A histone acetyltransferase contributes to the regulation of iron homeostasis.
Abstract
Iron homeostasis is essential for plant growth and development. Here, we report that a mutation in GENERAL CONTROL NONREPRESSED PROTEIN5 (GCN5) impaired iron translocation from the root to the shoot in Arabidopsis (Arabidopsis thaliana). Illumina high-throughput sequencing revealed 879 GCN5-regulated candidate genes potentially involved in iron homeostasis. Chromatin immunoprecipitation assays indicated that five genes (At3G08040, At2G01530, At2G39380, At2G47160, and At4G05200) are direct targets of GCN5 in iron homeostasis regulation. Notably, GCN5-mediated acetylation of histone 3 lysine 9 and histone 3 lysine 14 of FERRIC REDUCTASE DEFECTIVE3 (FRD3) determined the dynamic expression of FRD3. Consistent with the function of FRD3 as a citrate efflux protein, the iron retention defect in gcn5 was rescued and fertility was partly restored by overexpressing FRD3. Moreover, iron retention in gcn5 roots was significantly reduced by the exogenous application of citrate. Collectively, these data suggest that GCN5 plays a critical role in FRD3-mediated iron homeostasis. Our results provide novel insight into the chromatin-based regulation of iron homeostasis in Arabidopsis.
Iron is an essential trace nutrition element for plants; its availability affects root morphogenesis, photosynthesis, nitrogen fixation, respiration, and flower color and fertility (Guerinot and Yi, 1994; Walker and Connolly, 2008). Iron deficiency impairs fundamental processes and causes a decrease in chlorophyll production, thus influencing crop productivity and quality (Schikora and Schmidt, 2001; Curie and Briat, 2003). Conversely, iron in excess is toxic to plants. Therefore, to ensure the effective acquisition of iron from the soil and to avoid excess iron in cells, iron uptake and distribution are strictly controlled in plants (Guerinot and Yi, 1994). Once taken up into the root, iron must be moved to the central vascular cylinder, where it can be loaded into the xylem and translocated to the aerial part of the plant (Rogers, 2006).
At the molecular level, several genes involved in iron uptake and mobilization have been characterized, including FERRIC REDUCTASE DEFECTIVE3 (FRD3), FERRIC REDUCTASE OXIDASE2 (FRO2), ZIP family IRON REGULATED TRANSPORTER1 (IRT1), and other genes (Curie and Briat, 2003; Walker and Connolly, 2008). In Arabidopsis (Arabidopsis thaliana), FRO2 and IRT1 are two important genes responsible for iron uptake. IRT1 is the major transporter responsible for high-affinity iron uptake from the soil and is a key player in the regulation of plant iron homeostasis, as attested to by the severe chlorosis and lethality of the irt1 mutant (Korshunova et al., 1999; Varotto et al., 2002; Vert et al., 2002). Encoding a low-iron-inducible ferric chelate reductase, FRO2 is responsible for the reduction of iron at the root surface. When iron is loaded into the root xylem from the pericycle, iron distribution between the root and shoot is essential for tolerating fluctuations in iron availability (Long et al., 2010). Several studies revealed that FRD3, a multidrug and toxin efflux protein, facilitates iron chelation to citrate and the subsequent transport of iron citrate from the root to the shoot (Green and Rogers, 2004; Durrett et al., 2007).
Gene expression regulation is a crucial step for coping with iron deficiency or excess in plants. The tomato (Solanum lycopersicum) FE-EFFICIENCY REACTIONS (FER) gene, encoding a basic helix-loop-helix (bHLH) transcription factor, was the first gene shown to be involved in iron-deficiency responses and iron uptake (Ling et al., 2002). In Arabidopsis, FIT (for FER-like iron deficiency-induced transcription factor), a homolog of tomato FER, positively regulates the expression of the FRO2 and IRT1 genes (Colangelo and Guerinot, 2004; Yuan et al., 2005). In addition to FIT, the AtbHLH38, AtbHLH39, AtbHLH100, and AtbHLH101 proteins, which are classified into the Ib subgroup of bHLH transcription factors (Toledo-Ortiz et al., 2003), are required for the regulation of iron uptake and homeostasis in Arabidopsis (Wang et al., 2013).
With the discovery of the regulatory role of histone posttranslational modifications and their correlation with transcriptional states, studies have demonstrated that epigenetic mechanisms play important roles in iron homeostasis. For example, Shk1 binding protein1 (SKB1), which catalyzes the symmetric dimethylation of histone 4 arginine 3 (H4R3), is involved in iron homeostasis in Arabidopsis. Histone H4R3 dimethylation negatively regulates iron homeostasis by affecting several Ib subgroup bHLH genes and iron-uptake processes (Fan et al., 2014).
Histone acetylation and deacetylation are known to be important in the regulation of gene expression (Grunstein, 1997). Two enzymes, histone acetyltransferase (HAT) and histone deacetylase (HDAC), are responsible for histone acetylation and deacetylation, respectively (Jenuwein and Allis, 2001). The Arabidopsis genome encodes at least 12 HAT and 18 HDAC genes (Pandey et al., 2002). Although the functions of HATs and HDACs in plant development and reproduction have been reported extensively, their roles in iron homeostasis are poorly understood. In this study, we find that a mutation in GENERAL CONTROL NONREPRESSED PROTEIN5 (GCN5) impairs iron transportation from the root to the shoot and affects iron homeostasis in Arabidopsis. We also confirmed that GCN5 bound to the chromatin at FRD3 under iron-sufficient conditions and that the enrichment of GCN5 was enhanced under iron-deficient conditions. Therefore, we conclude that GCN5 regulates iron distribution pathways by affecting the acetylation level of its direct target FRD3. In addition, we propose that HISTONE DEACETYLASE7 (HDA7) may negatively regulate FRD3 expression through FRD3 deacetylation.
