyme regulation during growth of maize: I. Progressive desensitization of homoserine dehydrogenase during seedling growth. Plant Physiol 55: 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad G, King J (1995) l-O-Methylthreonine-resistant mutant of Arabidopsis defective in isoleucine feedback regulation. Plant Physiol 107: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer GJ, Somers DA, Matthews BF, Gengenbach BG (1994) Molecular genetics of the maize (Zea mays L.) aspartate kinase-homoserine dehydrogenase gene family. Plant Physiol 106: 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntz K, Christov V, Saalbach G, Saalbach I, Waddell D, Pickardt T, Schieder O, Wüstenhagen T (1998) Genetic engineering for high methionine grain legumes. Nahrung 42: 125–127 [DOI] [PubMed] [Google Scholar]
- Nguyen HC, Hoefgen R, Hesse H (2012) Improving the nutritive value of rice seeds: elevation of cysteine and methionine contents in rice plants by ectopic expression of a bacterial serine acetyltransferase. J Exp Bot 63: 5991–6001 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Paris S, Viemon C, Curien G, Dumas R (2003) Mechanism of control of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase by threonine. J Biol Chem 278: 5361–5366 [DOI] [PubMed] [Google Scholar]
- Paris S, Wessel PM, Dumas R (2002) Overproduction, purification, and characterization of recombinant aspartate semialdehyde dehydrogenase from Arabidopsis thaliana. Protein Expr Purif 24: 99–104 [DOI] [PubMed] [Google Scholar]
- Pechère JF, Capony JP (1968) On the colorimetric determination of acyl phosphates. Anal Biochem 22: 536–539 [DOI] [PubMed] [Google Scholar]
- Pfefferle W, Möckel B, Bathe B, Marx A (2003) Biotechnological manufacture of lysine. In Faurie R, Thommel J, Bathe B, Debabov VG, Huebner S, Ikeda M, Kimura E, Marx A, Möckel B, Mueller U, Pfefferle W, eds, Microbial Production of L-Amino Acids. Springer, Berlin, pp 59–112 [Google Scholar]
- Rébeillé F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103: 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton JM, Bonner PLR, Wallsgrove RM, Lea PJ (1988) Physical and kinetic-properties of lysine-sensitive aspartate kinase purified from carrot cell-suspension culture. Biochim Biophys Acta 953: 48–60 [Google Scholar]
- Rognes SE, Dewaele E, Aas SF, Jacobs M, Frankard V (2003) Transcriptional and biochemical regulation of a novel Arabidopsis thaliana bifunctional aspartate kinase-homoserine dehydrogenase gene isolated by functional complementation of a yeast hom6 mutant. Plant Mol Biol 51: 281–294 [DOI] [PubMed] [Google Scholar]
- Sarrobert C, Thibaud MC, Contard-David P, Gineste S, Bechtold N, Robaglia C, Nussaume L (2000) Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J 24: 357–367 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57: 241–254 [DOI] [PubMed] [Google Scholar]
- Stepansky A, Leustek T (2006) Histidine biosynthesis in plants. Amino Acids 30: 127–142 [DOI] [PubMed] [Google Scholar]
- Stiller I, Dancs G, Hesse H, Hoefgen R, Bánfalvi Z (2007) Improving the nutritive value of tubers: elevation of cysteine and glutathione contents in the potato cultivar White Lady by marker-free transformation. J Biotechnol 128: 335–343 [DOI] [PubMed] [Google Scholar]
- Tang G, Zhu-Shimoni JX, Amir R, Zchori IB, Galili G (1997) Cloning and expression of an Arabidopsis thaliana cDNA encoding a monofunctional aspartate kinase homologous to the lysine-sensitive enzyme of Escherichia coli. Plant Mol Biol 34: 287–293 [DOI] [PubMed] [Google Scholar]
- Ufaz S, Galili G (2008) Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol 147: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufaz S, Shukla V, Soloveichik Y, Golan Y, Breuer F, Koncz Z, Galili G, Koncz C, Zilberstein A (2011) Transcriptional control of aspartate kinase expression during darkness and sugar depletion in Arabidopsis: involvement of bZIP transcription factors. Planta 233: 1025–1040 [DOI] [PubMed] [Google Scholar]
- van Bochaute P, Novoa A, Ballet S, Rognes SE, Angenon G (2013) Regulatory mechanisms after short- and long-term perturbed lysine biosynthesis in the aspartate pathway: the need for isogenes in Arabidopsis thaliana. Physiol Plant 149: 449–460 [DOI] [PubMed] [Google Scholar]
- Wang X, Lopez-Valenzuela JA, Gibbon BC, Gakiere B, Galili G, Larkins BA (2007) Characterization of monofunctional aspartate kinase genes in maize and their relationship with free amino acid content in the endosperm. J Exp Bot 58: 2653–2660 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yoshioka Y, Kurei S, Machida Y (2001) Identification of a monofunctional aspartate kinase gene of Arabidopsis thaliana with spatially and temporally regulated expression. Genes Genet Syst 76: 189–198 [DOI] [PubMed] [Google Scholar]
- Zhu X, Tang G, Granier F, Bouchez D, Galili G (2001) A T-DNA insertion knockout of the bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase gene elevates lysine levels in Arabidopsis seeds. Plant Physiol 126: 1539–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Shimoni JX, Galili G (1998) Expression of an Arabidopsis aspartate Kinase/Homoserine dehydrogenase gene is metabolically regulated by photosynthesis-related signals but not by nitrogenous compounds. Plant Physiol 116: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.