RESULTS
The Mutation in GCN5 Caused Iron Retention in Roots and Affected Iron Homeostasis in Arabidopsis
Iron homeostasis comprises two successive steps: iron uptake at the root surface, and iron distribution in vivo (Curie and Briat, 2003). In this study, the Perl’s stain method, a qualitative measure of the ferric iron content in living tissues, was employed to screen 10 Arabidopsis transfer DNA insertion mutants (gcn5, hda2c, hda5, hda7, hda9, hda13, hda14, hda15, hda18, and hda19). After 1 d of iron-deficiency treatment, iron accumulated to higher levels in the roots of three single mutants (gcn5, hda9, and hda19) than in the wild type (Fig. 1A; Supplemental Fig. S1). Moreover, after treatment with trichostatin (TSA), an inhibitor of HDACs, iron accumulation throughout the roots was not observed in the gcn5 mutant (Fig. 1A). To better understand the underlying mechanism of action of GCN5 in iron homeostasis, we assessed the transcriptional response of GCN5 to iron deficiency in a wild-type background by using quantitative PCR (qPCR). Compared with the controls, the expression of GCN5 was higher after 1 d of iron-deficiency treatment, reaching a maximum at 3 d, then decreasing by 5 d (Fig. 1B), indicating that GCN5 is an iron deficiency-inducible gene. Collectively, these data indicate that GCN5 may play important positive regulatory roles in iron homeostasis.
Figure 1.
GCN5 is responsible for iron translocation and responses to iron deficiency. A, Iron retention is observed in the roots of the gcn5 mutant. Perl’s staining was performed to detect the ferric iron content in the root. Seedlings (Ws and gcn5 mutants) were grown on normal Murashige and Skoog (MS) plates for 7 d and then transferred to MS plates (0 µm iron-EDTA or 0 µm iron-EDTA and 0.5 µm TSA) for 1 d. TSA is an inhibitor of HDACs. B, GCN5 transcripts are induced in iron (Fe)-deficient conditions. Seven-day-old Ws seedlings on MS plates with iron were transferred onto MS plates without iron (0 µm iron-EDTA) or with iron (100 µm iron-EDTA) for 12 h or 1 to 5 d. Total RNA was isolated, and quantitative reverse transcription (qRT)-PCR showed the dynamic expression of the GCN5 transcript. The expression of β-ACTIN was used to normalize mRNA levels. Error bars represent sd values from at least three repetitions.
Illumina High-Throughput Sequencing Revealed 879 GCN5-Regulated Candidate Genes Involved in Iron Homeostasis
To obtain detailed information about the expression profiles of GCN5-regulated genes involved in iron-deficiency responses, we used Illumina high-throughput sequencing to compare the expression profiles of four samples, designated Ws (for Wassilewskija; wild type with sufficient iron), Ws-Fe (wild type with deficient iron), gcn5 (gcn5 mutant with sufficient iron), and gcn5-Fe (gcn5 mutant with deficient iron). The expression of GCN5 peaked at 3 d of iron-deficiency treatment, being up-regulated by more than 5-fold over the control. Thus, seedlings with 3 d of iron-deficiency treatment were collected for RNA sequencing (RNAseq). All sequencing reads were aligned to the reference genome of Arabidopsis, The Arabidopsis Information Resource 10 (http://www.arabidopsis.org/), using Bowtie2 software (Langmead and Salzberg, 2012), and only uniquely mapped reads were retained for the subsequent expression analysis by edgeR (Robinson et al., 2010). With two biological replicates for each sample, the correlation coefficients showed acceptable reproducibility, indicating the high reliability of our mRNA sequencing data (Supplemental Fig. S2). The accuracy of the transcriptome data was also validated by qPCR. (Primers are listed in Supplemental Table S1.) Among the 16 randomly selected genes, the qPCR results of 15 genes were completely consistent with the RNAseq data (Supplemental Fig. S3).
Under iron-deficient conditions, we found that 1,917 genes were significantly induced in the Ws genotype compared with the gcn5 mutant, designated Ws-Fe > gcn5-Fe. In addition, 1,278 genes displayed dramatic responses to iron-deficiency treatment in the wild-type background, designated Ws-Fe > Ws. Because GCN5 functioned as a positive regulator of transcription (Imoberdorf et al., 2006) and was up-regulated by iron-deficiency treatment, the GCN5-regulated genes involved in iron homeostasis should be included in the overlapping 884 genes of Ws-Fe > gcn5-Fe and Ws-Fe > Ws (Fig. 2A; Supplemental Table S2). Among the 884 overlapping genes, five genes belonged to the gcn5-Fe > gcn5 group, so these might not be regulated by GCN5. Therefore, 879 genes were considered to be GCN5-regulated candidate genes in iron homeostasis (Fig. 2A; Supplemental Table S2).
Figure 2.
Identification of potential GCN5 targets involved in iron homeostasis using RNAseq. A, The overlap (879 genes, underscored by a black line) in the Venn diagram represents potential GCN5 targets that are also involved in iron homeostasis. B, Distribution of GO categories (biological processes) for the 879 GCN5-regulated candidate genes.
Gene Ontology (GO) analysis of the 879 genes showed categories of significant enrichment. The most abundant category was metabolic processes (21%; including carbohydrate, glucuronoxylan, and lipid metabolic processes), followed by the response to abiotic or biotic stimulus category (11%; including responses to abscisic acid stimulus, auxin stimulus, high light intensity, and hydrogen peroxide). A significant fraction (8%) represented genes implicated in responses to stress, such as salt, water deprivation, cold, and heat stress. Up to 7% of genes were identified to be involved in transport, including nitrate, iron ion, lipid, and calcium ion transport. The other categories involved developmental processes (7%), signal transduction (2%), cell organization and biogenesis (2%), DNA-dependent transcription (1%), and electron transport or energy pathways (1%). In addition, a large group of genes (40%) involved in other biological processes could not be simply classified and were labeled as others (Fig. 2B).
Five Genes Are Direct Targets of GCN5 in Iron Homeostasis Regulation
To identify the direct target genes in GCN5-regulated iron homeostasis, we focused on the candidate genes related to iron transport, because the mutation in GCN5 resulted in iron retention in roots and the failure to transport this metal to the aerial parts of the plants. Through GO analysis of the 879 genes, we obtained all of the GO terms with P < 0.01 (data not shown) and selected two iron-binding or transportation-related GO terms (GO:0005506 and GO:0006826). Finally, we obtained 10 genes from the two GO terms based on their biological properties and described effects on iron transport (Green and Rogers, 2004; Feng et al., 2006) and their differential expression observed by RNAseq experiments (Table I). Seven-day-old seedlings of Ws and gcn5 genotypes subjected to iron-deficiency conditions for 3 d were collected for chromatin immunoprecipitation (ChIP) and gene expression assays (Fig. 3). ChIP-PCR analysis showed that GCN5 bound to the core promoter region of five genes in Ws to a significantly greater extent than in gcn5: FRD3 (At3G08040), MLP329 (At2G01530), EXOCYST SUBUNIT EXO70 FAMILY PROTEIN H2 (EXO70H2; At2G39380), REQUIRES HIGH BORON1 (BOR1; At2G47160), and CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE25 (CRK25; At4G05200; Fig. 3A). However, GCN5 was not enriched at the promoter of FRO6 (At5G49730), an important gene for iron homeostasis, indicating that this gene is an indirect target of GCN5 (Fig. 3A).
Table I. Potential GCN5-regulated, iron transport-related genes.
NA; Not available.
| Gene Identifier | Gene Name | Annotation |
|---|---|---|
| At5G49730 | FRO6 | Ferric reduction oxidase6 |
| At2G39380 | EXO70H2 | Exocyst subunit exo70 family protein H2 |
| At2G01530 | MYROSINASE-BINDING PROTEIN-LIKE PROTEIN329 (MLP329) | MLP-like protein329 |
| At2G47160 | BOR1 | HCO3− transporter family |
| At3G53420 | PIP2;1 | Plasma membrane intrinsic protein 2A |
| At3G08040 | FRD3 | Multidrug and toxin efflux family protein |
| At4G05200 | CRK25 | Cys-rich receptor-like protein kinase25 |
| At5G44610 | PCAP2 | Microtubule-associated protein18 |
| At5G51160 | NA | Ankyrin repeat family protein |
| At5G10580 | NA | Protein of unknown function, DUF599 |
Figure 3.
Identification of the direct targets of GCN5 and measurement of their acetylation states. Seven-day-old Ws and gcn5 mutant seedlings on MS plates with iron were separately transferred onto MS plates without iron (0 µm iron-EDTA) for 3 d. Nuclei were extracted from the cross-linked seedlings, sonicated, and immunoprecipitated with antibodies specific to GCN5, acetylation of histone 3 lysine 9 (H3K9ac), or H3K14ac. Primers were designed at the core promoter region. Error bars represent sd values from at least three repetitions. A, ChIP assays (anti-GCN5) identified the direct target of GCN5. B, ChIP assays (anti-H3K9ac or anti-H3K14ac) were used to examine the H3K9ac or H3K14ac levels of the five GCN5 target genes. C, qRT-PCR shows the dynamic expression of GCN5 target genes. Asterisks indicate significant differences (*, P < 0.05; and **, P < 0.01 by Student’s t test).
Previous studies demonstrated that GCN5 is responsible for H3K14ac and for facilitating H3K9ac and H3K27ac, which is expected to be required for the expression of a large number of genes (Vlachonasios et al., 2003; Earley et al., 2007; Benhamed et al., 2008). To further confirm the direct regulation of these five potential target genes by GCN5, we performed ChIP-PCR assays using antibodies against H3K9ac and H3K14ac (Fig. 3B). Consistent with the property of GCN5 as a HAT, the H3K9ac and/or H3K14ac levels of the five genes FRD3, EXO70H2, MLP329, BOR1, and CRK25 were significantly decreased due to the impairment of GCN5. Furthermore, the mRNA abundances of these five genes were significantly lower in gcn5 than in Ws under iron-deficiency conditions (Fig. 3C). Based on these findings, we propose that GCN5 directly binds to the promoters of these five genes, modulates their H3K9ac and/or H3K14ac levels, and in turn regulates their expression.
GCN5-Mediated Acetylation of FRD3 Determines the Dynamic Expression of FRD3
Iron retention in the roots of the gcn5 mutant was validated based on ion-coupled plasma mass spectrometry (ICP-MS) analysis. In addition, the gcn5 mutants showed nearly 2-fold higher levels of manganese and 5- to 6-fold higher levels of zinc in their roots than the wild type. Copper levels were unchanged in the mutant (Fig. 4A). These phenotypes of gcn5 are quite similar to those of frd3. Consistently, much less iron and manganese was detected in gcn5 shoots than in the wild type. The zinc content was somewhat lower in the gcn5 mutants than in the wild type (Fig. 4B). The micronutrient zinc plays an important role in the physiological and metabolic processes of plants. However, zinc can be toxic to the plant in excess concentrations, causing symptoms such as chlorosis (Marschner, 1986; Ramesh et al., 2004). According to the ICP-MS results, we believe that GCN5 also regulates zinc homeostasis in Arabidopsis. We propose that the misregulation of the distribution of other elements, such as zinc, also plays important roles in gcn5 phenotypes.
Figure 4.
gcn5 mutants accumulate more iron (Fe), manganese (Mn), and zinc (Zn) in the roots but not more copper (Cu). Pooled samples of 2-week-old roots (A) and shoots (B) from plants grown on MS plates were subjected to element analysis using ICP-MS. Experiments were performed at least twice. Asterisks indicate significant differences (*, P < 0.05; and **, P < 0.01 by Student’s t test).
Studies revealed that dynamic alteration in modifications of the histone H3 amino-tail were correlated with gene activation in response to abiotic stress (Bruce et al., 2007). Here, FRD3 was used for further investigation because the frd3 and gcn5 mutants showed a similar iron retention phenotype in roots (Green and Rogers, 2004). Seven-day-old seedlings of Ws were transferred to iron-deficient MS medium for 0, 1, 2, 3, 4, or 5 d, and samples were collected for further ChIP-PCR and gene expression analyses. ChIP-PCR revealed that GCN5 association with the FRD3 promoter exhibited up to approximately 2-fold enrichment from 1 to 4 d, compared with the control (0 d), but the enrichment of GCN5 at 5 d returned to levels similar to those of the control (Fig. 5A). The quantity of GCN5 associated with the FRD3 promoter was correlated with the status of iron, suggesting that GCN5 regulates iron homeostasis when environmentally soluble iron is limited. Furthermore, we investigated whether the enhanced association of GCN5 to FRD3 leads to dynamic changes in histone acetylation. We collected the seedlings at five time points for ChIP-PCR assays specific to H3K14ac. Compared with the control, the H3K14ac levels in the promoter region of FRD3 were clearly increased after 2 d of iron-deficiency treatment, reached a maximum at 3 d, and then were decreased at 4 d (Fig. 5B). As shown in Figure 5C, FRD3 transcripts were significantly up-regulated after 3 or 4 d of iron-deficiency treatment compared with the control.
Figure 5.
Regulation of FRD3 expression by GCN5. Seedlings grown on normal MS plates for 7 d were transferred to MS plates without iron (Fe) for 1 to 5 d. A, Nuclei were extracted, and ChIP assays were performed with antibodies specific to GCN5. The gene Arabidopsis CHALCONE SYNTHASE (AtCHS) was used as a negative control. B, Nuclei were extracted, and ChIP assays were performed with antibodies specific to H3K14ac. C, qRT-PCR was performed to detect FRD3 transcript levels. Asterisks indicate significant differences (*, P < 0.05; and **, P < 0.01 by Student’s t test).
qPCR analysis showed that the expression of FRD3 was approximately 3-fold lower in the gcn5 mutant than in the wild type (Supplemental Fig. S4A). ChIP assays also revealed a significantly greater association of GCN5 with the FRD3 promoter in the wild type than in the gcn5 mutant (Supplemental Fig. S4B). To confirm that GCN5 regulates FRD3 gene expression by modulating acetylation, we designed two pairs of primers for ChIP assays against H3K9ac or H3K14ac, which spanned the core promoter and the second exon, respectively. Consistent with the enrichment of GCN5, the H3K9ac and H3K14ac levels in both the promoter and gene body regions of FRD3 were significantly higher in the wild type than in the gcn5 mutant (Supplemental Fig. S4C). Collectively, these data provide further evidence that the acetylation of specific histone Lys residues, regulated by GCN5, is required for iron deficiency-induced FRD3 gene expression.
GCN5 Plays a Major Role in FRD3-Mediated Iron Homeostasis
To investigate the function of FRD3 in GCN5-regulated iron homeostasis, we generated FRD3 overexpression lines in the gcn5 mutant background. Owing to the low pollen fertility of the gcn5 mutant, we first transformed the 35S::FRD3 construct into Ws. Three independent transgenic plants (35S::FRD3/Ws 1 to 35S::FRD3/Ws 3) with significantly elevated FRD3 transcripts were selected to be crossed with gcn5 mutants. Finally, we obtained three homologous FRD3 overexpression lines in the gcn5 mutant background, which exhibited significantly higher expression levels of FRD3 than the gcn5 mutant, designated 35S::FRD3/gcn5 1 to 35S::FRD3/gcn5 3 (Supplemental Fig. S5).
FRD3 facilitates citrate release and the subsequent transport of iron citrate from the root to the shoot (Green and Rogers, 2004; Durrett et al., 2007). Iron retention was not detected in the three 35S::FRD3/gcn5 lines or the wild type, in contrast to the gcn5 mutant (Fig. 6A). In addition, iron retention in the gcn5 mutant was successfully rescued by the exogenous addition of citrate to the MS plates (Fig. 6A). Based on these data, we conclude that FRD3 is a critical direct target of GCN5 in iron distribution that regulates the citrate efflux in root tissues.
Figure 6.
Iron retention in roots caused by impairment of GCN5 affects chlorophyll content. A, Perl’s staining to detect the ferric iron content in roots. Seedlings (Ws, gcn5 mutants, and 35S::FRD3/gcn5 transgenic lines) were grown on normal MS plates for 7 d and then transferred to MS plates (0 µm iron-EDTA) for 1 d. The gcn5 mutant was transferred to MS plates (0 µm iron-EDTA and 3 mm sodium citrate) for 3 d. B, Impairment of GCN5 led to decreased total chlorophyll (Chl) content in new leaves. Ten-day-old seedlings (Ws, gcn5 mutants, and 35S::FRD3/gcn5 transgenic plants) were transferred to MS plates with 0 or 100 µm iron (Fe) for 3 d. In addition, gcn5 mutants were treated with 3 mm sodium citrate for 3 d. New leaves were collected, and total chlorophyll was quantified (n = 6; values are ±sd). Asterisks denote values that are significantly different (*, P < 0.05; and **, P < 0.01 by Student’s t test) compared with Ws. fw, Fresh weight.
Iron in the leaves and chloroplasts is tightly correlated with the content of chlorophyll (Jacobson, 1945). As shown in Figure 6B, gcn5 plants exhibited less chlorophyll in the new leaves relative to Ws, and this defect was enhanced after a 3-d iron-deficiency treatment. Moreover, the observed defect in the gcn5 mutants was partially rescued by the exogenous application of citrate or overexpression of FRD3 (Fig. 6B), suggesting that FRD3-mediated citrate efflux regulated by GCN5 plays an important role in chlorophyll synthesis.
Iron plays essential roles in pollen development (Lanquar et al., 2005; Roschzttardtz et al., 2011). The gcn5 mutant exhibited clear reproductive defects compared with the wild type, with shorter siliques and far fewer seeds (Table II). Electron microscopy analysis revealed that the 35S::FRD3/gcn5 or exogenous iron-treated gcn5 mutant showed enhanced pollen viability and compact stigma papillae compared with the gcn5 mutant (Fig. 7; Table III). Based on these results, we propose that GCN5-controlled iron homeostasis through the regulation of FRD3 expression also affects anther and stigma development.
Table II. Seed number and silique length in gcn5 mutants.
Asterisks denote values that are significantly different from the control (*, P < 0.05; and **, P < 0.01 by Student’s t test).
| Genotype | Seed No. | Silique Length |
|---|---|---|
| mm | ||
| gcn5 | 2.2 ± 1.4 | 4.3 ± 1.16 |
| gcn5 (+iron) | 4.1 ± 1.1** | 5.5 ± 1.08* |
Figure 7.
Reproductive growth of gcn5 is impaired. Observation was made of the anther and stigma of the gcn5 mutant with water (A and B) or exogenous iron (Fe) ethylene diamine diorthohydroxyphenyl acetic acid (C and D) treatment and normally grown 35S::FRD3/gcn5 plants (E and F). Iodine-potassium iodide staining was used to examine the pollen viability of gcn5 mutants (G) and 35S::FRD3/gcn5 plants (H).
Table III. Pollen viability in the gcn5 mutant and 35S::FRD3/gcn5 lines.
Asterisks denote values that are significantly different from the control (*, P < 0.05; and **, P < 0.01 by Student’s t test).
| Genotype | Fertile Pollen |
|---|---|
| % | |
| gcn5 | 16.88 ± 3.21 |
| 35S::FRD3/gcn5 1 | 30.27 ± 3.18** |
| 35S::FRD3/gcn5 2 | 24.37 ± 4.45* |
| 35S::FRD3/gcn5 3 | 25.8 ± 3.14** |
Nutrition-Deficiency Responses of the gcn5 Mutant under Nutrition-Replete Conditions
Iron homeostasis consists of two main processes: iron uptake and translocation in vivo. After suggesting one manner in which GCN5 regulates iron translocation, we investigated whether there were simultaneous influences in the iron-uptake processes. IRT1 and FRO2 are two important genes for iron uptake in Arabidopsis (Robinson et al., 1999; Vert et al., 2002). Consistent with other reports, these two genes were greatly induced by iron-starvation treatment in the wild-type background (Fig. 8A). However, both genes are significantly induced in the gcn5 mutant, even under nutrition-replete conditions, which was consistent with our RNAseq data. To clarify whether the induction of nutrition uptake-related genes was a common phenomenon in the gcn5 mutant plants, we analyzed the RNAseq data of normally grown Ws and gcn5 mutant samples. Among all of the 233 genes that exhibited at least 2-fold higher expression levels in the gcn5 mutant than in Ws, 50 genes were involved in nutrition homeostasis, including iron transporters, nitrate transporters, phosphate starvation-related genes, and other nutrition element-binding components (Table IV). In addition, the gcn5 mutant exhibited more and longer root hairs than Ws under normal conditions, which is a common morphology of malnourished plants (Fig. 8B). However, given that 18 out of the 233 genes were related to five important phytohormones, we could not rule out the possibility that this phenotype might be partially caused by a change in phytohormones. Nevertheless, these results suggested that, though grown under nutrition-replete conditions, gcn5 mutants exhibited nutrition-deficiency responses both in gene expression and morphogenesis phenotypes, which might be attributed to this mutant’s inefficient distribution of nutrition elements in vivo.
Figure 8.
Nutrition-deficiency responses of the gcn5 mutant under the nutrition-replete condition. A, Seven-day-old normally grown Ws seedlings were transferred to MS plates with or without iron (Fe) for 3 d. qRT-PCR was performed to detect the expression of two iron uptake-related genes (IRT1 and FRO2). B, Phenotypes of root hair zones for normally grown Ws and the gcn5 mutant. The gcn5 mutant exhibited more and longer root hairs than Ws under normal conditions.
Table IV. GO analysis of the 233 genes with higher expression in the gcn5 mutant than in Ws.
| GO Identifier | Gene No. | Annotation |
|---|---|---|
| Nutrition homeostasis (total 50) | ||
| GO:0005506 | 9 | Iron ion binding |
| GO:0046872 | 7 | Metal ion binding |
| GO:0016036 | 5 | Cellular response to phosphate starvation |
| GO:0000041 | 3 | Transition metal ion transport |
| GO:0015706 | 4 | Nitrate transport |
| GO:0046686 | 6 | Response to cadmium ion |
| GO:0005507 | 2 | Copper ion binding |
| GO:0008270 | 14 | Zinc ion binding |
| Hormone response (total 18) | ||
| GO:0009830 | 4 | Cell wall modification involved in abscission |
| GO:0009862 | 4 | Salicylic acid-mediated signaling pathway |
| GO:0009751 | 4 | Response to salicylic acid stimulus |
| GO:0009736 | 3 | Cytokinin-mediated signaling pathway |
| GO:0009867 | 3 | Jasmonic acid-mediated signaling pathway |
HDACs Negatively Regulate FRD3 Expression
Histone acetylation and deacetylation often coordinate to precisely regulate gene expression (Shahbazian and Grunstein, 2007). We found that FRD3 transcripts were significantly up-regulated (up to 3-fold) in two HDAC mutants (hda7 and hda14; Fig. 9A). Because HDA7 mainly catalyzes deacetylation in Lys-9 and Lys-14 sites of histone 3 in Arabidopsis (Cigliano et al., 2013), we designed five pairs of primers spanning the core promoter and gene body regions and examined the H3K9/14ac modifications at the FRD3 locus by performing ChIP-PCR experiments (Fig. 9B). The H3K14ac levels at the FRD3 locus did not drastically change, whereas the H3K9ac levels were much higher in the hda7 mutant than in the wild type, suggesting that HDA7 might negatively regulate FRD3 expression by reducing the H3K9ac levels of FRD3 in Arabidopsis.
Figure 9.
HDA7 negatively regulates FRD3 expression. A, Normally grown 10-d-old seedlings of the wild type (Columbia-0 [Col-0]) and nine HDAC mutants were collected. qRT-PCR was performed to detect the expression of FRD3 transcripts. B, Normally grown 10-d-old wild-type (Col-0) and hda7 seedlings were collected, and ChIP assays were performed using antibodies specific to H3K9ac or H3K14ac. The five primer sets (two-headed arrows) were numbered for FRD3. Asterisks denote values that are significantly different from the control (*, P < 0.05; and **, P < 0.01 by Student's t test).
DISCUSSION
Iron is essential for plant survival and growth. Recent studies have highlighted transcriptional responses to iron deficiency (Buckhout et al., 2009; Yang and Finnegan, 2010) and that the regulation of gene expression is accomplished by epigenetic mechanisms that modulate chromatin structure. Fan et al. (2014) reported that histone H4R3 dimethylation negatively regulated iron homeostasis by affecting several Ib subgroup bHLH genes and the iron-uptake processes. Here, our data show that histone acetylation is also important for iron distribution.
GCN5 Is Required for the Expression of Iron Deficiency-Responsive Genes
After iron is taken up by roots, its distribution is an important process for the mineral nutrition of plants (Curie and Briat, 2003). To date, several central genes responsible for iron distribution have been identified, but the underlying regulatory mechanism remains to be elucidated. In this study, we found that a gcn5 mutant exhibited iron retention in root tissues. Consistent with the up-regulation of GCN5, global H3K14ac levels clearly increased more in Ws than in the gcn5 mutants after iron-deficiency treatment (Supplemental Fig. S6). Moreover, the observed iron retention in the gcn5 mutants was rescued by TSA treatment. Thus, these data suggest that histone acetylation via GCN5 is an important mechanism for iron distribution in Arabidopsis.
GCN5 is required for the regulation of divergent sets of stress-response genes in yeast (Saccharomyces cerevisiae) and Arabidopsis (Grant et al., 1997; Benhamed et al., 2008). To understand the mechanisms underlying the involvement of GCN5 in iron-deficiency responses, we identified 1,278 genes that were up-regulated after iron-deficiency treatment, of which 879 are dependent on GCN5. GO analysis revealed a significant overrepresentation of genes involved in metabolic processes and responses to abiotic or biotic stimuli, including responses to abscisic acid and auxin stimulus, salt, water deprivation, and heat stresses. These data indicate that GCN5 is a general positive transcriptional regulator that regulates the expression of genes involved in a range of biological activities important for responding to iron-deficient conditions.
Our results show that iron retention in the gcn5 mutant was caused by inefficient iron transportation from the root to the shoot. Interestingly, we identified five iron transport-related targets of GCN5 using ChIP-PCR assays against GCN5 and H3K9/14ac: FRD3, MLP329, EXO70H2, BOR1, and CRK25. It is likely that these genes are downstream components jointly responsible for GCN5-dependent iron homeostasis regulation. FRD3 is well established as a citrate efflux protein, facilitating iron chelation to citrate and subsequent transport of iron citrate (Green and Rogers, 2004; Rogers, 2006; Durrett et al., 2007). BOR1 was the first boron transporter reported in Arabidopsis (Takano et al., 2002) and is required for efficient xylem loading and the preferential translocation of boron into young portions of plants under boron-deficient conditions (Noguchi et al., 1997; Takano et al., 2001). The other three genes are less well characterized and were predicted to have iron ion transport activity, the roles of which in iron homeostasis require further investigation.
FRD3 Is a Critical Target Gene in GCN5-Dependent Iron Homeostasis
Due to the poor solubility and high reactivity of iron, its translocation inside the plant body must be associated with suitable chelating molecules (Curie and Briat, 2003). Previous studies showed that FRD3 plays a dominant role in xylem iron transport. The iron retention phenotype, decreased expression, and histone acetylation levels of FRD3 in gcn5 mutants suggested that FRD3 might be a critical gene in GCN5-regulated iron homeostasis. In addition, both the frd3 and gcn5 mutants show iron-deficiency responses under normal conditions. ChIP assays further confirmed that the enrichment of GCN5 on the FRD3 core promoter significantly increased after iron-deficiency treatment, which was consistent with the increased H3K9/14ac levels. Notably, the iron retention in gcn5 mutants could be successfully rescued by the overexpression of FRD3 or exogenous citrate application. These results provide an epigenetic mechanism of iron homeostasis in which GCN5 regulates FRD3 expression by modulating its H3K9/14ac levels.
Iron availability affects chlorophyll synthesis and plant fertility (Guerinot and Yi, 1994). The frd3 mutant is chlorotic and almost completely sterile (Durrett et al., 2007; Roschzttardtz et al., 2011). Our results showed that pollen viability was also severely impaired and that the chlorophyll content in new leaves was decreased in gcn5 mutants. Moreover, the reduced chlorophyll content and pollen sterility in gcn5 mutants were partially restored by the overexpression of FRD3, which provides further evidence that FRD3 plays an important role in GCN5-regulated iron homeostasis. However, these defects of the gcn5 mutant were not fully rescued by FRD3 overexpression. Thus, to enable a comprehensive interpretation of the role of GCN5 in pollen development and chlorophyll synthesis, it is necessary to study the involvement of other target genes of GCN5 in these processes.
Complex Mechanisms Underlie the Regulation of Iron Homeostasis by HAT and HDAC
The antagonistic functions of the HAT/HDAC pair may operate on the acetylation of the same Lys residue (Jenuwein and Allis, 2001). In contrast to the decreased expression in gcn5 mutants, FRD3 expression increased in hda7 and hda14 mutants. It has been reported that GCN5 and HDA7 acted on the same sites of H3K9 and H3K14 but with opposite effects (Imoberdorf et al., 2006; Jin et al., 2011; Cigliano et al., 2013). Remarkably, the H3K9ac levels at the FRD3 locus were up-regulated in hda7 mutants, indicating that HDA7 might antagonistically act with GCN5 to regulate FRD3 expression. It should be noted that hda9 and hda19 mutants also exhibited iron retention in root tissues (Supplemental Fig. S1). Moreover, FRD3 expression was decreased in hda9 but did not change in hda19. Generally, the expression of direct targets of HDA9 will be increased in hda9. Thus, we hypothesize that HDA9 might regulate FRD3 expression and iron homeostasis indirectly. Collectively, these data indicate the complexity of the underlying regulation mechanisms for HAT and HDAC in iron homeostasis, which requires further investigation.
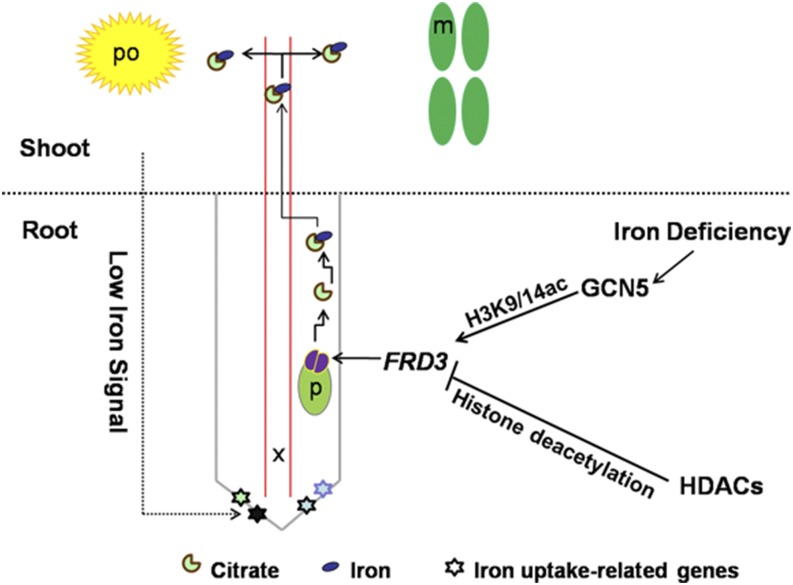
In conclusion, we revealed the roles of histone acetylation in the regulation of iron homeostasis and proposed a possible model for GCN5-regulated FRD3-dependent iron homeostasis in Arabidopsis (Fig. 10). Briefly, the impairment of GCN5 leads to iron retention in roots, impaired pollen viability, and compromised chlorophyll content. Iron deficiency-induced GCN5 acted as an epigenetic modulator to positively control the expression of FRD3 via H3K9/14ac modifications. In the gcn5 mutant, iron-deficiency signals from the shoot might induce the expression of IRT1 and FRO2 in roots even under normal conditions. The up-regulation of IRT1 and FRO2, the down-regulation of FRD3, and the increased production of root hairs jointly contributed to the accumulation of iron in roots, which might lead to iron toxicity in the root systems of the gcn5 mutant. In contrast, some HDACs, such as HDA7, may also participate in this process by negatively regulating FRD3 expression. This research into histone acetylation and iron homeostasis may help uncover prospects for improving iron nutrient utilization efficiency and crop yield.
Figure 10.
Model for acetylation regulation of iron homeostasis. The transcription of GCN5 is induced by iron deficiency. GCN5 and some HDACs (especially HDA7) might antagonistically act on the expression of FRD3 by modulating its acetylation levels. m, Mesophyll cell; p, pericycle; po, pollen grain; x, xylem.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) wild-type Columbia and Ws, the mutant gcn5-2 (Ws background), and hda7, hda5, hda9, hda13, hda14, hda15, hda18, hda19, and hda2c mutants (Columbia-0 background) were used in this study. Mutants were obtained in the homozygous state from the Arabidopsis Biological Resource Center or individual donors. Loss of function of these mutants was confirmed by PCR analysis. For seed germination, sterilized seeds were incubated at 4°C for 3 d; seeds were then sown on MS plates containing 1% (w/v) Suc and 0.6% (w/v) agar. Seedlings were grown under a 16-h-light/8-h-dark cycle at 22°C in a growth room.
Perl’s Stain for Ferric Iron
Arabidopsis roots were treated with Perl’s stain according to established histological methods for mammalian tissues. Briefly, equal amounts of solutions of 4% (v/v) HCl and 4% (w/v) potassium ferrocyanide were mixed immediately prior to use. The stain solution was vacuum infiltrated into 6- or 7-d-old Arabidopsis seedlings for approximately 15 min. Seedlings were rinsed in water, and Perl’s staining was observed immediately in whole roots.
RNA Isolation, Library Preparation, and Transcriptome Sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA concentration was measured using a NanoDrop 2000 spectrophotometer (ND-2000; Thermo Fisher Scientific). RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Paired-end sequencing libraries with average insert size of 200 bp were prepared with the TruSeq RNA Sample Preparation Kit version 2 (Illumina) and sequenced on HiSeq2000 (Illumina) according to the manufacturer’s standard protocols. Raw data obtained from Illumina sequencing were processed and filtered using the Illumina pipeline (http://www.Illumina.com) to generate FastQ files. Finally, approximately 16 GB high-quality 100-bp paired-end reads were generated from eight libraries (Supplemental Table S3). Genes with changes more than 2-fold and false discovery rate-adjusted P < 0.01 (edgeR) were defined as differentially expressed genes. The groups of genes from RNAseq in this study are shown in Supplemental Table S2.
Plasmid Construction and Plant Transformation
The complementary DNA clone of the FRD3 gene containing the full-length open reading frame was amplified by PCR-directed cloning based on annotation from The Arabidopsis Information Resource, with the following primer pair: FRD3-ORF-L and FRD3-ORF-R (Supplemental Table S1). The sequence-confirmed clone containing the full-length complementary DNA of target genes was digested by KpnI and XbaI and cloned into the binary expression vector pCAMBIA1300 (driven by the Cauliflower mosaic virus 35S promoter). Binary vectors were transferred into Agrobacterium tumefaciens strain GV3101. Transgenic plants were generated by a floral dip method and screened on solid plates containing 25 mg L−1 hygromycin.
ChIP
ChIP assays were performed as described previously (Fiil et al., 2008). Seeds of Arabidopsis were sterilized, kept for 3 d at 4°C, and grown in vitro under long-day conditions. After the proper growing period or after iron-deficiency treatments, seedlings were harvested and fixed in 1% (v/v) formaldehyde for 15 min in a vacuum and then neutralized by 0.125 m Gly. After washing with sterilized water, the samples were ground in liquid nitrogen. Nuclei pellets were suspended in a buffer containing 0.25 m Suc, 10 mm Tris-HCl, pH 8, 10 mm MgCl2, 1% (v/v) Triton X-100, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, and protease inhibitors (one minitablet per mL; Roche). The suspensions were transferred to microfuge tubes and centrifuged at 12,000g for 10 min. The pellets were suspended in 1.7 m Suc, 10 mm Tris-HCl, pH 8, 2 mm MgCl2, 0.15% (v/v) Triton X-100, 5 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, and protease inhibitors and centrifuged through a layer of the same buffer in microfuge tubes. The nuclear pellets were lysed in a buffer containing 50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% (w/v) SDS, and protease inhibitors. The lysed nuclei were sonicated four times for 15 s at 4°C followed by centrifugation. The supernatants containing chromatin fragments were diluted 10-fold with 1% (v/v) Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8, and 167 mm NaCl. Aliquots of the dilution were used for immunoprecipitation assays. The antibody of GCN5 was described by Benhamed et al. (2008). The H3K14ac and H3K9ac antibodies were purchased from Upstate Biotechnology. CHS was amplified as an endogenous control. Immunoprecipitated DNA was analyzed by PCR using the primer sets listed in Supplemental Table S1.
Elemental Analysis
Element levels were measured at the College of Resources and Environmental Sciences, China Agricultural University, in a VG AXIOM high-resolution ICP-MS device.
Chlorophyll Measurement
Ten-day-old seedlings of wild-type, gcn5, and 35S::FRD3/gcn5 plants grown on MS plates were transferred onto MS plates with 0 or 100 µm iron-EDTA. After 4 d, the two newest leaves were collected and their fresh weights were determined. Chlorophyll was extracted in 1 mL of 80% (v/v) acetone in the dark at room temperature (about 23°C) for at least 12 h until the leaves became blanched. The supernatant was then subjected to spectrophotometry at 470, 646, and 663 nm. The total chlorophyll content was calculated as described (Hartmut, 1983).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRP049039.
Supplemental Data
The following supplemental materials are available.
Supplementary Figure S1. Perl's stain to identify the mutants involved in iron homeostasis.
Supplementary Figure S2. The reproducibility of the RNA sequencing biological replicates.
Supplementary Figure S3. Sixteen genes selected to examine the accuracy of RNA sequencing using qRT-PCR.
Supplementary Figure S4. GCN5 regulates FRD3 under normal conditions.
Supplementary Figure S5. Expression of FRD3 in gcn5 mutant and 35S::FRD3/gcn5 lines.
Supplementary Figure S6. Examine the responses of histone acetylation to iron deficiency.
Supplemental Table S1. Gene-specific primer pairs used for PCR.
Supplemental Table S2. The groups of genes from RNA sequencing.
Supplemental Table S3. Summary of RNA sequencing data and reads mapping.
Supplementary Material
Acknowledgments
We thank Qixin Sun for providing constructive comments on the article and Shunong Bai for providing seeds.
Glossary
- bHLH
basic helix-loop-helix
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- qPCR
quantitative PCR
- RNAseq
RNA sequencing
- GO
Gene Ontology
- ChIP
chromatin immunoprecipitation
- ICP-MS
ion-coupled plasma mass spectrometry
- MS
Murashige and Skoog
- Ws
Wassilewskija
- TSA
trichostatin
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the National Basic Research Program of China 973 Program (grant no. 2012CB910900), the National Natural Science Foundation of China (grant no. 31471478), the 863 Project (grant no. 2012AA10A309), and the Chinese Universities Scientific Fund (grant no. 2014XJ019).
References
- Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, De Meyer B, Buysschaert C, Rombauts S, Villarroel R, et al. (2008) Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J 56: 493–504 [DOI] [PubMed] [Google Scholar]
- Bruce TJ, Matthes MC, Napier JA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173: 603–608 [Google Scholar]
- Buckhout TJ, Yang TJ, Schmidt W (2009) Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigliano RA, Cremona G, Paparo R, Termolino P, Perrella G, Gutzat R, Consiglio MF, Conicella C (2013) Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol 163: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Fan H, Zhang Z, Wang N, Cui Y, Sun H, Liu Y, Wu H, Zheng S, Bao S, Ling HQ (2014) SKB1/PRMT5‐mediated histone H4R3 dimethylation of Ib subgroup bHLH genes negatively regulates iron homeostasis in Arabidopsis thaliana. Plant J 77: 209–221 [DOI] [PubMed] [Google Scholar]
- Feng H, An F, Zhang S, Ji Z, Ling HQ, Zuo J (2006) Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant Physiol 140: 1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J (2008) Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. Cold Spring Harb Protoc 2008: pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Green LS, Rogers EE (2004) FRD3 controls iron localization in Arabidopsis. Plant Physiol 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature 389: 349–352 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmut K. (1983) Determinations of total carotenoids and chlorophylls b of leaf extracts in different solvents. Analysis 4: 142–196 [Google Scholar]
- Imoberdorf RM, Topalidou I, Strubin M (2006) A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol Cell Biol 26: 1610–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson L. (1945) Iron in the leaves and chloroplasts of some plants in relation to their chlorophyll content. Plant Physiol 20: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, et al. (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24: 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1986) Functions of mineral nutrients: macronutrients. In Marschner H, ed, Mineral Nutrition of Higher Plants. Academic Press, London, pp 226–235 [Google Scholar]
- Noguchi K, Yasumori M, Imai T, Naito S, Matsunaga T, Oda H, Hayashi H, Chino M, Fujiwara T (1997) bor1-1, an Arabidopsis thaliana mutant that requires a high level of boron. Plant Physiol 115: 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh SA, Choimes S, Schachtman DP (2004) Over-expression of an Arabidopsis zinc transporter in Hordeum vulgare increases short-term zinc uptake after zinc deprivation and seed zinc content. Plant Mol Biol 54: 373–385 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE. (2006) Role of FRD3 in iron translocation and homeostasis. In Iron Nutrition in Plants and Rhizospheric Microorganisms. Springer Verlag, New York, pp 327–339 [Google Scholar]
- Roschzttardtz H, Séguéla-Arnaud M, Briat JF, Vert G, Curie C (2011) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2001) Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol 125: 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M (2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420: 337–340 [DOI] [PubMed] [Google Scholar]
- Takano J, Yamagami M, Noguchi K, Hayashi H, Fujiwara T (2001) Preferential translocation of boron to young leaves in Arabidopsis thaliana regulated by the BOR1 gene. Soil Sci Plant Nutr 47: 345–357 [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ (2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM (2010) Regulation of phosphate starvation responses in higher plants. Ann Bot (Lond) 105: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.